Introduction
Globally, lung cancer is listed as one of the most
common and lethal types of cancer (1,2). In
total, ~85% of lung cancer cases are non-small cell lung carcinoma
(NSCLC) (3), with lung
adenocarcinoma being the most common subtype of NSCLC. Early-stage
lung cancer often presents with minimal or mild clinical symptoms;
therefore, ~75% of patients are already at an advanced stage at
initial diagnosis (4). When
diagnosed, pleural effusion is present in 20% of patients with lung
cancer (5) and the survival of
patients with lung cancer with malignant pleural effusion (MPE) is
5.5 months from diagnosis (6).
Furthermore, most patients with pleural effusion are not eligible
for surgical intervention due to the advanced stage of the
disease.
Mutations in the EGFR gene have been recognized as
key markers for individualized therapy and evaluating prognosis in
advanced lung cancer, given their close association with the
appropriateness of specific treatments, such as targeted therapy
(7). However, for numerous
patients with advanced lung cancer, sufficient tumor tissue for
molecular diagnosis cannot be obtained due to the inherent
invasiveness and sampling limitations of puncture biopsy or
surgical procedures. MPE cell blocks, often containing tumor cells
from patients, are recognized by pathologists as a valuable
resource for genetic testing, offering a unique opportunity to
directly analyze and understand the molecular characteristics of
malignancies (8). Pleural effusion
may be an alternative specimen to tumor tissue, as it can be safely
and repeatedly collected via thoracentesis (9). A previous study showed that the use
of pleural effusion cell blocks combined with immunohistochemistry
and molecular testing markedly improved the accuracy and
specificity of advanced lung cancer diagnosis compared with
standalone pleural effusion cytology (10).
The objective of the present study was to provide
clinical proof to confirm the diagnostic and predictive
significance of pleural effusion cell block technology in
diagnosing lung cancer and guiding decisions for patient-specific
clinical treatments. Immunohistochemical analysis was performed on
pleural effusion cell blocks from patients with advanced lung
cancer for histological classification, and an amplification
refractory mutation system (ARMS) was used to detect the EGFR
target gene mutation status in cell blocks from patients with
confirmed lung adenocarcinoma.
Materials and methods
Patients
The present study encompassed 231 patients with lung
cancer with pleural effusion complications (MPE) who presented at
Weifang No. 2 People's Hospital (Weifang, China) between July 2018
and December 2022. The population comprised of 129 male patients
and 102 female patients, aged 30-91 years old (mean age,
68.61±10.53 years). Samples of pleural effusion were collected from
each patient through the hospital medical system. No patient had
undergone targeted therapy. Data on clinicopathological features
were collected from the Weifang No. 2 People's Hospital health
record system. Patient overall survival (OS) information was
acquired via telephone follow-ups. The monitoring phase commenced
in July 2018 and concluded in February 2022.
Criteria for inclusion were as follows: i) Patients
diagnosed with lung cancer after admission on the basis of clinical
symptoms, bronchoscopy and pathological examination of cell blocks
using hematoxylin and eosin (H&E) staining and
immunohistochemistry; ii) patients with lung cancer who had not
received targeted therapy prior to pleural effusion collection;
iii) patients with sufficient pleural effusion material for the
preparation of cell blocks and subsequent experimental procedures
(at least 20 tumor cells in one H&E-stained section of the cell
block); and iv) comprehensive clinical data for patients provided
by the medical record system, encompassing the patient's age, sex,
clinical stage and smoking history.
The exclusion criteria were: i) Individuals
suffering from acute respiratory, heart or brain vessel disorders;
and ii) individuals with a background of or simultaneous presence
of other cancerous diseases, such as breast, colon or cervical
cancer.
The Ethics Committee of Weifang No. 2 People's
Hospital approved the present study (approval no. KY2023-015-01)
and all participants provided written informed consent for the use
of their tissues/data for scientific research purposes.
Reagents and instruments
The primary antibodies used in immunohistochemistry
were purchased from Fuzhou Maixin Biotechnology Development Co.,
Ltd. The secondary antibody and colorimetric reagents were obtained
from Roche Diagnostics (Shanghai) Co., Ltd.
A low-speed centrifuge (TDL-40B) was obtained from
Shandong Bio-Medical Technology Co., Ltd. The BenchMark GX
automated staining system was purchased from Roche Diagnostics
(Shanghai) Co., Ltd. The SLAN-96S fully automated medical
polymerase chain reaction (PCR) analysis system was purchased from
Shanghai Hongshi Medical Technology Co., Ltd.
Preparation of cell blocks
After collection of pleural fluid specimens, the
samples were promptly delivered to the Department of Pathology of
Weifang No. 2 People's Hospital. The sample was maintained at
ambient temperature for a duration of 15-30 min, following which
50-100 ml liquid was collected from the bottom of the container.
Samples underwent centrifugation at 1,000 x g for 5 min at room
temperature, followed by the extraction of the pellet for
cytological smear analysis by H&E staining. Following removal
of the supernatant, a solution of 4% neutral buffered formalin was
introduced to fix the pellet samples for 15 min at room
temperature. Subsequently, samples underwent centrifugation at
1,000 x g for 5 min at room temperature, after which the pellet was
moved to qualitative filter paper (Nanchang Yulu Experimental
Equipment Co., Ltd.) and then routinely dehydrated through a graded
ethanol series for 1-2 h at each concentration to remove water,
followed by clearing with xylene to achieve transparency for a
duration of 1-3 h. The slides were embedded in paraffin and
subsequently stained with H&E, with hematoxylin applied for 5
min and eosin applied for 30 sec, both at room temperature. An
optical microscope was used to observe the results.
Immunohistochemistry
The cell blocks were sectioned into 4-µm slices and
stained using the Roche BenchMark GX fully automated
immunohistochemistry system. Positive (antigen-expressing tissue)
and negative (replacement of primary antibody with negative
reagent) controls were routinely set up. The primary antibodies
used included antibodies against pan-cytokeratin (pCK), CK7, CK5/6,
napsin A, thyroid transcription factor-1 (TTF-1), P40, Wilms tumor
1 (WT-1), synaptophysin, chromogranin A (CgA), homeobox protein
CDX2, villin, CD56, Ki-67, desmin and calretinin (Table SI; Data SI). All cell blocks were
independently evaluated by two pathologists to ascertain whether
the cytomorphology matched the immunophenotyping results and to
assess whether the tumor cell content met the requirements for PCR
testing (a cell block H&E-stained section must contain at least
20 tumor cells). Tumor cells of lung adenocarcinoma exhibit
glandular/acinar patterns, while tumor cells of small cell
carcinoma are tightly arranged in a ‘stacked or regimented’
formation and tumor cells of squamous cell carcinoma are often
dispersed as single cells or in small clusters.
Genomic DNA extraction from
paraffin-embedded samples
After the tumor cell content was evaluated by
immunohistochemistry (number of tumor cells and percentage of tumor
cells relative to the total tissue area in the H&E-stained
section), 4-12 cell block sections were cut to a 5-µm thickness
using a disposable microtome blade and transferred into clean
1.5-ml centrifuge tubes. DNA extraction was performed using a
nucleic acid extraction kit (FFPE DNA; cat. no. 8.02.0017; Amoy
Diagnostics Co., Ltd.) following the manufacturer's instructions.
DNA purity and concentration were quantified using an Eppendorf UV
spectrophotometer (Eppendorf SE).
Detection of EGFR gene mutations
EGFR gene mutation status was evaluated by ADx-ARMS
using the human EGFR gene mutation detection kit (cat. no.
8.01.0131; Amoy Diagnostics Co., Ltd.), including primers and
fluorophore, following the manufacturer's guidelines. Using this
kit, the ARMS technique was used to identify 21 prevalent forms of
mutations in EGFR exons 18, 19, 20 and 21. The thermocycling
conditions used were as follows: 1 cycle at 95˚C for 5 min; 15
cycles at 95˚C for 25 sec, 64˚C for 20 sec and 72˚C for 20 sec; and
31 cycles at 93˚C for 25 sec, 60˚C for 35 sec and 72˚C for 20 sec.
Upon completion of the reaction, the SLAN-96S fluorescence
quantitative PCR instrument provided the amplification curve, and
the mutations were determined following the interpretation
principles outlined in the kit instructions. Each experiment
included a positive control from the kit and nuclease-free purified
water as a negative control.
Patient treatment
Patients with single EGFR mutations received EGFR
tyrosine kinase inhibitors as first-line treatment, as detailed in
Table I. Patients without EGFR
gene mutations were treated with chemotherapy, specifically
cisplatin (a platinum-based drug) at 75 mg/m² combined with
pemetrexed (an antimetabolite agent) at 500 mg/m² for 4-6 cycles,
followed by pemetrexed maintenance at 500 mg/m². Treatment
continued until disease progression or the occurrence of
intolerable adverse reactions. Based on the completeness of
follow-up data, prognostic analysis was performed only on 36 cases
with EGFR gene mutations and 65 cases without EGFR gene
mutations.
 | Table ITKI treatments (patients received only
one oral medication listed). |
Table I
TKI treatments (patients received only
one oral medication listed).
| EGFR mutation
type | Specific TKI | Generation | Dosage | Treatment
duration |
|---|
| L858R 19-del | Gefitinib | 1st Generation | 250 mg once
daily | Until disease
progression or unacceptable toxicity |
| | Erlotinib | 1st Generation | 150 mg once
daily | |
| | Icotinib | 1st Generation | 125 mg three times
daily | |
| | Afatinib | 2nd Generation | 40 mg once daily | |
| | Osimertinib | 3rd Generation | 80 mg once daily | |
| G719X,L861Q | Afatinib | 2nd Generation | 40 mg once daily | |
Statistical and prognostic
analyses
Statistical evaluation was performed using SPSS 25.0
software (IBM Corp.). The χ2 test was employed to
explore the associations between mutations in the EGFR gene and
clinicopathological features. Data are presented as n (%). The
analysis of prognosis was performed with GraphPad Prism 9.4.0
software (Dotmatics). Survival curves were generated using
Kaplan-Meier analysis. Statistical evaluation of the survival rates
for patients with EGFR mutations and those with wild-type EGFR was
performed using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Histological types of lung tumors in
the study group
H&E analysis and immunohistochemistry with
different antibodies was conducted on the cell blocks from pleural
effusions of 231 patients with advanced lung cancer for
histological classification. TTF-1, P40, CDX-2, Ki-67 and WT-1
exhibited nuclear staining, pCK, CK7, CK5/6, napsin A, villin,
desmin, CgA and synaptophysin showed cytoplasmic staining, CD56
displayed membrane staining, and calretinin showed either nuclear
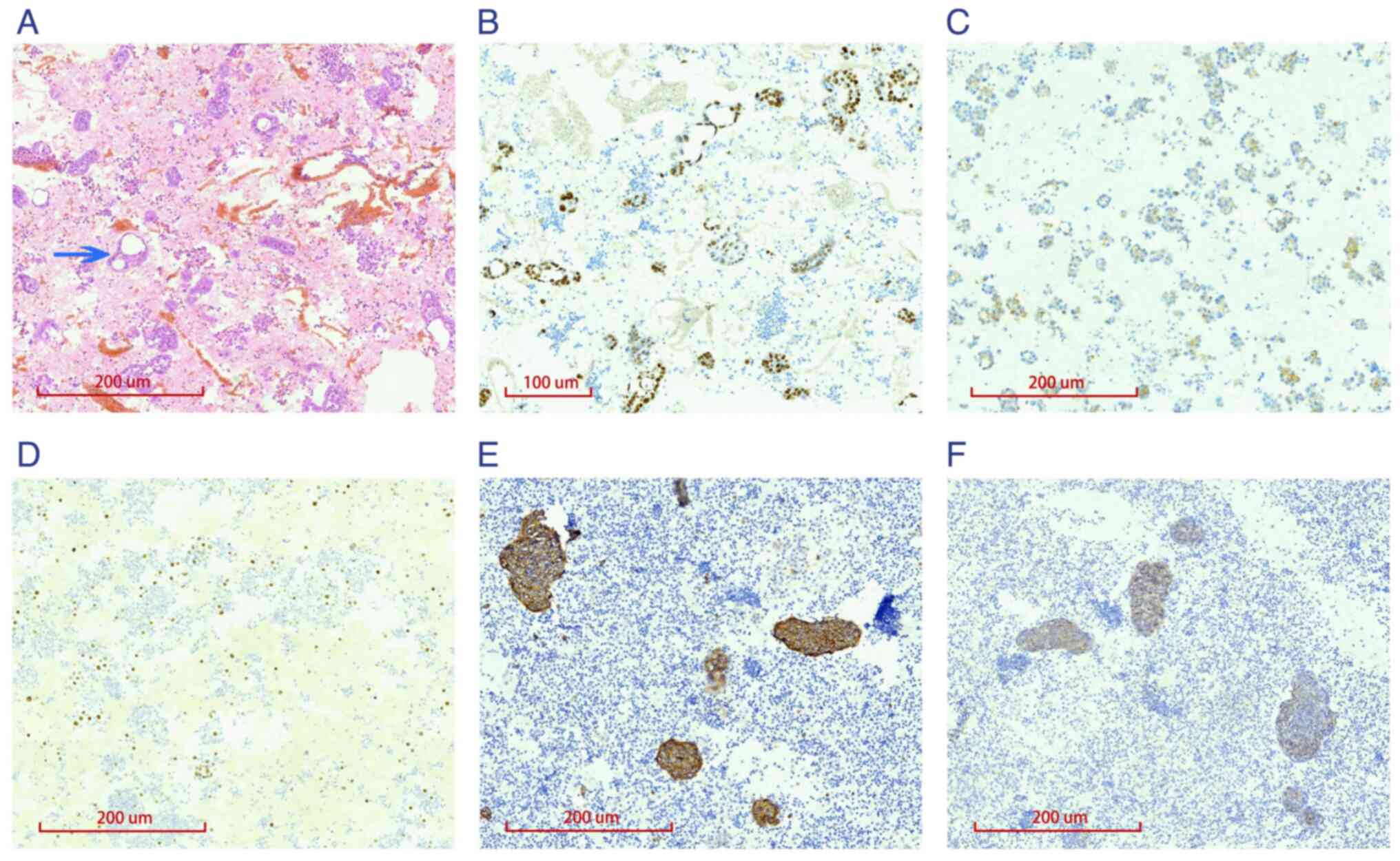
or cytoplasmic staining. Among the 231 cell blocks analyzed, 222
showed cancer cells arranged in glandular and rosette-like patterns
(Fig. 1A), with positive staining
for the lung adenocarcinoma markers TTF-1, napsin A and CK7
(Figs. 1B and C and S1A). Two cases were positive for the
squamous cell carcinoma markers P40 and CK5/6 (Figs. 1D and S2D). Seven cases showed positive
staining for the neuroendocrine markers synaptophysin, CD56 and CgA
(Figs. 1E and F and S2B), along with focal perinuclear
dot-like expression of cytokeratin (Fig. S2A), supporting a diagnosis of
small cell carcinoma. Additionally, the Ki-67 proliferation marker
was highly expressed in the small cell carcinoma cases (Fig. S2C). The remaining proteins
(calretinin, WT-1, villin, CDX2 and desmin) showed negative
staining in the tumor cells (Fig.
S1B-F).
EGFR gene mutations in the study
group
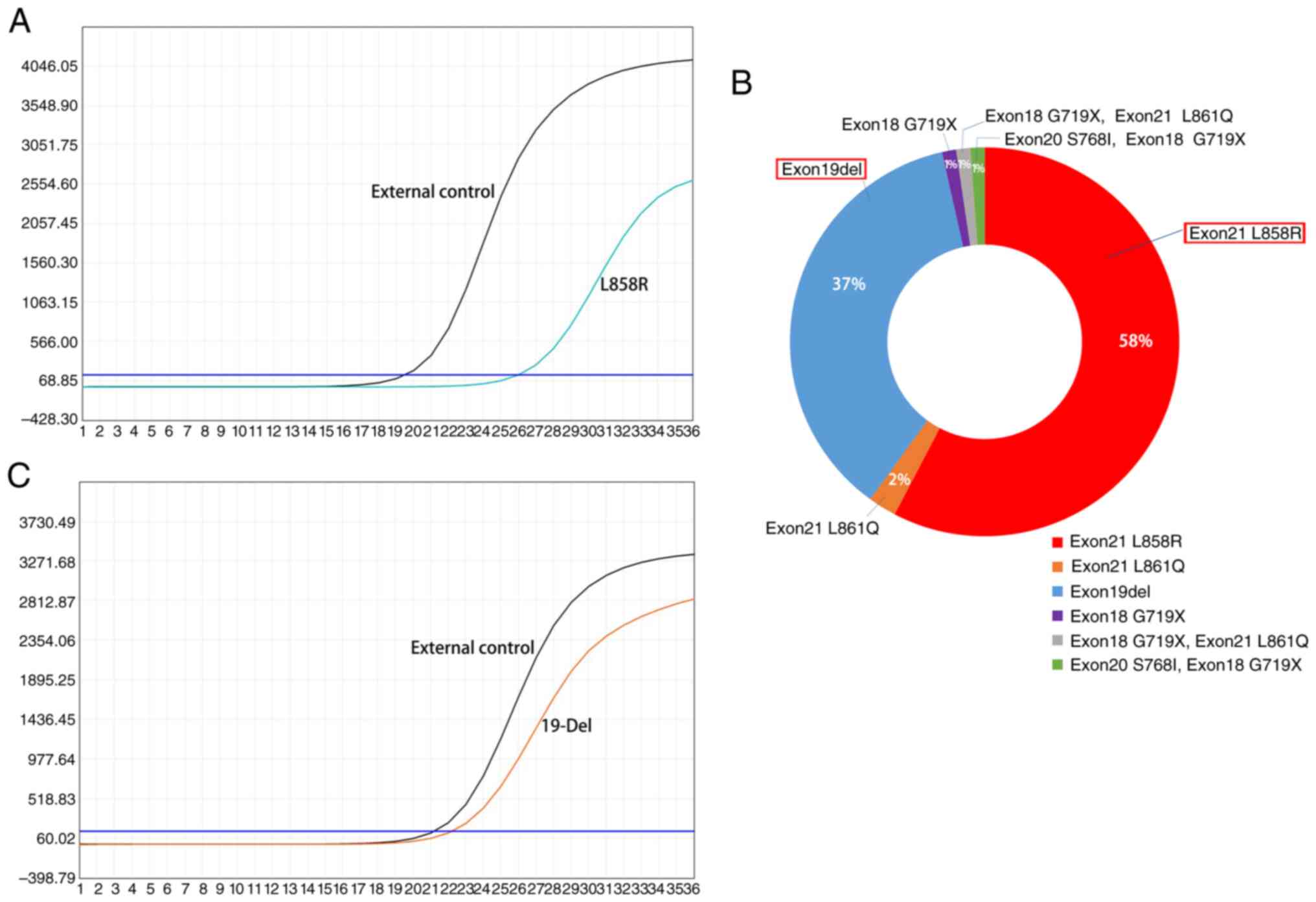
Among the 231 pleural effusion cell blocks from
patients with lung cancer, 161 underwent EGFR gene mutation
testing, all of which were obtained from patients with lung
adenocarcinoma. The remaining 70 patients did not receive the
testing for various reasons, such as financial constraints. Among
the 161 cases, 85 were positive for EGFR gene mutations (positive
rate, 52.8%). The detected genetic alterations encompassed 49
instances of the L858R mutation in exon 21, 31 instances of
deletions in exon 19, 2 instances of the L861Q mutation in exon 21
and a single case of the G719X mutation in exon 18. Another 2
patients showed complex mutations; 1 patient had the G719X mutation
in exon 18 alongside the L861Q mutation in exon 21, whereas the
other patient had the S768I mutation in exon 20 coupled with the
G719X mutation in exon 18 (Figs. 2
and S3).
Association between EGFR gene
mutations and clinicopathological characteristics in patients with
lung adenocarcinoma
Subsequently, the present study focused on the link
between the clinicopathological traits of 161 patients with lung
adenocarcinoma and EGFR mutations detected through pleural effusion
cell block analysis. There was a significantly greater occurrence
of EGFR gene mutations in female patients (32.3%) than in male
patients (20.5%) (P=0.001) (Table
II). Whereas EGFR gene mutations tended to show differences in
patients stratified by age and smoking history, the results did not
reach statistical significance (Table
II).
 | Table IIAssociation of EGFR gene mutations
with patient clinical parameters. |
Table II
Association of EGFR gene mutations
with patient clinical parameters.
| | EGFR gene
mutations | |
|---|
| Clinical
parameter | Patient number | Mutation | Wild-type | Mutation rate, % |
χ2-value | P-value |
|---|
| Sex | | | | | 11.682 | 0.001 |
|
Male | 83 | 33 | 50 | 20.50 | | |
|
Female | 78 | 52 | 26 | 32.30 | | |
| Age, years | | | | | 0.328 | 0.608 |
|
<65 | 48 | 27 | 21 | 16.77 | | |
|
≥65 | 113 | 58 | 55 | 36.03 | | |
| Smoking
history | | | | | 3.520 | 0.086 |
|
Smoker | 66 | 29 | 37 | 18.02 | | |
|
Non-smoker | 95 | 56 | 39 | 34.78 | | |
Prognostic comparison
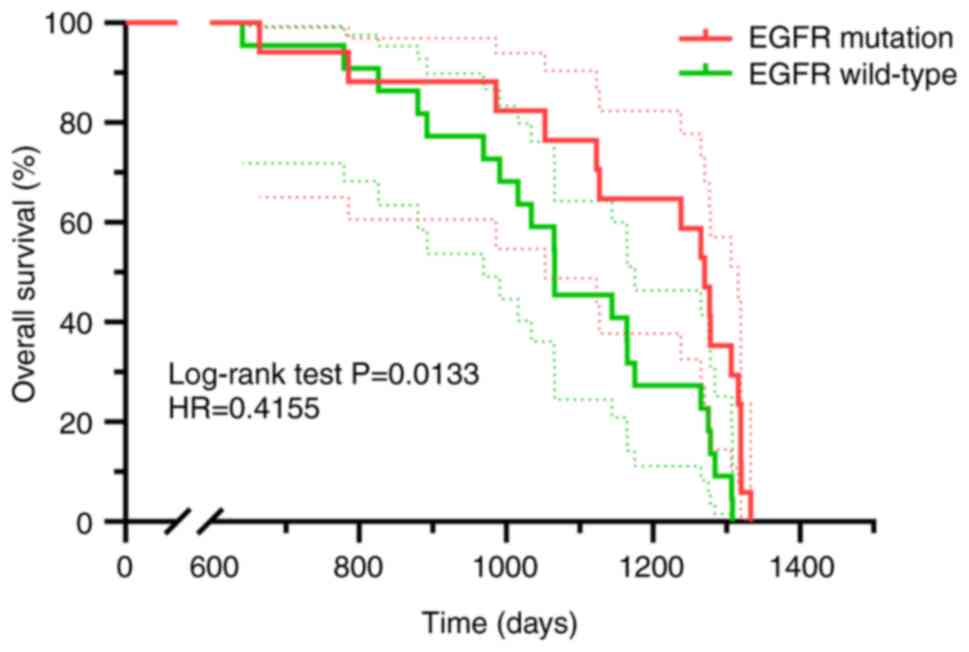
An observational analysis was performed on 36
patients exhibiting single EGFR mutations determined from pleural
effusion cell blocks and receiving EGFR tyrosine kinase inhibitor
(TKI) as their primary therapeutic intervention and 65 patients
without EGFR gene mutations determined from pleural effusion cell
blocks and treated with chemotherapy as first-line treatment. The
monitoring phase concluded in February 2022. Patients carrying EGFR
mutations exhibited a significantly extended OS rate compared with
patients with wild-type EGFR (hazard ratio=0.4155, P=0.0133;
Fig. 3).
Discussion
Over the last 20 years, studies have shown that
focusing on specific driver genes in therapy can extend the
duration of progression-free survival and OS in patients with lung
cancer. A study by Ramalingam et al (11) demonstrated that patients with EGFR
mutations (exon 19 deletion or L858R) who received osimertinib
treatment showed significantly extended OS. Alterations in the EGFR
gene frequently emerge as the primary driver of genetic changes in
lung cancer (12). Multiple
studies (13-15)
have demonstrated the superior efficacy and reduced toxicity of
targeted drugs compared with traditional chemotherapy, indicating
that targeted drug treatments provide sustained clinical benefits
and markedly improve prognostic outcomes for patients with lung
cancer. For patients with advanced NSCLC and sensitizing EGFR
mutations, EGFR TKIs serve as the primary treatment option
(16); however, sufficient tumor
tissue for genetic testing is not available for all patients with
advanced NSCLC.
The present study demonstrates that detecting
genetic mutations through MPE cell blocks is of value for
predicting patient response to targeted therapy. The ARMS-PCR
technique was employed to detect genetic mutations that drive tumor
progression in MPE cell blocks from patients with NSCLC lacking
tumor tissue samples. The findings showed a 52.8% detection rate of
EGFR mutations in MPE specimens, a rate similar to that observed
with tumor tissue samples (8,17).
The detected mutations included the L858R mutation in exon 21 and
the deletion mutation in exon 19 of EGFR, detected in 49 and 31
patients, respectively, constituting 95% of all detected EGFR
mutations. These proportions align with previous results in studies
of tumor tissues with reported rates of 85-90% (18-20).
The results of the present study also revealed the presence of
three less common mutation types (L861Q, G719X and S768I), with no
other rare EGFR mutations identified. These findings indicated that
the frequency of EGFR oncogenic driver gene mutations identified in
MPE may mirror that found in tumor tissues.
While a previous study has verified the diagnostic
precision of MPE in detecting oncogenic driver genes compared with
tumor tissue (21), to the best of
our knowledge there is almost no research on the efficacy of TKI
targeted therapy administered solely on the basis of the detection
of mutations through MPE analysis. The current study revealed that
patients with single EGFR mutations identified via MPE analysis and
treated initially with EGFR-TKIs experienced a significantly longer
OS rate compared with patients without EGFR mutations who received
chemotherapy as first-line treatment. This aligns with a previous
study on patients with EGFR sensitizing mutations detected on tumor
tissue who were administered EGFR-TKIs as an initial treatment
(11). Additionally, the findings
of the present study demonstrated variations in EGFR mutation rates
in MPE based on sex, with a notably higher incidence in female
patients than in male patients, consistent with the outcomes of
previous studies based on tumor tissue analysis, where the
detection rate of EGFR gene mutation was 52.8% in women and 31.6%
in men (22-24).
The present study has certain limitations. First,
only ARMS-PCR was used for detection of EGFR gene mutations, and
this technique has limitations in terms of detection sensitivity
and coverage (25,26), particularly in identifying
low-frequency mutations and rare mutations related to EGFR-TKI
resistance. This limitation affects the completeness of the
mutation spectrum in MPE samples. Upcoming initiatives ought to
focus on employing diverse mutation detection platforms, and
amalgamating advanced detection methods, such as next-generation
sequencing and droplet digital PCR, for a more detailed gene
mutation profile. Secondly, since this study only collected data
from malignant pleural effusion cell blocks of patients with lung
cancer, and the data analysis was based on comparisons with other
literature rather than a direct comparison with the patients' own
tumor tissue specimens, there are inherent differences in the
research subjects and tumor heterogeneity, which impose relative
limitations on the results. In future research, the study design
will be refined further by conducting advance planning and patient
recruitment to ensure standardized and consistent data
collection.
In conclusion, the findings of the present study
indicated that the OS rate of EGFR-TKI-treated patients, with EGFR
gene mutations identified solely via MPE cell block tests, may be
comparable with that of patients with similar mutations identified
on tumor tissues in previous studies. Furthermore, the frequency of
EGFR gene alterations identified in MPE paralleled that reported in
cancerous tissues in previous studies. Therefore, when sufficient
tumor tissue cannot be obtained, MPE is recommended as an
alternative specimen for the detection of EGFR mutations.
Supplementary Material
Supplementary methods
Immunohistochemical staining of
pleural effusion cell blocks from patients with lung cancer. (A)
Immunohistochemical staining showing positive cytokeratin 7
expression in adenocarcinoma. Immunohistochemical staining showing
(B) negative calretinin expression, (C) negative WT-1 expression,
(D) negative villin expression, (E) negative homeobox protein CDX2
expression and (F) negative desmin expression in tumor cells.
Immunohistochemical staining of
further markers in pleural effusion cell blocks from patients with
lung cancer. (A) Immunohistochemical staining showing perinuclear
dot-like positivity for pan-cytokeratin in small cell carcinoma.
(B) Immunohistochemical staining showing focal weak positivity (red
arrows) for chromogranin A in small cell carcinoma. (C)
Immunohistochemical staining showing a high Ki-67 proliferation
index in small cell carcinoma. (D) Immunohistochemical staining
showing positive cytokeratin 5/6 expression in squamous cell
carcinoma.
Representative detection images of the
other four types of EGFR gene mutations. (A) Exon 21 L861Q
mutation. (B) Exon 18 G719X mutation. (C) Compound mutation of exon
18 G719X and exon 21 L861Q. (D) Compound mutation of exon 20 S768I
and exon 18 G719X.
Ready-to-use primary antibodies
incubated at 37˚C for 32 min.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Health
Commission of Weifang, China (grant nos. WFWSJK-2023-122 and
2024-200) and the Medical and Health Science and Technology Project
of Shandong, China (grant no. 202401040090).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SZ, YuL and XC designed the present study. SZ, YuL
and TW conducted the experiments. ZC and YiL performed statistical
analysis, and SZ wrote the manuscript. XC and YiL conducted the
systematic literature search and supervised the writing of the
manuscript. SZ, YuL and XC confirm the authenticity of all the raw
data. All authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
ethical standards and was approved by the Ethics Committee of
Weifang No. 2 People's Hospital (approval no. KY2023-015-01;
Weifang, China). Written informed consent was obtained from all
patients for the use of their data and specimens in scientific
research.
Patient consent for publication
Written informed consent was obtained for
publication of the patient data and images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Huang S, Yang J, Shen N, Xu Q and Zhao Q:
Artificial intelligence in lung cancer diagnosis and prognosis:
Current application and future perspective. Semin Cancer Biol.
89:30–37. 2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lee E and Kazerooni EA: Lung cancer
screening. Semin Respir Crit Care Med. 43:839–850. 2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cho JH: Immunotherapy for non-small-cell
lung cancer: Current status and future obstacles. Immune Netw.
17:378–391. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chen Y, Mathy NW and Lu H: The role of
VEGF in the diagnosis and treatment of malignant pleural effusion
in patients with nonsmall cell lung cancer (review). Mol Med Rep.
17:8019–8030. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ferreiro L, Suárez-Antelo J,
Álvarez-Dobaño JM, Toubes ME, Riveiro V and Valdés L: Malignant
pleural effusion: Diagnosis and management. Can Respir J.
2020(2950751)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Russo A, Lopes AR, McCusker MG, Garrigues
SG, Ricciardi GR, Arensmeyer KE, Scilla KA, Mehra R and Rolfo C:
New targets in lung cancer (excluding EGFR, ALK, ROS1). Curr Oncol
Rep. 22(48)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mao L, Zhao W, Li X, Zhang S, Zhou C, Zhou
D, Ou X, Xu Y, Tang Y, Ou X, et al: Mutation spectrum of
EGFR from 21,324 Chinese patients with non-small cell lung
cancer (NSCLC) successfully tested by multiple methods in a
CAP-accredited laboratory. Pathol Oncol Res.
27(602726)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mohammed A, Hochfeld U, Hong S, Hosseini
DK, Kim K and Omidvari K: Thoracentesis techniques: A literature
review. Medicine (Baltimore). 103(e36850)2024.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Porcel JM: Diagnosis and characterization
of malignant effusions through pleural fluid cytological
examination. Curr Opin Pulm Med. 25:362–368. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ramalingam SS, Vansteenkiste J, Planchard
D, Cho BC, Gray JE, Ohe Y, Zhou C, Reungwetwattana T, Cheng Y,
Chewaskulyong B, et al: Overall survival with osimertinib in
untreated, EGFR-mutated advanced NSCLC. N Engl J Med.
382:41–50. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Su C: Emerging insights to lung cancer
drug resistance. Cancer Drug Resist. 5:534–540. 2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu YL, Saijo N, Thongprasert S, Yang JC,
Han B, Margono B, Chewaskulyong B, Sunpaweravong P, Ohe Y, Ichinose
Y, et al: Efficacy according to blind independent central review:
Post-hoc analyses from the phase III, randomized, multicenter,
IPASS study of first-line gefitinib versus carboplatin/paclitaxel
in Asian patients with EGFR mutation-positive advanced NSCLC. Lung
Cancer. 104:119–125. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shi Y, Zhang L, Liu X, Zhou C, Zhang L,
Zhang S, Wang D, Li Q, Qin S, Hu C, et al: Icotinib versus
gefitinib in previously treated advanced non-small-cell lung cancer
(ICOGEN): A randomised, double-blind phase 3 non-inferiority trial.
Lancet Oncol. 14:953–961. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hu X, Zhang L, Shi Y, Zhou C, Liu X, Wang
D, Song Y, Li Q, Feng J, Qin S, et al: The efficacy and safety of
Icotinib in patients with advanced non-small cell lung cancer
previously treated with chemotherapy: A single-arm, multi-center,
prospective study. PLoS One. 10(e0142500)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ettinger DS, Wood DE, Aisner DL, Akerley
W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, DeCamp M,
et al: NCCN guidelines® insights: Non-small cell lung
cancer, version 2.2023. J Natl Compr Canc Netw. 21:340–350.
2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang JL, Fu YD, Gao YH, Li XP, Xiong Q, Li
R, Hou B, Huang RS, Wang JF, Zhang JK, et al: Unique
characteristics of G719X and S768I compound double mutations of
epidermal growth factor receptor (EGFR) gene in lung cancer of
coal-producing areas of East Yunnan in Southwestern China. Genes
Environ. 44(17)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Batra U, Biswas B, Prabhash K and Krishna
MV: Differential clinicopathological features, treatments and
outcomes in patients with exon 19 deletion and exon 21 L858R EGFR
mutation-positive adenocarcinoma non-small-cell lung cancer. BMJ
Open Respir Res. 10(e001492)2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kutsuzawa N, Takahashi F, Tomomatsu K,
Obayashi S, Takeuchi T, Takihara T, Hayama N, Oguma T, Aoki T and
Asano K: Successful treatment of a patient with lung adenocarcinoma
harboring compound EGFR gene mutations, G719X and S768I, with
afatinib. Tokai J Exp Clin Med. 45:113–116. 2020.PubMed/NCBI
|
|
20
|
Suda K, Mitsudomi T, Shintani Y, Okami J,
Ito H, Ohtsuka T, Toyooka S, Mori T, Watanabe SI, Asamura H, et al:
Clinical impacts of EGFR mutation status: Analysis of 5780
surgically resected lung cancer cases. Ann Thorac Surg.
111:269–276. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yao Y, Peng M, Shen Q, Hu Q, Gong H, Li Q,
Zheng Z, Xu B, Li Y and Dong Y: Detecting EGFR mutations and
ALK/ROS1 rearrangements in non-small cell lung cancer using
malignant pleural effusion samples. Thorac Cancer. 10:193–202.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang H, Zhang W, Wang K and Li X:
Correlation between EML4-ALK, EGFR and clinicopathological features
based on IASLC/ATS/ERS classification of lung adenocarcinoma.
Medicine (Baltimore). 97(e11116)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang T, Zhang Y, Liu B, Hu M, Zhou N and
Zhi X: Associations between epidermal growth factor receptor
mutations and histological subtypes of lung adenocarcinoma
according to the IASLC/ATS/ERS classification in Chinese patients.
Thorac Cancer. 8:600–605. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li D, Ding L, Ran W, Huang Y, Li G, Wang
C, Xiao Y, Wang X, Lin D and Xing X: Status of 10 targeted genes of
non-small cell lung cancer in eastern China: A study of 884
patients based on NGS in a single institution. Thorac Cancer.
11:2580–2589. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
He C, Wei C, Wen J, Chen S, Chen L, Wu Y,
Shen Y, Bai H, Zhang Y, Chen X and Li X: Comprehensive analysis of
NGS and ARMS-PCR for detecting EGFR mutations based on 4467 cases
of NSCLC patients. J Cancer Res Clin Oncol. 148:321–330.
2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu T, Kang X, You X, Dai L, Tian D, Yan W,
Yang Y, Xiong H, Liang Z, Zhao GQ, et al: Cross-platform comparison
of four leading technologies for detecting EGFR mutations in
circulating tumor DNA from non-small cell lung carcinoma patient
plasma. Theranostics. 7:1437–1446. 2017.PubMed/NCBI View Article : Google Scholar
|