Introduction
Ovarian reserve, reflecting the female reproductive
capacity, refers to the quantity and quality of follicles at
various developmental stages (1,2). A
significant reduction in baseline antral follicle count (AFC) and
anti-Müllerian hormone (AMH) levels, coupled with an elevation in
basal follicle-stimulating hormone (FSH), indicates decreased
ovarian reserve (DOR) (3). Women
with DOR exhibit impaired ovarian function, manifested by
suboptimal responses to ovarian stimulation, decreased oocyte
retrieval and higher cycle cancellation rates, all contributing to
a lowered probability of successful conception (4). The impact of DOR on fertility has
become increasingly pronounced with the delay of reproductive age.
The etiology of DOR is complex, involving age, genetics, immune
factors and environmental exposures (5). For example, Biallelic germline BRCA1
frameshift mutations associated with isolated DOR (6); Dehydroepiandrosterone regulates the
balance of CD4+/CD8+ T cells to improve balance of CD4+/CD8+ T
cells with DOR (7); A history of
heavy smoking may increase risk of DOR (8). Despite advances, the precise
molecular mechanisms underlying DOR remain incompletely understood,
highlighting the need to unravel the molecular pathways implicated
in DOR and identify potential therapeutic targets.
Small extracellular vesicles (referred to as
exosomes in the present study) play a key role in intercellular
communication by transferring diverse molecules, including
proteins, nucleic acids and metabolites (9,10).
Increasing evidence highlights their function as central mediators
of extracellular signaling, with substantial implications for
biological processes, such as insulin signaling, lipolysis and
inflammation (11,12). Exosomes have been extensively
studied in the context of female reproductive disorders (13), such as polycystic ovary syndrome
(PCOS) (14,15), intrauterine adhesion (16), premature ovarian insufficiency
(POI) (17) and premature ovarian
failure (18). However, the
expression of exosome-derived circular RNAs (circRNAs) in DOR
follicles remains underexplored. Oxidative stress, caused by an
imbalance between pro-oxidants, such as reactive oxygen species
(ROS) and nitrogen species, and the body's antioxidant defenses,
can result in cellular damage to DNA, proteins and lipids (19). This imbalance is a recognized
contributor to fertility decline in both sexes (20) and plays a marked role in ovarian
aging (21). Huang et al
(22) observed a notable decrease
in the oxidative stress marker glutathione in follicular fluid from
individuals with DOR compared with that in healthy controls.
Despite these observations on oxidative stress in DOR, the precise
mechanisms by which circRNAs participate in regulation remain
poorly understood and are the subject of ongoing research.
To the best of our knowledge, no previous studies
have explored the alterations in exosomal circRNA profiles in the
follicular fluid of individuals with DOR. The present study
represents the first attempt to assess the variations in circRNA
expression within exosomes from the follicular fluid of patients
with DOR. The current study aimed to evaluate the influence of key
differentially expressed circRNAs on granulosa cells in DOR in
terms of cell viability and oxidative stress, in an effort to
identify potential therapeutic targets for DOR.
Materials and methods
Clinical samples
In total, 6 women diagnosed with DOR and 6
age-matched control women (median age, 32 years; range, 31-34
years) were recruited from Fujian Maternity and Child Health
Hospital (Fuzhou, China), with ethics approval granted by the
Ethics Committee of Fujian Maternity and Child Health Hospital
(approval no. 2023KYLLR01082). Controls were women who presented
for a routine fertility check-up or preconception consultation and
were found to have no significant infertility or known fertility
issues. All participants provided written informed consent. All
patients presented to the hospital between January 2020 and
December 2020. The subjects met the diagnostic criteria for DOR
based on AMH <1.1 ng/ml and AFC <5 on transvaginal ultrasound
(23,24). Exclusion criteria included any
history of medication affecting glucose or lipid metabolism, as
well as known conditions that cause hormonal, metabolic and
reproductive abnormalities, such as Cushing's syndrome,
endometriosis, congenital adrenal hyperplasia or androgen-secreting
tumors. Preovulatory follicular fluid was collected from
participants during oocyte retrieval, and samples were subsequently
stored at -80˚C for RNA extraction and exosome isolation. The
clinical characteristics of patients with DOR were obtained from
medical records.
Exosome isolation
To identify differentially expressed circRNAs in
exosomes of individuals with DOR and normal controls, exosomes were
isolated from the follicular fluid of both groups using
ultracentrifugation, as previously described (25). In brief, 500 µl follicular fluid
was centrifuged at 10,000 x g for 45 min at 4˚C. The supernatant
was filtered through a 0.45-µm membrane, and the filtrate was
collected and transferred to a new centrifuge tube.
Ultracentrifugation was then performed at 100,000 x g for 70 min at
4˚C. After removal of the supernatant, the pellet was resuspended
in ice-cold PBS. The sample was subjected to another round of
ultracentrifugation at 100,000 x g for 70 min at 4˚C. The resulting
precipitate was resuspended in 100 µl PBS. The isolated exosomes
were stored at -80˚C for further analysis.
Exosome analysis
Exosome isolation quality was assessed by diluting
the samples with PBS (w/v=1:100), followed by analysis using the
NanoSight LM10 instrument (Malvern Instruments, Ltd.) according to
the manufacturer's instructions. Size and concentration were
quantified through the Nanoparticle Tracking Analysis 2.0 (NTA 2.0)
software (Malvern Instruments, Ltd.).
Transmission electron microscopy
(TEM)
To examine the structural characteristics of the
isolated exosomes, 10 µl exosome suspension was carefully placed
onto a 300 mesh formvar/carbon-coated copper grid (MilliporeSigma).
The exosomes were allowed to adsorb to the grid for 1 min at room
temperature, after which excess liquid was removed to ensure
optimal sample preparation. The adsorbed exosomes were then
subjected to negative staining with 2.5% uranyl acetate for 5 min
and embedded with 1% methyl cellulose (Sigma-Aldrich, Shanghai,
China) on ice for 10 min. Following staining, the samples were
air-dried for several minutes at room temperature. Finally, the
exosomes were analyzed using TEM (FEI Tecnai 12; Philips Medical
Systems B.V.) at a magnification of x60,000 to observe their
structural features.
Western blot analysis
Total protein of samples was separated by
radio-immunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology), and then quantified using the BCA Protein Assay Kit
(Beyotime Institute of Biotechnology). A total of 20 µg protein
from each isolated exosome sample and follicular fluid [negative
control (NC)] were subjected to SDS-PAGE on 10% gels for protein
separation. The separated proteins were then transferred onto PVDF
membranes and incubated overnight in TBS-Tween (TBS-T) buffer,
containing Tris (15 mM, pH 7.8), NaCl (100 mM), Tween-20 (0.5%),
with 5% defatted dry milk at 4˚C. The membranes were subsequently
incubated with primary antibodies against CD9 (1:1,000; cat. no.
ab236630; Abcam) and CD63 (1:1,000; cat. no. A5271; ABclonal
Biotech Co., Ltd.) in TBS-T buffer with 5% defatted dry milk for 3
h at 25˚C. After washing with TBS-T buffer, the membranes were
incubated with HRP-conjugated secondary antibodies (goat
anti-mouse/rabbit IgG; cat. nos. SA00001-1& SA00001-2;
Proteintech Group, Inc.) at a 1:2,000 dilution for 1 h at 25˚C.
Protein bands were visualized using an electrochemiluminescence
detection system (Pierce ECL Western; Thermo Fisher Scientific,
Inc.). All experiments were repeated at least three times to ensure
the reliability and reproducibility of the results.
circRNA sequencing and analysis
circRNA sequencing was employed to identify
differentially expressed circRNAs in follicular fluid exosomes
between four DOR and four normal control samples, as previously
described (26). A concentration
≥50 ng/µl was used for each sample, and the EVs concentration was
determined by NTA software as the basis for standardization. Total
RNA was extracted from exosomes using TRIzol® (cat. no.
155976018; Invitrogen; Thermo Fisher Scientific, Inc.) as per the
manufacturer's protocol, with RNA quality assessed using the
optical density (OD)260/OD280 ratio between
1.8 and 2.0, adhering to establish quality control standards. For
library construction, RNA was processed with the TruSeq Stranded
Total RNA library preparation kit (cat. no. 20020599; Illumina,
Inc.). Post-preparation, library quality and quantity were
evaluated using the Bioanalyzer 2100 system (Agilent Technologies,
Inc.). The prepared library, at a concentration of 10 pM, was
converted into single-stranded DNA, which was then captured and
amplified in situ to form clusters (using a dilute NaOH for
denaturation and then hybiridzation and extension in ~35 cycles).
Sequencing was performed for 150 cycles in paired-end mode on the
Illumina HiSeq4000 sequencer (Illumina, Inc.) with the HiSeq
3000/4000 SBS Kit (150 cycles) (cat. no. TG-410-1002; Illumina,
Inc.), ensuring a Q30 quality score. Following trimming of 30
adapters with Cutadapt software (v3.4; National Bioinformatics
Infrastructure Sweden) and removal of low-quality sequences,
high-quality read fragments were used for circRNA analysis.
High-quality reads were aligned using the STAR software (v2.5.1b;
National Human Genome Research Institute of NIH), while circRNAs
were identified and analyzed with the DCC program (v0.4.4;
Karolinska Institutet), and annotation of the detected circRNAs was
performed using the Circ2Traits (http://gyanxet-beta.com/circdb/) and circBase
(https://www.circbase.org/) databases.
Data normalization and differential expression analysis of circRNAs
were conducted with the EdgeR software (v3.16.5; https://bioconductor.org/packages/edgeR), with
circRNAs considered differentially expressed when fold change was
≥2.0 and P<0.05. Heatmaps and clustering of differentially
expressed circRNAs were generated using the R ggplot2 package
(v2.1.0; https://www.rdocumentation.org/packages/ggplot2/versions/2.1.0)
(27). Functional predictions of
circRNAs were performed through Gene Ontology (GO; https://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; https://www.kegg.jp/) analyses on the associated
genes. These analyses were carried out using the Database for
Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov/home.jsp) (28). Terms with
Benjamini-Hochberg-adjusted P<0.05 and containing ≥5 annotated
genes were considered statistically significant. The raw data were
uploaded to the National Centre for Biotechnology Information
Sequence Read Archive database (BioProject accession no.
PRJNA1191187).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the follicular fluid
precipitate following centrifugation using TRIzol. RT of the RNA
into cDNA was carried out using a PrimeScript ™ RT reagent kit with
gDNA Eraser (Perfect Real Time) (cat. no. RR047Q; Takara
Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The resulting cDNA was subsequently amplified by
qPCR, utilizing the AceQ Universal SYBR qPCR Master Mix (cat. no.
Q511; Vazyme Biotech Co., Ltd.) and the ABI 7500 PCR system (cat.
no. 4351104; Applied Biosystems; Thermo Fisher Scientific, Inc.).
The amplification protocol included an initial denaturation step at
95˚C for 15 sec, followed by 45 cycles of amplification at 55-60˚C
for 15 sec, and a final extension step at 72˚C for 15 sec. Relative
RNA expression levels were calculated using the 2-ΔΔCq
method and using GAPDH as the reference gene (29). All reactions were performed in
triplicate to ensure reliability and reproducibility of the
results. The reference sequence accession nos. of circRNAs were
derived from the circBank database (http://www.circbank.cn/#/home). The circBank IDs of
hsa_circ_0000344, hsa_circ_0001126, hsa_circ_0005379,
hsa_circ_0005777 and hsa_circ_0007509 are hsa_RSF1_0004200,
hsa_PTPRA_0009800, hsa_GDI2_0006400, hsa_ARHGEF28_0005700 and
hsa_PPP4R1_0010200, respectively. Primer sequences are provided in
Table I.
 | Table IPrimer sequences used in the present
study. |
Table I
Primer sequences used in the present
study.
| Gene | Sequence
(5'-3') |
|---|
| GAPDH | F:
ACAGCAACAGGGTGGTGGAC |
| | R:
TTTGAGGGTGCAGCGAACTT |
|
hsa-circ-0000344 | F:
GCTTGGTGTATGTTGGTATCAGT |
| | R:
GAAGGCAGGCAGTATGGTATC |
|
hsa-circ-0001126 | F:
ACAAGAACAGCAAGCACCAAT |
| | R:
GCATCATCCATTACCACTCAGT |
|
hsa-circ-0005379 | F:
AGATCAACCGTGTTATGAAACCAT |
| | R:
TTGCCATTCACTGACATTATACCT |
|
hsa-circ-0005777 | F:
TTCGTGTGCTTCCAACTTGA |
| | R:
CACTGGGACTGGTTTCTGTAG |
|
hsa-circ-0007509 | F:
CGAAAGGCTTGTGCTGAATG |
| | R:
TCCAGGGCTGAAGGTATAATAATC |
Cell culture
The KGN ovarian granulosa cell line was obtained
from Shanghai Fuheng Biotechnology Co., Ltd., and cultured in
DMEM/F12 medium (cat. no. 11320033, Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum
(HyClone; Cytiva) and 0.5% penicillin-streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc.). The cells were maintained in a 5%
CO2 atmosphere at 37˚C with 100% humidity.
Cyclophosphamide (CTX; cat. no. HY-17420; MedChemExpress) may
destroy the follicular pool, leading to primary ovarian
insufficiency (30). To induce
cell damage, CTX was administered at concentrations of 0, 10, 15,
20, 25 and 30 µM for 24 h at 37˚C. The NC was treated without
CTX.
Cell transfection
To investigate the role of hsa_circ_0005379 in KGN
cells, short hairpin (sh)-hsa_circ_0005379 and NC
sh-circRNA(pLKO.1), as well as hsa_circ_0005379-overexpression (OE)
and NC plasmids (pcDNA3.1) were synthesized by GeneChem, Inc. KGN
cells were transfected for 5 h with the corresponding plasmids (500
µM) using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) at room temperature according to
the manufacturer's protocol, as previously described (31). The transfection efficiency was
detected by RT-qPCR 48 h after transfection. All subsequent
experiments were performed 48 h after transfection. The sequences
of sh-hsa_circ_0005379 and NC are presented in Table II.
 | Table IISequence of shRNAs and scramble
control. |
Table II
Sequence of shRNAs and scramble
control.
| Name | Sequence
(5'-3') |
|---|
|
hsa-circ-0005379-sh1 |
CTGTTGTGCTGACCTCTGA |
|
hsa-circ-0005379-sh2 |
GACAGCTTCATGGTCCAGA |
|
hsa-circ-0005379-sh3 |
TACCAGGATGTGCTGGTCA |
|
sh-hsa-circ-0005379-NC |
CCTAAGGTTAAGTCGCCCTCG |
Cell viability assay
Cell viability was assessed using Cell Counting
Kit-8 (CCK-8) (cat. no. A311; Vazyme Biotech Co., Ltd.) according
to the manufacturer's instructions, as previously described
(32). Briefly, 10 µl CCK-8
reagent were added to each well of a 96-well plate
(3x103 cells/well), followed by incubation for 1 h at
37˚C. Absorbance at 450 nm was measured using a BioTek
multifunctional microplate reader (Agilent Technologies, Inc.).
Cell apoptosis assay
A total of 1x106 cells were collected in
the logarithmic phase, washed with PBS and then digested with
trypsin (cat. no. C0205; Beyotime Institute of Biotechnology).
After treatment with 30 µM CTX, 1x105 KGN cells were
incubated with 5 µl Annexin V-APC and 5 µl PI at room temperature
for 15 min as per the manufacturer's instructions (Beyotime
Institute of Biotechnology). The apoptosis rate was then evaluated
by flow cytometry (FACSCalibur; BD Biosciences), and the data were
analyzed utilizing FlowJo software (version 10.0.7; TreeStar,
Inc.).
Biochemical assays of malondialdehyde
(MDA) and superoxide dismutase (SOD)
MDA and SOD levels of KGN cells were measured using
the MDA detection kit (cat. no. A003-1) and SOD detection kit (cat.
no. A001-3) from Nanjing Jiancheng Bioengineering Institute,
following the manufacturer's protocols.
ROS detection
ROS detection was performed using a ROS detection
kit (cat. no. MA0219; Dalian Meilun Biotechnology Co., Ltd.). KGN
cells were seeded in a six-well plate and treated with 30 µM CTX
for 24 h at room temperature. Each well received 1 ml
dichlorodihydrofluorescein diacetate (DCFH-DA; prepared at a
1:1,000 dilution in basal DMEM/F12) and incubated at 37˚C for 20
min to allow probe uptake. Following incubation, the cells were
washed with 1X PBS three times to remove excess probe. The cells
were then digested with 0.25% Trypsin (cat. no. C0205; Beyotime
Institute of Biotechnology), centrifuged at 300 x g for 5 min at
4˚C, and resuspended in 300 µl fresh ice-cold basal DMEM/F12
medium. ROS levels were quantified by flow cytometry (FACSCalibur;
BD Biosciences), with excitation and emission wavelengths set at
488 nm and 525 nm, respectively, as previously described (33). The data were analyzed utilizing
FlowJo software (version 10.0.7; TreeStar, Inc.). ROS mean
fluorescence intensity ratio was calculated as the fluorescently
stained cell number/total cell number.
Statistical analysis
All experiments were independently performed at
least 3 times. Data are presented as the mean ± standard deviation
and data analysis was performed using GraphPad Prism v9 software
(GraphPad; Dotmatics). Comparisons between two groups were
performed using unpaired Student's t-test. Comparisons among three
or more groups were conducted using one-way ANOVA followed by
Tukey's multiple comparisons test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Cohort characteristics and exosome
characterization
The clinical characteristics of patients with DOR
and controls were evaluated (Table
III), and no significant differences in age, BMI, duration of
infertility, estradiol, follicle-stimulating hormone, luteinizing
hormone, anti-mullerian hormone and cleaved zygotes ratio between
the two groups were observed. By contrast, significant differences
in AMH and AFC levels between the DOR and control groups were
detected. Significant decreases in AMH and AFC usually indicate a
significant decline in ovarian reserve function, which may be
accompanied by reduced fertility or an increased risk of
menopausal-related conditions. Exosomes were then isolated from the
follicular fluid of both DOR and control groups. TEM analysis was
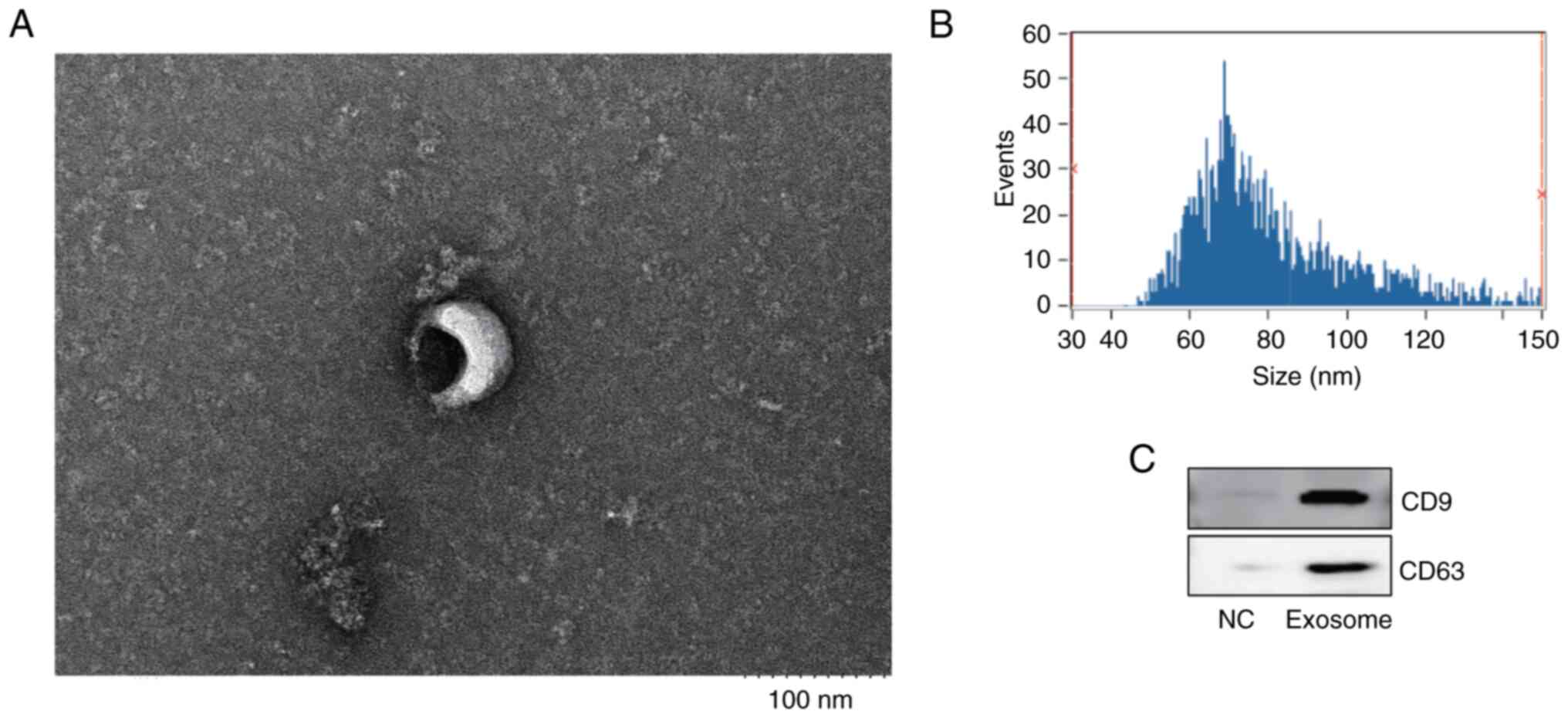
performed to examine exosome size and morphology (Fig. 1A). Nanoparticle tracking analysis
determined the exosome size and concentration, revealing that the
mean diameter was 81.18 nm and the concentration was
2.79x109 particles/ml (Fig.
1B). Additionally, the presence of the exosome markers CD9 and
CD63 was confirmed through western blot analysis of follicular
fluid exosomes (Fig. 1C).
 | Table IIIGeneral characteristics of the study
population. |
Table III
General characteristics of the study
population.
|
Characteristics | Normal control
group | Decreased ovarian
reserve group | P-value |
|---|
| Age, years | 31.50±1.73 | 32.50±1.29 | 0.390 |
| BMI | 20.41±3.10 | 22.18±4.38 | 0.530 |
| Duration of
infertility, years | 2.25±0.50 | 3.25±1.26 | 0.190 |
| Estradiol baseline,
pg/ml | 31.25±10.71 | 44.25±13.45 | 0.180 |
|
Follicle-stimulating hormone baseline,
mIU/ml | 5.98±0.54 | 8.27±3.70 | 0.270 |
| Luteinizing hormone
baseline, mIU/ml | 3.55±0.76 | 3.48±1.01 | 0.910 |
| Anti-mullerian
hormone baseline, ng/ml | 3.91±0.71 | 0.73±0.23 | <0.001 |
| Antral follicle
count | 10.75±2.36 | 4.00±2.45 | <0.010 |
| Cleaved zygotes,
% | 91.25±12.74 | 82.50±13.58 | 0.380 |
Expression profiling of circRNAs in
patients with DOR
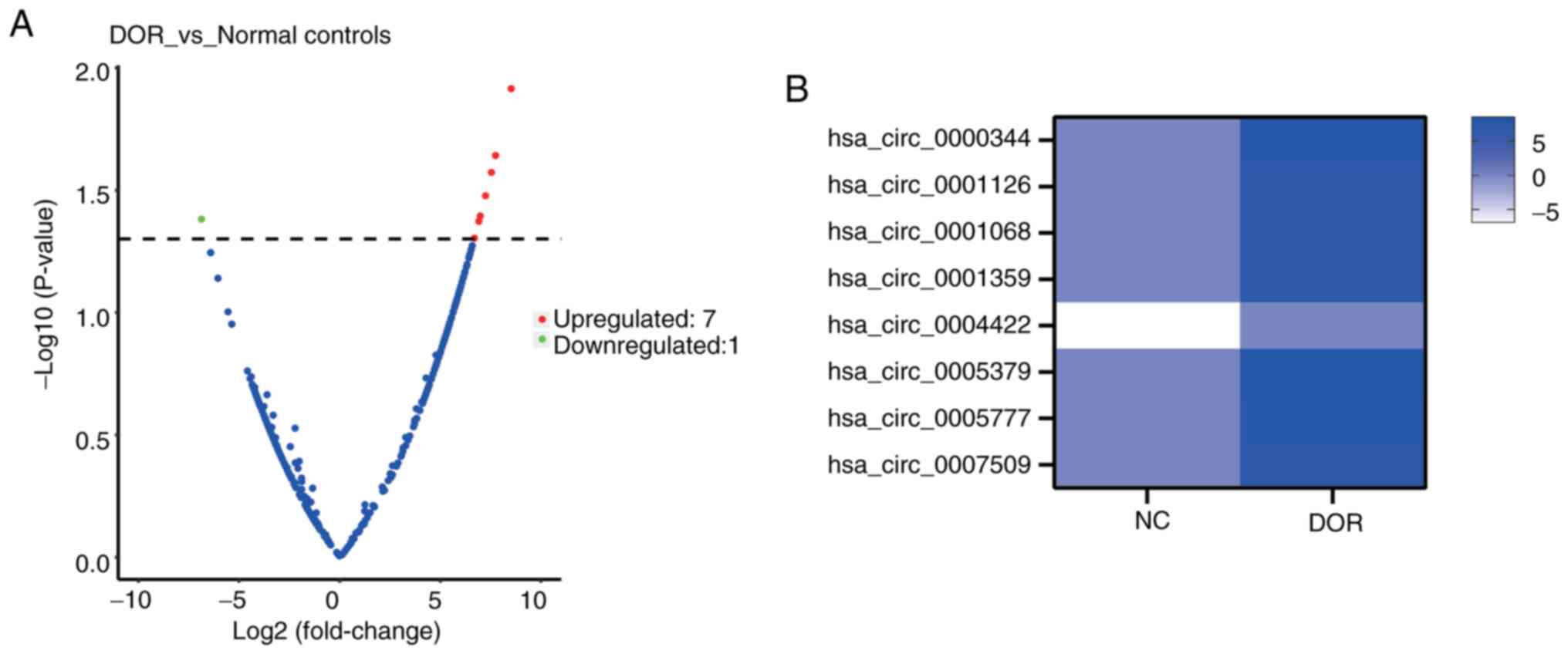
Differentially expressed circRNAs in exosomes were
identified and summarized in Table
IV. Information on the fold changes, P-values and the
corresponding host genes of all eight circRNAs is provided. As
illustrated in Fig. 2A, the
volcano plot revealed a clear separation between the DOR and
control groups, with seven circRNAs upregulated and one
downregulated in the DOR group. The heatmap in Fig. 2B displays the expression profiles
of these eight differentially expressed circRNAs.
 | Table IVDifferentially expressed circRNAs
between patients with decreased ovarian reserve and normal
controls. |
Table IV
Differentially expressed circRNAs
between patients with decreased ovarian reserve and normal
controls.
| circRNA ID | Log2 fold
change | P-value | Host gene |
|---|
|
hsa_circ_0000344 | 7.50 | 0.026802 | RSF1 |
|
hsa_circ_0001126 | 6.95 | 0.040373 | PTPRA |
|
hsa_circ_0001068 | 6.66 | 0.049757 | MGAT5 |
|
hsa_circ_0001359 | 6.88 | 0.042484 | PHC3 |
|
hsa_circ_0004422 | -6.90 | 0.041604 | - |
|
hsa_circ_0005379 | 8.49 | 0.012191 | GDI2 |
|
hsa_circ_0005777 | 7.71 | 0.022859 | RGNEF |
|
hsa_circ_0007509 | 7.21 | 0.033397 | PPP4R1 |
Functional enrichment analysis
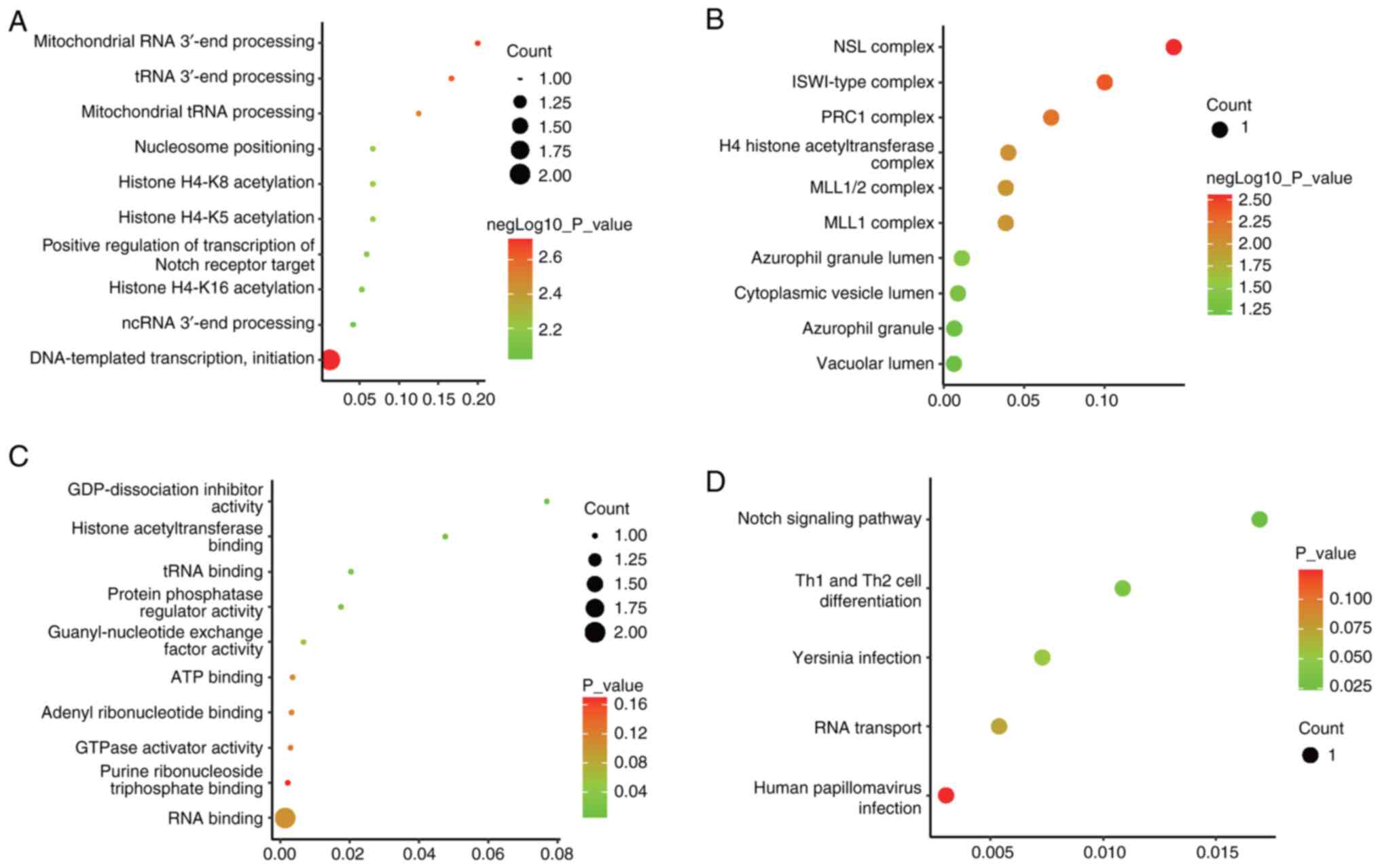
To explore the potential roles of differentially
expressed circRNAs, GO and KEGG analyses were performed, focusing
on the host genes associated with these circRNAs. The top 10
enriched terms related to molecular functions, cellular components
and biological processes are summarized in Fig. 3A-C. The leading biological
processes included ‘DNA-templated transcription, initiation’
‘mitochondrial RNA 3'-end processing’ and ‘tRNA 3'-end processing’
(Fig. 3A). For cellular
components, the most enriched terms were ‘NSL complex’, ‘ISWI-type
complex’ and ‘PRC1 complex’ (Fig.
3B). Regarding molecular functions, the most significant terms
were ‘GDP-dissociation inhibitor activity’, ‘histone
acetyltransferase binding’ and ‘tRNA binding’ (Fig. 3C). Additionally, five KEGG pathways
were enriched (Fig. 3D), with the
‘Notch signaling pathway’ and ‘Th1 and Th2 cell differentiation’
showing significant enrichment (P<0.05), which suggests
that these pathways may contribute to the pathogenesis of DOR.
Verification of selected circRNAs and
the function of hsa_circ_0005379
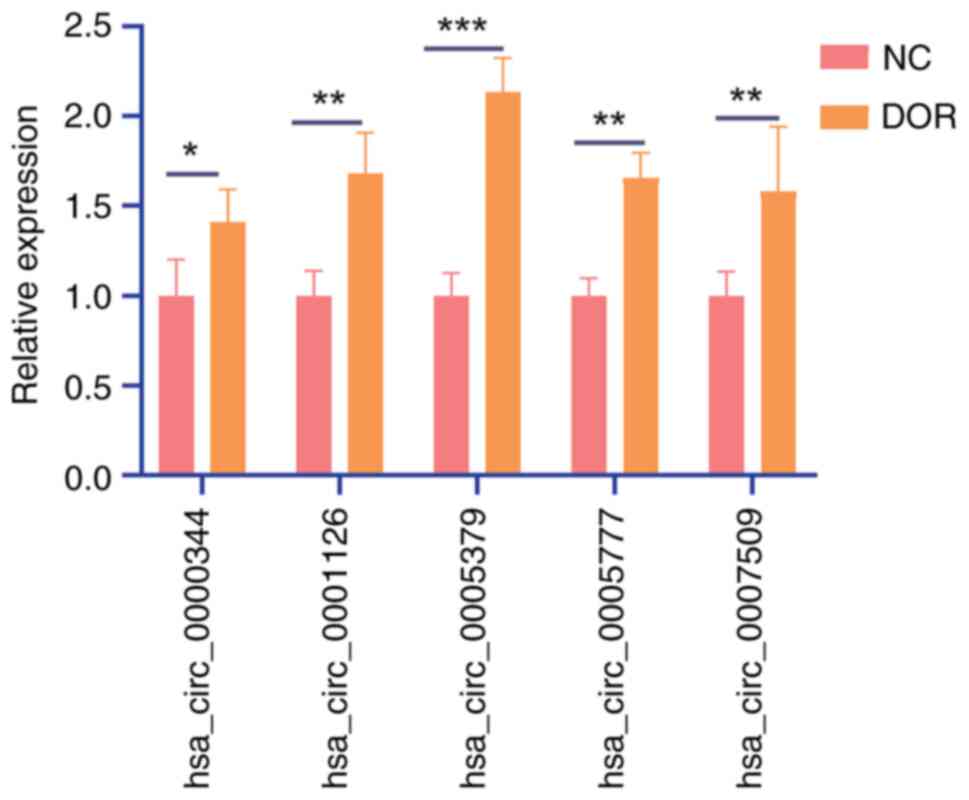
In total, 6 patients with DOR and 6 normal controls
were included for RT-qPCR validation. The top five most
significantly altered genes, based on the P-value from the RNA-seq
data, were selected for further validation (Table IV). Hsa_circ_0005379 was
identified as the most significantly upregulated circRNA in the DOR
group (Fig. 4) and was selected
for subsequent analysis. Ovarian granulosa KGN cells were treated
with varying concentrations of CTX (0, 10, 15, 20, 25 and 30 µM) to
induce cellular damage. The CCK-8 assay demonstrated a
dose-dependent decrease in KGN cell viability, with higher
concentrations of CTX producing a more pronounced effect (Fig. 5A). The 30 µM CTX treatment was
found to be the most effective in reducing cell viability. To
further investigate the biological role of hsa_circ_0005379, it was
selected as the optimal concentration for subsequent experiments.
Then, KGN cells were transfected with shRNAs or an overexpression
vector to explore the function of hsa_circ_0005379 (Fig. 5B and C). The results showed that sh1 had the
highest knockdown efficiency and was therefore used for subsequent
experiments (Fig. 5B). Following
CTX treatment and transfection of the overexpression vector, a
significant increase in the apoptosis rate was observed compared
with that in CTX-treated KGN cells transfected with the NC vector.
By contrast, knockdown of hsa_circ_0005379 with CTX treatment
resulted in a decrease in apoptosis compared with
sh-hsa_circ_0005379-NC+CTX group (Fig.
5D).
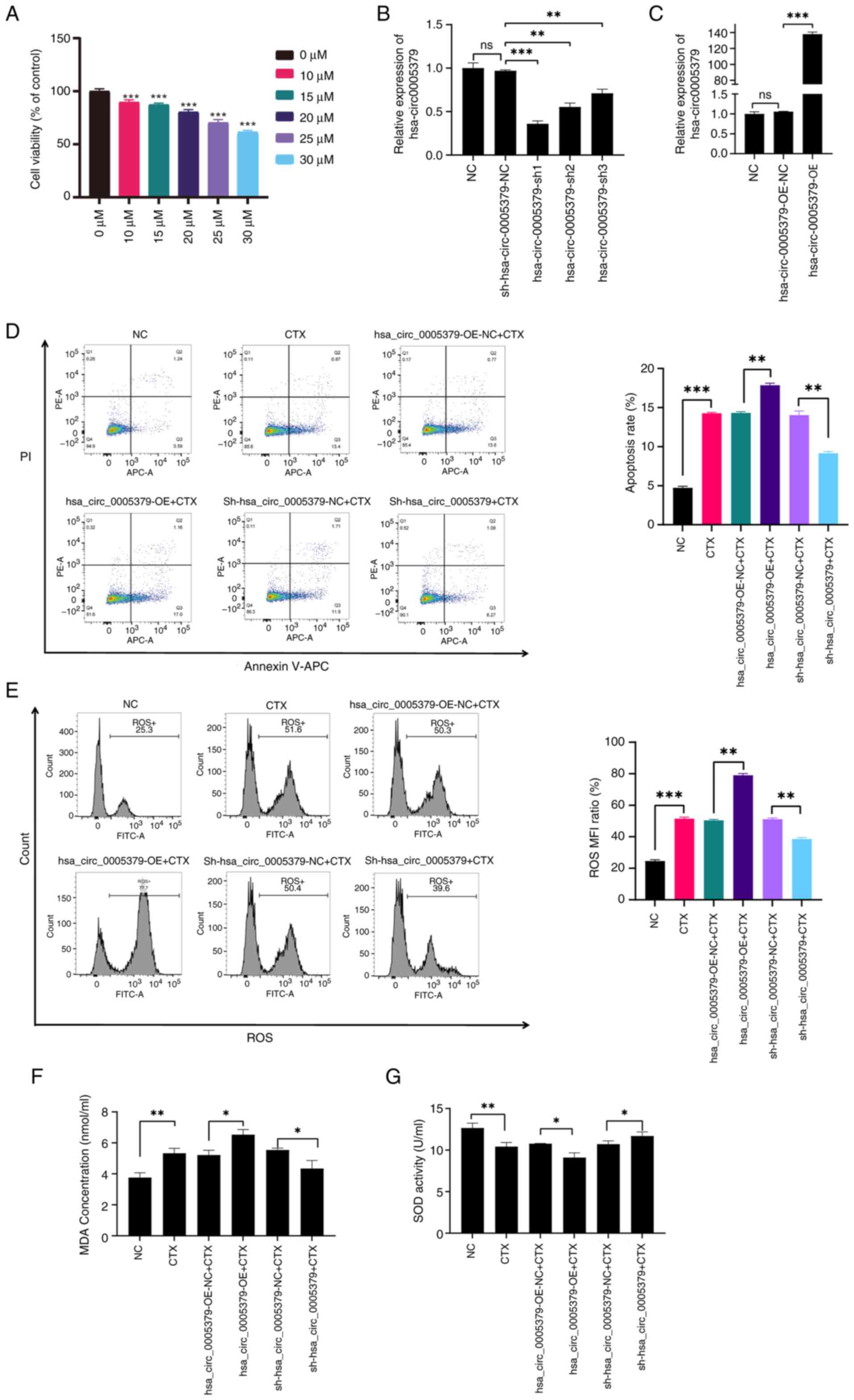 | Figure 5Function of hsa_circ_0005379 in
CTX-induced KGN ovarian granulosa cells. (A) Different gradient
concentrations (0, 10, 15, 20, 25 and 30 µM) of CTX were used to
induce ovarian granulosa cell damage, and Cell Counting Kit-8 assay
was used to detect KGN cell viability. (B) RT-qPCR analysis of
hsa_circ_0005379 expression after transfection with three shRNAs
and negative control sh-NC. (C) RT-qPCR analysis of
hsa_circ_0005379 expression after transfection with OE vector and
an empty OE-NC vector. (D) The apoptosis of sh-hsa_circ_0005379 or
hsa_circ_0005379-OE transfected KGN cells was detected using
annexin V/PI staining. (E) Flow cytometry was used to detect the
ROS level of cells after different treatments. The concentration of
(F) MDA and (G) SOD in KGN cells with different treatments was
determined using corresponding biochemistry detection kits. n=3,
*P<0.05, **P<0.01 and
***P<0.001 vs. 0 µΜ or as indicated. CTX,
cyclophosphamide; RT-qPCR, reverse transcription-quantitative PCR;
OE, overexpression; NC, negative control; sh, short hairpin; SOD,
superoxide dismutase; MDA, malondialdehyde; ROS, reactive oxygen
species; MFI, mean fluorescence intensity; ns, not significant. |
The effect of hsa_circ_0005379 on oxidative stress
in CTX-treated cells was further examined. The results indicated
increased levels of ROS and MDA in the hsa_circ_0005379-OE group,
whereas the sh-hsa_circ_0005379 group showed reduced levels of both
markers, compared with those in the corresponding NC groups
(Fig. 5E and F). By contrast, the antioxidant enzyme
SOD, which is involved in aging processes, displayed an inverse
pattern compared with that of ROS and MDA (Fig. 5G). These findings suggest that
silencing hsa_circ_0005379 may alleviate oxidative stress in
DOR.
Discussion
DOR is characterized by reduced oocyte quality and
quantity, leading to impaired ovarian endocrine function and
diminished fertility in women (34). The follicular microenvironment is
believed to play a critical role in oocyte maturation and
development (22). Therefore, the
present study investigated the follicular microenvironment
isolating exosomes (small vesicles containing biological molecules)
from the follicular fluid of patients with DOR. A subsequent
analysis of follicular exosomes using circRNA sequencing was
performed, thus conducting the first examination of the exosomal
circRNA profile in the follicular fluid of individuals with DOR, to
the best of our knowledge.
circRNAs, a class of single-stranded RNA molecules,
are characterized by covalently closed loops. These molecules are
widely distributed across various organisms, ranging from viruses
to mammals. Considerable progress has been made in understanding
the biogenesis, regulation, localization, degradation and
modification of circRNAs (35,36).
In the present study, eight differentially expressed circRNAs were
identified between patient with DOR and normal controls, followed
by enrichment analyses. GO biological process enrichment analysis
revealed the involvement of the identified circRNAs in numerous
biological processes, including ‘DNA-templated transcription,
initiation’. Notably, the enrichment of this process was also
observed in rats with DOR, induced by tripterygium glycoside tablet
suspension (37). Additionally,
two key pathways were found to be significantly enriched: ‘Notch
signaling pathway’ and ‘Th1 and Th2 cell differentiation’. Hughes
et al (38) reported that
elevated ovarian oocyte death triggered an increase in Notch
signaling and ovarian inflammation, and that Notch signaling
pathway activation in granulosa cells exacerbated apoptosis.
However, the specific role of the Notch pathway in DOR remains to
be further elucidated. In addition, increased oxidative stress
caused by smoking can further promote the onset and development of
cancer by amplifying the inflammatory response and activating
Notch-1 signaling (39).
Overexpression of long non-coding RNA NONHSAT098487.2 has been
shown to inhibit H2O2-induced oxidative
stress injury in cardiomyocytes by activating Notch signaling
pathway (40) and polystyrene
microplastics have been demonstrated to promote colon barrier
damage through oxidative stress-mediated overactivation of the
Notch signaling pathway (41).
These studies suggest an association between oxidative stress and
Notch pathway activation. Since oxidative stress is closely related
to DOR (42), future studies may
explore the association between DOR and the Notch pathway in terms
of oxidative stress generation. Furthermore, a recent study showed
that polysaccharides could collectively inhibit inflammation,
apoptosis and oxidative stress in asthmatic rats via regulation of
T helper (Th)1/Th2 and Th17/Treg cell immune imbalances (43). Dehydroepiandrosterone has been
shown to improve Th1 immune responses and regulate the balance of
the Th1/Th2 response in patients with DOR (7). These studies collectively suggest
that Th1/Th2 responses may be involved in the regulation of
oxidative stress in DOR.
During follicular development, the interaction
between oocytes and adjacent granulosa cells is essential for the
production of fertilizable oocytes and the regulation of ovarian
function (44-46).
Thus, diminished oocyte competence in women with DOR may stem from
dysregulated granulosa cell function (47). CTX exposure causes premature
ovarian insufficiency (48). In
the present study, a DOR cell model was established by exposing
granulosa cells to CTX, and this model was subsequently used to
investigate the role of hsa_circ_0005379.
Hsa_circ_0005379 has previously been identified as a
key regulator in neuroblastoma (49). In the present study, upregulation
of hsa_circ_0005379 was observed in exosomes isolated from the
follicular fluid of patients with DOR. Silencing hsa_circ_0005379
partially reversed granulosa cell apoptosis. A significant
association between oxidative stress and aging has been reported
(50), with oxidative stress also
being recognized as a major contributor to ovarian aging (21). Alterations in ROS, MDA and SOD
levels reflect changes in oxidative stress, providing an indication
of ovarian aging. Increased oxidative stress has been observed in
both animal models and patients with DOR (22,51).
In the present study, reduced hsa_circ_0005379 levels alleviated
oxidative stress in CTX-induced DOR cells, suggesting the potential
of hsa_circ_0005379 as a therapeutic target for DOR.
In addition, other circRNAs have been reported in
the literature to be associated with DOR. For example,
hsa_circ_0031584 has been indicated to be an important molecule
regulating the mitotic process of granulosa cells in DOR (52). The findings of the present study
were compared with studies on circRNAs in other female reproductive
diseases, noting conserved roles in oxidative stress regulation.
For example, Bu-Shen-Ning-Xin decoction has been shown to inhibit
oxidative stress by regulating circRNA_012284 expression in POI.
Human umbilical cord mesenchymal stem cell-derived exosomes
secreted circBRCA1, which directly sponged microRNA (miR)-642a-5p
to upregulate FOXO1, thereby preventing oxidative stress injuries
in granulosa cells and protecting ovarian function in rats with POI
(53). Furthermore, knockdown of
hsa_circ_0118530 was shown to suppress PCOS progression by
inhibiting oxidative stress and inflammation factor release
(54). In another study,
overexpression of circ_0097636 protected PCOS cell models from
dihydrotestosterone-induced oxidative stress by increasing sirtuin
3 expression (55).
Hsa_circ_0005777 is derived from the cyclization of
the host gene rho guanine nucleotide exchange factor (RGNEF);
circRGNEF promotes bladder cancer progression via regulation of the
miR-548/KIF2C axis (56).
Similarly, hsa_circ_0001126 originates from the protein tyrosine
phosphatase receptor type a (PTPRA) gene, and circPTPRA inhibits
RNA N6-methyladenosine recognition by interacting with insulin-like
growth factor 2 mRNA-binding protein 1, thereby suppressing bladder
cancer progression (57). Future
investigations involving small interfering RNA-mediated knockdown
or overexpression of these circRNAs are essential to further
elucidate their involvement in DOR pathogenesis.
The present study has certain limitations. The small
sample size represents an important constraint in this pilot
investigation. Due to the strict exclusion criteria (such as the
lack of history of medications known to influence glucose and lipid
metabolism) and the challenges in recruiting patients with
well-defined DOR, achieving a larger cohort was logistically
difficult within the study timeframe. In future studies, a
multicenter collaboration is necessary to recruit a larger, more
diverse cohort. Additionally, the exclusion of individuals with a
history of medications affecting glucose or lipid metabolism, as
well as conditions such as Cushing's syndrome, congenital adrenal
hyperplasia, androgen-secreting tumors and endometriosis, is rooted
in the need to minimize confounding variables. These factors could
independently alter metabolic or hormonal outcomes under
investigation, thereby obscuring the true effects of the
intervention or exposure being studied. For example, medications
influencing glucose/lipid metabolism (such as insulin, statins and
glucocorticoids) could mask or exaggerate metabolic changes
(58), compromising the assessment
of the study's primary endpoints. Furthermore, endocrine disorders
(such as Cushing's syndrome, congenital adrenal hyperplasia and
androgen-secreting tumors) directly disrupt hormonal or metabolic
pathways, potentially mimicking outcomes relevant to conditions
like PCOS or insulin resistance (59-61).
Endometriosis is known to alter follicular fluid composition and
inflammatory pathways, which could independently influence the
present study's endpoints, such as ROS levels and associated
pathways (20). By excluding these
factors, the internal validity of the present study was enhanced,
ensuring that the observed effects are more likely attributable to
the variables under investigation rather than pre-existing
conditions or treatments. Excluding certain groups of individuals
bolsters confidence in causal inferences by reducing confounding
factors, particularly in studies focusing on metabolic or endocrine
mechanisms (such as studies evaluating insulin sensitivity or
androgen levels). By contrast, the homogeneity of the study
population may limit external validity. For instance, the results
may not apply to individuals with overlapping conditions (such as
patients with PCOS or untreated congenital adrenal hyperplasia) or
those on common glucose/lipid-modifying therapies. This restricts
the findings to a narrower, ‘idealized’ population. If the excluded
conditions are rare (such as androgen-secreting tumors), their
omission may not markedly impact applicability. As a preliminary
study, the findings may be specific to idiopathic DOR. Future
research should include stratified analyses comparing subgroups
(patients with DOR with vs. without endometriosis) to evaluate the
broader applicability of these results, consistent with a recent
study on DOR heterogeneity (4).
In conclusion, the present study provided a
comprehensive profile of circRNA expression in exosomes isolated
from the follicular fluid of patients with DOR. Additionally, the
current study represents, to the best of our knowledge, the first
report on the role of hsa_circ_0005379 in DOR. The results
demonstrated that silencing hsa_circ_0005379 alleviated apoptosis
and oxidative stress in CTX-induced granulosa cells. However, the
underlying mechanisms through which hsa_circ_0005379 modulates
oxidative stress and contributes to DOR remain to be further
explored.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Fujian Natural
Science Foundation (grant no. 2021J05083), the Joint Funds for the
Innovation of Science and Technology of Fujian Province (grant no.
2023Y9384), the Fujian Provincial Health Technology Project (grant
no. 2025QNGGA010) and the Zhejiang Medical and Health Project
(grant no. 2023KY781).
Availability of data and materials
The raw RNA sequencing data generated in the present
study may be found in the National Center for Biotechnology
Information Sequence Read database under accession number
PRJNA1191187 or at the following URL: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1191187.
The other data generated in the present study may be requested from
the corresponding author.
Authors' contributions
PH, YF and DL were responsible for the
conceptualization, design and execution of the study. PH, YF, JC
and DL were responsible for drafting and revising the manuscript.
SC, JC and CJ were responsible for resources and investigation. PH,
SC, JC, CJ and BL analyzed and interpreted the data. SC and JC were
responsible for reviewing and editing the manuscript. PH, YF and DL
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Fujian Maternity and Child Health Hospital (approval
no. 2023KYLLR01082; Fuzhou, China). Written informed consent was
obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rotshenker-Olshinka K, Michaeli J, Srebnik
N, Samueloff A, Magen S, Farkash R and Eldar-Geva T: Extended
fertility at highly advanced reproductive age is not related to
anti-Mullerian hormone concentrations. Reprod Biomed Online.
45:147–152. 2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wen J, Huang K, Du X, Zhang H, Ding T,
Zhang C, Ma W, Zhong Y, Qu W, Liu Y, et al: Can Inhibin B Reflect
ovarian reserve of healthy reproductive age women effectively?
Front Endocrinol (Lausanne). 12(626534)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liang C, Zhang X, Qi C, Hu H, Zhang Q, Zhu
X and Fu Y: UHPLC-MS-MS analysis of oxylipins metabolomics
components of follicular fluid in infertile individuals with
diminished ovarian reserve. Reprod Biol Endocrinol.
19(143)2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cedars MI: Managing poor ovarian response
in the patient with diminished ovarian reserve. Fertil Steril.
117:655–656. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Park SU, Walsh L and Berkowitz KM:
Mechanisms of ovarian aging. Reproduction. 162:R19–R33.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Helbling-Leclerc A, Falampin M, Heddar A,
Guerrini-Rousseau L, Marchand M, Cavadias I, Auger N, Bressac-de
Paillerets B, Brugieres L, Lopez BS, et al: Biallelic Germline
BRCA1 Frameshift mutations associated with isolated diminished
ovarian reserve. Int J Mol Sci. 25(12460)2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhang J, Qiu X, Gui Y, Xu Y, Li D and Wang
L: Dehydroepiandrosterone improves the ovarian reserve of women
with diminished ovarian reserve and is a potential regulator of the
immune response in the ovaries. Biosci Trends. 9:350–359.
2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Oladipupo I, Ali T, Hein DW, Pagidas K,
Bohler H, Doll MA, Mann ML, Gentry A, Chiang JL, Pierson RC, et al:
Association between cigarette smoking and ovarian reserve among
women seeking fertility care. PLoS One. 17(e0278998)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ma X, Chen Z, Chen W, Chen Z and Meng X:
Exosome subpopulations: The isolation and the functions in
diseases. Gene. 893(147905)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gregory CD and Rimmer MP: Extracellular
vesicles arising from apoptosis: Forms, functions, and
applications. J Pathol. 260:592–608. 2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Quan M and Kuang S: Exosomal secretion of
adipose tissue during various physiological states. Pharmaceutical
Res. 37(221)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gallet R, Dawkins J, Valle J, Simsolo E,
de Couto G, Middleton R, Tseliou E, Luthringer D, Kreke M, Smith
RR, et al: Exosomes secreted by cardiosphere-derived cells reduce
scarring, attenuate adverse remodelling, and improve function in
acute and chronic porcine myocardial infarction. Eur Heart J.
38:201–211. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Esfandyari S, Elkafas H, Chugh RM, Park
HS, Navarro A and Al-Hendy A: Exosomes as biomarkers for female
reproductive diseases diagnosis and therapy. Int J Mol Sci.
22(2165)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yuan D, Luo J, Sun Y, Hao L, Zheng J and
Yang Z: PCOS follicular fluid derived exosomal miR-424-5p induces
granulosa cells senescence by targeting CDCA4 expression. Cell
Signal. 85(110030)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cao J, Huo P, Cui K, Wei H, Cao J, Wang J,
Liu Q, Lei X and Zhang S: Follicular fluid-derived exosomal
miR-143-3p/miR-155-5p regulate follicular dysplasia by modulating
glycolysis in granulosa cells in polycystic ovary syndrome. Cell
Commun Signal. 20(61)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yao Y, Chen R, Wang G, Zhang Y and Liu F:
Exosomes derived from mesenchymal stem cells reverse EMT via
TGF-beta1/Smad pathway and promote repair of damaged endometrium.
Stem Cell Res Ther. 10(225)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li Z, Zhang M, Zheng J, Tian Y, Zhang H,
Tan Y, Li Q, Zhang J and Huang X: Human umbilical cord mesenchymal
stem cell-derived exosomes improve ovarian function and
proliferation of premature ovarian insufficiency by regulating the
hippo signaling pathway. Front Endocrinol (Lausanne).
12(711902)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Qu Q, Liu L, Cui Y, Liu H, Yi J, Bing W,
Liu C, Jiang D and Bi Y: miR-126-3p containing exosomes derived
from human umbilical cord mesenchymal stem cells promote
angiogenesis and attenuate ovarian granulosa cell apoptosis in a
preclinical rat model of premature ovarian failure. Stem Cell Res
Ther. 13(352)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Burton GJ and Jauniaux E: Oxidative
stress. Best Pract Res Clin Obstet Gynaecol. 25:287–299.
2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Agarwal A, Aponte-Mellado A, Premkumar BJ,
Shaman A and Gupta S: The effects of oxidative stress on female
reproduction: A review. Reprod Biol Endocrinol.
10(49)2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang L, Tang J, Wang L, Tan F, Song H,
Zhou J and Li F: Oxidative stress in oocyte aging and female
reproduction. J Cell Physiol. 236:7966–7983. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Huang Y, Cheng Y, Zhang M, Xia Y, Chen X,
Xian Y, Lin D, Xie S and Guo X: Oxidative stress and inflammatory
markers in ovarian follicular fluid of women with diminished
ovarian reserve during in vitro fertilization. J Ovarian Res.
16(206)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang Q, Zhang D, Liu H, Fu J, Tang L and
Rao M: Associations between a normal-range free thyroxine
concentration and ovarian reserve in infertile women undergoing
treatment via assisted reproductive technology. Reprod Biol
Endocrinol. 22(72)2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Can S, Yang X, He Y, Wang C, Zou H, Fan Q,
Xu X, Cai G, Yunxia C and Xiaoqing P: Diminished ovarian reserve
associates with pregnancy and birth outcomes after IVF: A
retrospective cohort study. Hum Fertil (Camb).
27(2414813)2024.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lin Q, Qi Q, Hou S, Chen Z, Jiang N, Zhang
L and Lin C: Exosomal circular RNA hsa_circ_007293 promotes
proliferation, migration, invasion, and epithelial-mesenchymal
transition of papillary thyroid carcinoma cells through regulation
of the microRNA-653-5p/paired box 6 axis. Bioengineered.
12:10136–10149. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xiong S, Peng H, Ding X, Wang X, Wang L,
Wu C, Wang S, Xu H and Liu Y: Circular RNA expression profiling and
the potential role of hsa_circ_0089172 in Hashimoto's thyroiditis
via sponging miR125a-3p. Mol Ther Nucleic Acids. 17:38–48.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Gustavsson EK, Zhang D, Reynolds RH,
Garcia-Ruiz S and Ryten M: ggtranscript: An R package for the
visualization and interpretation of transcript isoforms using
ggplot2. Bioinformatics. 38:3844–3846. 2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sherman BT, Hao M, Qiu J, Jiao X, Baseler
MW, Lane HC, Imamichi T and Chang W: DAVID: A web server for
functional enrichment analysis and functional annotation of gene
lists (2021 update). Nucleic Acids Res. 50:W216–W221.
2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kalich-Philosoph L, Roness H, Carmely A,
Fishel-Bartal M, Ligumsky H, Paglin S, Wolf I, Kanety H, Sredni B
and Meirow D: Cyclophosphamide triggers follicle activation and
‘burnout’; AS101 prevents follicle loss and preserves fertility.
Sci Transl Med. 5(185ra62)2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mao X, Meng Q, Han J, Shen L, Sui X, Gu Y
and Wu G: Regulation of dynamin-related protein 1 (DRP1) levels
modulates myoblast atrophy induced by C26 colon cancer-conditioned
medium. Transl Cancer Res. 10:3020–3032. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang D, Yi S, Cai B, Wang Z, Chen M,
Zheng Z and Zhou C: Involvement of ferroptosis in the granulosa
cells proliferation of PCOS through the circRHBG/miR-515/SLC7A11
axis. Ann Transl Med. 9(1348)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sun L, Wang H, Yu S, Zhang L, Jiang J and
Zhou Q: Herceptin induces ferroptosis and mitochondrial dysfunction
in H9c2 cells. Int J Mol Med. 49(17)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lv Z, Lv Z, Song L, Zhang Q and Zhu S:
Role of lncRNAs in the pathogenic mechanism of human decreased
ovarian reserve. Front Genet. 14(1056061)2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and
Xu RH: Circular RNA: metabolism, functions and interactions with
proteins. Mol Cancer. 19(172)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Patop IL, Wust S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38(e100836)2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lu G, Zhu YY, Li HX, Yin YL, Shen J and
Shen MH: Effects of acupuncture treatment on microRNAs expression
in ovarian tissues from Tripterygium glycoside-induced diminished
ovarian reserve rats. Front Genet. 13(968711)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hughes CHK, Smith OE, Meinsohn MC,
Brunelle M, Gevry N and Murphy BD: Steroidogenic factor 1 (SF-1;
Nr5a1) regulates the formation of the ovarian reserve. Proc Natl
Acad Sci USA. 120(e2220849120)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Chiappara G, Di Vincenzo S, Cascio C and
Pace E: Stem cells, Notch-1 signaling, and oxidative stress: A
hellish trio in cancer development and progression within the
airways. Is there a role for natural compounds? Carcinogenesis.
45:621–629. 2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Feng G, Zhang H, Guo Q, Shen X, Wang S,
Guo Y and Zhong X: NONHSAT098487.2 protects cardiomyocytes from
oxidative stress injury by regulating the Notch pathway. Heliyon.
9(e17388)2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Shaoyong W, Jin H, Jiang X, Xu B, Liu Y,
Wang Y and Jin M: Benzo [a] pyrene-loaded aged polystyrene
microplastics promote colonic barrier injury via oxidative
stress-mediated notch signalling. J Hazard Mater.
457(131820)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zhou Z, Li Y, Ding J, Sun S, Cheng W, Yu
J, Cai Z, Ni Z and Yu C: Chronic unpredictable stress induces
anxiety-like behavior and oxidative stress, leading to diminished
ovarian reserve. Sci Rep. 14(30681)2024.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang B, Zeng M, Zhang Q, Wang R, Jia J,
Cao B, Liu M, Guo P, Zhang Y, Zheng X and Feng W: Ephedrae Herba
polysaccharides inhibit the inflammation of ovalbumin induced
asthma by regulating Th1/Th2 and Th17/Treg cell immune imbalance.
Mol Immunol. 152:14–26. 2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Buratini J, Dellaqua TT, Dal Canto M, La
Marca A, Carone D, Mignini Renzini M and Webb R: The putative roles
of FSH and AMH in the regulation of oocyte developmental
competence: From fertility prognosis to mechanisms underlying
age-related subfertility. Hum Reprod Update. 28:232–254.
2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Gilchrist RB, Lane M and Thompson JG:
Oocyte-secreted factors: Regulators of cumulus cell function and
oocyte quality. Hum Reprod Update. 14:159–177. 2008.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jin L, Yang Q, Zhou C, Liu L, Wang H, Hou
M, Wu Y, Shi F, Sheng J and Huang H: Profiles for long non-coding
RNAs in ovarian granulosa cells from women with PCOS with or
without hyperandrogenism. Reprod Biomed Online. 37:613–623.
2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Levi AJ, Raynault MF, Bergh PA, Drews MR,
Miller BT and Scott RT Jr: Reproductive outcome in patients with
diminished ovarian reserve. Fertil Steril. 76:666–669.
2001.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhang S, Zou X, Feng X, Shi S, Zheng Y, Li
Q and Wu Y: Exosomes derived from hypoxic mesenchymal stem cell
ameliorate premature ovarian insufficiency by reducing
mitochondrial oxidative stress. Sci Rep. 15(8235)2025.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Zhang L, Zhou H, Li J, Wang X, Zhang X,
Shi T and Feng G: Comprehensive Characterization of Circular RNAs
in Neuroblastoma Cell Lines. Technol Cancer Res Treat.
19(1533033820957622)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Khan F, Chen Y, Hartwell HJ, Yan J, Lin
YH, Freedman A, Zhang Z, Zhang Y, Lambe AT, Turpin BJ, et al:
Heterogeneous oxidation products of fine particulate isoprene
Epoxydiol-derived methyltetrol sulfates increase oxidative stress
and inflammatory gene responses in human lung cells. Chem Res
Toxicol. 36:1814–1825. 2023.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Li F, Wang Y, Xu M, Hu N, Miao J, Zhao Y
and Wang L: Single-nucleus RNA Sequencing reveals the mechanism of
cigarette smoke exposure on diminished ovarian reserve in mice.
Ecotoxicol Environ Saf. 245(114093)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chen P, Li W, Liu X, Wang Y, Mai H and
Huang R: Circular RNA expression profiles of ovarian granulosa
cells in advanced-age women explain new mechanisms of ovarian
aging. Epigenomics. 14:1029–1038. 2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Zhu X, Li W, Lu M, Shang J, Zhou J, Lin L,
Liu Y, Xing J, Zhang M, Zhao S, et al: M6A demethylase
FTO-stabilized exosomal circBRCA1 alleviates oxidative
stress-induced granulosa cell damage via the miR-642a-5p/FOXO1
axis. J Nanobiotechnology. 22(367)2024.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Jia C, Wang S, Yin C, Liu L, Zhou L and Ma
Y: Loss of hsa_circ_0118530 inhibits human Granulosa-like tumor
cell line KGN cell injury by sponging miR-136. Gene.
744(144591)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wang S, Wang Y, Qin Q, Li J, Chen Q, Zhang
Y, Li X and Liu J: Berberine protects against
Dihydrotestosterone-induced human ovarian granulosa cell injury and
ferroptosis by regulating the circ_0097636/MiR-186-5p/SIRT3
pathway. Appl Biochem Biotechnol. 196:5265–5282. 2024.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yang C, Li Q, Chen X, Zhang Z, Mou Z, Ye
F, Jin S, Jun X, Tang F and Jiang H: Circular RNA circRGNEF
promotes bladder cancer progression via miR-548/KIF2C axis
regulation. Aging (Albany NY). 12:6865–6879. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Xie F, Huang C, Liu F, Zhang H, Xiao X,
Sun J, Zhang X and Jiang G: CircPTPRA blocks the recognition of RNA
N6-methyladenosine through interacting with IGF2BP1 to
suppress bladder cancer progression. Mol Cancer.
20(68)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
She J, Tuerhongjiang G, Guo M, Liu J, Hao
X, Guo L, Liu N, Xi W, Zheng T, Du B, et al: Statins aggravate
insulin resistance through reduced blood glucagon-like peptide-1
levels in a microbiota-dependent manner. Cell Metab. 36:408–421.e5.
2024.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Ferriere A and Tabarin A: Cushing's
syndrome: Treatment and new therapeutic approaches. Best Pract Res
Clin Endocrinol Metab. 34(101381)2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Auer MK, Nordenstrom A, Lajic S and Reisch
N: Congenital adrenal hyperplasia. Lancet. 401:227–244.
2023.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Macut D, Ilic D, Mitrovic Jovanovic A and
Bjekic-Macut J: Androgen-Secreting ovarian tumors. Front Horm Res.
53:100–107. 2019.PubMed/NCBI View Article : Google Scholar
|