Introduction
Colorectal cancer (CRC) is a major global public
health concern, responsible for high rates of both mortality and
morbidity (1). Colorectal polyps
(CPs) are widely recognized as the predominant precursors of CRC
(2), due to their propensity to
undergo malignant transformation (3). Modern medical research has confirmed
several risk factors associated with CPs, including aging, high
protein consumption (particularly red meat), a diet rich in fats
and deficient in fiber, smoking and excessive alcohol consumption
(4,5).
Notably, the intestinal microbiota has gained
recognition as an important contributor that mechanistically links
various risk factors to the development of CRC (6-8).
More specifically, the intestinal microbiota and its metabolites
have been demonstrated to induce epigenetic modifications in host
cells, with these metabolites playing an important role as
signaling molecules in intercellular communication (9). Fusobacterium nucleatum, a
commonly found microorganism prevalent in CRC, has been
demonstrated to increase gene methylation levels and induce
microsatellite instability (10,11).
Furthermore, trimethylamine is primarily produced by Escherichia
coli and induces DNA methylation, which is closely linked to
CRC (12). Bilophila
wadsworthia and Pyramidobacter spp. are additional
examples of microorganisms enriched in CRC, which have been
reported to promote carcinogenesis through the production of
genotoxic hydrogen sulfide within the gastrointestinal tract
(13-15).
Conversely, specific gut bacteria including
Faecalibacterium, Bifidobacterium, Roseburia,
Lactobacillus and Eubacterium can ferment dietary
fibers into short-chain fatty acids (SCFAs). These SCFAs possess
gut-protective properties and are inversely correlated with CRC;
SCFAs such as acetate, propionate and butyrate provide protection
against CRC by regulating gut inflammation and the immune system
through multiple mechanisms (15).
Moreover, both butyrate and acetate can function as histone
deacetylase inhibitors, exerting an influence on the epigenetic
alterations that regulate the development of CRC (16).
Although colonoscopy is still considered the
standard procedure for diagnosing CRC, its invasive nature and high
cost limits its use in widespread screening (17). The progress in metabolomic
profiling technology has enabled several studies to showcase the
potential of fecal metabolites in noninvasive CRC diagnosis
(18,19). Our previous study revealed a
structural imbalance in the gut microbiota composition of patients
with CPs compared with that in healthy individuals, characterized
by decreased beneficial bacteria and increased harmful species
(20). However, the signatures of
fecal metabolites in patients with CPs and healthy individuals have
yet to be fully understood.
In the present study, the gut metabolome was
profiled in patients diagnosed with CPs and was compared with that
of healthy individuals. The objective was to define fecal metabolic
profiles that could differentiate patients with CPs from healthy
individuals.
Materials and methods
Participants
The study cohort used in the present study included
patients described in our previous study (20). The present study involved
conducting untargeted metabolomics analysis on fecal samples
obtained from patients with CPs and healthy individuals at the
Shanghai Jiading District Hospital of Chinese Medicine (Shanghai,
China) between March and August 2024. All participants were
enrolled consecutively upon providing written informed consent.
Patients undergoing colonoscopy and diagnosed with CPs and healthy
individuals without CPs were included. A total of 30 patients with
CPs were classified as the CP group, whereas 30 healthy individuals
were classified as the healthy control (HC) group. Participants in
each group were matched based on age, sex and body weight during
enrollment. The CP group was selected according to the following
inclusion criteria: Aged >18 years, and a diagnosis of CP
confirmed by colonoscopy and histopathology. The exclusion criteria
for the CP group were as follows: i) Use of antibiotics or
probiotics within the past 2 months; ii) a previous history of CRC.
The inclusion criteria of the HC group was: Aged >18 years
without CP confirmed by colonoscopy examination. The exclusion
criteria for the HC group were as follows: i) Presence of any
underlying diseases, including autoimmune diseases such as
ankylosing spondylitis and systemic lupus erythematosus, infectious
diseases, cachexia, organ failure, respiratory diseases and
cardiovascular diseases; ii) history of surgery or chronic drug
use; iii) use of antibiotics or probiotics within the past 2
months.
Sample processing
Fecal samples were collected by the participants
after enrolling in the study. Immediately after collection, the
samples were placed on dry ice and stored at -80˚C until further
use. Subsequently, the samples were sent to Shanghai Bioprofile
Biotechnology Co., Ltd. for untargeted metabolomics analysis. These
samples were mixed with 400 µl cold (4˚C) methanol acetonitrile
(v/v; 4:1) and homogenized with a tissue crusher. The mixture was
then sonicated (53 kHz) for 20 min in an ice bath. Following this,
the mixture was incubated at -20˚C for 1 h, and centrifuged at 4˚C
for 20 min at 16,000 x g. Next, the supernatants were harvested and
were vacuum dried.
Ultra-high performance liquid
chromatography (UHPLC)-mass spectrometry (MS)/MS analysis
UHPLC (LC-30AD; Shimadzu Corporation) coupled with
Q-Exactive™ Plus (Thermo Fisher Scientific, Inc.) was used for
metabolomics profiling analysis.
Samples were placed in a 4˚C autosampler and
underwent liquid chromatography separation using an ACQUITY
UPLC® HSS T3 column (2.1x100 mm; 1.8 µm; Waters
Corporation). The injection volume was 4 µl and the column
temperature was 40˚C. The flow rate was set at 0.3 ml/min, and the
mobile phase consisted of two components: i) A (0.1% formic acid in
water); and ii) B (0.1% formic acid in acetonitrile). The gradient
started at 0% buffer B for 2 min, increased linearly to 48% over 4
min, was further increased to 100% over 4 min and held for 2 min,
then decreased to 0% buffer B within 0.1 min, followed by a 3 min
re-equilibration period.
Electrospray ionization (ESI) in both positive and
negative modes was used for separate acquisition of MS data. The
heated-ESI source was configured with the following conditions: i)
Spray voltage at 3.8 kV (positive) and 3.2 kV (negative); ii)
capillary temperature at 320˚C; iii) sheath gas (nitrogen) flow at
30 arbitrary units (AU) with temperature at 300˚C and with
nebulizer pressure at 100 pounds per square inch; iv) aux gas flow
at 5 AU; v) probe heater temperature at 350˚C; and vi) S-Lens radio
frequency level at 50. The instrument was set to acquire full MS
data over the m/z range of 70-1,050 Da. The full MS scans were
performed at a resolution of 70,000 at m/z 200 and 17,500 at m/z
200 for MS/MS scan. The maximum injection time was set at 100 msec
for MS and 50 msec for MS/MS. The isolation window for MS/MS was
set to 2 m/z and the normalized collision energy (stepped) was set
to 20, 30 and 40 V for fragmentation.
Data preprocessing and filtering
The raw MS data underwent processing using MS-DIAL
(version 4.9) (21) to align
peaks, correct retention time and extract peak areas. Metabolites
were identified based on accurate mass (with a mass tolerance of
<10 parts per million) and MS/MS data (with a mass tolerance of
<0.01 Da) through matching against The Human Metabolome Database
(https://hmdb.ca/), MassBank (https://massbank.eu/MassBank), Global Natural Product
Social Molecular Networking (https://gnps.ucsd.edu) library databases and Baipu
Metabolite Standard Library (in-house database of Shanghai
Bioprofile Biotechnology Co., Ltd.). Only variables with >50%
non-zero measurement values in at least one group were retained
from the extracted-ion features.
Metabolomics data analysis
R programming language (v4.0.3) and R packages were
used for all multivariate data analyses and modeling (https://www.r-project.org/). Metabolomics data were
mean-centered using Pareto scaling (22). Models were constructed using
principal component analysis (PCA), orthogonal partial least
squares (OPLS)-discriminant analysis (DA) and partial least squares
(PLS)-DA. All models were evaluated and tested for overfitting
using permutation tests. The descriptive performance of the models
was assessed based on the cumulative R2X (with a perfect model
having R2X, 1) and R2Y (with a perfect model having R2Y, 1) values,
while their predictive performance was evaluated using cumulative
Q2 (with a perfect model having Q2, 1) and a permutation test
(n=200). A permuted model should be unable to predict the classes:
The R2 and Q2 values at the Y-axis intercept should be lower than
those of the non-permuted model.
Discriminating metabolites that differentiate
patients with CPs from HCs were identified utilizing a
statistically significant threshold of the variable influence on
projection (VIP) values derived from the OPLS-DA model, along with
two-tailed unpaired Student's t-test (P-value) applied to the
normalized raw data at the univariate analysis level. The VIP score
represents the contribution of the variable to discriminating
between sample classes, calculated as the weighted sum of squares
of PLS weights for each variable. The mean VIP value is 1, and VIP
values >1 and P<0.05 are considered significant, indicating a
strong discriminatory ability and serving as criteria for biomarker
selection. Fold change was determined as the logarithm ratio of the
average mass response (area) between two arbitrary classes.
Additionally, the identified differential metabolites were
subjected to cluster analyses using R package (v4.0.3) and a
heatmap was generated. Receiver operating characteristic (ROC)
analysis was performed and the area under the curve (AUC) was
calculated to assess the predictive capability of potential
bacterial markers for distinguishing between the CP and HC
groups.
Statistical analysis and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis
To identify the disrupted biological pathways, KEGG
pathway analysis was conducted on the differential metabolite data
using the KEGG database (http://www.kegg.jp). KEGG enrichment analyses were
performed using Fisher's exact test, and multiple testing
correction was applied using the false discovery rate method.
Pathways enriched at P<0.05 level were considered statistically
significant. The correlations between change in metabolites were
estimated with Pearson correlation tests using R package
(v4.0.3).
Results
Demographic and clinical
characteristics of study participants
No significant differences were found between the CP
and HC groups in terms of age, sex, height, weight or BMI, as
previously shown (20). The
histological types of CPs comprised the following: i) Tubular
adenoma (63.3%); ii) tubular adenoma/serrated polyps (3.3%); iii)
tubular adenoma/hyperplastic polyps (16.7%); iv) tubular
adenoma/hyperplastic polyps/serrated polyps (3.3%); v) serrated
adenoma (3.3%); and vi) hyperplastic polyps (10.0%).
Metabolomics analysis
After filtering and pre-processing data, a total of
62,209 peaks were normalized and utilized for subsequent analyses.
The unsupervised PCA revealed that the quality control study pools
formed a distinct cluster at the center of the corresponding study
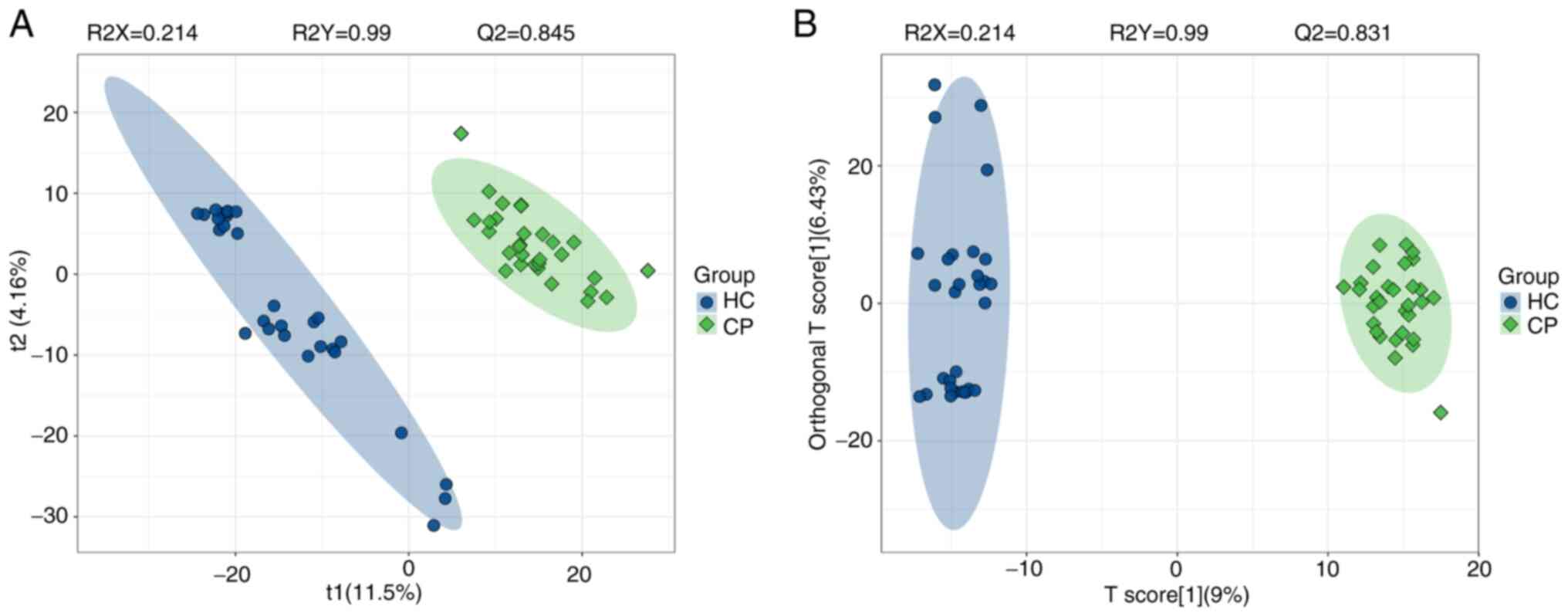
samples (Fig. S1). PLS-DA, a
supervised multivariate analysis, effectively distinguished CP and
HC samples into well-defined groups using the untargeted data,
supported by strong model statistics (R2Y, 0.990; R2X, 0.214; Q2,
0.845; Fig. 1A). Similarly,
supervised multivariate analysis using OPLS-DA also discriminated
CP and HC groups with good model statistics (R2Y, 0.990; R2X,
0.214; Q2, 0.831; Fig. 1B).
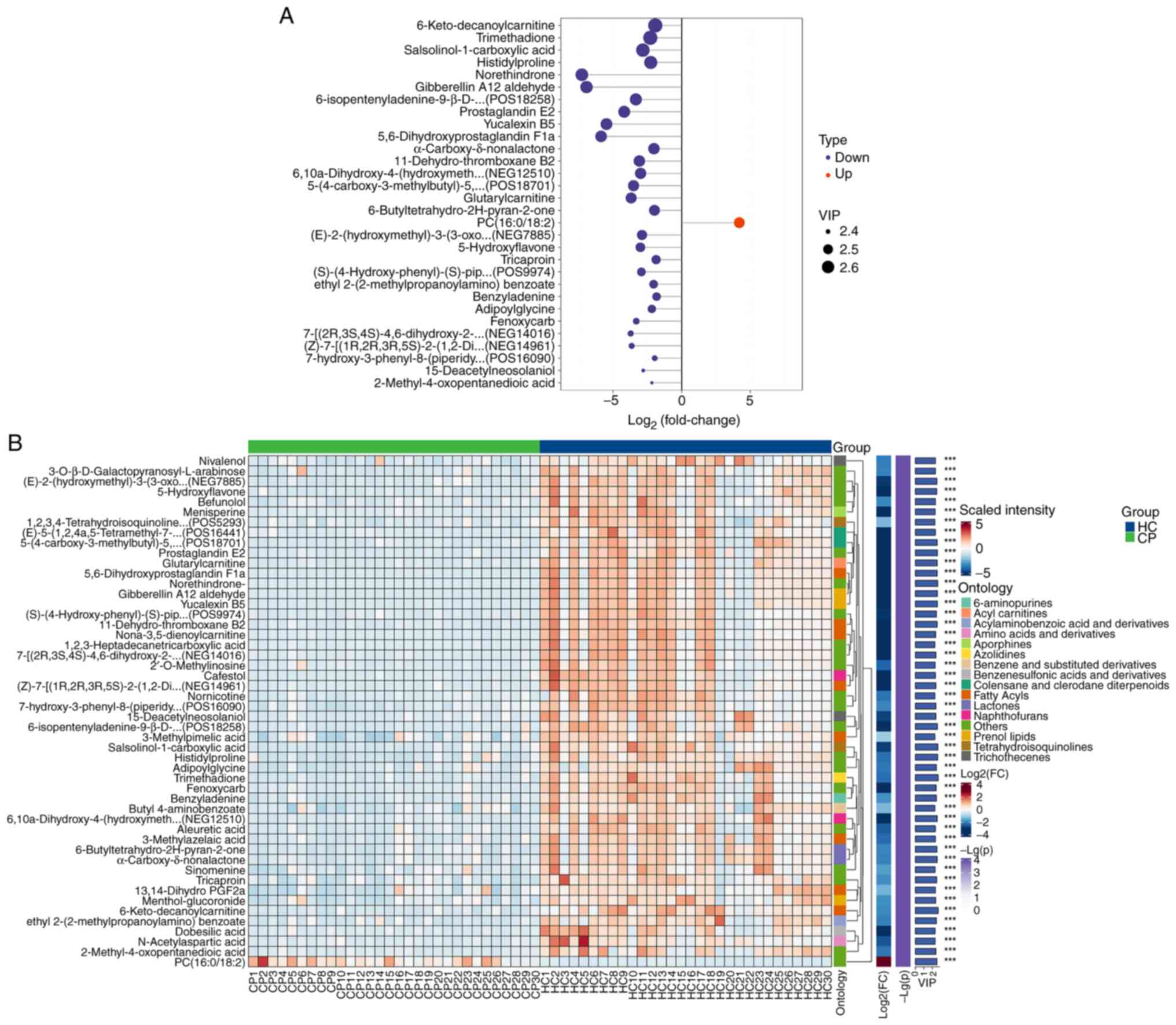
Next, differential abundance analysis of metabolites
was performed, and a total of 300 metabolites were identified
(Table SI). Fig. 2A displays the top 30 metabolites
with statistically significant differences (based on P-value)
between the CP and HC groups, with a VIP score >1 and P<0.001
(Table SII). Specifically, the
log2 fold change, VIP and P-value for prostaglandin A1
were -7.453520969, 2.161393135 and 4.88x10-8,
respectively (Table SI).
Correlation analysis showed a positive correlation between these
identified metabolites (Fig. S2;
Table SIII). Specifically,
6-keto-decanoylcarnitine, trimethadione, salsolinol-1-carboxylic
acid, histidyl-proline, norethindrone, gibberellin A12 aldehyde,
6-isopentenyladenine-9-β-D-glucopyranoside, prostaglandin E2,
yucalexin B5 and 5,6-dihydroxyprostaglandin F1a were significantly
less abundant in the CP group compared with in the HC group
(Fig. 2A). By contrast,
phosphatidylcholine (PC; 16:0/18:2) exhibited a significantly
greater abundance in the CP group compared with in the HC group
(Fig. 2A). The hierarchical
clustering heatmap (Fig. 2B)
visually illustrates the distribution of metabolites that
significantly differ between the CP (green) and HC (blue)
groups.
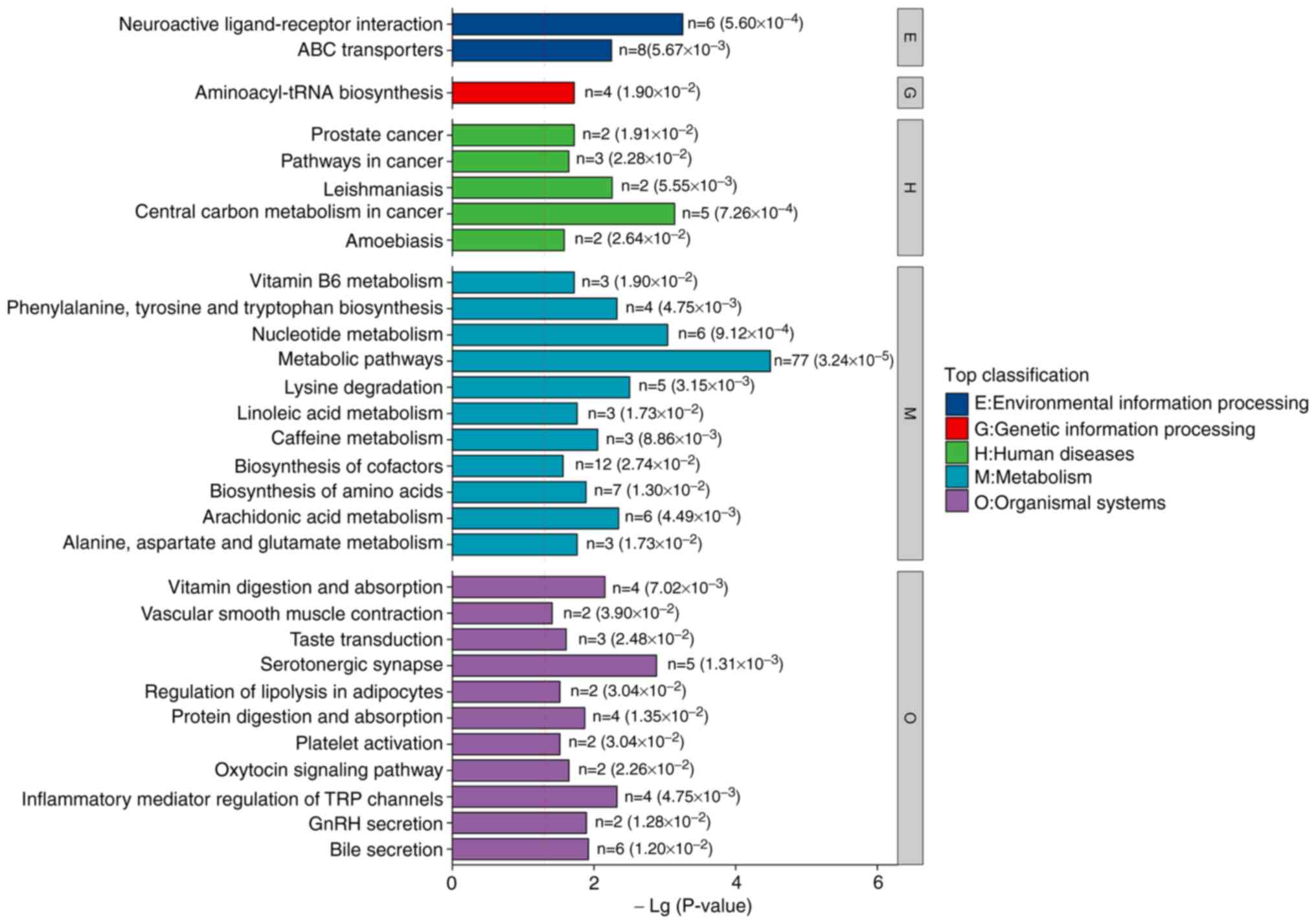
KEGG pathway analysis identified multiple perturbed
metabolic pathways between the CP and HC groups. The perturbed
pathways (Fig. 3) encompassed
‘neuroactive ligand-receptor interaction’, ‘aminoacyl-tRNA
biosynthesis’, ‘central carbon metabolism in cancer’, and
‘phenylalanine, tyrosine and tryptophan biosynthesis’. These
findings indicated that metabolic pathways, as well as individual
metabolites, may be altered in patients with CPs.
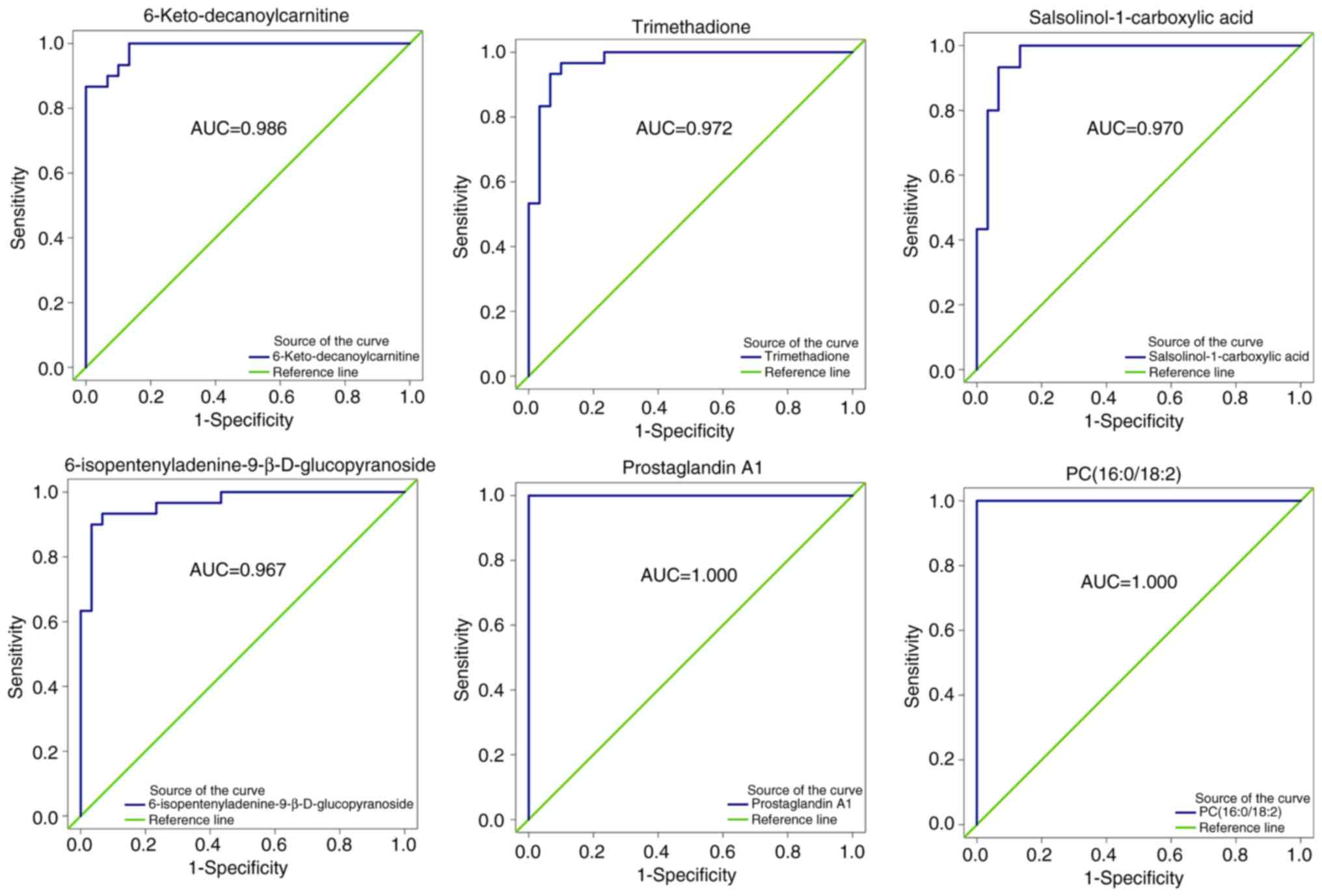
ROC analysis
All 300 metabolites with differential abundance
underwent ROC analysis. Fig. 4
summarizes the ROC analysis results for six significantly abundant
metabolites. 6-Keto-decanoylcarnitine, trimethadione,
salsolinol-1-carboxylic acid and
6-isopentenyladenine-9-β-D-glucopyranoside were ranked in the top
30 by VIP score and with AUC values in the top 5% (≥0.967),
indicating accurate performance for the CP status. In addition, PC
(16:0/18:2) and prostaglandin A1 achieved an AUC of 1 indicating
perfect discrimination.
Discussion
Accumulating evidence has suggested that the gut
microbiota and its metabolites have important implications in the
development of CRC (23,24). Consequently, exploring the
potential of using the gut microbiome and its metabolites as
screening tools for early CRC detection holds great promise.
Notably, the fecal metabolome reflects the complex interactions
among genetic, dietary and environmental factors (25). Thus, conducting metabolomics
research using fecal samples may enable the identification of
metabolic biomarkers linked to CRC. In the present study, metabolic
analysis of fecal samples obtained from patients with CPs and
healthy individuals was conducted to identify metabolites
associated with pathophysiological changes. These findings
highlight the potential of fecal metabolites as non-invasive
biomarkers for the early detection of CPs.
The observed increase in PC (16:0/18:2) in the CP
group highlighted the role of phospholipids in CRC development; PCs
are essential components of cell membranes and are involved in
various signaling pathways, such as the CDP-choline pathway and the
phosphatidylethanolamine methylation pathway (26). Elevated levels of specific PC
species, such as PC (16:0/16:1), have been reported in CRC tissues,
suggesting that these lipids may serve as biomarkers for early
detection (27). The findings in
the present study further supported the notion that dysregulation
of lipid metabolism, particularly in the context of PCs, may be a
key feature of CPs and could potentially contribute to their
progression to malignancy. By contrast, the reduced abundance of
metabolites such as 6-keto-decanoylcarnitine, trimethadione and
prostaglandin E2 in the CP group suggests a disruption in
inflammatory pathways. Prostaglandin E2 in particular is known to
play a dual role in cancer biology, acting both as a
proinflammatory mediator and a regulator of immune responses
(28,29). The decreased levels of
prostaglandin E2 in the current study may reflect an altered
inflammatory and immune response in the gut microenvironment of
patients with CP, which may influence the progression of CPs to
CRC.
Pathway enrichment analysis revealed significant
alterations in aminoacyl-tRNA biosynthesis and aromatic amino acid
metabolism in the CP group. Aminoacyl-tRNA synthetases, which are
critical for protein synthesis, have been implicated in various
types of cancer, including CRC (30); the dysregulation of these enzymes
in patients with CP suggests that translational control mechanisms
may be perturbed early in the carcinogenic process. Furthermore,
the observed changes in aromatic amino acid metabolism,
particularly phenylalanine, tyrosine and tryptophan, are
noteworthy. These amino acids are not only essential for protein
synthesis, but also serve as precursors for various signaling
molecules, including neurotransmitters and immune modulators
(31). The dysregulation of these
pathways may contribute to the disruption of the gut barrier
function and immune homeostasis, both of which are critical factors
in CRC development (32). The
potential of fecal metabolites as non-invasive tools for the
detection of CPs is further supported by the performance of the
identified biomarkers in ROC analysis, with AUC values >0.96.
Notably, PC (16:0/18:2) and prostaglandin A1 achieved perfect
discrimination (AUC, 1), highlighting their potential utility in
clinical settings. Nonetheless, a pending further validation study
in larger cohorts is necessary to validate the candidate
biomarker.
The current study has several limitations. Firstly,
the single-center design and relatively small sample size may limit
the generalizability of the findings. Multicenter studies with
larger cohorts are needed to validate these results. Secondly, the
complex interplay between diet, gut microbiota and host metabolism
poses a challenge in distinguishing between host- and
microbial-derived metabolites. Future studies incorporating dietary
assessments and isotope labeling techniques may elucidate the
origins of these metabolites and their role in CP pathogenesis.
Furthermore, functional validation is necessary in future studies
to confirm the biological relevance of the metabolic alterations
identified.
In conclusion, the present study provides compelling
evidence that fecal metabolomics profiling can serve as a
non-invasive tool for the detection of CPs. The identified
metabolic alterations, particularly in lipid and amino acid
metabolism, offer new insights into the molecular mechanisms
underlying CP development. These findings pave the way for further
research into the role of gut metabolites in CRC pathogenesis and
their potential as therapeutic targets.
Supplementary Material
Principal component analysis plot
showing the spread of the study samples and the high
reproducibility of the QC pools. PC1 and PC2 denote the first and
second components, respectively. HC, healthy control; QC, quality
control; PC, principal component.
Heatmap of correlation for the top 10
variable influence projection-ranked differential metabolites.
Differential metabolites between the
CP group and the HC group.
Top 30 differentially abundant
metabolites between the CP group and the HC group.
Correlation for the differential
metabolites.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Natural Science Research
project of Jiading District, Shanghai (grant nos. JDKW-2022-0030
and JDKW-2023-0051).
Availability of data and materials
The UHPLC-MS/MS data generated in the present study
may be found in the National Genomics Data Center repository under
BioProject number: PRJCA040951 and accession number: OMIX010385 or
at the following URL: ngdc.cncb.ac.cn/omix/release/OMIX010385.
Authors' contributions
DD, ZG, and YJ conceived and designed the study. DD,
YY, HZ, and HS interpreted the analysis results and wrote the
manuscript. DD, YY, YX, HW, and LZ collected the samples, and
contributed to the acquisition and analysis of data. DD, ZG and YJ
confirm the authenticity of all the raw data. All authors revised
the manuscript, and read and approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval was obtained from the Ethics
Committee of the Shanghai Jiading District Hospital of Chinese
Medicine (approval no. 2022-002; Shanghai, China) and written
informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Favoriti P, Carbone G, Greco M, Pirozzi F,
Pirozzi RE and Corcione F: Worldwide burden of colorectal cancer: A
review. Updates Surg. 68:7–11. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Barberis E, Joseph S, Amede E, Clavenna
MG, La Vecchia M, Sculco M, Aspesi A, Occhipinti P, Robotti E,
Boldorini R, et al: A new method for investigating
microbiota-produced small molecules in adenomatous polyps. Anal
Chim Acta. 1179(338841)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Øines M, Helsingen LM, Bretthauer M and
Emilsson L: Epidemiology and risk factors of colorectal polyps.
Best Pract Res Clin Gastroenterol. 31:419–424. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kordahi MC, Stanaway IB, Avril M, Chac D,
Blanc MP, Ross B, Diener C, Jain S, McCleary P, Parker A, et al:
Genomic and functional characterization of a mucosal symbiont
involved in early-stage colorectal cancer. Cell Host Microbe.
29:1589–1598.e6. 2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zitvogel L, Galluzzi L, Viaud S, Vétizou
M, Daillère R, Merad M and Kroemer G: Cancer and the gut
microbiota: An unexpected link. Sci Transl Med.
7(271ps1)2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Vipperla K and O'Keefe SJ: Diet,
microbiota, and dysbiosis: A ‘recipe’ for colorectal cancer. Food
Funct. 7:1731–1740. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wong CC and Yu J: Gut microbiota in
colorectal cancer development and therapy. Nat Rev Clin Oncol.
20:429–452. 2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Woo V and Alenghat T: Epigenetic
regulation by gut microbiota. Gut Microbes.
14(2022407)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Baba Y, Hara Y, Toihata T, Kosumi K,
Iwatsuki M, Iwagami S, Miyamoto Y, Yoshida N, Komohara Y and Baba
H: Relationship between gut microbiome Fusobacterium nucleatum and
LINE-1 methylation level in esophageal cancer. Esophagus.
20:704–712. 2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mima K, Nishihara R, Qian ZR, Cao Y,
Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al:
Fusobacterium nucleatum in colorectal carcinoma tissue and patient
prognosis. Gut. 65:1973–1980. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xu R, Wang Q and Li L: A genome-wide
systems analysis reveals strong link between colorectal cancer and
trimethylamine N-oxide (TMAO), a gut microbial metabolite of
dietary meat and fat. BMC Genomics 16 Suppl. 7(S4)2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hale VL, Jeraldo P, Mundy M, Yao J, Keeney
G, Scott N, Cheek EH, Davidson J, Greene M, Martinez C, et al:
Synthesis of multi-omic data and community metabolic models reveals
insights into the role of hydrogen sulfide in colon cancer.
Methods. 149:59–68. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yazici C, Wolf PG, Kim H, Cross TL,
Vermillion K, Carroll T, Augustus GJ, Mutlu E, Tussing-Humphreys L,
Braunschweig C, et al: Race-dependent association of sulfidogenic
bacteria with colorectal cancer. Gut. 66:1983–1994. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Davies J, Mayer MJ, Juge N, Narbad A and
Sayavedra L: Bacteroides thetaiotaomicron enhances H2S production
in Bilophila wadsworthia. Gut Microbes.
16(2431644)2024.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fusco W, Lorenzo MB, Cintoni M, Porcari S,
Rinninella E, Kaitsas F, Lener E, Mele MC, Gasbarrini A, Collado
MC, et al: Short-chain fatty-acid-producing bacteria: Key
components of the human gut microbiota. Nutrients.
15(2211)2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Doubeni CA, Corley DA, Quinn VP, Jensen
CD, Zauber AG, Goodman M, Johnson JR, Mehta SJ, Becerra TA, Zhao
WK, et al: Effectiveness of screening colonoscopy in reducing the
risk of death from right and left colon cancer: A large
community-based study. Gut. 67:291–298. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Coker OO, Liu C, Wu WKK, Wong SH, Jia W,
Sung JJY and Yu J: Altered gut metabolites and microbiota
interactions are implicated in colorectal carcinogenesis and can be
non-invasive diagnostic biomarkers. Microbiome.
10(35)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Li Z, Deng X, Luo J, Lei Y, Jin X, Zhu J
and Lv G: Metabolomic comparison of patients with colorectal cancer
at different anticancer treatment stages. Front Oncol.
11(574318)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Deng D, Zhao L, Song H, Wang H, Cao H, Cui
H, Zhou Y and Cui R: Microbiome analysis of gut microbiota in
patients with colorectal polyps and healthy individuals. Sci Rep.
15(7126)2025.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tsugawa H, Cajka T, Kind T, Ma Y, Higgins
B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O and Arita M:
MS-DIAL: Data-independent MS/MS deconvolution for comprehensive
metabolome analysis. Nat Methods. 12:523–526. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
van den Berg RA, Hoefsloot HC, Westerhuis
JA, Smilde AK and van der Werf MJ: Centering, scaling, and
transformations: Improving the biological information content of
metabolomics data. BMC Genomics. 7(142)2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Eng C, Yoshino T, Ruíz-García E, Mostafa
N, Cann CG, O'Brian B, Benny A, Perez RO and Cremolini C:
Colorectal cancer. Lancet. 404:294–310. 2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Louis P, Hold GL and Flint HJ: The gut
microbiota, bacterial metabolites and colorectal cancer. Nat Rev
Microbiol. 12:661–672. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zierer J, Jackson MA, Kastenmüller G,
Mangino M, Long T, Telenti A, Mohney RP, Small KS, Bell JT, Steves
CJ, et al: The fecal metabolome as a functional readout of the gut
microbiome. Nat Genet. 50:790–795. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
van der Veen JN, Kennelly JP, Wan S, Vance
JE, Vance DE and Jacobs RL: The critical role of
phosphatidylcholine and phosphatidylethanolamine metabolism in
health and disease. Biochim Biophys Acta Biomembr. 1859:1558–1572.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Pakiet A, Kobiela J, Stepnowski P,
Sledzinski T and Mika A: Changes in lipids composition and
metabolism in colorectal cancer: A review. Lipids Health Dis.
18(29)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Finetti F, Travelli C, Ercoli J, Colombo
G, Buoso E and Trabalzini L: Prostaglandin E2 and cancer: Insight
into tumor progression and immunity. Biology (Basel).
9(434)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Karpisheh V, Nikkhoo A, Hojjat-Farsangi M,
Namdar A, Azizi G, Ghalamfarsa G, Sabz G, Yousefi M, Yousefi B and
Jadidi-Niaragh F: Prostaglandin E2 as a potent therapeutic target
for treatment of colon cancer. Prostaglandins Other Lipid Mediat.
144(106338)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhou Z, Sun B, Huang S, Yu D and Zhang X:
Roles of aminoacyl-tRNA synthetase-interacting multi-functional
proteins in physiology and cancer. Cell Death Dis.
11(579)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Dodd D, Spitzer MH, Van Treuren W, Merrill
BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP,
Fischbach MA and Sonnenburg JL: A gut bacterial pathway metabolizes
aromatic amino acids into nine circulating metabolites. Nature.
551:648–652. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Platten M, Nollen EAA, Röhrig UF,
Fallarino F and Opitz CA: Tryptophan metabolism as a common
therapeutic target in cancer, neurodegeneration and beyond. Nat Rev
Drug Discov. 18:379–401. 2019.PubMed/NCBI View Article : Google Scholar
|