Introduction
Venous thromboembolism (VTE) arises from the
development and detachment of venous thrombi, encompassing deep
venous thrombosis (DVT) and pulmonary embolism (PE). Its insidious
clinical presentation elevates VTE (mainly PE) above myocardial
infarction and stroke, emerging as a primary cause of sudden
mortality in patients (1). Early
diagnosis poses a notable challenge in VTE management (2), as current diagnostic tests are
predominantly indirect and non-specific. Consequently, researchers
have dedicated years to investigating VTE-related biomarkers
(3). However, the development of
early and universally applicable biomarkers holds paramount
importance for the initial management of VTE.
Emerging evidence highlights the pivotal role of
inflammatory and immune responses in thrombosis (4). Given this mechanistic link between
inflammation and thrombosis, recent studies have intensified
efforts to identify novel inflammatory biomarkers for VTE, which
could elucidate its pathophysiological underpinnings (5-7).
Pyroptosis is a type of programmed cell death characterized by
rapid cytomembrane pore formation and the subsequent release of a
plethora of proinflammatory mediators such as interleukin-1β and
interleukin-18(8). These events
trigger the activation of endothelial cells, the coagulation system
and consequent thrombotic processes (9). Considered as the initiating factor in
VTE, pyroptosis holds promise for early diagnostic applications
(10). This newly recognized
programmed cell death, associated with a robust inflammatory
response and thrombotic inflammation linked to innate immunity, has
garnered attention (11). While
preliminary studies suggest a potential significance of pyroptosis
in venous thrombosis (3,10,12),
the precise relationship between pyroptosis and VTE remains
unresolved. The present study investigates the potential
association between VTE and pyroptosis by integrating VTE
transcriptomic data from the GEO database and employing multi-omics
analytical approaches to identify pyroptosis-related biomarkers.
Through differential expression analysis, weighted gene
co-expression network analysis (WGCNA) co-expression network
construction, and gene set enrichment analysis (GSEA) scoring to
assess pyroptosis activity, least absolute shrinkage and selection
operator (LASSO) and support vector machine recursive feature
elimination (SVM-RFE) machine learning algorithms were applied to
screen characteristic genes, ultimately validating diagnostic
efficacy using receiver operating characteristic (ROC) curve
analysis. The primary aim of the present study was to identify
early diagnostic biomarkers by exploring the interplay between
pyroptosis and VTE.
Materials and methods
Data collection
Sequencing data from GSE19151 and GSE48000 (13,14)
were stemmed from the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/).
The microarray data from GSE19151, comprising 70 VTE samples and 63
control samples, was utilized as the training cohort (13). The GSE48000 dataset, consisting of
107 VTE samples and 25 control samples, served as the validation
cohort. A total of 52 pyroptosis-related genes (PRGs) were sourced
from literature (14).
Single sample GSEA (ssGSEA)
Utilizing the background set of PRGs (15), the ssGSEA analysis was used for
calculating the pyroptosis score of the samples in the GSE19151
training set through the R package-GSEA (version 1.46.0) (16). Subsequently, this score was
employed as a phenotypic trait to identify the gene modules
exhibiting the highest correlation with the pyroptosis scores.
WGCNA
WGCNA was employed using the R package-WGCNA
(17) on all genes derived from
blood samples in the GSE19151 training set. Initially, clustering
analysis was utilized to identify and remove sample outliers.
Subsequently, the optimal soft-threshold power was determined to
construct a network ensuring a scale-free index (R2) of
0.85 and an average connectivity close to 0. A systematic
intergenic cluster tree was drawn according to the coefficient of
dissimilarity between genes. Moreover, the minimum gene count per
module was set at 100 and modules were merged when the threshold
reached 0.3. VTE and Pyroptosis Score were considered as phenotypic
traits to identify gene modules exhibiting the highest correlation
coefficients with respect to VTE and Pyroptosis Score. Statistical
significance was set at P<0.05. Lastly, gene significance (GS),
module membership (MM) and pyroptosis scores were computed to
assess the relationship between key modules and VTE. Genes
extracted from these key modules were designated as hub genes
identified through WGCNA analysis.
Identification of VTE differentially
expressed PRGs (DE-PRGs)
In the GSE19151 training set, differential genes
between VTE and control groups were filtrated using the R
package-limma (18). Differential
genes were filtered based on criteria that |log2FC|≥1
and P<0.05. Visualization of the results was carried out using
volcano and heat maps using the R package-ggplot2(19). The intersected genes of
differentially expressed genes (DEGs) and hub genes were defined as
differentially expressed DE-PRGs.
Function analyses of DE-PRGs
To elucidate the biological functions and signaling
pathways associated with the key differentially expressed module
genes, Gene Ontology annotation analysis and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway enrichment analysis were executed
by means of R package-clusterProfiler on DE-PRGs in dataset
GSE19151. The results were visualized using the R package-ggplot2,
with statistical significance set at P<0.05.
Protein-protein interaction (PPI)
network
The PPI network was constructed to depict gene
interactions at the protein level. The STRING database (https://cn.string-db.org/) was utilized to identify
known proteins and predict protein relationships. Subsequently, the
top 20 genes from each of the six algorithms (maximal clique
centrality, closeness, maximum neighborhood component, degree,
radiality and edge percolated component) were selected, and the
overlapping genes among the top 20 genes were designated as
candidate genes. The PPI network was visualized using Cytoscape
(version 3.9.1) (https://cn.string-db.org/).
Identification of biomarkers
associated with pyroptosis in VTE
Feature genes were selected form the candidate genes
using the LASSO algorithm and SVM-RFE model. The LASSO model was
applied using R package-glmnet, while the SVM-RFE model was
established by the R package-e1071(20). The feature genes with the least
error were taken for the analysis results of LASSO and SVM-RFE
models. Subsequently, candidate biomarkers for VTE were obtained
via overlapping feature genes obtained from both algorithms.
Furthermore, the biomarkers were screened from candidate biomarkers
with a ROC value >0.7. The ROC curve was plotted using the R
package-pROC and the area under the curve (AUC) was computed to
assess the diagnostic capability of biomarkers.
GSEA
GSEA analysis was conducted to explore the
underlying biological pathways of biomarkers using the R
package-clusterProfiler. Initially, the correlation between the
biomarkers and other genes in GSE19151 training set was calculated.
All genes were ranked based on their correlations and considered as
the test set for analysis. Subsequently, the C2: KEGG signaling
pathway set acquired from the MSigDB database (https://ngdc.cncb.ac.cn/databasecommons/database/id/1077)
was regarded as a background set to recognize these sorted genes
enrichment in the background set.
A pre-experiment: Reverse
transcription-quantitative PCR (RT-qPCR)
A pilot study was conducted with approval and
oversight from the Ethics Committee of the First Affiliated
Hospital of Kunming Medical University (approval no. 2023L198) from
January 2024, to December 2024. All participants provided written
informed consent. As a preliminary investigation, to ensure the
research direction and minimize confounding factors, the case group
consisted of individuals with idiopathic VTE (all five cases were
lower extremity DVT complicated with PE, excluding infections and
immune-related diseases). The control group comprised healthy
individuals undergoing routine physical examinations. Given the
substantial budget required for a clinical trial, initially 10
samples (5 VTE cases and 5 controls) were used for
pre-experiments.
The inclusion criteria for the Case group was: i)
Lower extremity DVT complicated with PE, diagnosed by
ultrasonography and computed tomographic pulmonary angiography, ii)
aged between 18 and 80 years, iii) no history of thyroid, heart,
liver, kidney and metabolic disorders, iv) no history of
trauma-surgery, rheumatoid immune disorders, oral contraceptive use
or pregnancy, v) Fracture-free.
The inclusion criteria for the Control group was: i)
Healthy individuals after normal physical examination and no
history of VT, ii) aged between 18 and 80 years, iii) no history of
thyroid, heart, liver, kidney or metabolic disorders, iv) no
history of trauma-surgery, rheumatoid immune disorders, oral
contraceptive use or pregnancy, v) fracture-free.
RT-qPCR analysis
RT-qPCR processing mainly includes 2 parts:
Amplification and dissolution curve preparation, and Cq value
recording. Expression levels between two groups were normalized to
the internal reference GAPDH and calculated using the
2-ΔΔCq method (21). A
total of 5 pairs of frozen blood samples were processed for RT-qPCR
analysis as follows: A volume of 3,000 µl blood was transferred
into 15 ml centrifuge tubes and mixed with 3 ml peripheral blood
mononuclear cell (PBMC) separation solution (Wuhan Servicebio
Technology Co., Ltd.) to isolate PBMC cells. Subsequently, 1 ml
TRIzol (Thermo Fisher Scientific, Inc.) reagent was added to the
tubes, thoroughly homogenized and incubated on ice for 10 min to
ensure complete cell lysis. Following this, 300 µl chloroform was
added into the tubes which were shaken vigorously for 30 sec and
allowed to stand at room temperature for 10 min to separate the
liquid phases. The samples were then centrifuged at 1,225 x g and
4˚C for 15 min for RNA stratification. Equal volumes of ice-cold
isopropyl alcohol (Chengdu Kelong Chemical Co., Ltd.) was added to
precipitate the RNA, followed by centrifugation at 1,224 x g and
4˚C for 10 min. Afterward, RNA was cleaned with 1 ml 75% ethanol
(Chronchem), air-dried and centrifuged at 765 x g, 4˚C for 5 min,
with this washing step repeated twice before removing the
supernatant and drying the RNA. Finally, RNA was dissolved in
RNase-free water (Wuhan Servicebio Technology Co., Ltd.) and the
concentration was detected by NanoPhotometer N50 (Implen). RT of
mRNA was performed using SweScript First Strand cDNA synthesis kit
(Wuhan Servicebio Technology Co., Ltd.). Sequential addition of
various reagents and solutions was carried out on ice (Table I), followed by a brief
centrifugation step (centrifuged at 765 x g, 4˚C for 1 min). RT was
carried out on PCR apparatus (Bio-Rad Laboratories, Inc.) under the
following conditions: 25˚C for 5 min, 50˚C for 15 min, 85˚C for 5
sec and hold at 4˚C. The cDNA product was diluted 5-20 times with
RNase/DNase-free ddH2O. Then the qPCR reaction was
performed using the reaction system outlined in Table II. Subsequently, 40 cycles of
reaction were carried out on a CFX96 real-time quantitative
fluorescent PCR instrument (Bio-Rad Laboratories, Inc.) under the
following conditions: Pre-denaturation: 95˚C for 1 min;
denaturation: 95˚C for 20 sec, annealing: 55˚C for 20 sec,
extension: 72˚C for 30 sec. The primer sequences (Beijing Tsingke
Biotech Co., Ltd.) are provided in Table III. Despite the issue of
insufficient experimental budget, western blotting (WB) experiments
were carried out on PBMC cells from another three pairs of
samples.
 | Table IReverse transcription reagents of the
SweScript First Strand cDNA synthesis kit (Wuhan Servicebio
Technology Co., Ltd.). |
Table I
Reverse transcription reagents of the
SweScript First Strand cDNA synthesis kit (Wuhan Servicebio
Technology Co., Ltd.).
| Component | Volume |
|---|
| 5x Reaction Buffer,
µl | 4 |
| Primer, µl | 1 |
| SweScript RT I
Enzyme Mix, µl | 1 |
| Total RNA, µg | 0.0001-5 |
| Nuclease-free
water | Add to 20 µl |
 | Table IIQuantitative PCR reaction system. |
Table II
Quantitative PCR reaction system.
| Component | Volume, µl |
|---|
| cDNA | 3 |
| 2x Universal Blue
SYBR Green qPCR | 5 |
| Master Mix | |
| Forward primer (10
µM) | 1 |
| Reverse primer (10
µM) | 1 |
 | Table IIIPrimer sequences. |
Table III
Primer sequences.
| Primer | Forward sequence
(5'-3') | Reverse sequence
(5'-3') |
|---|
| RPL31 |
GACACCAGGCTCAACAAAGC |
GCATCTTCCCACACCAACAA |
| RPL34 |
GGTGTAGGGCGGTGTTTCTC |
CGTCGGTATGTCAAACGCTG |
| RPL9 |
GCTGCGTCTACTGCGAGAAT |
GTGATTGAAGTCCCTCCGCA |
| RPS27L |
AGTGGCATGATTTACCCGCA |
AGGCACCAGAACCACTCAAC |
| HINT1 |
GGCAAGAAATGTGCTGCTGA |
TTTGCCGACCTCCAAGAACA |
| GAPDH |
ATGGGCAGCCGTTAGGAAAG |
AGGAAAAGCATCACCCGGAG |
Statistical analysis
Bioinformatics analysis in the present study was
conducted using R software (version 4.2.3) (16). With the help of ‘R software’ limma
package, the integration and comparative analysis of the two
datasets were executed. Statistical significance was typically
defined as P<0.05. The aforementioned data was processed using
Graphpad prism 6 (Dotmatics) statistical software package to obtain
P-values. When two groups were compared, the Mann-Whitney U Test
was used as normality could not be tested due to the small sample
size, which is shown as Mean ± SD or Median ± quartile spacing. The
quartile spacing, the upper quartile to the lower quartile
(depending on the distribution type of the sample). P<0.05
indicated a statistically significant difference.
Results
Ascertaining hub genes
Correlations between the pyroptosis score and PRGs
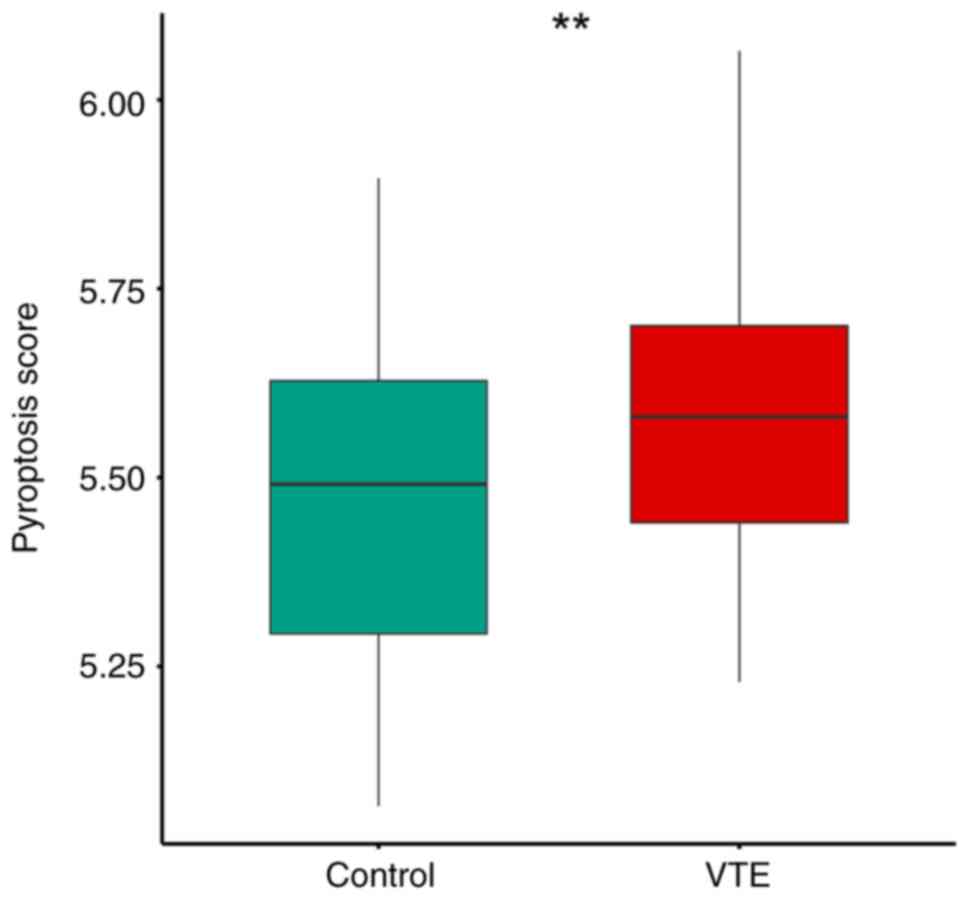
modules were calculated by ssGSEA. The results indicated a
significantly higher pyroptosis score in the VTE group compared
with the control group (P<0.05), suggesting a substantial impact
of pyroptosis on the onset and progression of VTE (Fig. 1). The optimal soft threshold power
was determined to be 11, meeting the criteria of R2
reaching 0.85 and mean connectivity approaching 0. The hierarchical
clustering tree analysis showed distinct co-expression blocks for
the filtered genes, resulting in the identification of a total of
10 modules. Following the generation of the WGCNA network, the
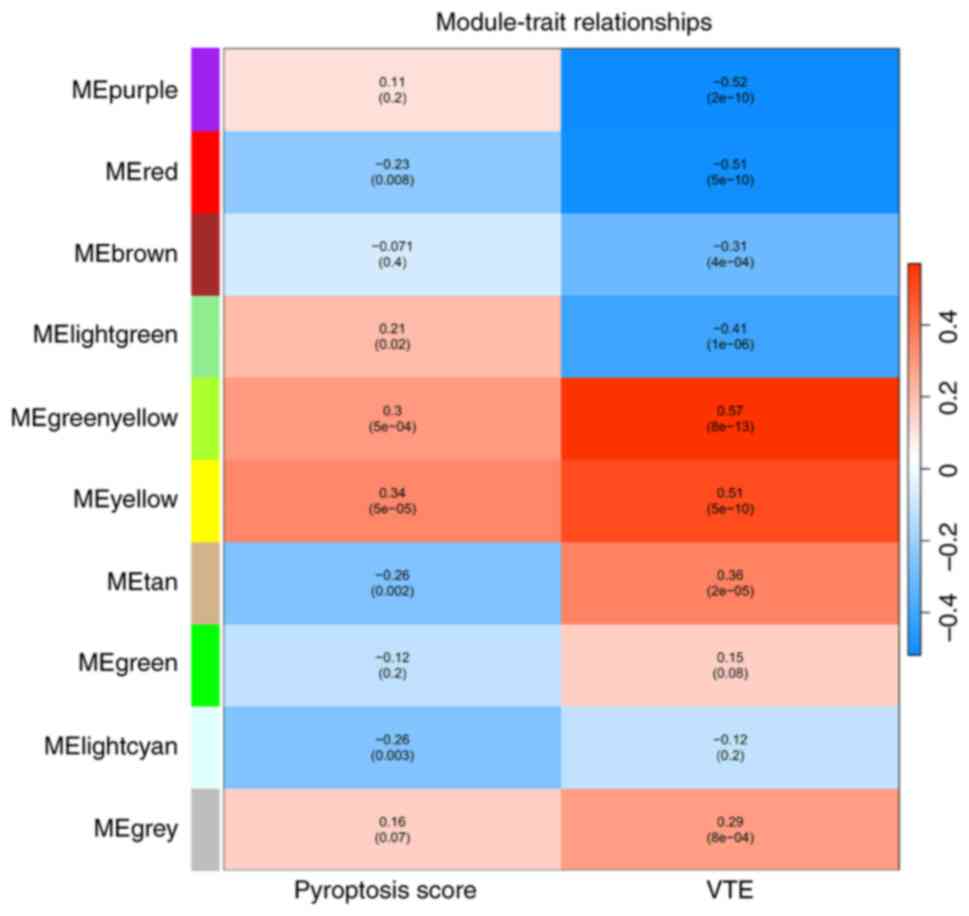
relationships between modules and traits were depicted in a heatmap
(Fig. 2). The results revealed
that in the column of VTE group, the MEgreenyellow module exhibited
correlations with VTE progression, comprising 941 genes that may
modulate the onset of VTE. In the Pyroptosis score category, the
MEyellow module demonstrated robust associations with pyroptosis,
consisting of 1739 genes. Subsequent analysis of module membership
and gene significance identified 155 genes in the MEgreenyellow
block and 464 genes in the MEyellow module as key candidates,
totaling 619 genes that functioned as hub genes in the WGCNA
analysis.
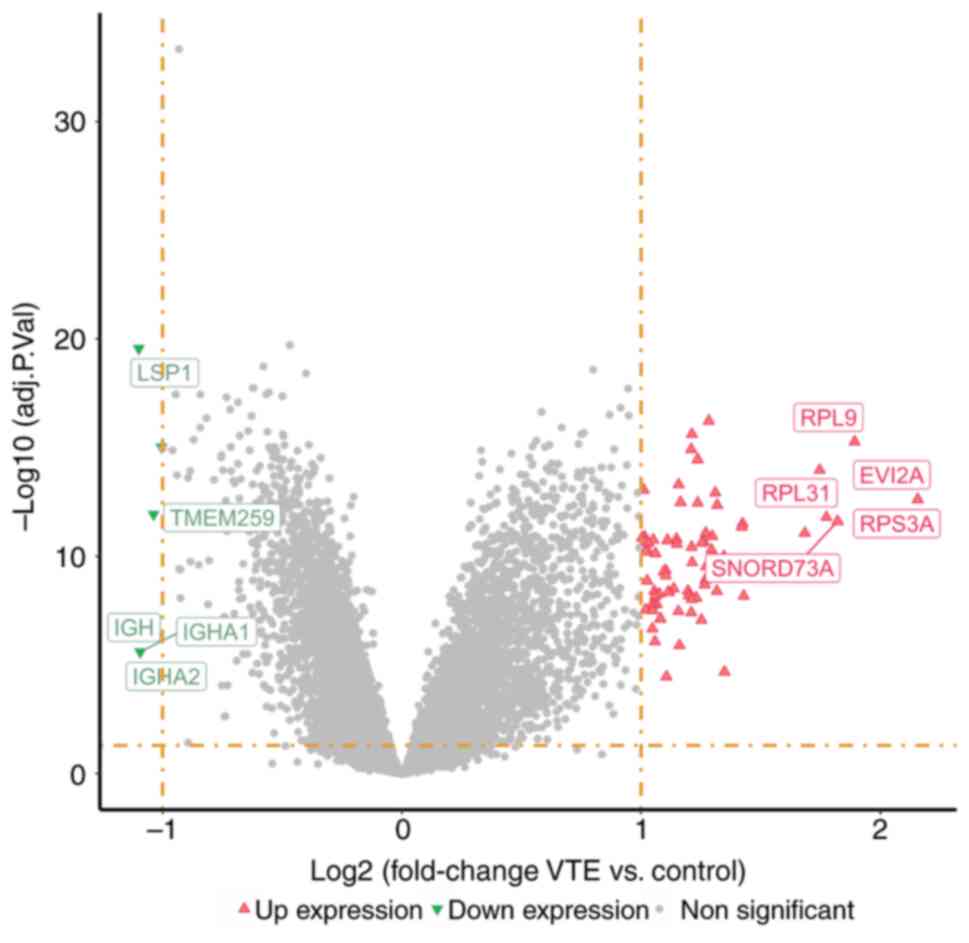
Acquiring DEGs
DEGs between VTE samples and normal samples were
analyzed. The volcano plot (Fig.
3) revealed 91 genes as DEGs, with 85 DEGs showing
significantly elevated expression levels and 6 DEGs displaying
significantly reduced expression levels.
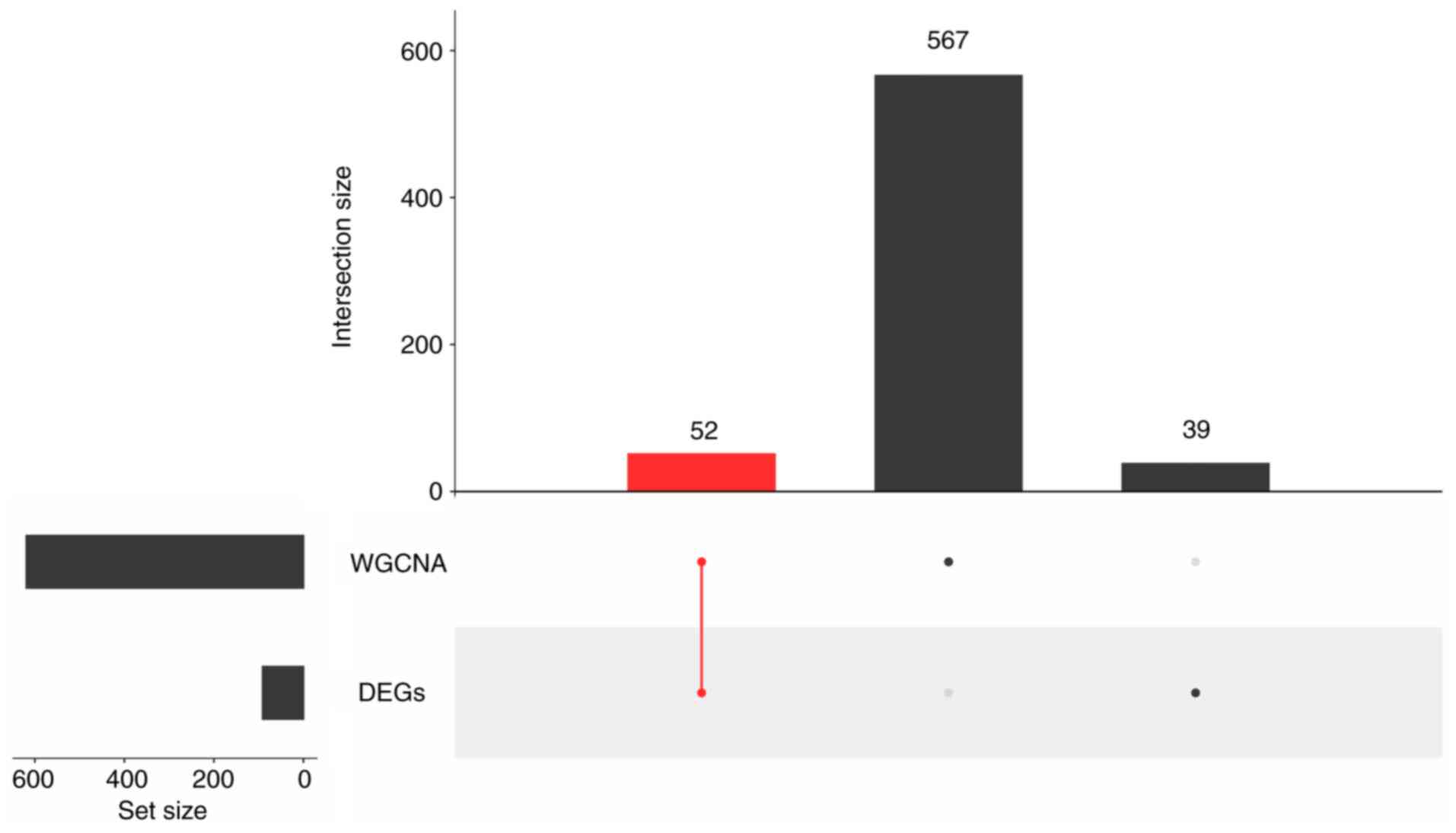
Filtering DE-PRGs
A total of 52 DE-PRGs were filtered by the feat of
intersection of 91 DEGs and 619 hub genes (Fig. 4). It meant that these 52 DE-PRGs
met with both hub genes and DEGs.
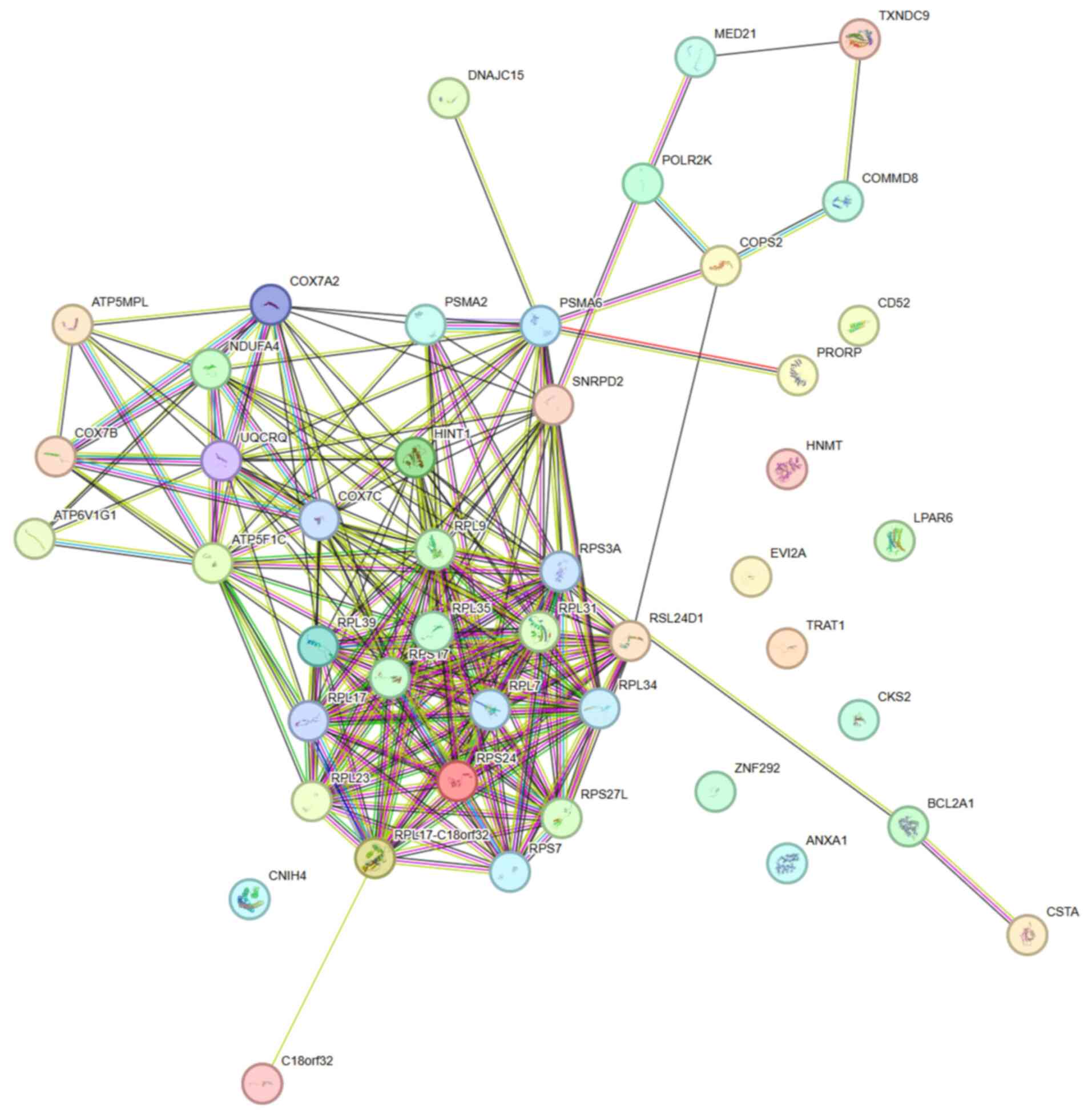
PPI network of DE-PRGs
Based on the 52 DE-PRGs, a PPI network that
contained 46 nodes and 227 edges was constructed that demonstrated
the interaction among 46 key DE-PRGs at the protein level (Fig. 5). The correlation (or importance)
of each gene was positively related with the number of its edges.
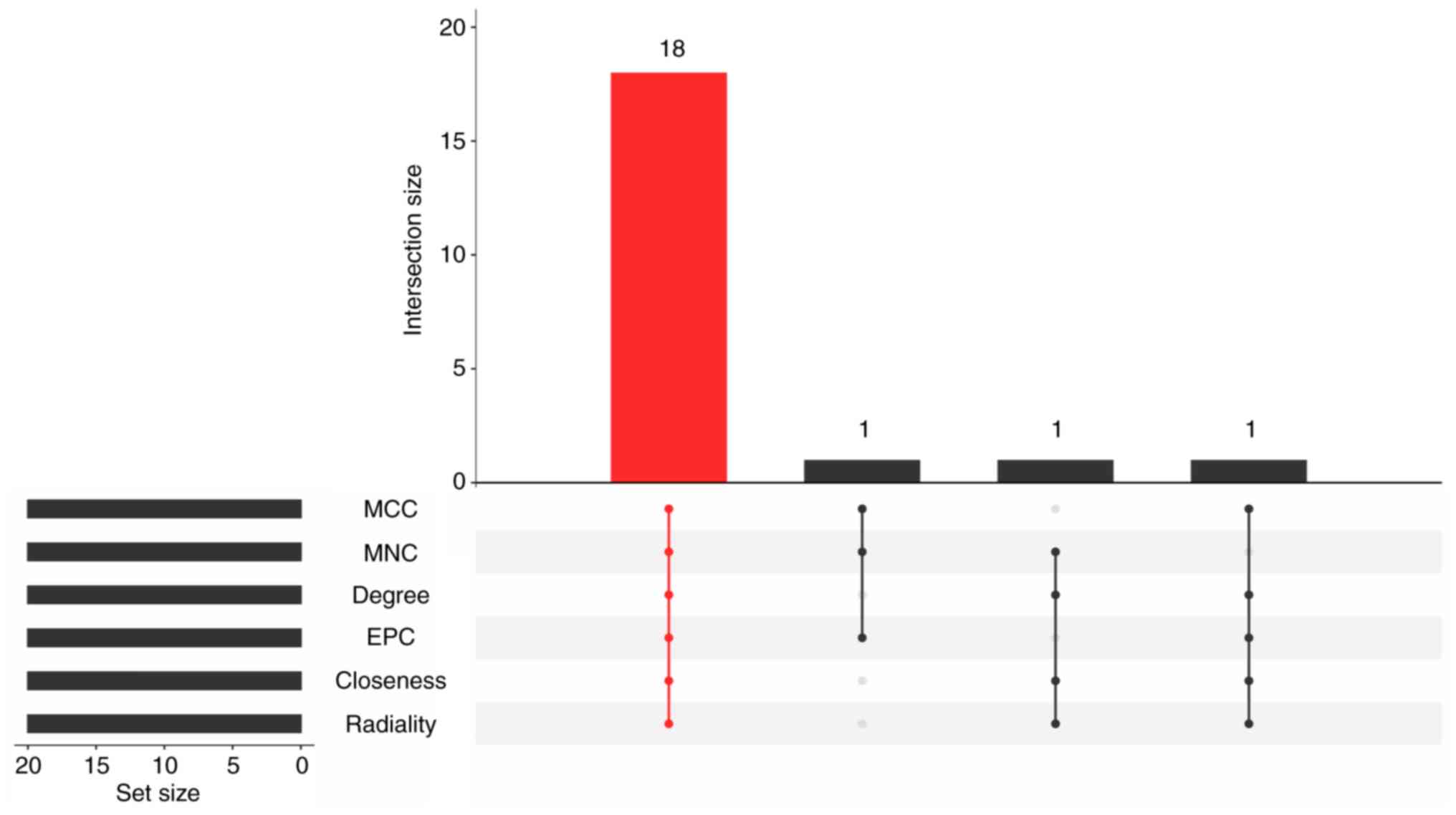
Through the utilization of six algorithms, 18 candidate genes were
identified by selecting the top 20 genes from each algorithm and
determining their intersection (Fig.
6).
A total of five key biomarkers are
associated with pyroptosis in VTE
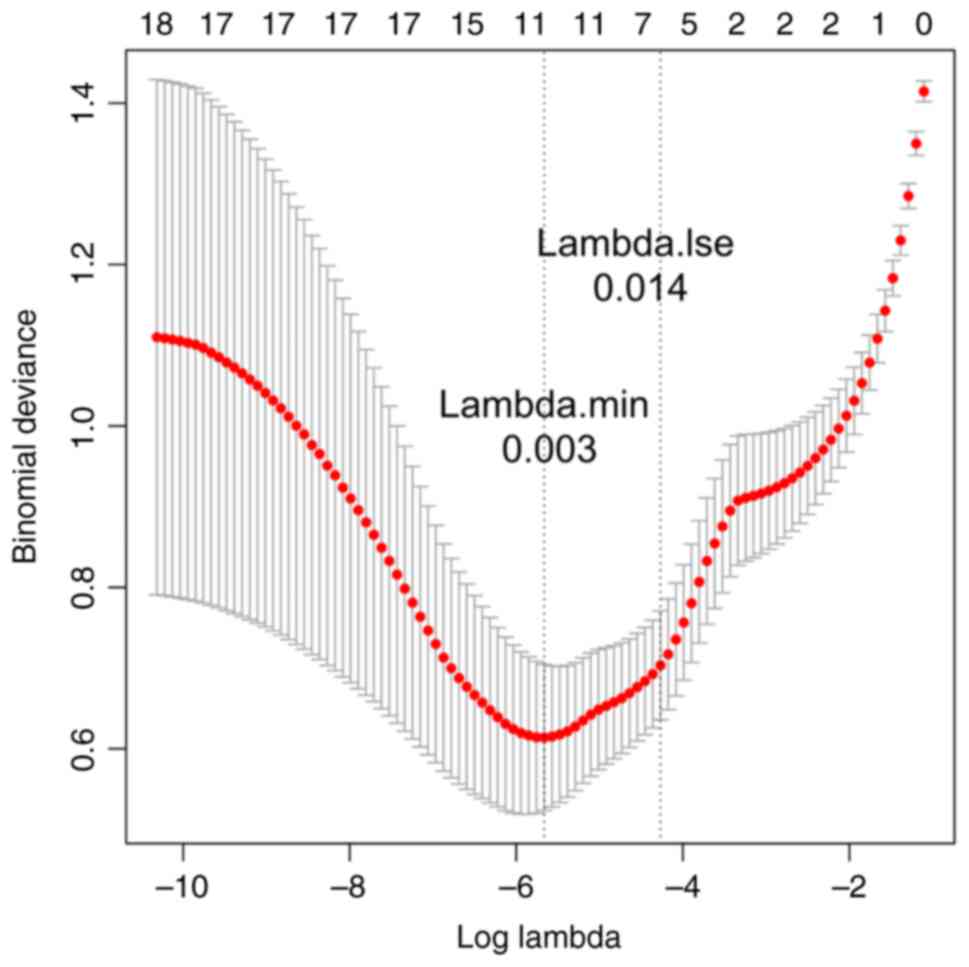
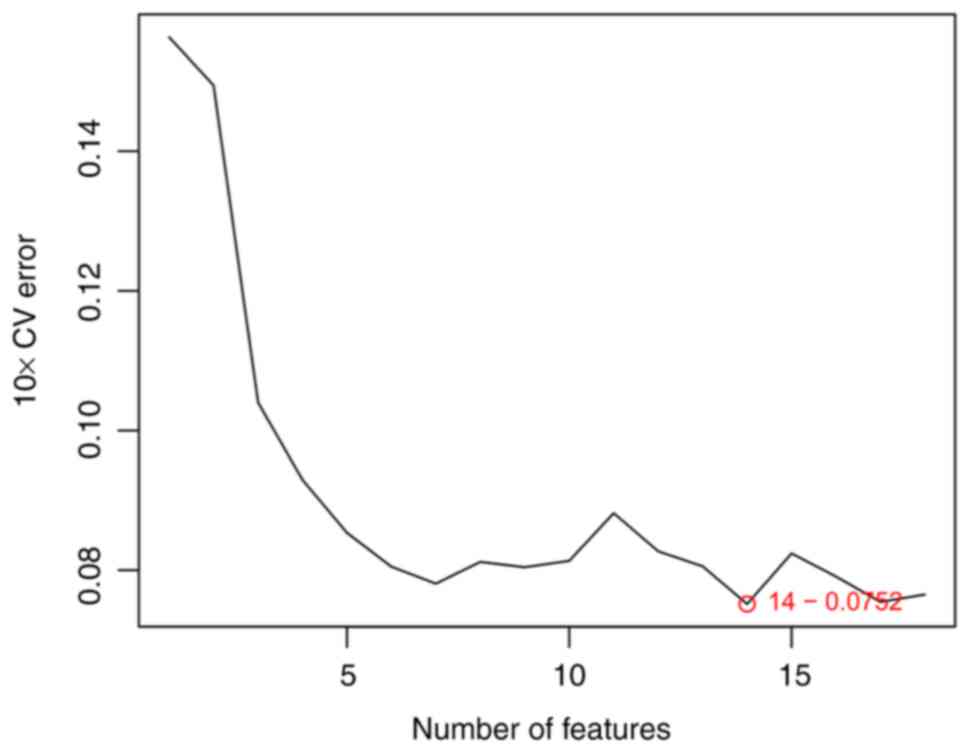
A total of 11 feature genes were further verified by
LASSO logistic regression algorithm (Fig. 7), while 14 feature genes were
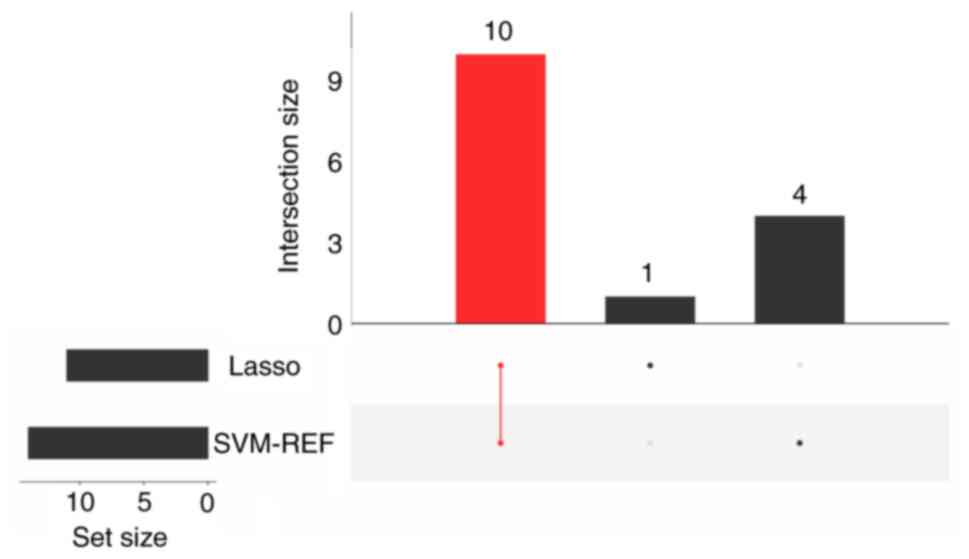
confirmed via the SVM-RFE algorithm (Fig. 8). A total of 10 candidate
biomarkers (RPL31, RPL34, RPS17, RPL9, RPL17, RPS27L,
RPL17-C18orf32, HINT1, SNRPD2 and UQCRQ) were identified upon their
intersection (Fig. 9). The
diagnostic value of these 10 candidate biomarkers was diagnosed and
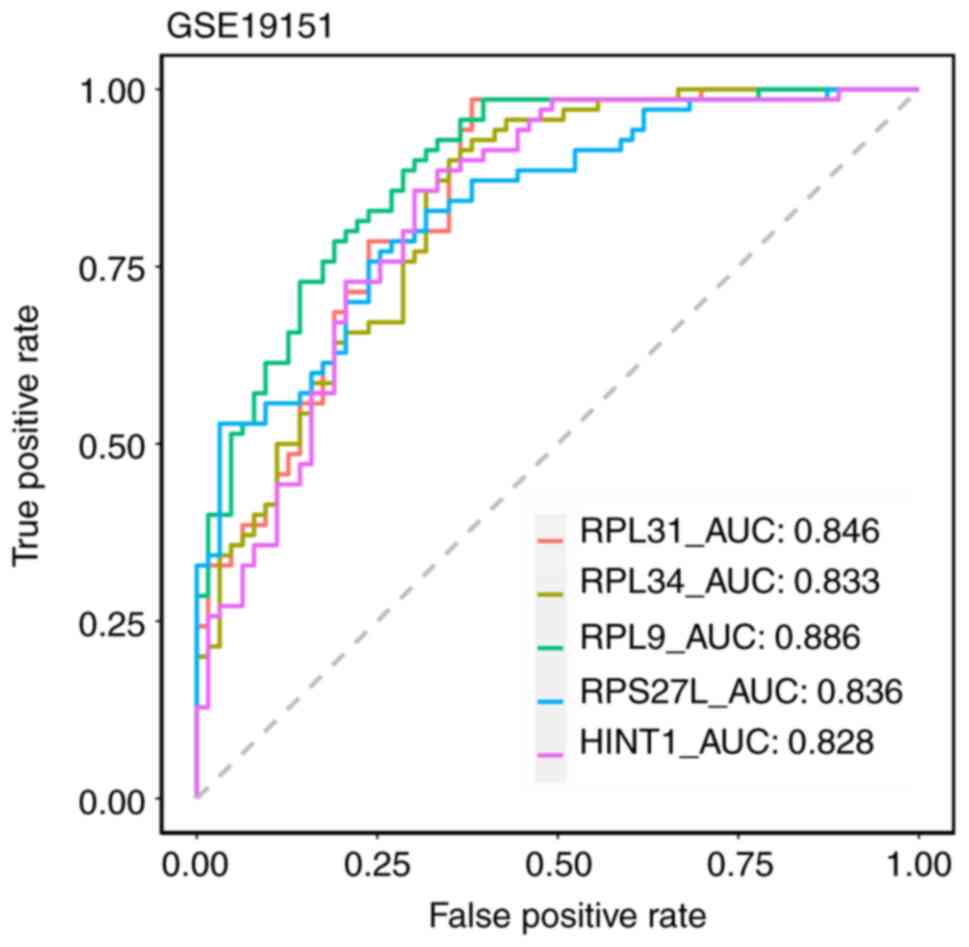
further refined through ROC analysis. Notably, five key biomarkers
(RPL31, RPL34, RPL9, RPS27L and HINT1) were screened under the
condition of ROC >0.7. The AUC values for these five biomarkers
was >0.8 (Fig. 10). Moreover,
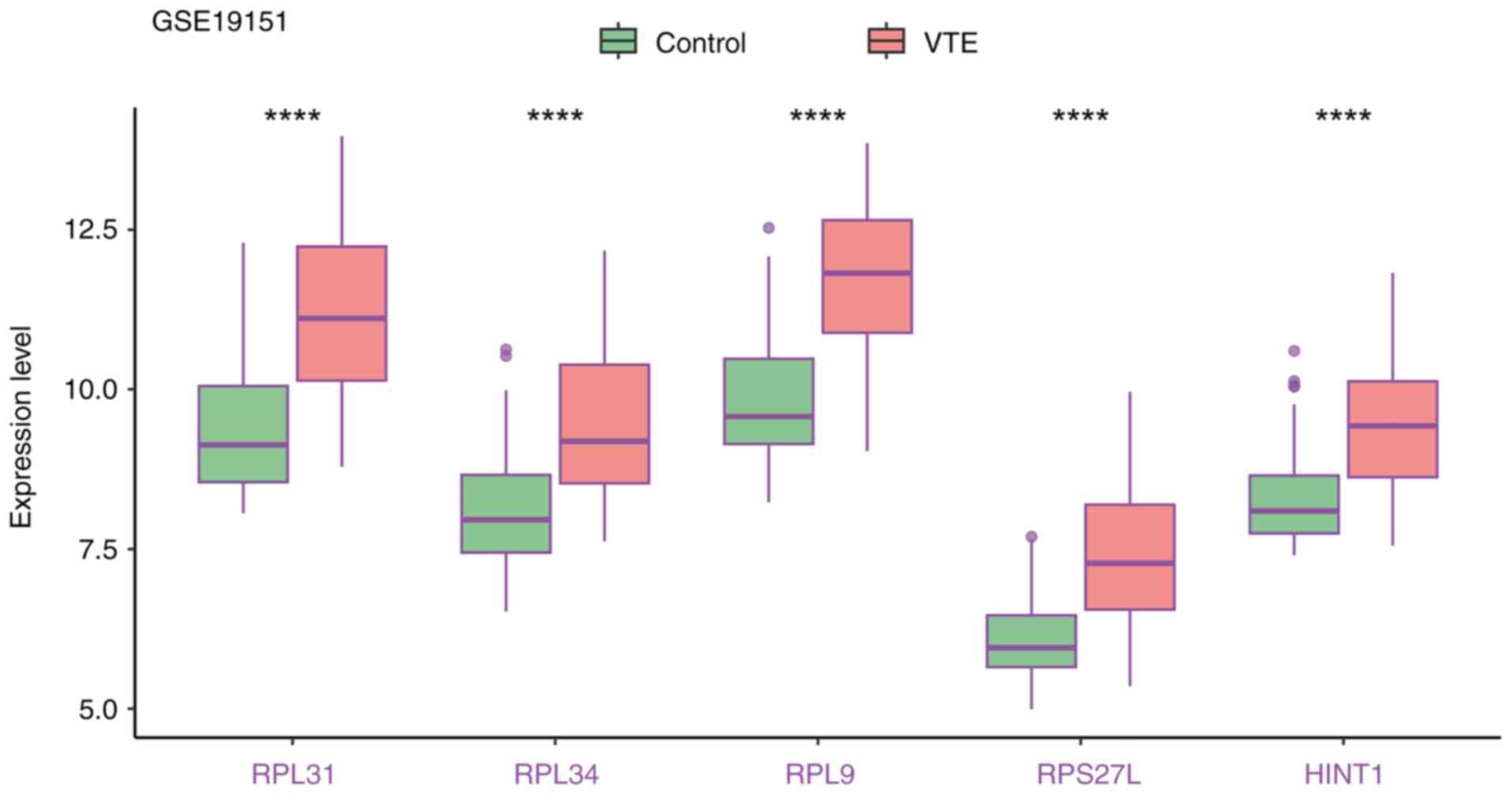
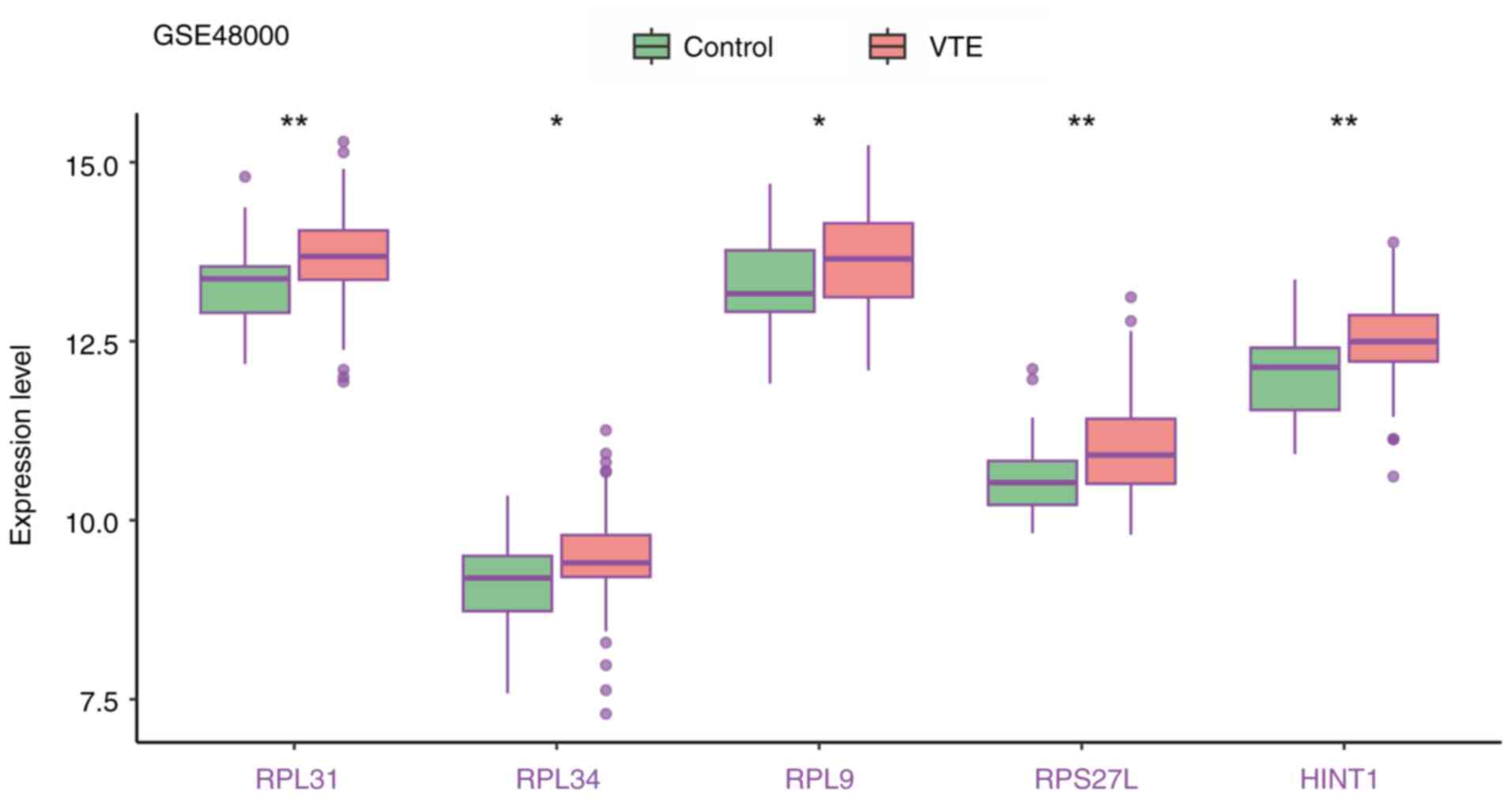
the expression levels of these five biomarkers in the training and
validation cohort were significantly increased in VTE samples
(Figs. 11 and 12).
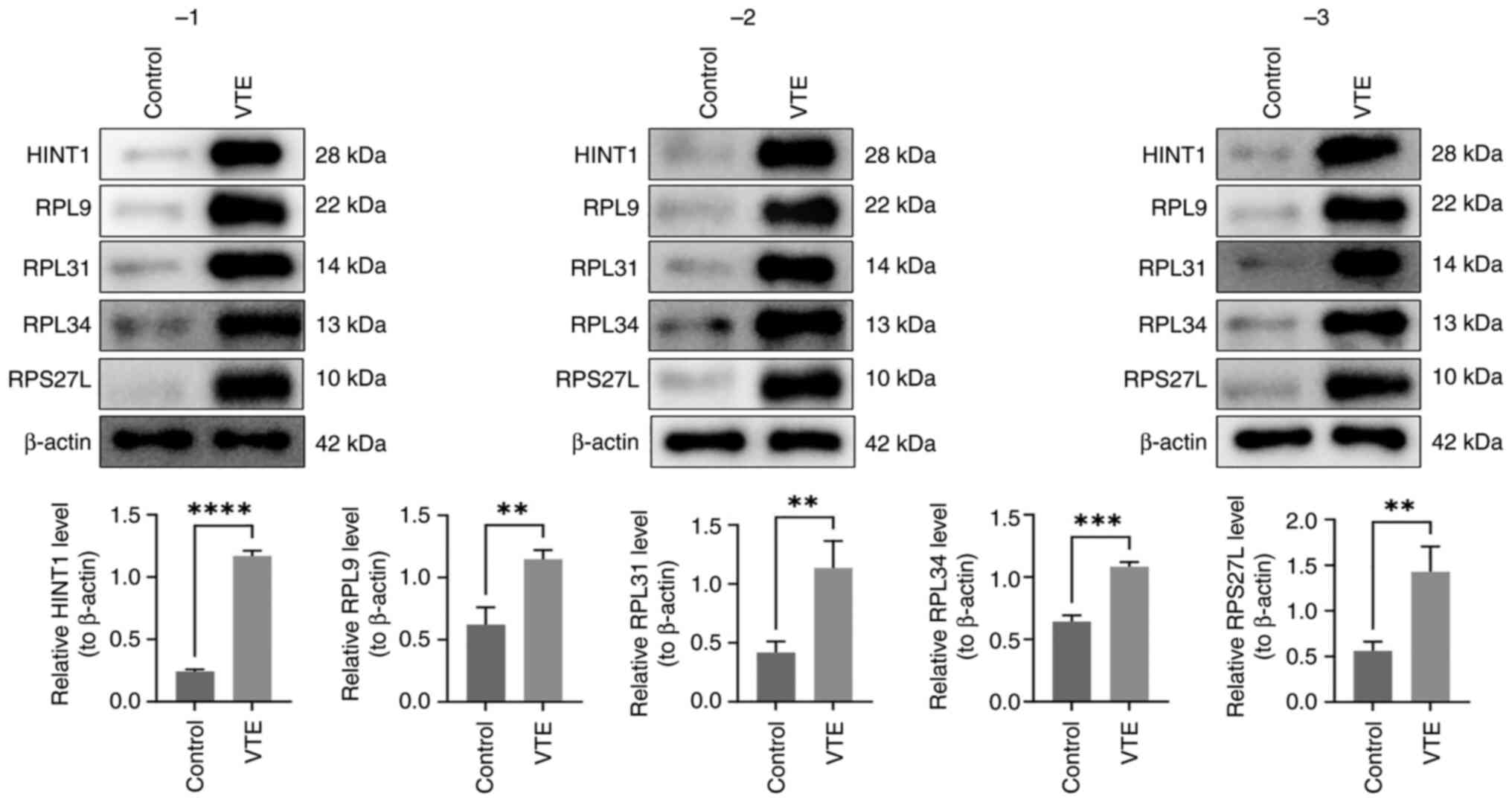
Expression levels of the five
biomarkers are all observably increased in VTE
RT-qPCR results demonstrated significantly elevated
expression levels of RPL34, RPS27L and HINT1 in VTE samples
compared with control samples. While the expression levels of RPL31
and RPL9 were higher in the VTE group, statistical significance was
not observed (Table IV).
Importantly, the expression trends of these genes were consistent
with those in the GSE19151 and GSE48000 datasets. Western blotting
analysis in three replicates revealed that all five biomarkers
significantly increased in the VTE group (Fig. 13).
 | Table IVReverse transcription-quantitative
PCR results for the five biomarkers. |
Table IV
Reverse transcription-quantitative
PCR results for the five biomarkers.
| Biomarkers | Control | Venous
thromboembolism | P-value |
|---|
| RPL31 | 1±0.4534 | 3.6857±3.8089 | 0.1561 |
| RPL34 | 1±0.8099 | 8.3609±3.4477 | 0.0030a |
| RPL9 | 1±0.4192 | 1.4419±0.8531 | 0.3920 |
| RPS27L | 1±0.8207 | 4.3979±2.8337 | 0.0360a |
| HINT1 | 1±0.5200 | 2.1421±0.9540 | 0.0466a |
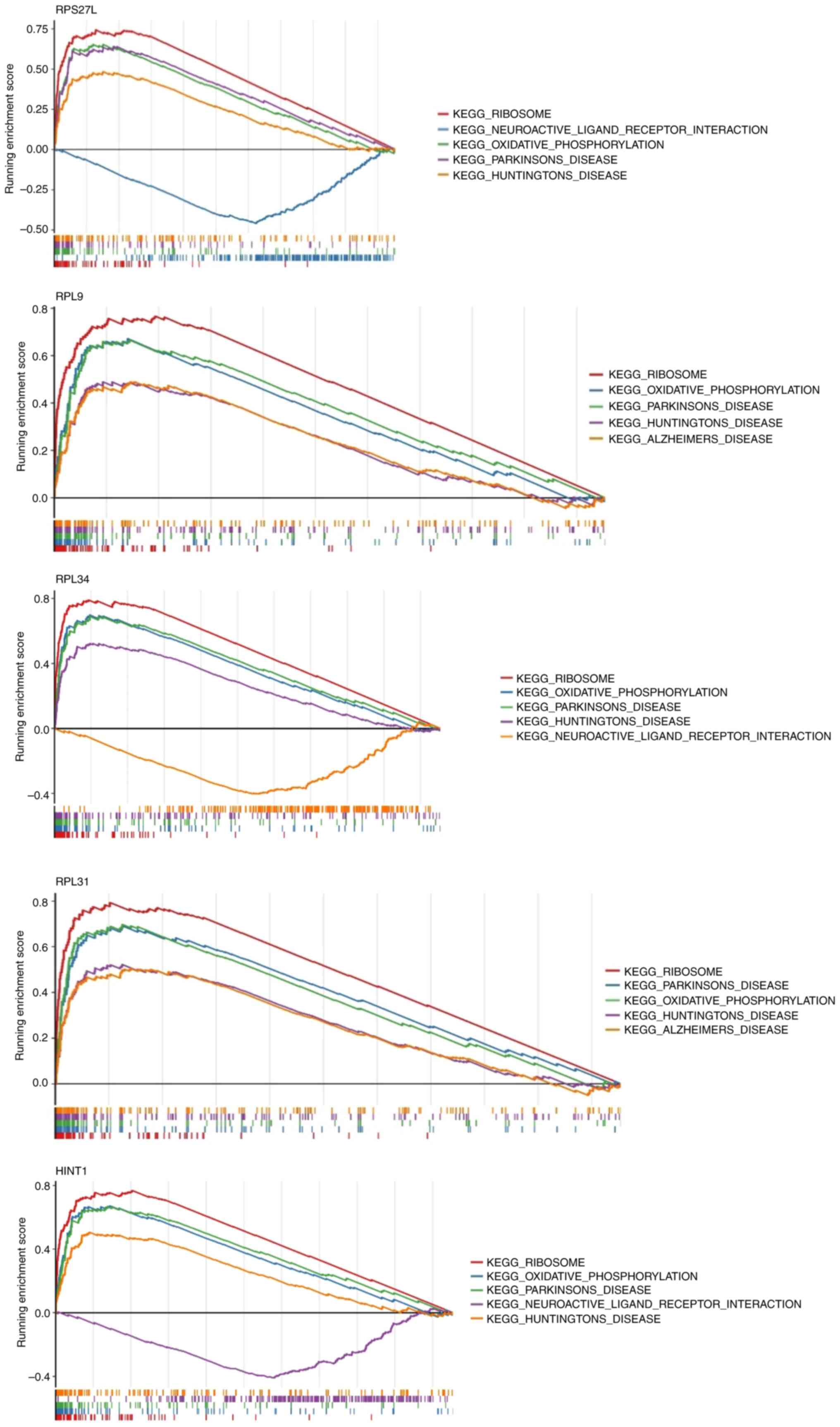
A total of five biomarkers are
associated with the pathways that may cause pyroptosis
GSEA was employed to elucidate the pathways
influenced by pyroptosis in VTE. The results revealed that ribosome
and oxidative phosphorylation signaling pathways were highly
enriched in these five biomarkers. These pathways may contribute to
the initiation of VTE by regulating
pyroptosis-inflammation-coagulation axis (Fig. 14).
Discussion
VTE, encompassing PE and DVT, is a life-threatening
condition. Due to its often-asymptomatic presentation, VTE
supersedes myocardial infarction and stroke as a leading cause of
sudden mortality. Globally, VTE results in a mortality every 37
sec, with millions of diagnoses and >840,000 mortalities
annually, a figure that continues to rise. Early diagnosis poses a
considerable challenge in VTE management, as it relies on clinical
manifestations supported by laboratory and imaging investigations.
However, these diagnostic modalities have inherent limitations that
can lead to missed or incorrect diagnoses in the early stages of
the condition. Therefore, the development of early and universally
applicable biomarkers holds paramount importance for its initial
management. Emerging research implicates thrombotic inflammation,
particularly pyroptosis, a more recently discovered form of
programmed cell death, elicits a strong inflammatory response and
thrombotic inflammation related to innate immunity (10), in VTE pathogenesis, though its
molecular mechanisms remain unclear. Through integrated
bioinformatics analysis, five pyroptosis-related biomarkers (RPL31,
RPL34, RPL9, RPS27L and HINT1) associated with ribosomal
biosynthesis and mitochondrial oxidative phosphorylation were
identified, suggesting their potential role in VTE via
mitochondrial dysfunction. The present study provides the first
systematic evidence connecting these biomarkers to pyroptosis in
VTE, offering novel targets for diagnosis and therapeutic
intervention.
Pyroptosis is characterized by quick formation of
cytomembrane pores and subsequent release of massive
proinflammatory mediators, culminating in the activation of
endothelial cells, the coagulation system and ensuing thrombotic
occurrences. In detail, pyroptosis leads to the release of tissue
factors from immune cells, initiating the coagulation cascade and
facilitating thrombus formation. Activation of the inflammasome and
subsequent pyroptosis carries out a central role in the development
and progression of VTE (12).
Therefore, pyroptosis serves as a key determinant in the occurrence
of VTE, preceding its clinical manifestation. Through an analysis
of the interplay between pyroptosis and VTE, five key biomarkers
(RPL31, RPL34, RPL9, RPS27L and HINT1, all associated with
ribosomal proteins and oxidative phosphorylation) (3,22)
have been identified as associated with the pathogenesis of both
conditions. The expression levels of these five biomarkers in the
training cohort (containing 70 VTE samples as well as 63 control
samples) and validation cohort (containing 107 VTE samples and 25
control samples) were significantly increased in the VTE samples.
Upon RT-qPCR analysis, RPL34, RPS27L and HINT1 exhibited
significantly increased expression levels in VTE samples compared
with controls (P<0.05), while RPL31 and RPL9 displayed increased
expression in the VTE group without statistical significance.
Although the RT-qPCR analysis conducted on 5 sample pairs was
limited in scale and yielded slightly differing results from the
bioinformatics analysis, the expression trends were consistent with
those of the GSE19151 and GSE48000 datasets.
A study conducted by Ma et al (3) on catheter-associated venous
thrombosis utilized similar methodologies and datasets to identify
12 VTE-related genes, four of which overlapped with the genes
identified in the present study (RPL31, RPL34, RPL9 and RPS27L).
However, Ma et al's (3)
study mainly focused on catheter-associated venous thrombosis.
Although the present study's methods and databases were similar
with that used by Ma et al (3), the present study focused on
systematically exploring early diagnostic markers for VTE from the
perspective of cell pyroptosis, rather than solely investigating
catheter-related thrombosis. Therefore, the present study attempted
to avoid the limitation of a single applicable population as much
as possible. Additionally, research by Zhang et al (22) on COVID-19 and VTE highlighted Hint1
and RPL34 as promising diagnostic markers for COVID-19 and VTE.
However, these studies did not specifically investigate pyroptosis
and may not have universal applicability to VTE in a broader
context. The principle of RT-qPCR is to combine reverse
transcription of RNA with chain amplification of complementary DNA,
and then detect the changes in yield in each cycle of amplification
in real time through changes in fluorescence signal. Finally,
precise quantitative analysis of the starting template is carried
out to detect the expression levels of genes in cells. Due to the
involvement of several of PCR cycles, there may be technical bias
in the experimental results. This requires a reasonable sample size
as much as possible, or at the same time, use another experimental
techniques (such as western blotting) for supporting verification.
It should be noted that although this bias did exist, the levels of
all five biomarkers were elevated in the VTE group, although this
elevation only showed significant differences in three biomarkers
(RPL34, RPS27L and HINT1).
In the present study, GSEA highlighted a significant
enrichment of the ribosome and oxidative phosphorylation signaling
pathways in the five identified biomarkers. It is hypothesized that
these biomarkers may contribute to the onset of VTE by modulating
the oxidative phosphorylation signaling pathway. This hypothesis
was also partially confirmed in Ma et al's (3) and Zhang et al's (22) study. Ma et al's (3) study showed that the expression levels
of RPL31, RPL34, RPL9 and RPS27L in VTE were higher than normal
whole blood tissue samples, NDUFB11 is highly expressed in
catheter-related VTE during continuous blood purification, which
may lead to the formation of venous thrombosis through the
oxidative phosphorylation pathway. While Zhang et al's
(22) study identified HINT1,
RPL34 and NDUFA4 as diagnostic markers for COVID-19 and VTE.
Structurally, these biomarkers are associated with biosynthesis
(ribosomal proteins RPL31, RPL34, RPL9 and RPS27L) and signal
transduction (HINT1). Through rigorous bioinformatics analysis, the
present study has systematically identified five key biomarkers
associated with pyroptosis in the context of VTE.
Ribosomal proteins are components of ribosomes
involved in protein translation and ribosome assembly (23), key for the growth and viability of
all cell types (24). Meanwhile,
ribosomal proteins have other functions involved in DNA repair,
cell development regulation and differentiation. Certain ribosomal
protein genes exhibit elevated expression levels in tumor tissues
such as gastric cancer, colorectal cancer, and pancreatic cancer
(25-27).
In the eukaryotic cells, the ribosome is divided into the 60S and
40S subunits, with RPL31, RPL34 and RPL9 belonging to the 60S
subunit, while RPS27L is a member of the 40S subunit. Specifically,
RPL31 regulates a variety of physiological and pathological
processes in the cytoplasm (25).
RPL31 is implicated in ribosome self-assembly, protein synthesis,
cell proliferation, DNA repair and tumorigenesis. RPL34, besides
its role as a ribosomal protein, harbors a zinc finger motif and
has been associated with various cellular processes (26). RPL9 has been associated with the
progression of colorectal carcinoma (27) and to inflammatory processes
(28). RPS27L, an evolutionarily
conserved 84-amino acid ribosomal protein within the 40S small
subunit of the ribosome, differs from its family member RPS27 by
only three amino acids (R5K, L12P, K17R) at the N-terminus
(29). The study by Xiong et
al demonstrated that neddylation stabilizes RPS27L and RPS27,
thereby promoting the survival of cancer cells (30). HINT1 is a highly conserved protein
prevalent in mammalian tissues across evolution, primarily
localized in the nucleus and cytoskeleton. HINT1 engages in
intracellular and extracellular signal transduction by interacting
with protein kinase C, participating in a spectrum of PPIs that
encompass nociception and mast cell activation (31,32).
However, the expression, functional roles and prognostic
implications of the aforementioned five biomarkers in VTE remain
unexplored in prior studies.
The comprehensive application of multiple
statistical methods serves as a key aspect in the initial discovery
of these biomarkers. LASSO regression, a contraction estimation
method, aims to minimize the sum of squared residuals while
ensuring that the absolute sum of regression coefficients falls
below a specified constant. This approach results in some
regression coefficients being precisely zero, facilitating the
creation of an interpretable model. SVMs represent a binary
classification model that employs a learning strategy focused on
maximizing the margin, addressing convex quadratic programming
optimization. However, the ROC curve stands as a fundamental metric
for assessing the discriminative performance of medical diagnostic
experiments and predictive models. A key attribute of the ROC curve
is its AUC, where a value closer to 1 signifies superior
discriminatory ability. In the present study, the AUC values for
all five biomarkers fall within the range of 0.8 to 0.9, indicating
excellent recognition ability.
The present study is the first to systematically
identify five PRGs as potential biomarkers for VTE through
integrated multi-omics analysis, revealing the key role of
ribosomal stress and mitochondrial dysfunction in VTE pathogenesis
and providing novel insights into thrombotic inflammation. However,
several limitations should be noted: i) The relatively small sample
size and exclusive reliance on peripheral blood transcriptome data
may not fully reflect tissue-specific mechanisms; ii)
bioinformatics predictions require further experimental validation
using in vitro models (such as endothelial cell pyroptosis
assays) and animal models (such as murine venous thrombosis
experiments); iii) the clinical translational value of candidate
biomarkers needs evaluation in prospective cohorts.
Based on these findings, the present study
recommends the following directions for future research: i)
Application of single-cell sequencing to characterize pyroptosis
pathway activation in specific cell subpopulations (such as,
platelets and endothelial cells) from patients with VTE; ii)
development of gene-targeting strategies (such as, small
interfering RNA or inhibitors) against RPL31/RPL34 to elucidate
their regulatory mechanisms in thrombus formation; iii)
establishment of multicenter clinical cohorts to validate the
utility of these biomarkers for early VTE diagnosis and risk
stratification.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Foundation of
Applied Basic Research Program of Yunnan Province [Kunming, China;
grant no. 2019FE001(-216)].
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SH designed the present study and wrote the original
draft. JX interpreted data and translated the manuscript. CY and SD
collected and organized the data. SD was responsible for
statistical analysis and made substantial contributions to
conception and design. HG searched for literature and conducted the
bioinformatic analysis. All authors read and approved the final
version of the manuscript. SH and SD confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
The present study was approved by the First
Affiliated Hospital of Kunming Medical University ethics committee
(approval no. 2023L198).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Di Nisio M, van Es N and Büller HR: Deep
vein thrombosis and pulmonary embolism. Lancet. 388:3060–3073.
2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lutsey PL and Zakai NA: Epidemiology and
prevention of venous thromboembolism. Nat Rev Cardiol. 20:1–15.
2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ma Y and Guo S: High expression of NADH
ubiquinone oxidoreductase subunit B11 induces catheter-associated
venous thrombosis on continuous blood purification. Medicine
(Baltimore). 102(e36520)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Colling ME, Tourdot BE and Kanthi Y:
Inflammation, infection and venous thromboembolism. CircRes.
128:2017–2036. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Maaroof A and Wahab Z: Enhancing venous
thromboembolism risk prediction through novel biomarkers and
inflammatory indicators. J Appl Hematol. 15:313–318.
2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zeng J, Feng J, Luo Y, Wei H, Ge H, Liu H,
Zhang J, Li X, Pan P, Xie X, et al: Inflammatory biomarkers as
predictors of symptomatic venous thromboembolism in hospitalized
patients with AECOPD: A multicenter cohort study. J Atheroscler
Thromb. 32:439–457. 2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu Y, Guo B, Meng Z, Fan Y, Xie Y, Gao L
and Ma R: Coagulation and inflammatory indicators in pneumonia
patients with venous thromboembolism: A propensity-score matching
study. J Inflamm Res. 18:5627–5635. 2025.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Swanson KV, Deng M and Ting JP: . The
NLRP3 inflammasome: molecular activation and regulation to
therapeutics. Nat Rev Immunol. 19:477–489. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Bakirci EM, Topcu S, Kalkan K, Tanboga IH,
Borekci A, Sevimli S and Acikel M: The role of the nonspecific
inflammatory markers in determining the anatomic extent of venous
thromboembolism. Clin Appl Thromb. 21:181–185. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Potere N, Abbate A, Kanthi Y, Carrier M,
Toldo S, Porreca E and Di Nisio M: Inflammasome signaling,
thromboinflammation, and venous thromboembolism. JACC Basic Transl
Sci. 8:1245–1261. 2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Johnson AG, Wein T, Mayer ML, Duncan-Lowey
B, Yirmiya E, Oppenheimer-Shaanan Y, Amitai G, Sorek R and
Kranzusch PJ: Bacterial gasdermins reveal an ancient mechanism of
cell death. Science. 375:221–225. 2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang Y, Cui J, Zhang G, Wu C, Abdel-Latif
A, Smyth SS, Shiroishi T, Mackman N, Wei Y, Tao M and Li Z:
Inflammasome activation promotes venous thrombosis through
pyroptosis. Blood Adv. 5:2619–2623. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Lu S, Lijuan R, Tang QH, Liu QL and
Xian-Lan Z: Bioinformatics analysis and identification of genes and
molecular pathways involved in venous thromboembolism (VTE). Ann
Vasc Surg. 74:389–399. 2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen H, Luo H, Wang J, Li J and Jiang Y:
Identification of a pyroptosis-related prognostic signature in
breast cancer. BMC Cancer. 22(429)2022.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W and Zhan L: ClusterProfiler 4.0: A universal
enrichment tool for interpreting omics data. Innovation (Camb).
2(100141)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang Z, Zhang Z, Zhang K, Zhou Q, Chen S,
Zheng H, Wang G, Cai S, Wang F and Li S: Multi-omics
characterization of a glycerolipid metabolism-related gene
enrichment score in colon cancer. Front Oncol.
12(881953)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9(559)2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cheng Q, Chen X, Wu H and Du Y: Three
hematologic/immune system-specific expressed genes are considered
as the potential biomarkers for the diagnosis of early rheumatoid
arthritis through bioinformatics analysis. J Transl Med.
19(18)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tan Z, Li F, Chen Q, Chen H, Xue Z, Zhang
J, Gao Y, Liang L, Huang T, Zhang S, et al: Integrated bulk and
single-cell RNA-sequencing reveals SPOCK2 as a novel biomarker gene
in the development of congenital pulmonary airway malformation.
Respir Res. 24(127)2023.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wu X, Qin K, Iroegbu CD, Xiang K, Peng J,
Guo J, Yang J and Fan C: Genetic analysis of potential biomarkers
and therapeutic targets in ferroptosis from coronary artery
disease. J Cell Mol Med. 26:2177–2190. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang L, Qin J and Li P: Bioinformatics
analysis of potential common pathogenic mechanisms for COVID-19 and
venous thromboembolism. Cytokine. 181(156682)2024.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fatica A and Tollervey D: Making
ribosomes. Curr Opin Cell Biol. 14:313–318. 2002.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Roberts E, Sethi A, Montoya J, Woese CR
and Luthey-Schulten Z: Molecular signatures of ribosomal evolution.
Proc Natl Acad Sci U S A. 105:13953–1398. 2008.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wu F, Liu Y, Hu S and Lu C: Ribosomal
protein L31 (RPL31) inhibits the proliferation and migration of
gastric cancer cells. Heliyon. 9(e13076)2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wei F, Ding L, Wei Z, Zhang Y, Li Y,
Qinghua L, Ma Y, Guo L, Lv G and Liu Y: Ribosomal protein L34
promotes the proliferation, invasion and metastasis of pancreatic
cancer cells. Oncotarget. 51:85259–85272. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Baik IH, Jo GH, Seo D, Ko MJ, Cho CH, Lee
MG and Lee YH: Knockdown of RPL9 expression inhibits colorectal
carcinoma growth via the inactivation of Id-1/NF-κB signaling axis.
Int J Oncol. 49:1953–1962. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Watanabe M, Toyomura T, Wake H, Nishinaka
T, Hatipoglu OF, Takahashi H, Nishibori M and Mori S:
Identification of ribosomal protein L9 as a novel regulator of
proinflammatory damage-associated molecular pattern molecules. Mol
Biol Rep. 49:2831–2838. 2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xiong X, Zhao Y, He H and Sun Y: Ribosomal
protein S27-like and S27 interplay with p53-MDM2 axis as a target,
a substrate and a regulator. Oncogene. 30:1798–1811.
2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xiong X, Cui D, Bi Y, Sun Y and Zhao Y:
Neddylation modification of ribosomal protein RPS27L or RPS27 by
MDM2 or NEDP1 regulates cancer cell survival. The FASEB J.
34:13419–13429. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Genovese G, Ghosh P, Li H, Rettino A,
Sioletic S, Cittadini A and Sgambato A: The tumor suppressor HINT1
regulates MITF and β-catenin transcriptional activity in melanoma
cells. CellCycle. 11:2206–2215. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
32
|
Dillenburg M, Smith J and Wagner CR: The
many faces of histidine triad nucleotide binding protein 1 (HINT1).
ACS Pharmacol. Transl. Sci. 6:1310–1322. 2023.PubMed/NCBI View Article : Google Scholar
|