Introduction
Choledocholithiasis (CDL), the presence of
gallstones in the common bile duct (CBD), affects approximately
10-15% of patients with cholelithiasis over 10 years (1). In the U.S., gallstone disease
contributes to over 1.2 million emergency department visits
annually (2). The incidence of CDL
is rising, likely due to an aging population and increased use of
diagnostic imaging (3). Prompt and
accurate diagnosis is critical, as undetected CDL can lead to
complications such as cholangitis, pancreatitis, or biliary
obstruction, while unnecessary endoscopic retrograde
cholangiopancreatography (ERCP) may cause iatrogenic harm (4).
While ERCP remains the gold standard for diagnosing
and treating CDL, its invasive nature demands judicious patient
selection (5). The American
Society for Gastrointestinal Endoscopy (ASGE) revised its risk
stratification criteria in 2019, classifying patients into low,
intermediate, or high-risk groups based on clinical, biochemical,
and imaging data (6,7). However, these criteria have shown
inconsistent accuracy, particularly in high-volume settings,
raising concerns about overuse of ERCP and missed diagnoses
(8).
Artificial intelligence (AI) transforms medicine by
enabling data-driven, personalized care (9). In gastroenterology, AI has been
applied in real-time endoscopic image analysis, polyp detection,
colorectal neoplasia and malignant biliary strictures, and disease
risk stratification (10-12).
Supervised machine learning models (MLMs), such as
gradient-boosting machines (GBMs), learn from labeled input-output
pairs by building and correcting sequential decision trees
(10,13). Deep learning (DL) models, including
artificial neural networks (ANNs) and convolutional neural networks
(CNNs), can integrate diverse clinical inputs such as biochemical
studies and imaging findings, and continuously refine predictions
through iterative learning (10,13).
Unsupervised models, which learn from unlabeled data, are
increasingly used to uncover hidden disease subtypes and patterns
(10,13,14).
Data-driven ML and DL algorithms in diagnosing and
early predicting CDL may improve their therapeutic implementations
and decision-making. Applying AI to CDL diagnosis may reduce
unnecessary ERCP, lower complication rates, and improve outcomes by
enabling noninvasive, individualized risk assessment. However,
evidence of their diagnostic performance remains fragmented. This
systematic review and meta-analysis aim to evaluate AI-assisted
tools' sensitivity, specificity, and overall diagnostic accuracy in
predicting CDL and compare their performance with guideline-based
approaches such as those from the ASGE.
Materials and methods
Search strategy
Two independent investigators systematically
searched MEDLINE, EMBASE, PubMed, and Web of Science from inception
to March 2, 2025. Keywords included ‘Artificial intelligence’,
‘machine learning’, ‘computer-aided diagnosis’, ‘biliary
obstruction’, ‘choledocholithiasis’, and ‘endoscopic retrograde
cholangiopancreatography or ECRP’ with Boolean operators used to
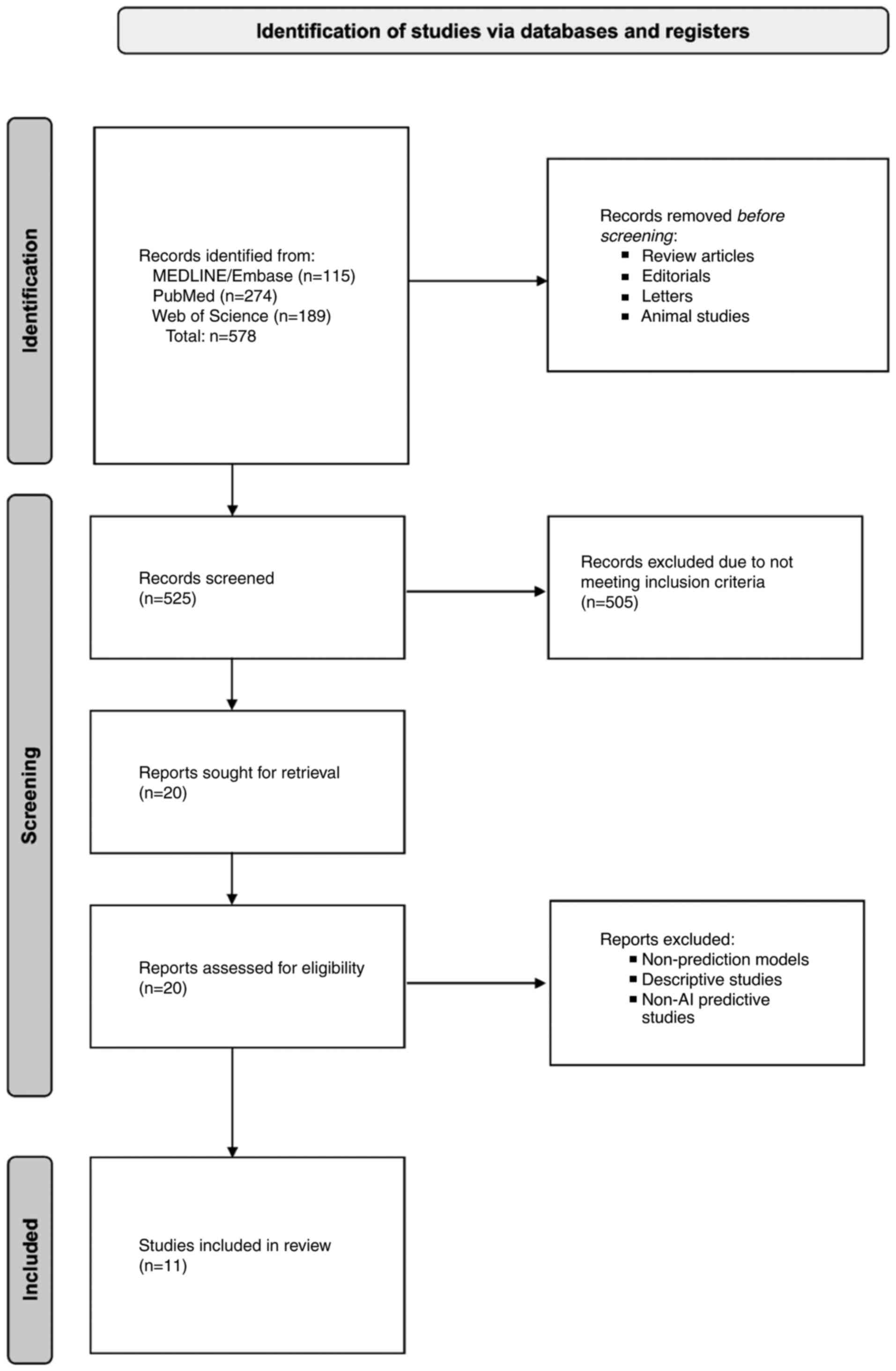
optimize search results. A PRISMA diagram was designed according to
the PRISMA 2020 guidelines, as shown in Fig. 1, including our meta-analysis
studies (15-25).
Our meta-analysis focused on the diagnostic accuracy of AI models,
which does not fall within the scope of PROSPERO registration, as
it primarily registers reviews of interventional studies;
therefore, it was not registered.
Inclusion and exclusion criteria
Studies were eligible if they: i) Evaluated AI or
MLMs for CDL diagnosis or prediction, ii) involved human subjects
without age restrictions, and iii) were observational, cohort,
retrospective, or prospective studies. Studies predicting
recurrence or procedural complexity were excluded since they did
not directly evaluate diagnostic performance for initial CDL
detection, which was our primary outcome. Specifically, exclusion
criteria included: i) Models not aimed at diagnostic prediction,
ii) studies not using AI or machine learning (ML) tools, iii)
models focused on predicting procedural complexity or recurrence
prediction after stone removal rather than initial diagnostic
performance for CDL, and iv) studies lacking detailed model
diagnostic accuracy metrics such as sensitivity, specificity,
positive predictive value, negative predictive value, to limit
heterogeneity. All metrics are defined based on the reference
standard, ERCP or intraoperative cholangiography (IOC)
confirmation.
Data extraction
Two reviewers independently extracted data on study
design, model type, comparator (e.g., ASGE guidelines), cohorts,
and performance metrics. Extracted values included sensitivity,
specificity, PPV, LR+, true/false positives and negatives, and
accuracy. LR+ and 95% confidence intervals (CI) were calculated
from raw data where needed. Validation cohort data were prioritized
over training data. Discrepancies were resolved by consensus. Only
clearly reported data was included. Each study was assessed for
data originality to avoid duplication.
Study quality assessment
Reviewers independently assessed the risk of bias
and applicability concerns using the Quality Assessment of
Diagnostic Accuracy Studies 2 (QUADAS-2) tool, designed for
diagnostic accuracy studies. This tool evaluates four bias domains
(patient selection, index test, reference standard, and
flow/timing) and three applicability domains. All studies were
included regardless of bias ratings, though heterogeneity due to
bias and applicability concerns was acknowledged and considered in
data synthesis and interpretation.
Statistical analysis
All statistical analyses were performed using R
Studio (Version 2020). A bivariate diagnostic random-effects
meta-analysis was conducted, with variance components estimated
using the restricted maximum likelihood (REML) method to minimize
bias. Python and the Clopper-Pearson (exact binomial) method were
used to calculate 95% CI for sensitivity when raw data were
available. Heterogeneity was assessed by calculating the
I2 and τ² statistics within the bivariate
random-effects framework, following the Zhou and Dendukuri approach
for diagnostic test accuracy (DTA) meta-analysis (26). We also visually inspected forest
plots and summary receiver operating characteristic (SROC) curves
to detect threshold effects or outliers. Because moderate
heterogeneity was present in several outcomes, we used a
random-effects model for all pooled estimates and interpreted
summary estimates with caution, emphasizing the direction rather
than the absolute magnitude of effect. SROC curves and
corresponding area under the curve (AUC) values were generated to
evaluate pooled AI model performance and to compare against 2019
ASGE guidelines. P<0.05 was considered to indicate a
statistically significant difference. Using RStudio, pooled
sensitivity, specificity plots, likelihood ratio positive (LR+) and
Deeks' asymmetry test funnel plots for publication bias were also
created.
Results
Study characteristics
Our systematic search across four databases yielded
578 articles. After screening titles, abstracts, and full texts
using predefined criteria, 20 studies were assessed for
eligibility, and 11 studies were included in the final
meta-analysis (Fig. 1). We
excluded Huerta-Reyna et al for using AI-assisted tools
outside the scope of our review (27) and Shi et al focusing on
recurrence prediction post-ERCP (28). Studies by Huang et al and
Wang et al which assessed the complexity of stone extraction
rather than CDL prediction, were also excluded (29,30).
Li et al's study focused on visualization enhancement of
biliary pathology, rather than stone prediction, which was
similarly omitted (31). Akabane
et al's large multi-center study was excluded due to missing
sensitivity, specificity, and other key diagnostic metrics
(32). Additionally, studies
applying AI solely to imaging without biochemical or clinical data
were excluded (31,33,34).
The 11 included studies encompassed over 7,000
patients evaluated with MLMs. Most studies benchmarked MLM
performance against the ASGE guidelines (2010 or 2019 versions)
(7,35), while several also compared their
models to logistic regression, support vector machines (SVMs), and
k-nearest neighbors (KNN) algorithms, enhancing data robustness.
Table I summarizes AI models,
comparators, patient numbers, sensitivity, specificity, mean age,
sex ratio, PPV, NPV, TP, TN, FN, accuracy, AUC, number of input
variables, and CBD diameter cut-off values. Input variables varied,
incorporating laboratory markers (total bilirubin (TB), alanine
aminotransferase (AST), alanine transaminase (ALT), imaging
features (CBD dilation, ductal stones), and clinical
characteristics. Seven studies compared their models against ASGE
criteria and additional classifiers such as logistic regression,
CART, and KNN.
 | Table IDetailed data characteristics of each
study, including sensitivity, specificity, AI model type,
comparator group and total number of patients. |
Table I
Detailed data characteristics of each
study, including sensitivity, specificity, AI model type,
comparator group and total number of patients.
| First author,
year | AI model | Comparator | Patient number | Sens., % | Spec., % | 95% CI Sens. | Accuracy | AUC | Variables | CBD cutoff | F/M | Mean age,
years | PPV | NPV | (Refs.) |
|---|
| Blum, 2024 | RF, LR, KNN,
XGBoost | ASGE | 222 | 86 | 72 | 0.794; 0.923 | 0.79 | 0.83 | 9 | 6 | 1.13:1 | 63.6 | 77 | 82 | (15) |
| Cohen, 2021 | MLR, ΚΝΝ | ASGE | 316 | 40.8 | 90.3 | 0.339; 0.477 | 0.72 | 0.738 | 4 | NM | 1.08:1 | 13.8 | 71.4 | 72.1 | (16) |
| Dalai, 2021 | RF | ASGE | 52 | 61.4 | 100 | 0.789; 0.896 | 0.77 | 0.791 | 9 | 9 | 1.22:1 | 46 | 100 | 32 | (17) |
| Jovanovic,
2014 | ANN | LR | 291 | 92.74 | 68.42 | 0.753; 0.847 | 0.92 | 0.844 | 10 | 7 | 1.49:1 | 63 | 92.34 | 69.64 | (18) |
| Mena-Camilo,
2024 | CNN | LR, LDA, ASGE | 292 | 96.77 | 92.86 | 0.946; 0.990 | 0.93 | 0.927 | 12 | NM | 1.94:1 | 46 | 92.86 | 72.35 | (19) |
| Floan Sachs,
2023 | Extra-Trees ML | Pediatric DUCT
Score | 1,597 | 91.3 | 93.4 | 0.881; 0.945 | 0.72 | 0.935 | 9 | NM | 2.86:1 | 13.9 | 75 | 98 | (20) |
| Steinway, 2023 | GBM | ASGE, ESGE | 1,378 | 70.3 | 72.3 | 0.673; 0.731 | 0.72 | 0.79 | 8 | 6 | 1.58:1 | 43 | 78.1 | 63.4 | (21) |
| Stojadinovic,
2015 | CART | LR | 157 | 80.9 | 94.7 | 0.581; 0.946 | 0.93 | 0.939 | 3 | 4 | 1.44:1 | 57 | 70.8 | 96.3 | (22) |
| Tranter-Entwistle,
2020 | GBM | No comparison | 1,315 | 37 | 96 | 0.458; 0.597 | 0.85 | 0.840 | 9 | NM | 1.33:1 | 70 | 67 | 87 | (23) |
| Vukicevic,
2016 | ANN | LR, DT, NB, SVM,
KNN | 303 | 88.2 | 95.8 | 0.844; 0.920 | 0.92 | 0.934 | 8 | 7 | 2.12:1 | 57 | 78.9 | 97.8 | (24) |
| Zhang, 2023 | Model Arts AI | ASGE, ESGE | 1,199 | 97 | 97 | 0.957; 0.983 | 0.97 | ~0.97 | 11 | 6 | 1.07:1 | 66 | 97.1 | 50 | (25) |
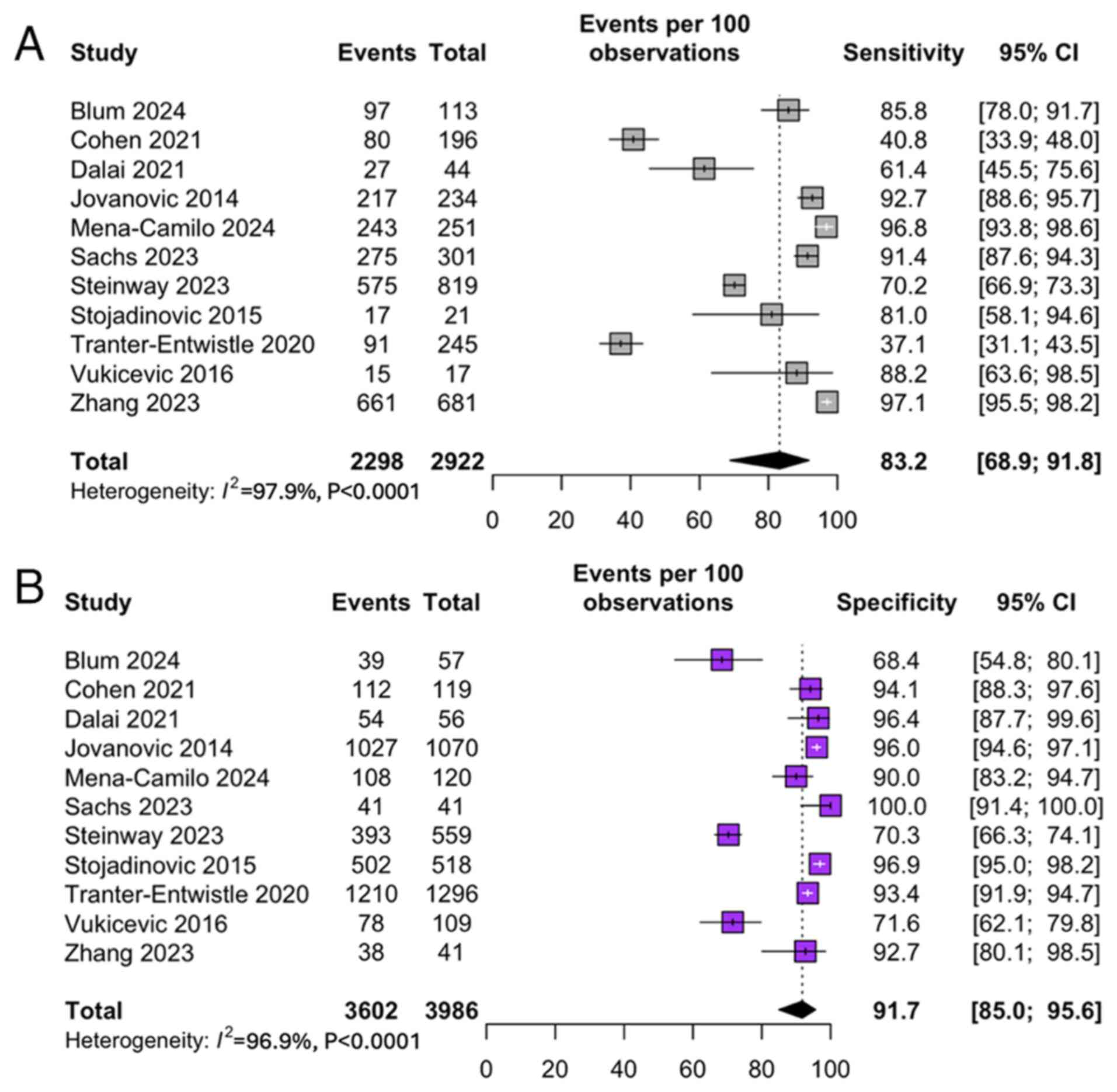
Sensitivity and specificity of AI
models
The pooled sensitivity and specificity for MLM
predicting CDL were 83.2% [95% CI: 68.9; 91.8] (Fig. 2A) and 91.1% [95% CI: 84.7; 95.0]
(Fig. 2B), respectively. These
values, calculated using a bivariate random-effects model, reflect
consistently high diagnostic performance across diverse study
settings. Individual studies, such as Blum et al and Floan
Sachs et al reported sensitivities >90% (15,20).
In contrast, others, like Cohen et al exhibited trade-offs
between sensitivity and specificity depending on the model
threshold chosen (16).
Positive predictive value and
likelihood ratio
The pooled positive likelihood ratio (LR+) from the
common-effect model was 8.39 [95% CI: 4.4; 9.5], indicating that
patients testing positive via an MLM algorithm were approximately
seven times more likely to truly have CDL compared to those who
tested negative (Fig. 3A). The
pooled PPV was 87.7 [95% CI: 77.4; 93.7] based on the calculated
PPV from the included studies (Fig.
3B).
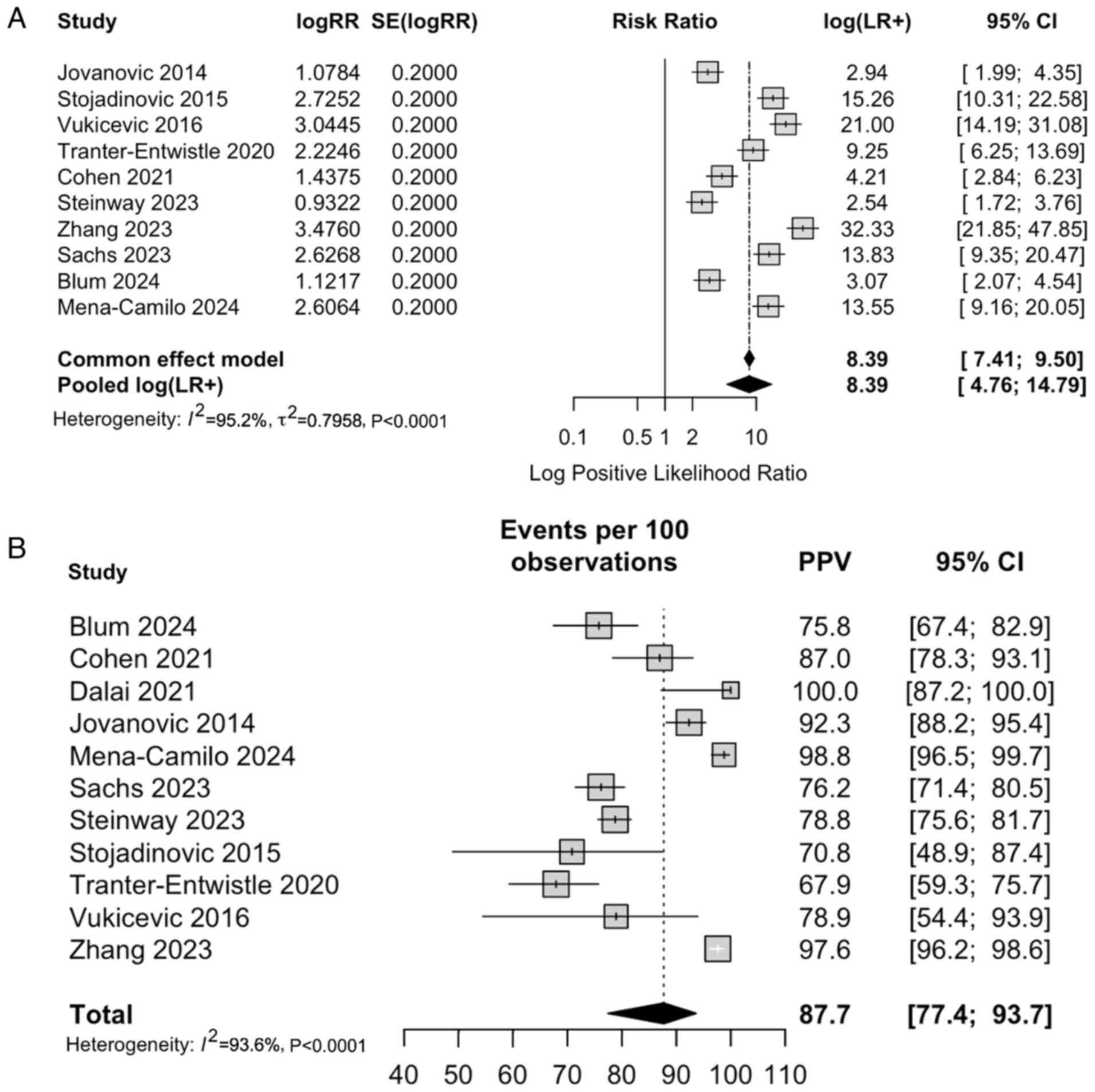 | Figure 3(A) Forest plot of LR+ for AI-based
diagnostic models across the 11 included studies (>7,000
patients), illustrating their ability to correctly ‘rule in’ CDL
when the test result is positive. The pooled LR⁺ from the
common-effect model was 8.39 (95% CI: 7.41-9.50), indicating that
AI-assisted tools substantially increase the post-test probability
of disease compared to pre-test estimates. (B) Forest plot
summarizing PPV estimates from studies evaluating AI-based
diagnostic tools for CDL. Pooled PPV was 87.7% (95% CI: 77.4-93.7),
demonstrating strong predictive performance in clinical practice.
Cis for individual studies highlight variability related to patient
selection, disease prevalence, and input variable differences, but
overall estimates indicate robust diagnostic accuracy across
diverse populations and model architectures. CDL,
choledocholithiasis; CI, confidence interval; LR+, positive
likelihood ratio; PPV, positive predictive value. AI, artificial
intelligence. |
SROC analysis
SROC curves and AUC values were generated to assess
pooled diagnostic performance across MLMs (Fig. 4A). The primary SROC curve indicated
excellent discriminative ability, confirming AI models' robust and
consistent performance across diverse study populations and
clinical settings.
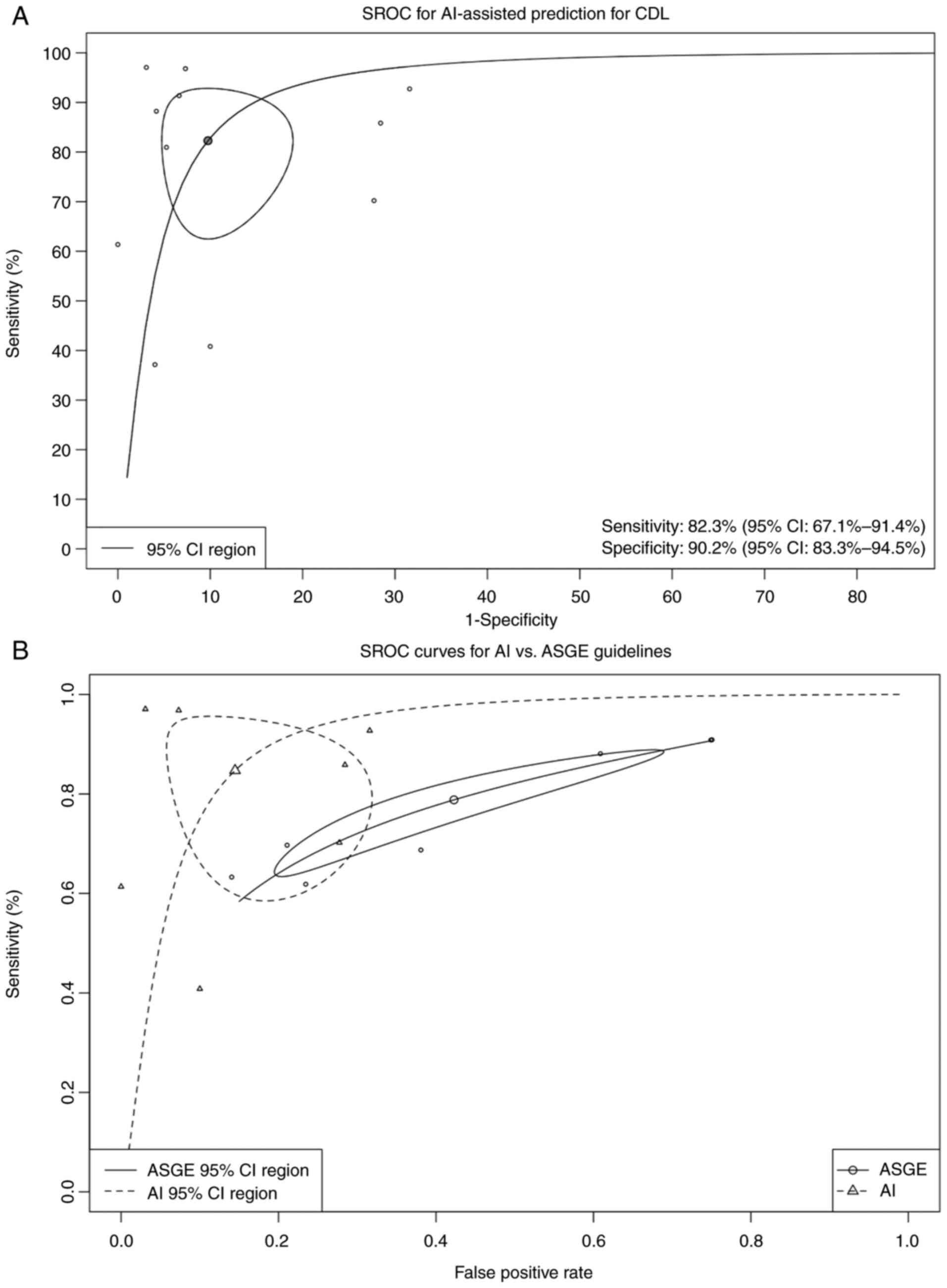 | Figure 4(A) Bivariate random-effects SROC
curve of AI-assisted diagnostic tools derived from 11 studies
involving over 7,000 individuals. The pooled AUC was 0.94 (95% CI:
0.83-0.98), indicating high discriminative performance across
diverse clinical populations. Threshold effect testing showed no
significant heterogeneity (P>0.05). Using Deeks' test, funnel
plot analysis for publication bias did not demonstrate significant
asymmetry (t=1.84, P=0.13). (B) Comparative SROC curves of
AI-assisted models vs. the 2019 ASGE high-risk criteria. AI models
demonstrated significantly higher pooled diagnostic accuracy with
an AUC of 0.91 (97.5% CI: 0.76-0.96) compared to ASGE criteria (AUC
0.76; 97.5% CI: 0.72-0.80). Mixed-effects meta-regression
(generalized linear mixed model, likelihood ratio test P=0.019)
confirmed the superior performance of AI models. Sensitivity and
specificity estimates for AI models were 86.1% (95% CI: 68.5-94.7)
and 93.3% (95% CI: 71.4-98.7), respectively, compared to ASGE
guideline performance of 78.4% (95% CI: 67.5-86.4) and 57.5% (95%
CI: 37.3-75.6). CDL, choledocholithiasis; SROC, summary receiver
operating characteristic; AI, artificial intelligence; AUC, area
under the curve; CI, confidence interval; ASGE, American Society
for Gastrointestinal Endoscopy. |
Subgroup analysis: AI-models vs. ASGE
guidelines
Seven studies directly compared their MLM to the
2019 ASGE risk stratification criteria. Pooled sensitivity and
specificity values were used to generate SROC curves for both
models. As shown in Fig. 4B, MLMs
consistently achieved higher AUC values and diagnostic accuracy
than ASGE high-risk criteria. This suggests that AI may enhance
precision and reduce reliance on subjective guideline
interpretation in pre-ERCP decision-making.
In bivariate random-effects meta-analysis, MLMs
achieved an AUC of 0.915 (95% CI: 0.763-0.961) compared to 0.766
(95% CI: 0.7;-0.7) for ASGE guidelines. The absolute difference in
AUC (ΔAUC=0.148) reflects a clinically meaningful improvement
favoring AI tools. Although the 95% CI for this difference (-0.002
to 0.205) includes zero, the P-value (0.053) indicates a trend
toward statistical significance. The pooled sensitivity for AI
models was 82.3%, with a false positive rate of 9.8%. Heterogeneity
across studies ranged from 52.4% (Zhou and Dendukuri method) to
97.8% (Holling unadjusted).
Quality assessment for bias
QUADAS-2 tool evaluation identified ‘some concerns’
in several studies, mainly in patient selection (convenience
sampling, exclusion of cholangitis, etc.) and flow/timing (delayed
or unclear index/reference timing) domains (Fig. 5). Specifically, Tranter-Entwistle
et al faced issues with missing clinical data, 36%
misclassification, and manual diagnosis reassignment (23). Temporal alignment of index and
reference tests was inconsistently reported. Steinway et al
lacked clarity regarding blinding details and applied a
retrospective cohort design, excluding cholangitis, potentially
limiting generalizability (21).
Mena-Camilo et al despite ERCP-confirmed diagnoses,
demonstrated significant class imbalance (86% positive) and heavy
reliance on imputed clinical data, raising data integrity concerns
(19). QUADAS-2 revealed moderate
risk biases likely inflated accuracy estimates in some models by
limiting negative cases or excluding diagnostically challenging
scenarios. Therefore, while AI models demonstrated strong pooled
performance, conclusions should be interpreted with caution given
these study-level limitations.
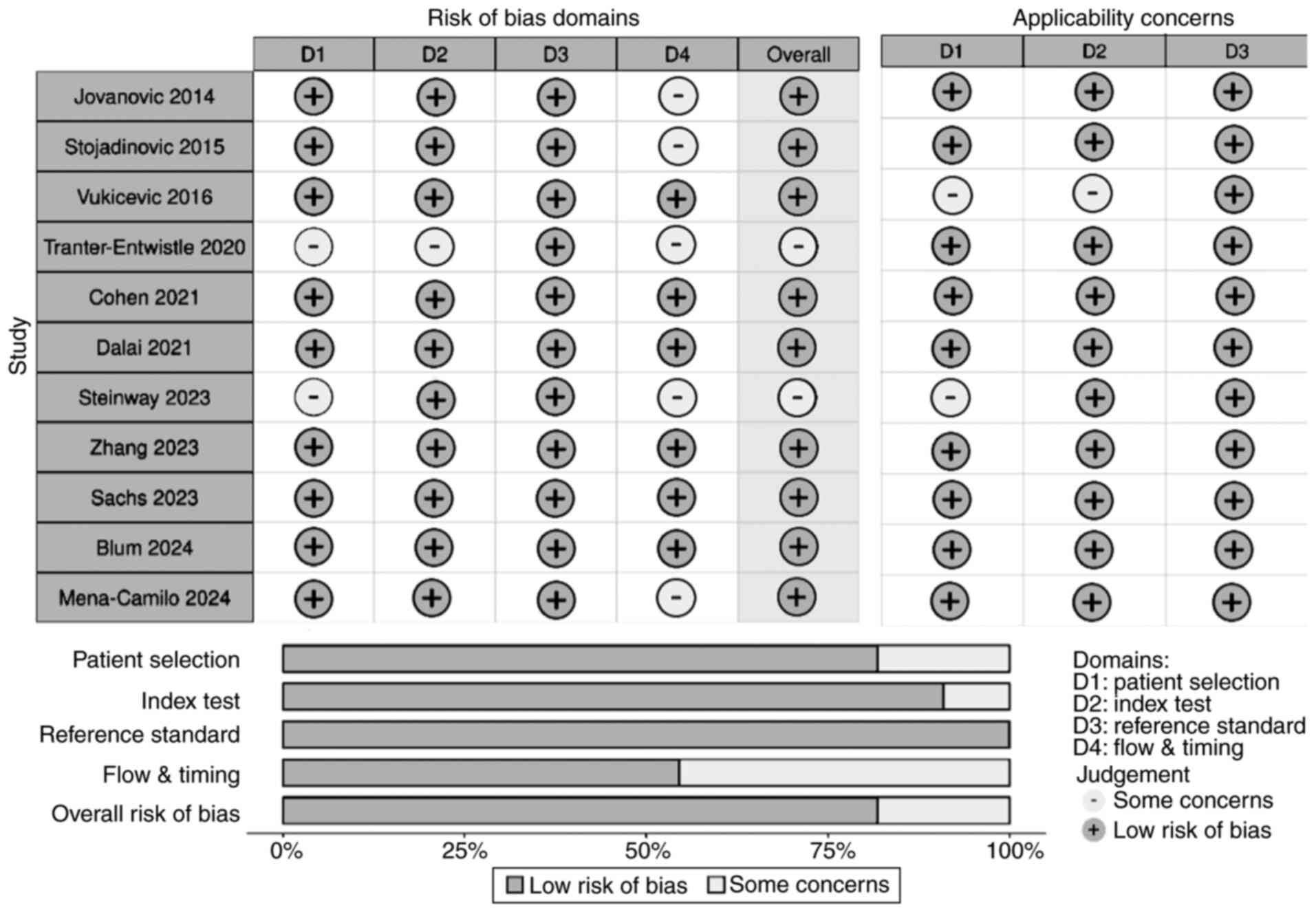 | Figure 5The Quality Assessment of Diagnostic
Accuracy Studies 2 tool was used to evaluate risk of bias and
applicability concerns for the 11 included studies (>7,000
patients). The round-shaped plot (top) summarizes the judgment of
each study across four bias domains, patient selection, index test,
reference standard, and flow/timing, and three applicability
domains, using grey (low risk), and white (some concerns). No areas
of high risk were noted. The summary plot (bottom) depicts the
overall proportion of studies rated at low risk or with some
concerns within each domain. Overall, most studies demonstrated low
risk in the index test and reference standard domains, while
patient selection and flow/timing presented the most frequent
concerns, primarily due to retrospective designs, incomplete
clinical data, or unclear temporal alignment of index and reference
tests. Applicability concerns were minimal for most studies,
reflecting consistent patient populations and appropriate test
applications. These results indicate an overall moderate to high
quality of included studies, supporting the robustness of the
meta-analysis findings. |
Discussion
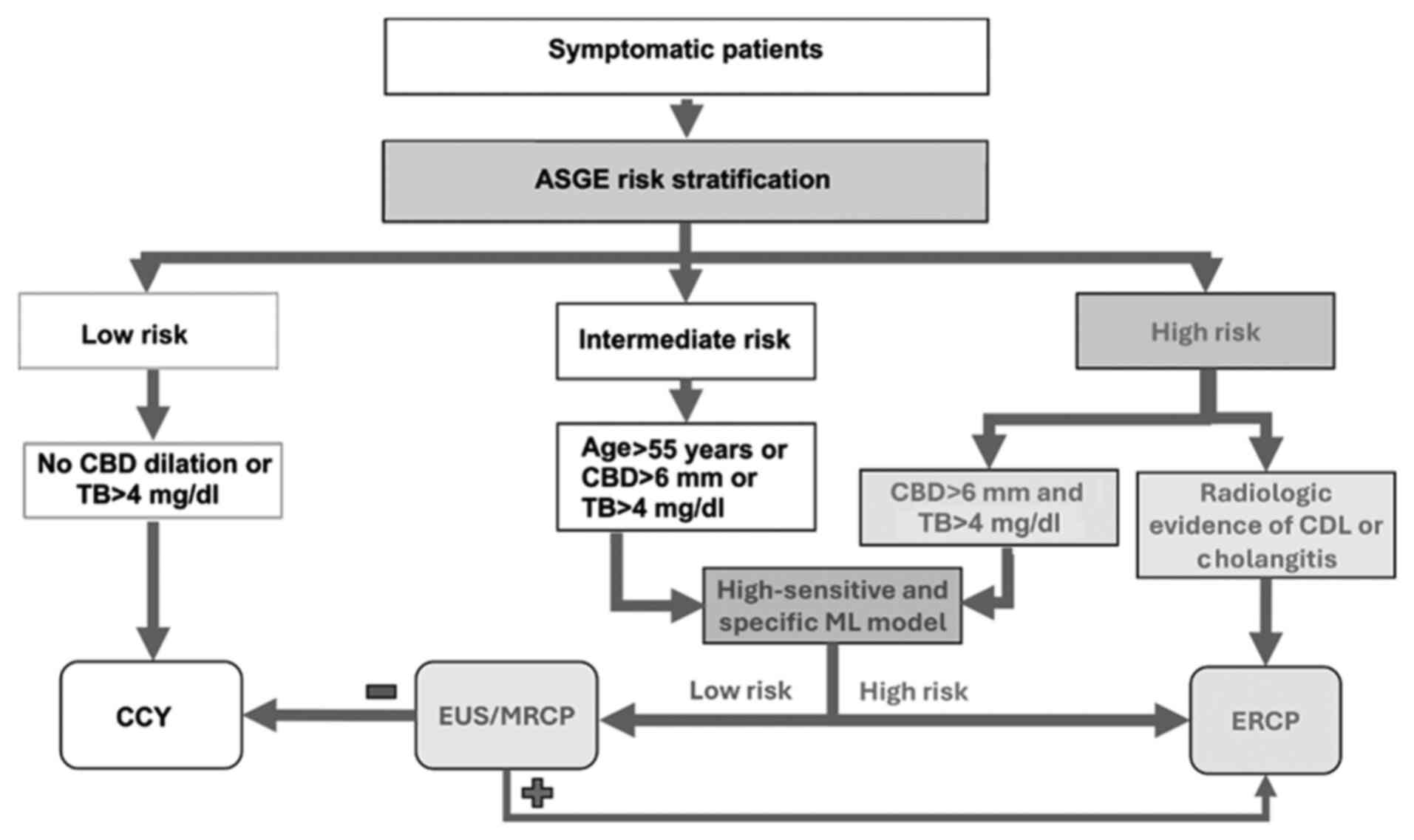
Accurate stratification of patients with suspected
CDL remains clinically challenging. To improve specificity and
reduce unnecessary ERCPs, the 2019 ASGE guidelines revised
high-risk criteria to include only patients with both TB >4
mg/dl and CBD dilation on imaging (Fig. 6) (6). This change, supported by prior
studies showing improved specificity with these combined features
(36,37), raised specificity from 55% under
the 2010 criteria to approximately 80% (38). The 2019 update also incorporated
computed tomography (CT) alongside ultrasonography (USG) for risk
stratification (6). Per the
current guidelines, in the absence of definitive radiologic
evidence of CDL, ERCP is recommended for patients with a suggestive
clinical presentation, elevated liver enzymes, CBD dilation >6
mm, TB >4 mg/dl, or signs of cholangitis (Fig. 6). Those not meeting high-risk
criteria are advised to undergo magnetic resonance
cholangiopancreatography (MRCP) or EUS (Fig. 6) (6). While MRCP is a sensitive and specific
tool (39), its overuse may
prolong hospital stays and increase costs (40). Despite refinements, current
guidelines lack tools for quantitative CDL risk estimation. Our
meta-analysis is the first comprehensive synthesis of AI-assisted
prediction models for CDL, highlighting their potential for
non-invasive risk stratification and optimized ERCP selection
(Fig. 6).
The reviewed studies employed diverse MLMs,
including traditional and DL approaches like ANNs, CNNs, and GBMs,
and supervised learning algorithms such as classification and
regression trees (CART), logistic regression, KNN, Naïve Bayes
(NB), and Scikit-learn frameworks Several models demonstrated
superior performance: Stojadinovic et al found that CART
outperformed logistic (22);
Tranter-Entwistle et al reported that GBM outperformed
random forest and KNN (23); Dalai
et al tested GLM, SVM, and random forest, with the latter
performing best and included in our meta-analysis (17). Cohen et al utilized a
KNN-based model (16), while Zhang
et al developed the ModelArts ExeML tool (Huawei),
outperforming logistic regression, LDA, QDA, NB, KNN, and Ranger
(25). Floan Sachs et al
applied Extra-Trees to a 9-feature pediatric model, outperforming
the 3-feature Pediatric DUCT score (20). Blum et al compared logistic
regression, XGBoost, random forest, and KNN, with random forest
performing best in adults with suspected CDL (15).
ANNs were among the earliest tools. Jovanovic et
al introduced an ANN-based model (18), while Vukicevic et al
enhanced it by automating configuration using genetic algorithms,
outperforming logistic regression, decision trees, NB, SVMs, and
KNN (24). Mena-Camilo et
al applied a CNN-based model that exceeded logistic regression
and LDA in sensitivity, supporting DL's role in non-invasive CDL
diagnosis (19).
Our meta-analysis of 7,000+ subjects across 11
studies showed strong predictive performance of AI-assisted models,
with a pooled sensitivity of 83.2%, specificity of 91.1% and a PPV
of 87.7% [77.4; 93.7]. Despite heterogeneity in models, patient
populations, and predictive variables, consistently high accuracy
supports AI as a non-invasive diagnostic tool and underscores its
adaptability, especially where guidelines fall short. A key source
of heterogeneity is in training/validation cohorts, impacting
generalizability. Jovanovic et al excluded patients with
prior CCY, cholangitis, and primary sclerosing cholangitis,
achieving high AUC (0.884) and sensitivity (92.7%) but lower
specificity (68.7%) due to few ERCP-negative cases and low NPV
(18).
Other studies also assessed high-suspicion cohorts
undergoing ERCP or IOC, excluding cholangitis cases (15,21).
Dalai et al focused on Hispanic/Latino patients with prior
CCY (17). Again, low
ERCP-negative rates skewed the sample distribution. Blum et
al applying similar criteria, confirmed stones in only 50% of
cases (15). Mena-Camilo et
al trained a CNN regardless of CCY status (19); Zhang et al used MRCP or IOC
for diagnosis, excluding cholangitis (25). Tranter-Entwistle et al found
CDL in 19% of acute biliary cases, with their GBM model showing low
sensitivity (37%) and high specificity (96%) due to missing data
imputation (23). In elective
surgical populations, Stojadinovic et al found 37% CDL
prevalence; their CART model exceeded logistic regression in
sensitivity (90.9% vs. 57.2%) but not specificity (92.3% vs. 94.7%)
(22). Similarly, Vukicevic et
al's ANN achieved 88.2% sensitivity and 95.8% specificity among
CCY patients with IOC (24).
Although CDL is typically adult-onset, pediatric
cases are rising with obesity (41-43).
Cohen et al evaluated suspected pediatric CDL using
IOC/ERCP, prioritizing PPV by setting 90% specificity, which
yielded only 40.8% sensitivity and 71.5% accuracy, marking it an
outlier (16). Floan Sachs et
al used pediatric elective CCY cases, with CDL confirmed via
MRCP, ERCP, and/or IOC. Their 9-feature model outperformed the
3-feature Pediatric DUCT score (AUC 0.957 vs. 0.935; sensitivity
91.3% vs. 78.3%), while maintaining specificity (20), estimating only 8.7% of cases
missed, compared to 21.7% with the 3-feature model. These results
support ML's role in optimizing ERCP in children, with performance
thresholds that are unable to meet institutional needs.
Input variables ranged from 3 to 53 (Table I), typically combining biochemical
laboratory data and imaging. USG and CT were common, while MRCP was
underused. Dalai et al showed models incorporating CT/MRCP
had higher AUC than USG alone (17). Cohen et al excluded CBD
stone presence on USG due to its high predictive value; univariate
analysis identified (AST), CBD diameter, and ductal stone presence
on USG as key predictors (16).
Common biochemical values in all studies included TB, AST, ALT, and
alkaline phosphatase (ALP). Few used follow-up biochemical values.
Steinway et al found no benefit from serial labs (21). Stojadinovic et al emphasized
cystic duct diameter (>4 mm), elevated ALP, and multiple small
gallstones (‘dangerous’ stones) as key predictors, though their
elective CCY cohort may not reflect acute cases (22). Jovanovic et al (18) found lipase and CBD diameter to be
influential, with lipase and AST negatively, and CBD diameter
positively associated with CDL, supporting the theory that
pancreatitis may follow stone passage rather than concurrent CDL
(6,18,22).
Seven studies directly compared MLMs to the 2019
ASGE guidelines; others used published ASGE performance data for
benchmarking. MLMs consistently outperformed current guidelines,
particularly in high-suspicion cases (Table I) (18,19,21).
Most models achieved high sensitivity, except Tranter-Entwistle
et al (23) and Cohen et
al (16), which prioritized
specificity above 90%, reducing sensitivity. Most models reported
>90% accuracy, though accuracy can be prevalence-dependent.
Large sample sizes strengthen these conclusions. Integrating MLMs
may enhance individualized CDL risk stratification.
Steinway et al's GBM achieved sensitivity and
specificity above 70%, outperforming ASGE (61.9/62.8%) and European
Society of Gastrointestinal Endoscopy (ESGE) (46.9/86.3%), avoiding
22% of unnecessary ERCPs and identifying 48% of true positives
missed by ESGE guidelines (21).
Dalai et al's best-performing model (77.3% sensitivity, 75%
specificity) outperformed the ASGE's high-likelihood group, which
had higher sensitivity (90.3%) but lower specificity (25%)
(17). Threshold tuning raised
sensitivity to 97.7%, illustrating MLM flexibility. Blum et
al found Random Forest best among four classifiers (AUC 0.83,
sensitivity 94%), exceeding ASGE (15). Zhang et al reported ASGE's
high sensitivity (98.5%) but poor specificity, while their
ModelArts ExML tool achieved AUCs of 0.77-0.81(25). Jovanovic et al's ANN model
(AUC 0.884) surpassed logistic regression (AUC 0.752), despite
excluding several ASGE criteria (18). Cohen et al's model
prioritized specificity in pediatrics, achieving 40.8% sensitivity
but outperforming ASGE in specificity (16).
This meta-analysis showed higher accuracy for MLMs
(AUC 0.91) vs. ASGE 2019 guidelines (AUC 0.72). Unlike ASGE's fixed
risk categories, MLMs may provide individualized risk
stratification. While ASGE guidelines remain a valuable risk
stratification tool, their broad criteria often underperform in
real-world populations (6). AI
models offer personalization, especially in low-resource settings
or special populations like pediatrics (16,20).
High-specificity models reduce unnecessary ERCPs; high-sensitivity
models expedite referrals. From a therapeutic standpoint, these
tools may refine patient selection for ERCP, particularly in
intermediate/high-risk patients lacking definitive findings. In
such cases, MLMs could guide decisions between MRCP and ERCP. The
findings also open avenues for developing real-time, bedside
decision-support tools trained on multicenter data and embedded
within electronic medical records. Though AI tool thresholds may
vary, high sensitivity supports timely referral in community
hospitals, while high specificity aids triage in tertiary centers.
ASGE guidelines recommend direct CCY for low-risk patients, yet
some harbor occult stones (6).
While the benefit of pre-CCY ERCP is debated, AI may identify
low-risk patients needing further evaluation (6,44-46).
These findings are consistent with broader AI applications in
gastroenterology, including colorectal polyp and neoplasia
detection or classification (11,12,47,48),
highlighting the potential of AI-based tools to improve diagnostic
accuracy and efficiency. However, extrapolation across clinical
settings remains challenging, underscoring the need for external
validation and standardized performance reporting. Integrating AI
into clinical workflows may reduce unnecessary testing, optimize
resources, and improve outcomes (49).
This study supports the diagnostic potential of AI
tools for CDL, but several limitations exist. We identified 11
distinct AI models using varied input data: labs, imaging, clinical
features, introducing heterogeneity that hinders direct comparisons
and pooled estimates. A key limitation is the lack of prospective
validation; most studies were retrospective, single-centered,
limiting generalizability. Homogeneous populations and limited
external validation further constrain applicability. Comparisons
were inconsistent, using differing ASGE guidelines, impairing
uniform benchmarking. Future studies should prioritize standardized
comparisons using current ASGE criteria and diverse cohorts. Some
studies lacked key statistics (e.g., CI, PPV, NPV), and unclear
training-validation cohort separation may have skewed performance.
In several cases, unclear differentiation between training and
validation cohorts may have distorted performance outcomes. These
issues underscore the need for prospective validation and
standardized reporting to enable clinical adoption.
In conclusion, this study highlights that ML tools
demonstrate promising diagnostic potential for CDL detection and
may enhance current or future guideline-based risk stratification.
While pooled sensitivity and specificity were high, heterogeneity,
reliance on retrospective data, and a somewhat limited prospective
validation constrain definitive conclusions. AI-based tools may
complement existing guidelines via a non-invasive accurate
alternative in refining patient selection for ERCP, enhancing both
the safety and efficiency of care, especially in acute and
resource-limited settings. Future work should prioritize validation
in prospective multicenter studies, ensure model generalizability,
and support integration into clinical workflows before clinical
implementation.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
PGD, SGD and AB conceived and designed the study.
PGD and SGD performed literature search, data analysis, and
interpretation of data and results. PGD and SGD confirm the
authenticity of all the raw data. PGD, SGD and AB prepared the
draft manuscript and critically revised the manuscript. AB
supervised the study. All authors reviewed the results, and read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
Panagiotis G. Doukas: https://orcid.org/0000-0001-8050-963X. Sotirios G.
Doukas: https://orcid.org/0000-0002-1840-659X.
References
|
1
|
Tazuma S, Unno M, Igarashi Y, Inui K,
Uchiyama K, Kai M, Tsuyuguchi T, Maguchi H, Mori T, Yamaguchi K, et
al: Evidence-based clinical practice guidelines for cholelithiasis
2016. J Gastroenterol. 52:276–300. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Unalp-Arida A and Ruhl CE: Burden of
gallstone disease in the United States population: Prepandemic
rates and trends. World J Gastrointest Surg. 16:1130–1148.
2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li S, Guizzetti L, Ma C, Shaheen AA, Dixon
E, Ball C, Wani S and Forbes N: Epidemiology and outcomes of
choledocholithiasis and cholangitis in the United States: Trends
and Urban-rural variations. BMC Gastroenterol.
23(254)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Molvar C and Glaenzer B:
Choledocholithiasis: Evaluation, treatment, and outcomes. Semin
Intervent Radiol. 33:268–276. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Morales SJ, Sampath K and Gardner TB: A
review of prevention of Post-ERCP pancreatitis. Gastroenterol
Hepatol (N Y). 14:286–292. 2018.PubMed/NCBI
|
|
6
|
ASGE Standards of Practice Committee.
Buxbaum JL, Abbas Fehmi SM, Sultan S, Fishman DS, Qumseya BJ,
Cortessis VK, Schilperoort H, Kysh L, Matsuoka L, et al: ASGE
guideline on the role of endoscopy in the evaluation and management
of choledocholithiasis. Gastrointest Endosc. 89:1075–1105.e15.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
ASGE Standards of Practice Committee.
Maple JT, Ben-Menachem T, Anderson MA, Appalaneni V, Banerjee S,
Cash BD, Fisher L, Harrison ME, Fanelli RD, et al: The role of
endoscopy in the evaluation of suspected choledocholithiasis.
Gastrointest Endosc. 71:1–9. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tunruttanakul S, Chareonsil B, Verasmith
K, Patumanond J and Mingmalairak C: Evaluation of the American
society of gastrointestinal endoscopy 2019 and the european society
of gastrointestinal endoscopy guidelines' performances for
choledocholithiasis prediction in clinically suspected patients: A
retrospective cohort study. JGH Open. 6:434–440. 2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Rajpurkar P, Chen E, Banerjee O and Topol
EJ: AI in health and medicine. Nat Med. 28:31–38. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kröner PT, Engels MM, Glicksberg BS,
Johnson KW, Mzaik O, van Hooft JE, Wallace MB, El-Serag HB and
Krittanawong C: Artificial intelligence in gastroenterology: A
state-of-the-art review. World J Gastroenterol. 27:6794–824.
2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Byrne MF, Chapados N, Soudan F, Oertel C,
Linares Pérez M, Kelly R, Iqbal N, Chandelier F and Rex DK:
Real-time differentiation of adenomatous and hyperplastic
diminutive colorectal polyps during analysis of unaltered videos of
standard colonoscopy using a deep learning model. Gut. 68:94–100.
2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Korbar B, Olofson AM, Miraflor AP, Nicka
CM, Suriawinata MA, Torresani L, Suriawinata AA and Hassanpour S:
Deep learning for classification of colorectal polyps on
Whole-slide images. J Pathol Inform. 8(30)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
An Q, Rahman S, Zhou J and Kang JJ: A
comprehensive review on machine learning in healthcare industry:
Classification, restrictions, opportunities and challenges. Sensors
(Basel). 23(4178)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Trezza A, Visibelli A, Roncaglia B, Spiga
O and Santucci A: Unsupervised learning in precision medicine:
Unlocking personalized healthcare through AI. Appl Sci.
14(9305)2024.
|
|
15
|
Blum J, Hunn S, Smith J, Chan FY and
Turner R: Using artificial intelligence to predict
choledocholithiasis: Can machine learning models abate the use of
MRCP in patients with biliary dysfunction? ANZ J Surg.
94:1260–1265. 2024.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Cohen RZ, Tian H, Sauer CG, Willingham FF,
Santore MT, Mei Y and Freeman AJ: Creation of a pediatric
choledocholithiasis prediction model. J Pediatr Gastroenterol Nutr.
73:636–641. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Dalai C, Azizian J, Trieu H, Rajan A, Chen
F, Dong T, Beaven S and Tabibian JH: Machine learning models
compared to existing criteria for noninvasive prediction of
endoscopic retrograde cholangiopancreatography-confirmed
choledocholithiasis. Liver Res. 5:224–231. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jovanovic P, Salkic NN and Zerem E:
Artificial neural network predicts the need for therapeutic ERCP in
patients with suspected choledocholithiasis. Gastrointest Endosc.
80:260–268. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mena-Camilo E, Salazar-Colores S,
Aceves-Fernández MA, Lozada-Hernández EE and Ramos-Arreguín JM:
Non-invasive prediction of choledocholithiasis using 1D
convolutional neural networks and clinical data. Diagnostics.
14(1278)2024.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Floan Sachs G, Ourshalimian S, Jensen AR,
Kelley-Quon LI, Padilla BE, Shew SB, Lofberg KM, Smith CA, Roach
JP, Pandya SR, et al: Machine learning to predict pediatric
choledocholithiasis: A western pediatric surgery research
consortium retrospective study. Surgery. 174:934–939.
2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Steinway SN, Tang B, Telezing J, Ashok A,
Kamal A, Yu CY, Jagtap N, Buxbaum JL, Elmunzer J, Wani SB, et al: A
machine learning-based choledocholithiasis prediction tool to
improve ERCP decision making: A proof-of-concept study. Endoscopy.
56:165–171. 2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Stojadinovic MM and Pejovic T: Regression
tree for choledocholithiasis prediction. Eur J Gastroenterol
Hepatol. 27:607–613. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tranter-Entwistle I, Wang H, Daly K,
Maxwell S and Connor S: The challenges of implementing artificial
intelligence into surgical practice. World J Surg. 45:420–428.
2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vukicevic AM, Stojadinovic M, Radovic M,
Djordjevic M, Cirkovic BA, Pejovic T, Jovicic G and Filipovic N:
Automated development of artificial neural networks for clinical
purposes: Application for predicting the outcome of
choledocholithiasis surgery. Comput Biol Med. 75:80–89.
2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang H, Gao J, Sun Z, Zhang Q, Qi B,
Jiang X, Li S and Shang D: Diagnostic accuracy of updated risk
assessment criteria and development of novel computational
prediction models for patients with suspected choledocholithiasis.
Surg Endosc. 37:7348–7357. 2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou Y and Dendukuri N: Statistics for
quantifying heterogeneity in univariate and bivariate meta-analyses
of binary data: The case of Meta-analyses of diagnostic accuracy.
Stat Med. 33:2701–2717. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Huerta-Reyna R, Guevara-Torres L,
Martínez-Jiménez MA, Armas-Zarate F, Aguilar-García J,
Waldo-Hernández LI and Martínez-Martínez MU: Development and
validation of a predictive model for choledocholithiasis. World J
Surg. 48:1730–1738. 2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Shi Y, Lin J, Zhu J, Gao J, Liu L, Yin M,
Yu C, Liu X, Wang Y and Xu C: Predicting the recurrence of common
bile duct stones after ERCP treatment with automated machine
learning algorithms. Dig Dis Sci. 68:2866–2877. 2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Huang L, Lu X, Huang X, Zou X, Wu L, Zhou
Z, Wu D, Tang D, Chen D, Wan X, et al: Intelligent difficulty
scoring and assistance system for endoscopic extraction of common
bile duct stones based on deep learning: Multicenter study.
Endoscopy. 53:491–498. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang Z, Yuan H, Lin K, Zhang Y, Xue Y, Liu
P, Chen Z and Wu M: Artificial intelligence-empowered assessment of
bile duct stone removal challenges. Expert Systems Applications.
258(125146)2024.
|
|
31
|
Li D, Du B, Shen Y and Ge L: Artificial
Intelligence-assisted visual sensing technology under duodenoscopy
of gallbladder stones. J Sensors. 2021(5158577)2021.
|
|
32
|
Akabane S, Iwagami M, Bell-Allen N,
Navadgi S, Kawahara T and Bhandari M: Machine learning-based
prediction for incidence of endoscopic retrograde
cholangiopancreatography after emergency laparoscopic
cholecystectomy: A retrospective, multicenter cohort study. Surg
Endosc. 39:1770–1777. 2025.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hou JU, Park SW, Park SM, Park DH, Park CH
and Min S: Efficacy of an artificial neural network algorithm based
on Thick-slab magnetic resonance cholangiopancreatography images
for the automated diagnosis of common bile duct stones. J
Gastroenterol Hepatol. 36:3532–3540. 2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sun K, Li M, Shi Y, He H, Li Y, Sun L,
Wang H, Jin C, Chen M and Li L: Convolutional neural network for
identifying common bile duct stones based on magnetic resonance
cholangiopancreatography. Clin Radiol. 79:553–558. 2024.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Singhvi G, Ampara R, Baum J and Gumaste V:
ASGE guidelines result in Cost-saving in the management of
choledocholithiasis. Ann Gastroenterol. 29:85–90. 2016.PubMed/NCBI
|
|
36
|
Kang J, Paik KH, Lee JC, Kim HW, Lee J,
Hwang JH and Kim J: The efficacy of clinical predictors for
patients with intermediate risk of choledocholithiasis. Digestion.
94:100–105. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
He H, Tan C, Wu J, Dai N, Hu W, Zhang Y,
Laine L, Scheiman J and Kim JJ: Accuracy of ASGE high-risk criteria
in evaluation of patients with suspected common bile duct stones.
Gastrointest Endosc. 86:525–532. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jacob JS, Lee ME, Chew EY, Thrift AP and
Sealock RJ: Evaluating the revised american society for
gastrointestinal endoscopy guidelines for common bile duct stone
diagnosis. Clin Endosc. 54:269–274. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lee H, Song T, Park DH, Lee SS, Seo DW,
Lee SK, Kim MH, Jun JH, Moon JE and Song YH: Diagnostic performance
of the current Risk-stratified approach with computed tomography
for suspected choledocholithiasis and its options when negative
finding. Hepatobiliary Pancreat Dis Int. 18:366–372.
2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Anand G, Yeh HC, Khashab M, Kim KJ, Lennon
AM, Shin EJ, Canto M, Okolo PI, Kalloo AN and Singh VK: Mo1582
Patterns of MRCP Utilization prior to ERCP among patients at high
risk for choledocholithiasis. Gastrointestinal Endoscopy.
73:AB393–AB394. 2011.
|
|
41
|
Tuna Kirsaclioglu C, Çuhacı Çakır B,
Bayram G, Akbıyık F, Işık P and Tunç B: Risk factors, complications
and outcome of cholelithiasis in children: A retrospective,
single-centre review. J Paediatr Child Health. 52:944–949.
2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Frybova B, Drabek J, Lochmannova J, Douda
L, Hlava S, Zemkova D, Mixa V, Kyncl M, Zeman L, Rygl M and Keil R:
Cholelithiasis and choledocholithiasis in children; risk factors
for development. PLoS One. 13(e0196475)2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Murphy PB, Vogt KN, Winick-Ng J, McClure
JA, Welk B and Jones SA: The increasing incidence of gallbladder
disease in children: A 20year perspective. J Pediatr Surg.
51:748–752. 2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lascia AD, Tartaglia N, Pavone G, Pacilli
M, Ambrosi A, Buccino RV, Petruzzelli F, Menga MR, Fersini A and
Maddalena F: One-step versus two-step procedure for management
procedures for management of concurrent gallbladder and common bile
duct stones. Outcomes and cost analysis. Ann Ital Chir. 92:260–207.
2021.PubMed/NCBI
|
|
45
|
Jones M, Johnson M, Samourjian E, Slauch K
and Ozobia N: ERCP and laparoscopic cholecystectomy in a combined
(one-step) procedure: A random comparison to the standard
(two-step) procedure. Surg Endosc. 27:1907–1912. 2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
de Medeiros KS, Aragão Fernandes AC, Fulco
Gonçalves G, Villarim CVO, Costa E Silva LC, de Sousa VMC, Meneses
Rêgo AC and Araújo-Filho I: Cholecystectomy before, simultaneously,
or after ERCP in patients with acute cholecystitis: A protocol for
systematic review and/or meta analysis. Medicine (Baltimore).
101(e30772)2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ebigbo A, Messmann H and Lee SH:
Artificial intelligence applications in Image-based diagnosis of
early esophageal and gastric neoplasms. Gastroenterology.
169:396–415.e2. 2025.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sharma P and Hassan C: Artificial
intelligence and deep learning for upper gastrointestinal
neoplasia. Gastroenterology. 162:1056–1066. 2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Alhejaily AG: Artificial intelligence in
healthcare (review). Biomed Rep. 22(11)2024.PubMed/NCBI View Article : Google Scholar
|