Introduction
Osteoarthritis (OA) is a prevalent degenerative
joint disorder that affects the quality of life of >344 million
individuals worldwide, especially postmenopausal women. This places
a substantial burden on healthcare systems as well as a
socioeconomic stress (1-3).
The pathogenesis of OA involves multifactorial factors, including
mechanical stress, inflammation, hormonal imbalance and metabolic
dysregulation. However, a complete understanding of its etiology
and pathological progression is yet to be elucidated. This has led
to a lack of effective clinical interventions among patients with
early-stage OA (4,5).
Among the multifactorial factors involved, estrogen
serves a crucial role in female patients (6). A number of studies demonstrate that
estrogen, particularly estradiol (E2), is a protective factor
against OA as it reduces the release of inflammatory mediators
(such as IL-1β, TNF-α and IL-6) and promotes cartilage matrix
synthesis, which can mitigate the onset and progression of OA
(3,7-9).
Clinical evidence also indicates that estrogen replacement therapy
can delay cartilage degeneration and reduce the risk of OA among
menopausal women, which highlights its anti-inflammatory and
cartilage-protective properties (10,11).
However, estrogen-dependent uterine fibroids (UFs)
are the most common pelvic tumors in women of childbearing age,
affecting >70% of the global population of women (9,12,13).
UFs cause symptoms including excessive menstrual bleeding, anemia,
pelvic pressure and pain. In addition, UFs elevate the risk of
endometrial cancer and compromises the quality of life of women
(9,14-16).
As typical estrogen-dependent tumors, estrogen binds to estrogen
receptors (ERα/ERβ) on myometrial cells, activating proliferation
pathways in fibroid stem cells, such as the β-catenin pathway. This
promotes the proliferation of smooth muscle cells and inhibits
apoptosis (17). Furthermore,
estrogen also enhances the expression of various cytokines, such as
TGF-β3 and activin A, which leads to an increase in the deposition
of extracellular matrix (ECM) in the fibroid. This serves a
critical role in the proliferation and enlargement of fibroids
(17,18). These dual, yet opposing, roles of
estrogen in OA and UFs indicate the complexity of the hormone.
Inflammation is a common risk factor for UFs and OA.
In OA, inflammatory cytokines, such as IL-1β, induce cartilage
matrix degradation through the upregulation of matrix
metalloproteinases and the suppression of cartilage-specific
proteins, such as collagen II, aggrecan and cartilage oligomeric
matrix protein (19). Furthermore,
TGF-β, which is normally protective, is often dysregulated,
exacerbating cartilage degradation under inflammatory conditions
(20). Additionally, UF tissues
have increased levels of inflammatory cytokines (such as MMPs, TNFs
and ILs) that promote chronic inflammation and the excessive
deposition of ECM (21).
Therefore, inflammation-driven ECM remodeling represents a shared,
yet divergent, pathway in OA and UF pathogenesis.
The complex association between OA and UF is yet to
be fully elucidated. A number of studies suggest a potential
inverse association between UFs and the risk of OA progression
(22-24).
However, a cross-sectional study by Kovari et al (25) reports a higher incidence of UFs in
individuals with advanced OA. This raises the possibility that OA
may influence the development of UFs through shared hormonal or
inflammatory pathways. For instance, both conditions have been
linked to hormonal imbalances, such as increased estrogen activity,
which can influence the growth and inflammation of tissues.
Inflammation-related pathways, including those involving cytokines
such as IL-1β, TNF-α and TGF-β, are commonly seen in both OA and
UF. These cytokines contribute to cartilage degradation in OA and
to the proliferation of smooth muscle cells in uterine fibroids.
Additionally, the activation of angiogenic factors, such as VEGF,
in both conditions may further promote tissue growth and fibrosis.
However, an investigation by Yan et al (26) demonstrates that there is no causal
link between the concentration of E2 in serum from female patients
and the risk of developing OA. The apparent contradictions in the
evidence may be attributed to the vulnerability of conventional
epidemiological approaches to confounders and reverse causality,
rendering the causal link between UF and OA ambiguous (27). However, conducting extensive
randomized controlled trials to investigate the potential genetic
association between UF and OA is impractical and unethical
(28).
Therefore, Mendelian Randomization (MR) was used in
the present study to reduce the biases introduced by confounding
factors and reverse causation (29). The association between the selected
exposures inferred from genetic polymorphisms and their effects
were investigated in the present MR analysis. The unaltered
characteristics of the genetic polymorphisms after conception
suggested that their use as instrumental variables (IVs)
strengthened the validity of the conclusions in the present study
(30). Additionally, the
widespread availability of genetic information in public databases
facilitated the strategy used in the present study (29).
Materials and methods
Study summary
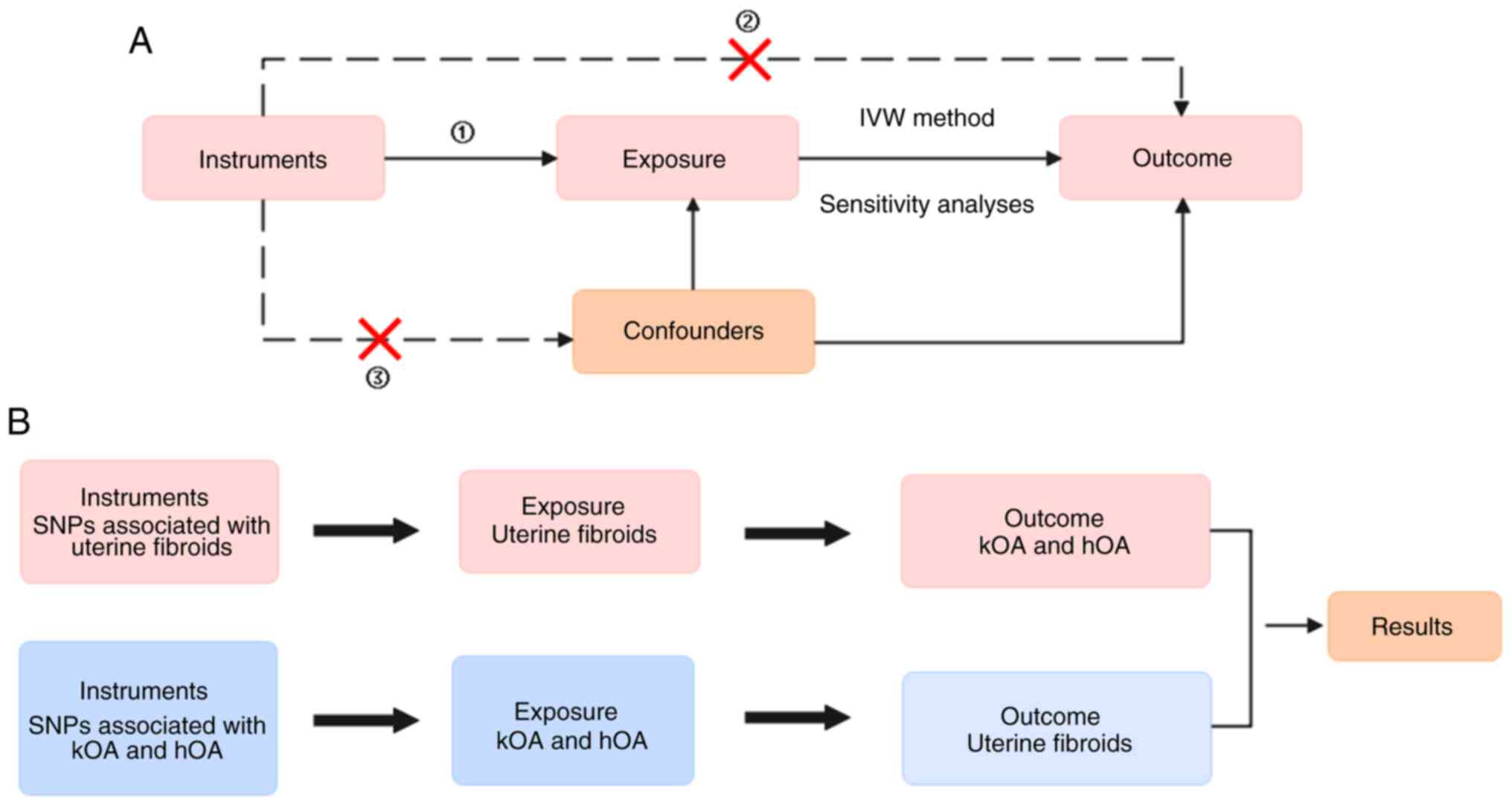
Fig. 1 presents the
design used in the present study of the bidirectional MR
analysis.
Data source and selection of
single-nucleotide polymorphisms (SNPs)
Publicly available data on hip OA (hOA) and knee OA
(kOA) were acquired from a genome-wide study that included
>400,000 European patients (31). The genome-wide association study
(GWAS) database of UF (https://opengwas.io/datasets/ebi-a-GCST90018934) was
obtained from an independent GWAS that included 258,718 European
individuals (32). The detailed
sample characteristics are presented in Table SI.
To satisfy the MR study criteria (33,34),
SNPs with strong associations with UF were selected from the GWAS
database (P<5x10-8). Linkage disequilibrium (LD)
analysis was conducted on these nucleotide polymorphisms to cluster
SNPs (r2 <0.001), and the genetic variants for pairs
in LD were pruned. To minimize bias from sample overlap,
instruments (such as those with an F-statistic >10 for the
instrument-exposure association) were used. Additionally, SNPs were
annotated using the SNiPA SNP annotator online platform (https://snipa.helmholtz-muenchen.de/snipa3/) to find
available proxies for unmatchable SNPs in the GWAS database. The
Phenoscanner database (http://www.phenoscanner.medschl.cam.ac.uk/) was used
to identify SNPs associated with exposure and links to confounding
factors were investigated (P<1x10-5). Table SII was produced by systematically
selecting SNPs from the GWAS database based on the aforementioned
criteria. The selected SNPs were then summarized in Table SII, where their chromosomal
positions, associated genes, effect alleles, non-effect alleles,
effect allele frequencies, β values, P-value, F values and
confounders are shown. These tables directly correspond to the
instrumental variables selected for the bidirectional MR analysis,
which aimed to evaluate causal relationships between UF and OA.
Finally, the filtered SNPs were used as IVs for MR analysis. The
F-statistic was calculated as: F=R2 x (sample
size-2)/(1-R2), where R² is the explained variance in
the exposure by each IV (35,36).
MR analysis
MR analyses were conducted using the version 4.3.1
of TwoSampleMR package (MRC Biostatistics Unit; https://github.com/MRCIEU/TwoSampleMR)
(36), including inverse variance
weighted (IVW), simple median, MR-Egger, weighted mode and weighted
median. If the P-value of the Egger intercept was >0.05, no
potential pleiotropic effects were suggested (37). The MR-pleiotropy residual sum and
outlier method was used to assess the presence of horizontal
pleiotropic outliers. Subsequently, the degree of heterogeneity was
quantified using Cochran's Q statistic (38). ‘Leave-one-out’ analysis, the
sequential exclusion of each SNP, was used to support the
sensitivity and reliability of the outcomes (29). As the results were binary
variables, the β effect estimates were converted to odds ratios
(OR), which were interpreted as the odds of an outcome per unit
increase in exposure (29). The
P-values were adjusted using the Benjamini-Hochberg method
(39).
Cell culture conditions
For the chondrocytes isolated from rats (cat. no.
CP-R087; Procell Life Science & Technology Co., Ltd.), the
culture medium used was rat Articular Chondrocyte Complete Medium
(cat. no. CM-R092; Procell Life Science & Technology Co.,
Ltd.), which primarily contains DMEM/F12, 10% fetal bovine serum
(FBS), 1% penicillin-streptomycin (P-S) solution (cat. no. C0222;
Beyotime Institute of Biotechnology), transferrin and selenium. For
the ELT3 cells (cat. no. 4616; BioVector NTCC Inc.), the complete
culture medium mainly contained DMEM/F12 (cat. no. PM150312;
Procell Life Science & Technology Co., Ltd.), 10% FBS (cat. no.
164250; Procell Life Science & Technology Co., Ltd.), 1% P-S
solution (cat. no. C0222; Beyotime Institute of Biotechnology),
ferrous sulfate and sodium selenite. All the cells were incubated
at 37˚C in a humidified atmosphere containing 5%
CO2.
Cell viability analysis
Using the Cell Counting Kit-8 (CCK-8) assay (cat.
no. C0037; Beyotime Institute of Biotechnology) and a Calcein
AM/propidium iodide (PI) kit (cat. no. CA1630; Beijing Solarbio
Science & Technology Co., Ltd.), the viability of chondrocytes
was evaluated. Chondrocytes were seeded in 6-well plates at a
density of 1-1.5x104 cells/well and pretreated with 10
ng/ml IL-1β for 2 h at 37˚C (OA group). Afterward, they were
incubated at 37˚C for 24 h either alone or with ELT3 cells
(0.4-0.8x104 cells/well) in the OA + UF cell (UFC) group
using a 6-well Transwell co-culture system with a 0.4-µm pore size
(cat. no. 3460; Corning, Inc.). Subsequently, 10 µl of CCK-8
reagent was added to each well, followed by a 2 h incubation at
37˚C. The absorbance at 450 nm was recorded using a Rayto RT-6100
microplate reader.
For live/dead staining, chondrocytes were cultured
under the aforementioned conditions and then treated with Calcein
AM and a PI kit, according to the manufacturer's protocol. Live
cells were stained green with Calcein AM for 20 min at room
temperature and protected from light, while dead cells were stained
red with PI for 5 min under the same conditions. Stained cells were
visualized under a fluorescence microscope and quantified using
Image-Pro (version 6.0; Media Cybernetics, Inc.). All experiments
were performed in triplicate. Data are presented as the mean ±
standard deviation (SD). To avoid phenotypic loss, only passages
1-3 of primary cells were used.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the chondrocytes using
the RNA Easy Fast Tissue/Cell kit (Tiangen Biotech Co., Ltd.). The
extracted RNA was reverse-transcribed into cDNA using the HiScript
III RT SuperMix for qPCR (+gDNA wiper) kit (cat. no. R323-01;
Vazyme Biotech Co., Ltd.) following the manufacturer's protocol..
The cDNA served as a template for qPCR analysis using TB Green
PreMix Ex Taq (cat. no. RR420A; Takara Bio, Inc.), with the
following thermocycling conditions: 95˚C for 30 sec, followed by 40
cycles of 95˚C for 5 sec and 60˚C for 30 sec. Gene expression
levels were determined by calculating the cycle threshold values,
normalized to the internal control GAPDH, and analyzed using the
2-ΔΔCq method (40).
The primer sequences used for the target genes are listed in
Table I.
 | Table IPrimer sequences used in reverse
transcription-quantitative PCR. |
Table I
Primer sequences used in reverse
transcription-quantitative PCR.
| Target genes | Forward primer
sequence (5'-3') | Reverse primer
sequence (5'-3') |
|---|
| Collagen II |
GAGTGGAAGAGCGGAGACTACTG |
GTCTCCATGTTGCAGAAGACTTTCA |
| Aggrecan |
CTAGCTGCTTAGCAGGGATAACG |
GATGACCCGCAGAGTCACAAAG |
| GAPDH |
GAAGGTCGGTGTGAACGGATTTG |
CATGTAGACCATGTAGTTGAGGTCA |
Statistical analysis
Experimental data are presented as the mean ± SD and
were visualized using GraphPad Prism (version 6.02; Dotmatics).
Statistical comparisons between groups were carried out using the
unpaired Student's t-test. All experiments were conducted in
triplicate. P<0.05 was considered to indicate a statistically
significant difference.
Results
Genetic associations between UFs and
hOA
In total, 49 SNPs were initially extracted from the
GWAS database for UF. However, one SNP that was unmatchable in the
GWAS for UF lacked an available proxy on the SNipA SNP annotator
online platform and was removed from the analysis. Using the
Phenoscanner database, two SNPs associated with confounding
factors, such as age, body mass index and ankylosing spondylitis,
were removed. Therefore, 46 genetically independent variants were
selected as IVs for UFs for use in the subsequent MR analyses
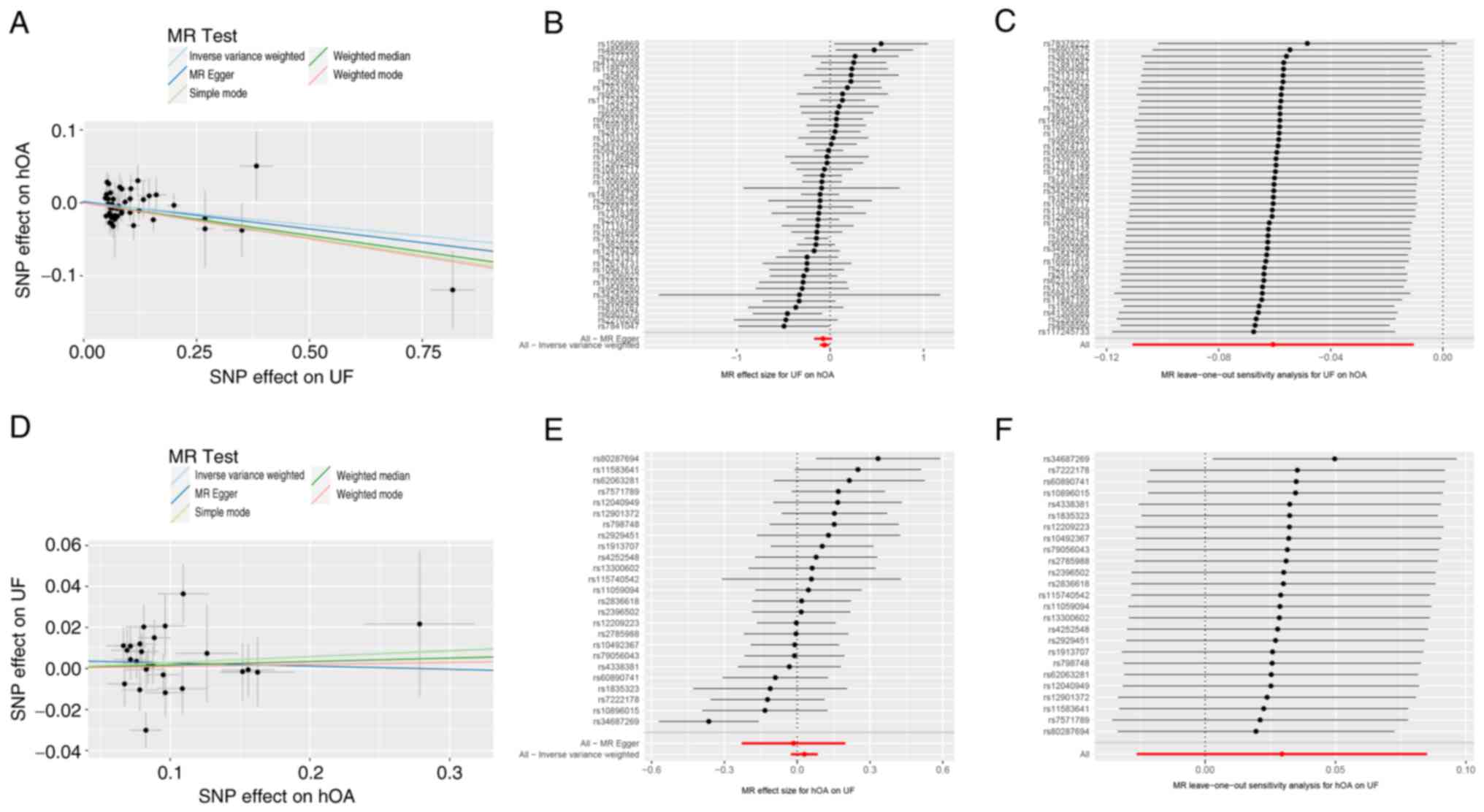
(Table SII). As shown in Table II, no heterogeneity was observed
in the associations between the selected IVs of UF and hOA using
the Cochran's Q test (Q=55.24; P=0.14); therefore, IVW was used as
the primary random-effects model. The IVW results indicated a
significant inverse genetic association between UFs and hOA (OR,
0.941; 95% CI, 0.895-0.990; adjusted P=0.042). Additionally, the
weighted median (OR, 0.914; 95% CI, 0.854-0.979; adjusted P=0.045)
and weighted mode (OR, 0.906; 95% CI, 0.833-0.985; adjusted
P=0.042) were in line with the IVW method (Fig. 2A and B; Table
II). However, MR-Egger analysis did not indicate a genetic
association (OR, 0.928; 95% CI, 0.843-1.021; adjusted P=0.161) nor
the presence of horizontal pleiotropy in the IVs (intercept P=0.73)
(Fig. 2A and B; Table
II), which indicated uncertainty regarding the robustness of
the association estimates.
 | Table IIMR results of UF and hOA genetic
association. |
Table II
MR results of UF and hOA genetic
association.
| A, 46 SNPs (Exp,
UF; Out, hOA) |
|---|
| | Heterogeneity
test | Pleiotropy
test |
|---|
| Methods | OR (95% CI) | P-value | Adjusted
P-value | Cochran's Q | P-value | Intercept | P-value |
|---|
| IVW | 0.941 (0.895,
0.990) | 0.018 | 0.042 | 55.24 | 0.14 | | |
| Weighted
median | 0.914 (0.854,
0.979) | 0.011 | 0.045 | | | | |
| MR-Egger | 0.928 (0.843,
1.021) | 0.131 | 0.161 | | | 0.002 | 0.73 |
| Simple mode | 0.909 (0.797,
1.037) | 0.161 | 0.161 | | | | |
| Weighted mode | 0.906 (0.833,
0.985) | 0.025 | 0.042 | | | | |
| B, 25 SNPs (Exp,
hOA; Out, UF) |
| | Heterogeneity
test | Pleiotropy
test |
| Methods | OR (95% CI) | P-value | Adjusted
P-value | Cochran's Q | P-value | Intercept | P-value |
| IVW | 1.030 (0.974,
1.089) | 0.300 | 0.891 | 35.71 | 0.06 | | |
| Weighted
median | 1.017 (0.953,
1.086) | 0.612 | 0.891 | | | | |
| MR-Egger | 0.985 (0.795,
1.220) | 0.891 | 0.891 | | | 0.004 | 0.68 |
| Simple mode | 1.029 (0.909,
1.163) | 0.657 | 0.891 | | | | |
| Weighted mode | 1.010 (0.907,
1.125) | 0.859 | 0.891 | | | | |
A ‘leave-one-out’ analysis was conducted to assess
the influence of each UF-associated SNP on the overall association
with hOA, which provided a sensitivity evaluation of the results
(Fig. 2C). The analysis revealed
that the removal of rs11,7245,733 had a significant impact on the
association, suggesting that this SNP plays a critical role in the
observed relationship between UF and hOA.. The funnel plots are
presented in Fig. S1A.
In the reverse MR analysis, 27 SNPs were extracted
from the hOA GWAS database. However, two SNPs associated with
confounding factors were removed, and 25 genetically independent
variants were selected as the IVs of hOA (Table SII). As shown in Table II, the IVW method did not find a
genetic association between hOA and UFs (OR, 1.030; 95% CI,
0.974-1.089; adjusted P=0.891), and Cochran's Q test did not reveal
any significant heterogeneity (Q=35.71; P=0.06). Additionally, the
MR-Egger analysis corroborated the IVW findings (OR, 0.985; 95% CI,
0.795-1.220; adjusted P=0.891) (Fig.
2D and E; Table II). The MR-Egger analysis also
revealed no evidence of horizontal pleiotropy in the IVs (intercept
P=0.68) (Fig. 2D and E; Table
II).
A ‘leave-one-out’ analysis was carried out to assess
the influence of each hOA-associated SNP on UF, which provided a
sensitivity evaluation and consistency of the findings. The
findings indicated that none of the SNPs exhibited a discernible
effect on the pooled results (Fig.
2F). The funnel plots are presented in Fig. S1B.
Genetic associations between UFs and
kOA
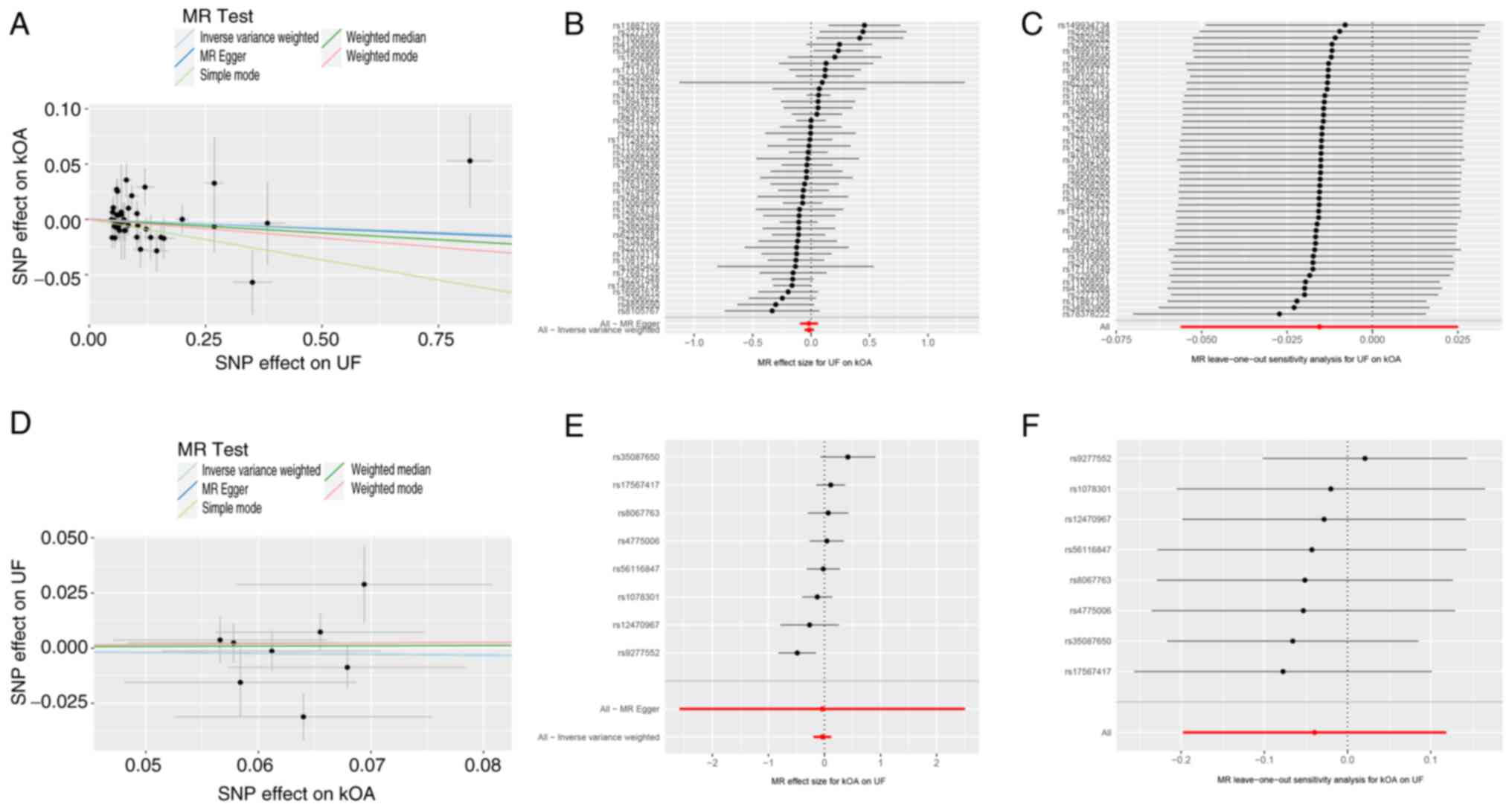
As aforementioned, 46 genetically independent
variants were identified and selected as IVs for the UF analysis
(Table SII). In the primary
analysis using the IVW method, no genetic association was found
between UFs and kOA (OR, 0.985; 95% CI, 0.945-1.025; adjusted
P=0.569). Cochran's Q test indicated the absence of significant
heterogeneity in the results (Q=57.20; P=0.10) (Fig. 3A and B; Table
III). The results of the MR-Egger analysis were consistent with
those of the IVW analysis (OR, 0.983; 95% CI, 0.910-1.062; adjusted
P=0.662), which indicated the absence of horizontal pleiotropy in
the IVs (intercept P=0.956) (Fig.
3A and B; Table III). Furthermore, the result of
the ‘leave-one-out’ analysis indicated that the genetic association
between UF and kOA was not associated to a single SNP (Fig. 3C). The funnel plots are presented
in Fig. S1C.
 | Table IIIMR results of UF and kOA genetic
association. |
Table III
MR results of UF and kOA genetic
association.
| A, 46 SNPs (Exp,
UF; Out, kOA) |
|---|
| | Heterogeneity
test | Pleiotropy
test |
|---|
| Methods | OR (95% CI) | P-value | Adjusted
P-value | Cochran's Q | P-value | Intercept | P-value |
|---|
| IVW | 0.985 (0.945,
1.025) | 0.455 | 0.569 | 57.20 | 0.10 | | |
| Weighted
median | 0.976 (0.921,
1.034) | 0.409 | 0.569 | | | | |
| MR-Egger | 0.983 (0.910,
1.062) | 0.662 | 0.662 | | | 0.0002 | 0.956 |
| Simple mode | 0.930 (0.835,
1.035) | 0.191 | 0.569 | | | | |
| Weighted mode | 0.967 (0.886,
1.054) | 0.454 | 0.569 | | | | |
| B, 8 SNPs (Exp,
kOA; Out, UF) |
| | Heterogeneity
test | Pleiotropy
test |
| Methods | OR (95% CI) | P-value | Adjusted
P-value | Cochran's Q | P-value | Intercept | P-value |
| IVW | 0.961 (0.820,
1.126) | 0.624 | 0.975 | 13.18 | 0.07 | | |
| Weighted
median | 1.016 (0.872,
1.182) | 0.841 | 0.975 | | | | |
| MR-Egger | 0.960 (0.075,
12.247) | 0.975 | 0.975 | | |
7.68x10-5 | 0.999 |
| Simple mode | 1.033 (0.833,
1.283) | 0.774 | 0.975 | | | | |
| Weighted mode | 1.033 (0.855,
1.249) | 0.744 | 0.975 | | | | |
In the reverse MR analysis, the genetic associations
between kOA and UFs were analyzed using the same aforementioned
methodology. In total, 10 SNPs were extracted from the GWAS for
UFs, and eight variants were identified as IVs (Table SII). The subsequent analysis
revealed the absence of a genetic association of kOA on UFs using
the IVW method (OR, 0.961; 95% CI, 0.820-1.126; adjusted P=0.975),
which aligned with the result obtained from the MR-Egger method
(OR, 0.960; 95% CI, 0.075-12.247; adjusted P=0.975) (Fig. 3D and E; Table
III). The MR-Egger analysis showed no evidence of horizontal
pleiotropy among the IVs (intercept P=0.999) (Fig. 3D and E; Table
III). Furthermore, Cochran's Q test revealed no heterogeneity
(Q=13.18; P=0.07) (Table III).
Additionally, the results of the ‘leave-one-out’ analysis indicated
that the genetic association of kOA on UF was not attributable to a
single SNP (Fig. 3F). The funnel
plots are presented in Fig.
S1D.
Investigating the role of UFCs in OA
chondrocyte degeneration
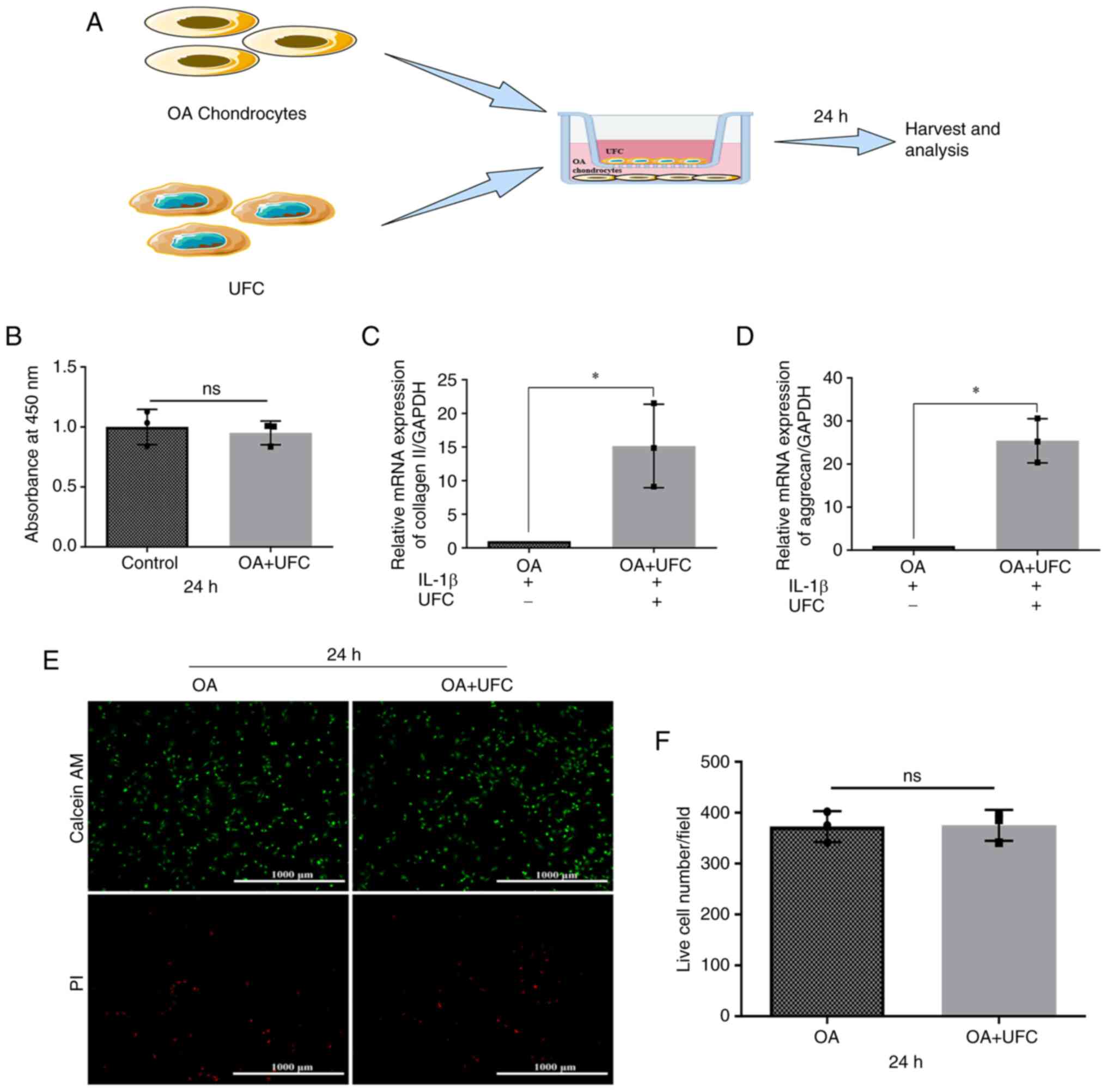
A Transwell co-culture system was used to
investigate the effects of UFCs on chondrocytes of OA. Rat
chondrocytes were cultured in the lower chamber and pre-treated
with IL-1β for 2 h to mimic an inflammatory cartilage environment.
UFCs were then cultured in the upper chamber for another 24 h
(Fig. 4A). The CCK-8 assay
indicated that there was no significant difference in the viability
of chondrocytes after exposure to UFCs compared with those in the
control group not exposed to UFCs (Fig. 4B). This was further confirmed by
live/dead cell staining (Fig. 4E
and F). Furthermore, it was
demonstrated that the mRNA levels of Collagen II and Aggrecan were
increased in the chondrocytes treated with UFCs compared with the
chondrocytes that were not co-cultured with UFCs (Fig. 4C and D). These results suggested that UFCs
reduced the levels of markers of chondrocyte degeneration in
vitro, which indicated a possible biological interaction that
warrants further basic mechanistic investigation.
Discussion
In the present study, a bidirectional MR analysis
was carried out using population-based genetic datasets to
investigate the potential genetic association between OA of
weight-bearing joints and the risk of UF. Analysis using the IVW
method indicated an inverse genetic association between genetic
liability to UF and hOA, whereas this association was not observed
in the MR-Egger analysis. This discrepancy highlights the need for
cautious interpretation of the findings of the present study. The
IVW method estimates the association between genetically predicted
exposure and outcome through a weighted regression of SNP-specific
Wald ratios (βoutcome/βexposure), which may
be more sensitive to undetected pleiotropy or confounding factors
(29). The MR-Egger method tests
for directional pleiotropy and provides an assessment of potential
associations, but the strength of evidence is reduced when
accounting for pleiotropy or other unmeasured confounders (35,41).
Therefore, the inconsistency between these two analyses introduced
uncertainty regarding the robustness and interpretation of these
associations. Given the potential limitations in the robustness of
these association inferences from MR analysis, in vitro
co-culture experiments were carried out. These in vitro
experiments used OA chondrocytes and UFCs to provide additional
biological insights. In the present study, IL-1β-stimulated rat
chondrocytes were co-cultured with UFCs to mimic an inflammatory
cartilage environment. The experiments demonstrated that the
presence of UFCs in the co-culture system significantly inhibited
the IL-1β-induced reduction in Aggrecan and Collagen II mRNA
expression in chondrocytes, compared with the OA group. These
results suggest a potential interaction between UFCs and
IL-1β-stimulated chondrocytes, but further mechanistic studies are
needed to validate these findings.
Numerous studies have investigated the potential
associations of various exposures on OA, but the results have been
unfavorable (26,28,42).
For example, a previous MR analysis demonstrates that elevated
serum testosterone and dihydrotestosterone levels may increase the
risk of total hip arthroplasty (26). IVs linked to genetic
predispositions for osteoporosis are associated with an increased
risk of OA, and increasing bone mineral density serves as an
effective strategy to prevent OA (42). Furthermore, MR analysis reveals an
association between OA and bladder cancer (28). However, the association between UFs
and OA is yet to be elucidated. UFs, the most common benign tumor
among women of reproductive age, have an etiology that is still not
completely understood (9). An
observational study by Kovari et al (25) reports a notably higher prevalence
of UFs in patients diagnosed with hOA, which contradict the MR
findings of the present study. One possible explanation for this
discrepancy may involve estrogen (specifically, E2). Previous
studies reveal that elevated serum E2 levels are associated with an
increased risk of UFs, while emerging evidence suggests that E2 may
exert anti-degenerative effects on cartilage, potentially
influencing OA onset or progression (7-9).
Systemic E2 levels in patients with UFs may
contribute to protection against OA via estrogen receptor signaling
(43). However, this mechanism
alone cannot explain the results of the present study, which
demonstrated in vitro that UFCs restrained the IL-1β-induced
reduction of Aggrecan and Collegen II in chondrocytes at the mRNA
level. Given that UFCs do not secrete E2, the observed reduction in
IL-1β-induced reduction of Aggrecan and Collagen II in chondrocytes
may involve alternative mechanisms, such as paracrine
anti-inflammatory signaling or modulation of receptor sensitivity
in chondrocytes (44). Integrating
these findings with the findings of the MR analysis in the present
study, we hypothesize that the inverse association between UF and
hOA likely reflects indirect biological interactions mediated by
alterations in the tumor microenvironment instead of direct effects
of the tumor itself. For example, changes in hormonal levels,
immune cell modulation and inflammatory cytokine profiles within
the tumor microenvironment may influence cartilage metabolism or
immune homeostasis in anatomically adjacent tissues, such as the
hip joint (7,44). This localized paracrine mechanism
may partially account for the anatomical specificity observed in
the MR findings of the present study, in which UF was inversely
associated with hOA, an anatomically adjacent site, but not with
kOA. This specificity may be attributed to the proximity of the
uterus to the hip, which potentially facilitates local crosstalk or
signaling gradients that may not extend to more distant joints.
The findings of the present study indicated novel
evidence of a potential genetic link between UF and hOA, suggesting
that the association between UF pathophysiology and joint
degeneration may be more complex than previously considered
(45,46). However, as the simplified in
vitro model used in the present study cannot fully recapitulate
the complex biological environment of OA, the proposed hypothesis
is speculative. Further comprehensive mechanistic experiments
involving in vivo models and clinical samples are required
to validate these interactions.
However, there were a number of limitations in the
present study that should be considered. Firstly, the present study
was limited by the relatively small number of SNPs. Therefore, to
enhance the validity of the MR analyses, larger GWAS datasets
should be used and a broader array of SNPs should be incorporated
as IVs in future investigations. Secondly, the background of the
patients included in the present study were restricted to those
with European ancestry, and the instruments identified within
European populations may not be applicable to non-European groups.
Therefore, additional MR analyses that include a broader range of
ethnic groups are required. Furthermore, the weak instruments used
in MR analyses can lead to biased and inconsistent association
estimates. Therefore, to assess the strength of the IVs used in the
present study, F-statistics for each SNP was calculated. The
results of the present study showed that the F-statistics for the
instruments exceeded the commonly accepted threshold of 10,
indicating that the instruments were strong and the risk of weak
instrument bias was minimized. However, despite the apparent
strength of the instruments, the possibility of a weak instrument
bias affecting the association estimates cannot be excluded. Weak
instrument bias could lead to attenuated or distorted estimates,
undermining the validity of the conclusions of the present study.
Although, precautions to mitigate this risk were taken in the
present study, further research using alternative IVs or additional
methodological approaches may strengthen the robustness of the
findings of the present study. Finally, the MR analysis combined
with the cellular experiments used in the present study assumed a
unidirectional association between exposure and outcome. However,
biological systems may include complex feedback loops, potentially
affecting the interpretation of results (47).
In conclusion, the novelty of the present study was
in the integration of the bidirectional MR with in vitro
co-culture experiments, an approach that, to the best of our
knowledge, has not previously been used to investigate the
association between UF and hOA. Using this combined strategy the
present study proposed a novel microenvironment-mediated mechanism
that may explain the possible inverse association and its
anatomical specificity. Furthermore, a possible inverse genetic
association between UF and the risk of hOA was demonstrated in the
present study. This was further suggested by the findings of the
in vitro experiments, which demonstrated that Aggrecan and
Collagen II mRNA levels were increased in IL-1β-induced
chondrocytes in the presence of UFCs, compared with that in the OA
group. Furthermore, we hypothesize that this association
potentially reflects indirect effects mediated by the tumor
microenvironment instead of the direct effects of fibroids
themselves. However, due to the discrepancies across the MR
analytical approaches, further detailed mechanistic studies are
warranted to validate the hypothesis of the present study, which
may potentially provide a framework for integrating genetic
epidemiology with experimental biology when investigating complex
disease interactions.
Supplementary Material
Funnel plot. (A) Funnel plot of the
estimated associations of (A) UF on hOA, (B) hOA on UF, (C) UF on
kOA and (D) kOA on UF. UF, uterine fibroids; OA, osteoarthritis;
hOA, hip OA; kOA, knee OA; IV, instrumental variables; MR,
mendalian randomization; SEIV, standard error of IVs.
Baseline profiles of the cohort used
in the present study.
Characteristics of SNPs used as
genetic instruments in UF and hOA.
Acknowledgements
Not applicable.
Funding
Funding: The present work was supported by the National Natural
Science Foundation of China Key Programme (grant no. 32130052) and
the National Natural Science Foundation of China Major Research
Plan (grant no. 91949203).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XM, SM, TW and YZ conceived and designed the present
study. XM, JM, XJ, HZ and SM collected, analyzed and interpreted
the data. XM and SM drafted the manuscript. XM, JM, XJ, TW and YZ
revised the manuscript. All authors read and approved the final
version of the manuscript. XM and YZ confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jaramillo Quiceno GA, Sarmiento Riveros
PA, Ochoa Perea GA, Vergara MG, Rodriguez Muñoz LF, Arias Perez RD,
Piovesan NO and Muñoz Salamanca JA: Satisfactory clinical outcomes
with autologous matrix-induced chondrogenesis in the treatment of
grade IV chondral injuries of the knee. J ISAKOS. 8:86–93.
2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sacitharan PK: Ageing and Osteoarthritis.
Subcell Biochem. 91:123–159. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Atasoy-Zeybek A, Showel KK, Nagelli CV,
Westendorf JJ and Evans CH: The intersection of aging and estrogen
in osteoarthritis. NPJ Women's Health. 3(15)2025.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nelson AE, Smith MW, Golightly YM and
Jordan JM: ‘Generalized osteoarthritis’: A systematic review. Semin
Arthritis Rheum. 43:713–720. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Loughlin J: Translating osteoarthritis
genetics research: Challenging times ahead. Trends Mol Med.
28:176–182. 2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cho HJ, Morey V, Kang JY, Kim KW and Kim
TK: Prevalence and risk factors of spine, shoulder, hand, hip, and
knee osteoarthritis in community-dwelling koreans older than age 65
years. Clin Orthop Relat Res. 473:3307–3314. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nevitt MC, Cummings SR, Lane NE, Hochberg
MC, Scott JC, Pressman AR, Genant HK and Cauley JA: Association of
estrogen replacement therapy with the risk of osteoarthritis of the
hip in elderly white women. Study of osteoporotic fractures
research group. Arch Intern Med. 156:2073–2080. 1996.PubMed/NCBI
|
|
8
|
Fan DX, Yang XH, Li YN and Guo L:
17β-Estradiol on the Expression of G-Protein coupled estrogen
receptor (GPER/GPR30) mitophagy, and the PI3K/Akt signaling pathway
in ATDC5 chondrocytes in vitro. Med Sci Monit. 24:1936–1947.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yang Q, Ciebiera M, Bariani MV, Ali M,
Elkafas H, Boyer TG and Al-Hendy A: Comprehensive review of uterine
fibroids: Developmental origin, pathogenesis, and treatment. Endocr
Rev. 43:678–719. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Dennison EM: Osteoarthritis: The
importance of hormonal status in midlife women. Maturitas.
165:8–11. 2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lu Z, Zhang A, Wang J, Han K and Gao H:
Estrogen alleviates post-traumatic osteoarthritis progression and
decreases p-EGFR levels in female mouse cartilage. BMC
Musculoskelet Disord. 23(685)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Al-Hendy A, Myers ER and Stewart E:
Uterine fibroids: Burden and unmet medical need. Semin Reprod Med.
35:473–480. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Baird DD, Dunson DB, Hill MC, Cousins D
and Schectman JM: High cumulative incidence of uterine leiomyoma in
black and white women: Ultrasound evidence. Am J Obstet Gynecol.
188:100–107. 2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kho PF and Mortlock S: Endometrial Cancer
Association Consortium; International Endometriosis Genetics
Consortium. Rogers PAW, Nyholt DR, Montgomery GW, Spurdle AB, Glubb
DM and O'Mara TA: Genetic analyses of gynecological disease
identify genetic relationships between uterine fibroids and
endometrial cancer, and a novel endometrial cancer genetic risk
region at the WNT4 1p36.12 locus. Hum Genet. 140:1353–1365.
2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Stewart EA, Cookson CL, Gandolfo RA and
Schulze-Rath R: Epidemiology of uterine fibroids: A systematic
review. BJOG. 124:1501–1512. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Stewart EA, Laughlin-Tommaso SK, Catherino
WH, Lalitkumar S, Gupta D and Vollenhoven B: Uterine fibroids. Nat
Rev Dis Primers. 2(16043)2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Borahay MA, Asoglu MR, Mas A, Adam S,
Kilic GS and Al-Hendy A: Estrogen receptors and signaling in
fibroids: Role in pathobiology and therapeutic implications. Reprod
Sci. 24:1235–1244. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tal R and Segars JH: The role of
angiogenic factors in fibroid pathogenesis: Potential implications
for future therapy. Hum Reprod Update. 20:194–216. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kapoor M, Martel-Pelletier J, Lajeunesse
D, Pelletier JP and Fahmi H: Role of proinflammatory cytokines in
the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 7:33–42.
2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Blaney Davidson EN, van der Kraan PM and
van den Berg WB: TGF-beta and osteoarthritis. Osteoarthritis
Cartilage. 15:597–604. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ciebiera M, Włodarczyk M, Wrzosek M,
Męczekalski B, Nowicka G, Łukaszuk K, Ciebiera M,
Słabuszewska-Jóźwiak A and Jakiel G: Role of transforming growth
factor β in uterine fibroid biology. Int J Mol Sci.
18(2435)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pang H, Chen S, Klyne DM, Harrich D, Ding
W, Yang S and Han FY: Low back pain and osteoarthritis pain: A
perspective of estrogen. Bone Res. 11(42)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wright VJ, Schwartzman JD, Itinoche R and
Wittstein J: The musculoskeletal syndrome of menopause.
Climacteric. 27:466–472. 2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Felson DT, Lawrence RC, Dieppe PA, Hirsch
R, Helmick CG, Jordan JM, Kington RS, Lane NE, Nevitt MC, Zhang Y,
et al: Osteoarthritis: New insights. Part 1: the disease and its
risk factors. Ann Intern Med. 133:635–646. 2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kovari E, Kaposi A, Bekes G, Kiss Z,
Kurucz R, Mandl P, Balint GP, Poor G, Szendroi M and Balint PV:
Comorbidity clusters in generalized osteoarthritis among female
patients: A cross-sectional study. Semin Arthritis Rheum.
50:183–191. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yan YS, Qu Z, Yu DQ, Wang W, Yan S and
Huang HF: . Sex steroids and osteoarthritis: A mendelian
randomization study. Front Endocrinol (Lausanne).
12(683226)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hill HA, Schoenbach VJ, Kleinbaum DG,
Strecher VJ, Orleans CT, Gebski VJ and Kaplan BH: A longitudinal
analysis of predictors of quitting smoking among participants in a
self-help intervention trial. Addict Behav. 19:159–173.
1994.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang X, Wen Z, Xing Z, Zhou X, Yang Z,
Dong R and Yang J: The causal relationship between osteoarthritis
and bladder cancer: A Mendelian randomization study. Cancer Med.
13(e6829)2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Chen D, Xu H, Sun L and Li Y, Wang T and
Li Y: Assessing causality between osteoarthritis with urate levels
and gout: A bidirectional Mendelian randomization study.
Osteoarthritis Cartilage. 30:551–558. 2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Bowden J and Holmes MV: Meta-analysis and
mendelian randomization: A review. Res Synth Methods. 10:486–496.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tachmazidou I, Hatzikotoulas K, Southam L,
Esparza-Gordillo J, Haberland V, Zheng J, Johnson T, Koprulu M,
Zengini E, Steinberg J, et al: Identification of new therapeutic
targets for osteoarthritis through genome-wide analyses of UK
Biobank data. Nat Genet. 51:230–236. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sakaue S, Kanai M, Tanigawa Y, Karjalainen
J, Kurki M, Koshiba S, Narita A, Konuma T, Yamamoto K, Akiyama M,
et al: A cross-population atlas of genetic associations for 220
human phenotypes. Nat Genet. 53:1415–1424. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Skrivankova VW, Richmond RC, Woolf BAR,
Yarmolinsky J, Davies NM, Swanson SA, VanderWeele TJ, Higgins JPT,
Timpson NJ, Dimou N, et al: Strengthening the reporting of
observational studies in epidemiology using mendelian
randomization: The STROBE-MR Statement. JAMA. 326:1614–1621.
2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Cao Z, Wu Y, Li Q, Li Y and Wu J: A causal
relationship between childhood obesity and risk of osteoarthritis:
Results from a two-sample Mendelian randomization analysis. Ann
Med. 54:1636–1645. 2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ni J, Zhou W, Cen H, Chen G, Huang J, Yin
K and Sui C: Evidence for causal effects of sleep disturbances on
risk for osteoarthritis: A univariable and multivariable Mendelian
randomization study. Osteoarthritis Cartilage. 30:443–450.
2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Palmer TM, Lawlor DA, Harbord RM, Sheehan
NA, Tobias JH, Timpson NJ, Davey Smith G and Sterne JA: Using
multiple genetic variants as instrumental variables for modifiable
risk factors. Stat Methods Med Res. 21:223–242. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bowden J, Davey Smith G and Burgess S:
Mendelian randomization with invalid instruments: effect estimation
and bias detection through Egger regression. Int J Epidemiol.
44:512–525. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bowden J, Del Greco MF, Minelli C, Davey
Smith G, Sheehan NA and Thompson JR: Assessing the suitability of
summary data for two-sample mendelian randomization analyses using
MR-egger regression: The role of the I2 statistic. Int J Epidemiol.
45:1961–1974. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Inkster AM, Konwar C, Peñaherrera MS,
Brain U, Khan A, Price EM, Schuetz JM, Portales-Casamar É, Burt A,
Marsit CJ, et al: Profiling placental DNA methylation associated
with maternal SSRI treatment during pregnancy. Sci Rep.
12(22576)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Livak KJ and Schmittgen TD: . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bowden J, Davey Smith G, Haycock PC and
Burgess S: Consistent estimation in mendelian randomization with
some invalid instruments using a weighted median estimator. Genet
Epidemiol. 40:304–314. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Qu Y, Chen S, Han M, Gu Z, Zhang Y, Fan T,
Zeng M, Ruan G, Cao P, Yang Q, et al: Osteoporosis and
osteoarthritis: A bi-directional Mendelian randomization study.
Arthritis Res Ther. 25(242)2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhao Z, Niu S, Chen J, Zhang H, Liang L,
Xu K, Dong C, Su C, Yan T, Zhang Y, et al: G protein-coupled
receptor 30 activation inhibits ferroptosis and protects
chondrocytes against osteoarthritis. J Orthop Translat. 44:125–138.
2024.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bulun SE, Yin P, Wei J, Zuberi A, Iizuka
T, Suzuki T, Saini P, Goad J, Parker JB, Adli M, et al: UTERINE
FIBROIDS. Physiol Rev. 105:1947–1988. 2025.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kiely M, Lord B and Ambs S: Immune
response and inflammation in cancer health disparities. Trends
Cancer. 8:316–327. 2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Haycock PC, Burgess S, Wade KH, Bowden J,
Relton C and Davey Smith G: Best (but oft-forgotten) practices: The
design, analysis, and interpretation of Mendelian randomization
studies. Am J Clin Nutr. 103:965–978. 2016.PubMed/NCBI View Article : Google Scholar
|