Introduction
Achalasia is a rare motility disorder of the
esophagus characterized by dysphagia, chest pain, and weight loss
(1,2). It has an annual incidence of one in
100,000 individuals and a prevalence of 10 in 100,000 individuals,
and occurs equally in male and female patients (3). Although the exact cause remains
largely unknown, a combination of genetic and environmental
factors, such as viral infection-mediated immune reactions, may
lead to ganglionitis and the loss of esophageal neurons (1,4,5).
Achalasia is defined by a combination of symptoms, endoscopic
findings, esophagography, and esophageal high-resolution manometry
(HRM) (2,6).
Although the underlying causes and variations in
esophageal motility disorders remain largely unknown, HRM has
emerged as an essential tool for defining and detecting these
disorders (2,6). The Chicago classification using HRM
is helpful for the diagnosis and pretreatment evaluation of
esophageal achalasia (7). The
integrated relaxation pressure (IRP) indicates the elevation of the
lower esophageal sphincter (LES) pressure for several seconds after
swallowing (8). In the Chicago
classification, this value serves as the starting point for
developing the algorithm, indicating an abnormal motility disorder
(7). However, relying solely on
the IRP in the algorithm is often inconsistent with the clinical
diagnosis (9); therefore, a
comprehensive assessment by a physician, incorporating other
modalities, is usually necessary. The IRP value, essential for
diagnosis, is dynamic and influenced by factors such as
diaphragmatic activity, LES function, intrabolus pressure, and
catheter positioning (10,11). An accurate interpretation requires
a comprehensive assessment that extends beyond the numerical values
(11). Interpreting HRM poses
significant challenges and requires specialized expertise for an
accurate diagnosis. This exclusive reliance on specialists for this
diagnostic procedure has led to delays in identifying esophageal
motility disorders, such as achalasia.
Artificial intelligence (AI) can alleviate issues
associated with the clinical interpretation of HRM. Due to the
rarity of this disease, few studies have assessed AI in patients
with achalasia. Kou et al (12) first demonstrated a deep
learning-based model for diagnosing achalasia using HRM images,
employing a variational autoencoder (VAE). Surdea-Blaga et
al (13) demonstrated the
classification of swallowing disorders using the DenseNet201
convolutional neural network (CNN) architecture combined with the
automated Chicago classification, yielding 86% accuracy without
human intervention. Although AI-based tools are emerging in the
medical field, a black-box problem arises: they do not reveal how
or why they were concluded. In AI-based achalasia, whether AI
focuses on the same area as clinicians remains unclear.
In this study, we validated an automated diagnostic
system using AI to assess HRM. In addition, we utilized a gradient
class activation map (Grad-CAM) to evaluate the features of HRM,
where the AI focused on distinguishing achalasia from other
esophageal motor disorders.
Materials and methods
Data collection and ethical
approval
The research implementation system was conducted at
a single center. This retrospective study was approved by the
Nagasaki University Hospital Research Ethics Committee. This
retrospective study was approved by the ethics review board and the
requirement for informed consent was waived.
Data pre-processing
A total of 211 HRM images were de-identified to
protect patient confidentiality and were collected between December
2018 and August 2022 at the Nagasaki University Hospital. Typical
HRM images obtained from patients with achalasia who underwent POEM
were used as achalasia images, whereas HRM images from asymptomatic
individuals were used as normal controls. Images that were unclear
or diagnostically ambiguous were excluded from the analyses. Before
submitting the images for model training, each image was cropped to
remove the patient identification, machine settings, annotations,
and scale bars. High-resolution esophageal manometry was performed
using the Starlet system (Star Medical, Tokyo, Japan).
We selected tracings from ten test swallows for
analysis (n=22 patients; 211 images): achalasia type I, 67 images;
achalasia type II, 30 images; achalasia type III, 9 images;
hypercontractile esophagus, 20 images; and normal esophageal
motility, 97 images. We used the raw data, which contained only the
markers mentioned above (vertical white lines) preceding each wet
swallow. The manometry software enabled the storage of 60 s long
images of the recordings, which represented the raw images. The
region of interest (wet swallowing) was marked with a white
vertical line during the procedure. For each patient, we created a
folder with ten images, with each image representing a test
swallow.
Each set is assigned a diagnostic category. We used
five diagnostic categories as follows: normal, type I achalasia,
type II achalasia, type III achalasia, and hypercontractile
esophagus, based on the Chicago v3.0 classification for esophageal
motility disorders (7).
Developing a deep learning
algorithm
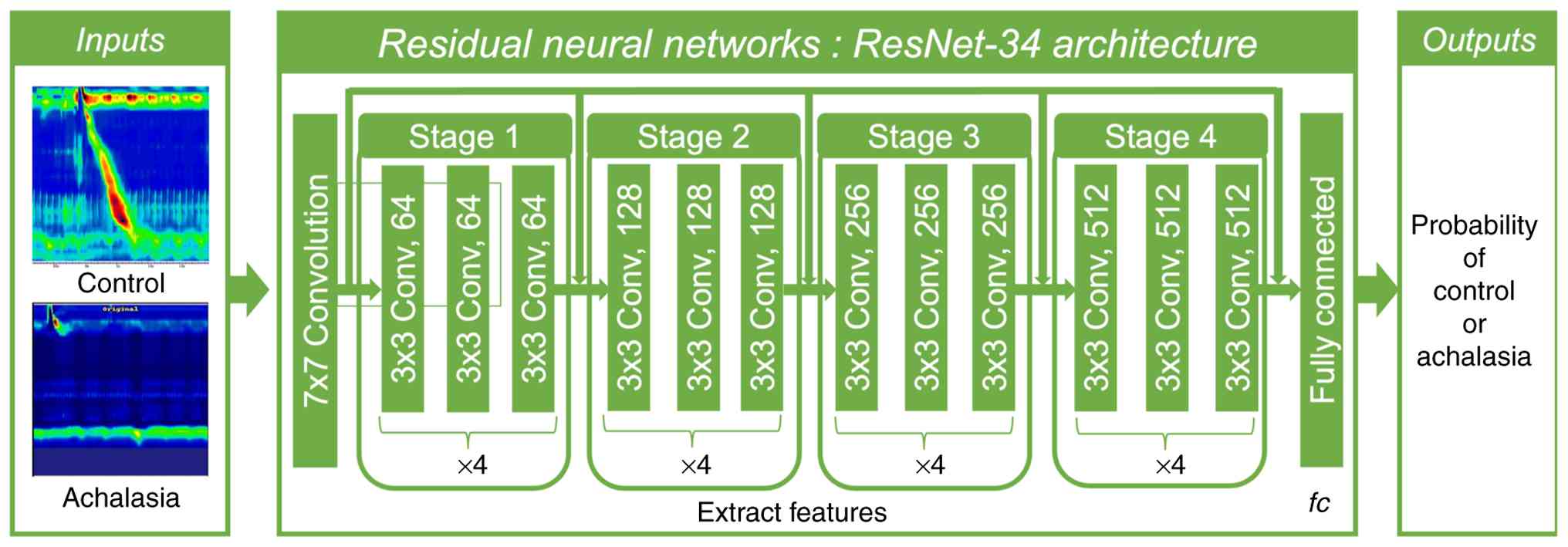
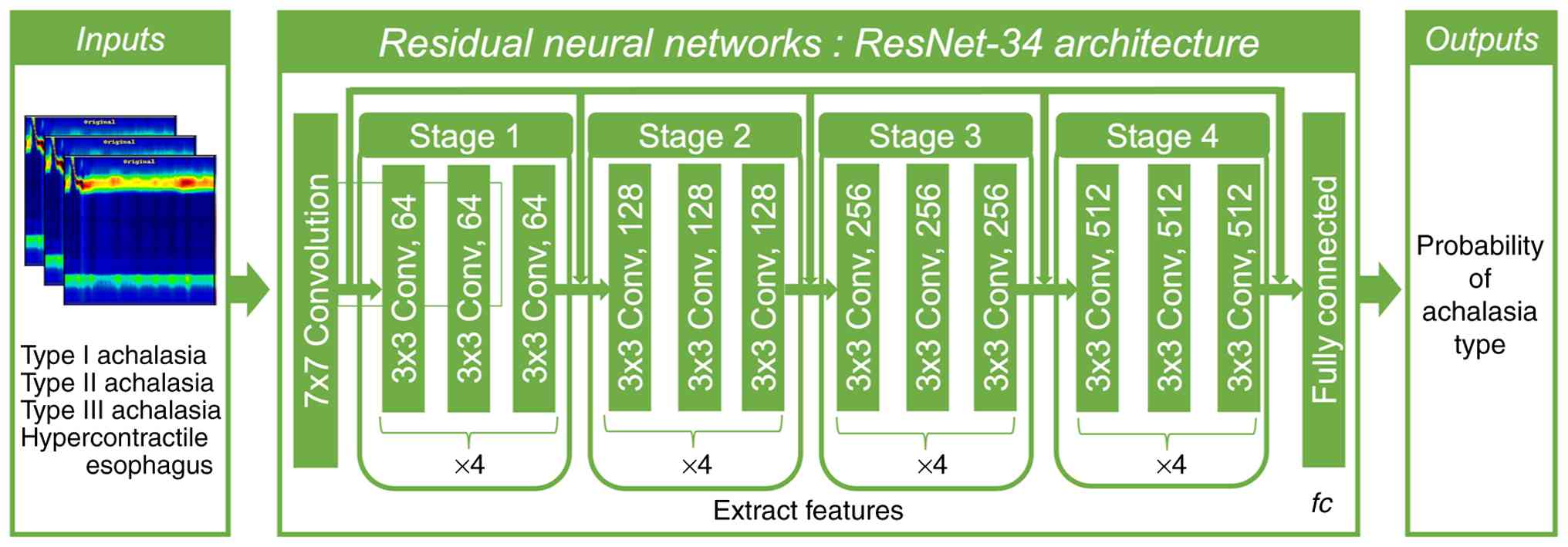
Achalasia was predicted by using the CNN. The
ResNet-34 network was used as the backbone of the encoder network
to encode the downsampling layer of the image. The model was
trained using Python 3.8 (Python Software Foundation, Beaverton,
OR, USA), Pytorch 1.8.1 (The Linux Foundation), and an Nvidia RTX
A6000 graphics card. The achalasia prediction training process
utilized a mean-squared error loss function and an Adam optimizer
with a batch size of 4; the learning rate decreased linearly from
5e-4 to 1e-6.
Grad-CAM method
Grad-CAM was used to reconstruct the image and
localize intensive areas for feature extraction. This method
identifies the image features used by a CNN to perform the
decision-making process, thereby enabling an understanding of the
predictive features determined by the trained model. We used
Grad-CAM to identify key abnormal features in the dataset. Regions
with high feature extraction intensities were assumed to be key
points in identifying anomalies.
Dimensions reduction analysis
Principal component analysis (PCA) and
three-dimensional t-distributed stochastic neighbor embedding
(3D-tSNE) were used for feature analysis of the entire image
(14). PCA was used to reduce the
dimensions. A TensorBoard Embedding Projector, which offers 3D-tSNE
views, was used to compute the top ten principal components. These
components were projected onto a combination of three components.
The most popular nonlinear dimensionality reduction technique is
t-SNE (15).
Statistical analysis
Testing accuracy, sensitivity, and specificity, as
calculated from the confusion matrix, were used to evaluate the
models. The receiver operating characteristic (ROC) curve, which is
a graphical plot highly correlated with the confusion matrix, was
also used as an evaluation index. Diagnostic accuracy based on the
confusion matrix was calculated as follows: (true positives + true
negatives)/(true positives + true negatives + false positives +
false negatives). All statistical analyses were performed using the
R statistical software (version 4.1.1; The R Foundation for
Statistical Computing, Vienna, Austria). Statistical significance
was set at P<0.05.
Results
All patients with achalasia included in the study
were treatment-naïve and had no prior interventions, such as
balloon dilation, Heller myotomy, or peroral endoscopic myotomy
(POEM) (Table I). The training
process for the achalasia-2CNN is illustrated in Fig. 1. First, we developed an achalasia
prediction model using HRM with CNN (Fig. 1). The ResNet-34 model, which has
already been proven to be versatile, was used for transfer
learning. Two hundred images were divided into training and
validation datasets, and a CNN based primarily on the PyTorch
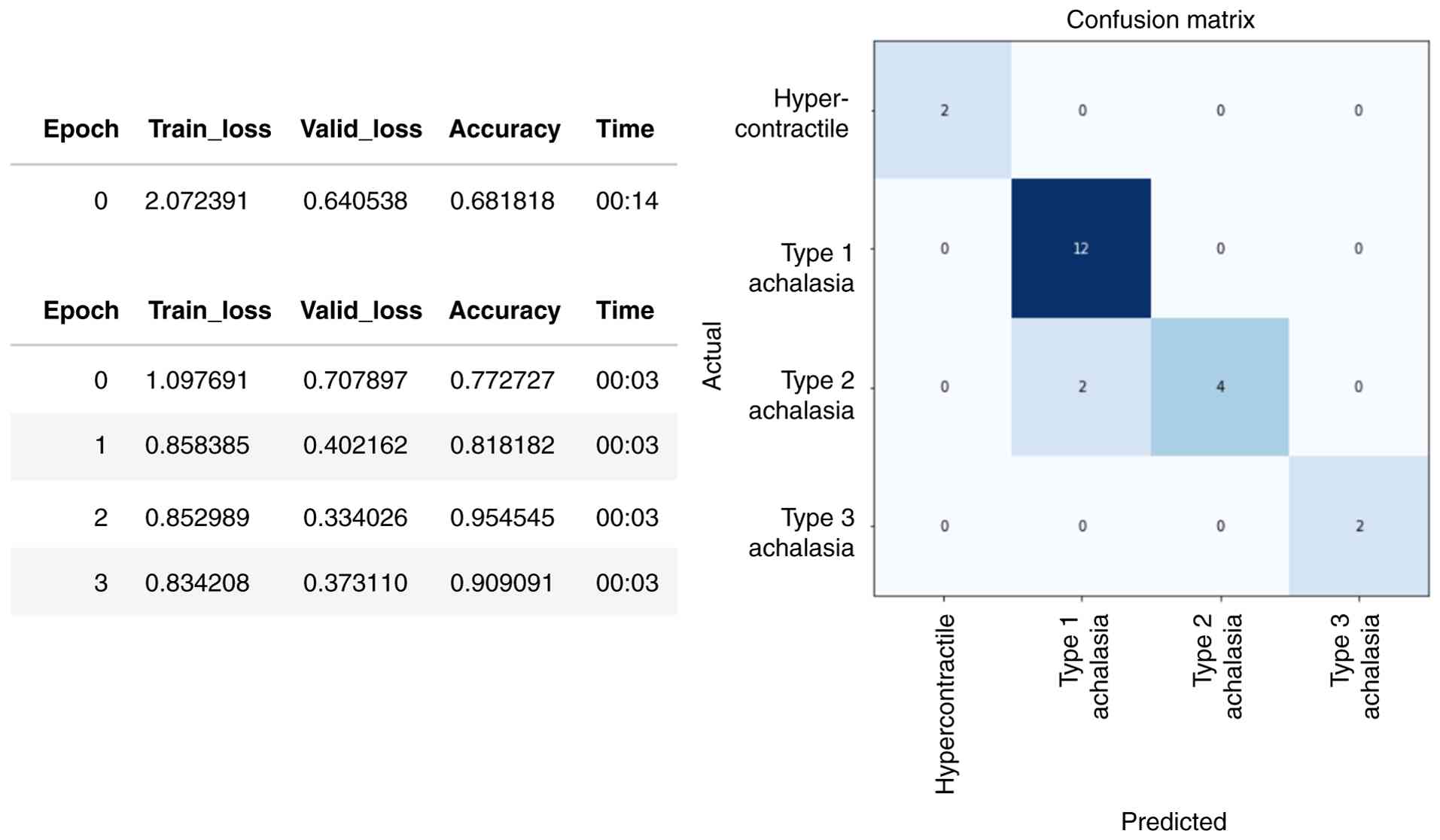
framework was used. With training, the accuracy of the training and
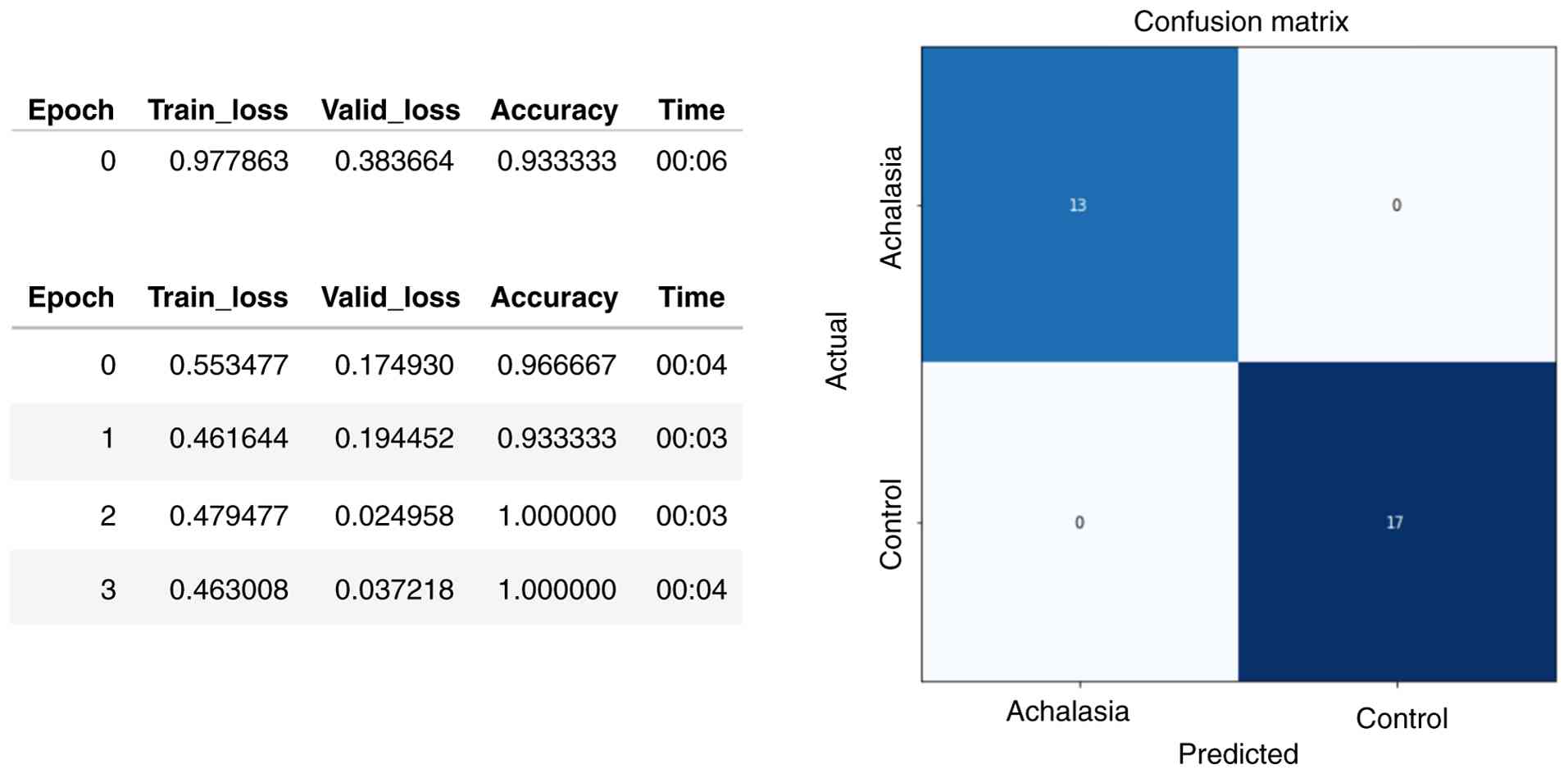
validation sets increased, and the loss value decreased. The
accuracy of the validation set was highest in the fourth epoch of
training (Fig. 2, left panel).
Fig. 2 (right panel) shows that
the confusion matrix of the achalasia-CNN optimal model was 100%
accurate for the external test set, with both sensitivity and
specificity reaching 100%. The receiver operating characteristic
(ROC) curve, area under the curve (AUC), sensitivity, and
specificity are shown in Table SI
and Fig. S1.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
|
Characteristics | Achalasia
(n=12) | Normal (n=9) |
|---|
| Age, years | 41 (16-89) | 72 (50-82) |
| Sex (male) | 7(58) | 6(67) |
| Eckardt score
(normal, ≤3) | 7 (1-10) | 0 (0-0) |
| Prior treatment
(yes) | 0 (0) | 0 (0) |
| IRP, mmHg (normal,
≤25 mmHg) | 23.6
(0.6-55.0) | 10.4
(3.6-18.5) |
| Maximum DCI,
mmHg·sec·cm (normal range, 450-8,000 mmHg·sec·cm) | 796.1
(397.0-493,490.0) | 1,701.1
(238.1-3,713.1) |
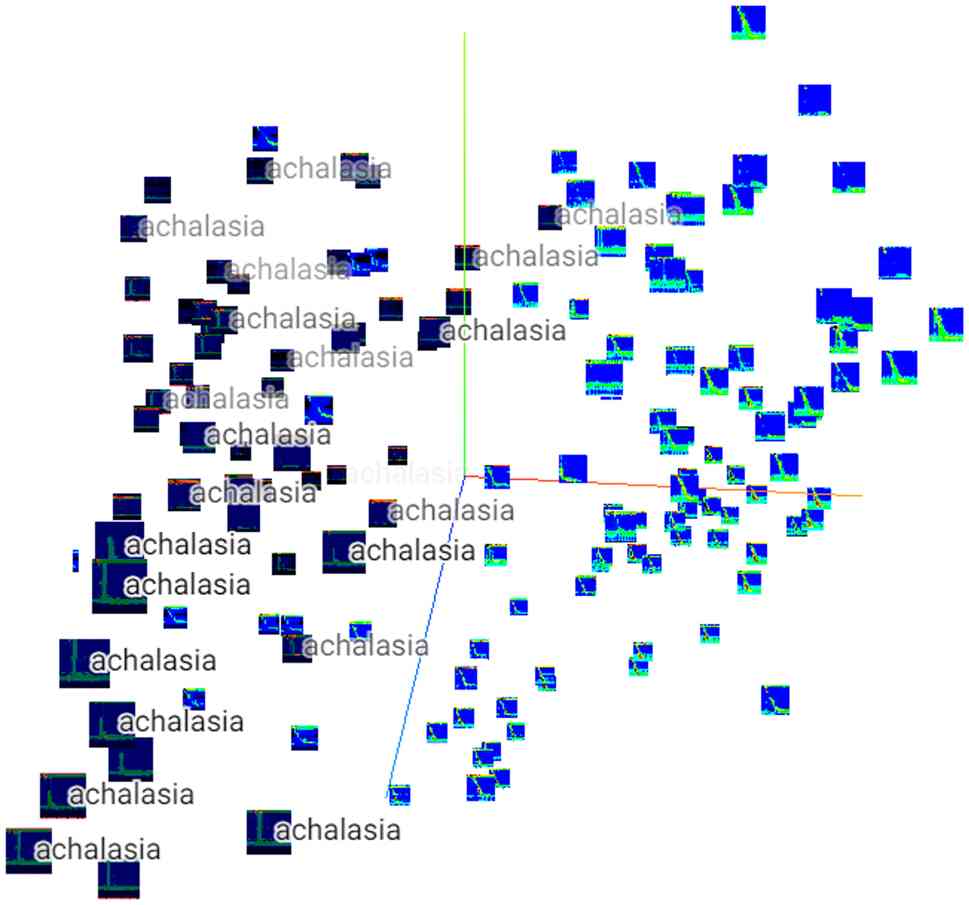
Owing to the nearly 100% correct response rate, and
despite the small dataset, PCA was performed on the characteristics
of the dataset. The HRM of patients with achalasia was distributed
mainly on the left side of Fig. 3,
indicating that the features of each HRM image differed between the
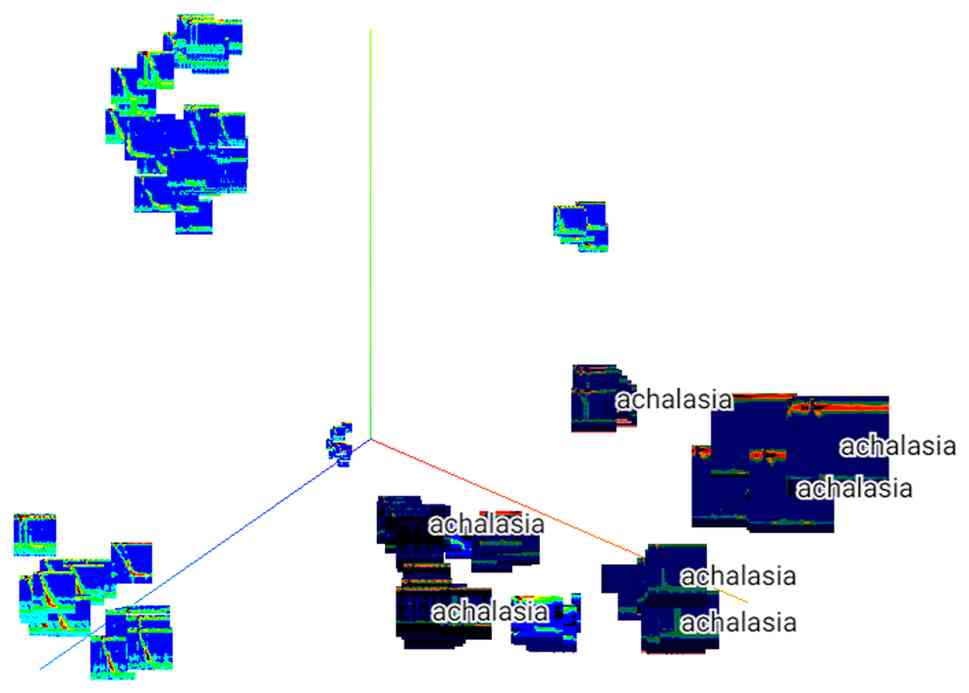
controls and patients with achalasia. Similarly, a 3D-tSNE
analysis, a dimensionality removal method, was performed (Fig. 4), which showed that the achalasia
group could be further subdivided into several groups, suggesting
the possibility of extracting features using an achalasia-type
classification.
The difference in HRM features between the controls
and patients with achalasia, even in the absence of a large number
of HRM images, suggests that deep learning may be predominantly
predictive of the disease. Thus, predictors of each type of
achalasia were developed using the same methods.
Next, we developed an achalasia-type prediction
model using HRM with deep learning. The confusion matrix in
Fig. 5 shows the predictions for
each type. Of the 22 validation datasets, two were incorrectly
classified as type 1 for type 2 achalasia; however, the others were
correctly classified (Fig. 6).
These results demonstrate that deep learning can achieve high
accuracy rates in HRM diagnosis, even with small datasets. The
comprehensive performance metrics are presented in Table SII and Fig. S2.
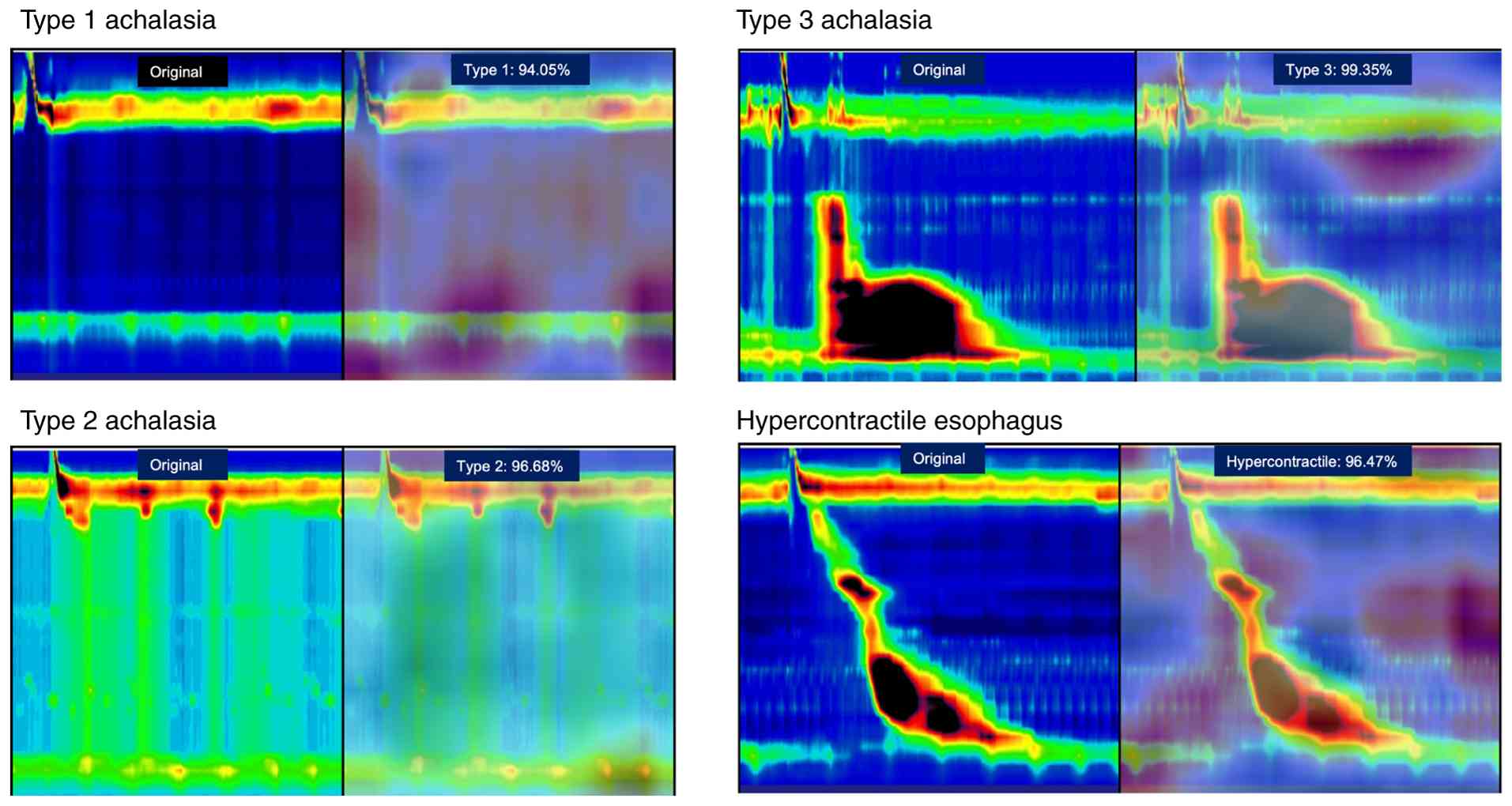
The legends visualized using Grad-CAM are shown in
Fig. 7. The images were reviewed
by two achalasia diagnosticians, primarily based on the diagnosis,
with high-signal images in the heatmap of the LES pressure region,
which is a crucial area for achalasia diagnosis.
When reexamining the areas of focus of Grad-CAM, we
found that pan-esophageal pressurization, which is continuous with
upper esophageal sphincter contraction, is a feature of type 2
achalasia (Fig. S3).
Interestingly, these features are not found in type 1 achalasia,
suggesting that they may be imaging features of type 2
achalasia.
Discussion
In recent years, multimodal therapy combining
surgical resection with chemotherapy, chemoradiotherapy, and
immunotherapy has become widely adopted for malignant esophageal
diseases such as esophageal cancer, leading to significant
improvements in patient prognosis (16-19).
Furthermore, advances in endoscopic diagnosis have enabled earlier
detection of esophageal squamous cell carcinoma (ESCC), and
endoscopic submucosal dissection (ESD) for early lesions has
markedly improved prognosis (20,21).
In contrast, benign esophageal motility disorders,
such as achalasia, require precise functional evaluation and have
often been overlooked due to the complexity of diagnostic tools and
a limited comprehensive understanding of these conditions (22,23).
HRM serves as a highly valuable functional imaging examination
tool, and its diagnostic accuracy and progression are pivotal for
future elucidation and treatment of the underlying causes of
esophageal motility disorders (1,9,24).
Once achalasia is accurately diagnosed using HRM, patients can
experience marked symptom improvement through appropriate
therapeutic interventions, such as peroral endoscopic myotomy
(POEM) or Heller-Dor surgery (1,25,26).
However, the diagnosis of esophageal motility
disorders using HRM can be highly challenging. Even in achalasia,
cases with normal IRP values (9 out of 12 patients, within the
normal range of ≤25 mmHg) were observed in our cohort, and such
patients cannot always be accurately diagnosed, even when the
Chicago Classification is applied. Several studies have reported
the presence of achalasia with normal IRP values. Proposed
mechanisms include impaired esophagogastric junction opening due to
fibrotic changes of the LES (27),
as well as advanced age or pronounced esophageal tortuosity and
dilatation (28). In addition,
when using the Starlet catheter, Type I achalasia may present with
lower IRP values than expected (29). Therefore, diagnosing esophageal
motility disorders based on HRM topography, rather than relying
solely on numerical indices such as IRP, is essential. However,
describing and quantifying the image features that serve as
diagnostic criteria remains challenging. The present results
demonstrate that AI-assisted diagnosis can be a valuable approach
for identifying achalasia.
Rare esophageal motility disorders, such as distal
esophageal spasm (DES) and esophagogastric junction outflow
obstruction (EGJOO), have been reported in previous studies
(30). Achalasia is characterized
by a selective loss of inhibitory myenteric neurons in the distal
esophagus and LES, resulting in impaired esophagogastric junction
(EGJ) relaxation and loss of normal peristalsis (31). High-resolution manometry shows an
elevated IRP with absent peristalsis, with type I-III subtypes
distinguished by the presence of panesophageal pressurization or
premature spastic contractions. In contrast, EGJ outflow
obstruction is defined in the Chicago Classification as an elevated
median IRP with preserved- or only weakly impaired-peristalsis, and
is considered a heterogeneous manometric pattern that may reflect
early or variant achalasia, mechanical obstruction (e.g., hiatal
hernia, stricture, malignancy), or post-surgical changes rather
than a single primary motility disorder (32). Accordingly, the current Chicago
Classification v4.0 requires not only HRM findings but also
supportive evidence from endoscopy, timed barium esophagram, or
functional lumen imaging probe (FLIP) to distinguish clinically
relevant EGJOO from incidental manometric abnormalities (33). Distal esophageal spasm (DES) is
thought to result from an imbalance between excitatory and
inhibitory innervation confined mainly to the distal esophagus; HRM
in DES demonstrates premature contractions (distal latency <4.5
s) in ≥20% of swallows with normal EGJ relaxation, typically in
patients with dysphagia or non-cardiac chest pain (34). Thus, while all three conditions
share abnormalities in inhibitory neural control of the esophagus,
achalasia is defined by impaired EGJ relaxation with absent
peristalsis, EGJOO by impaired EGJ relaxation with preserved
peristalsis, and DES by premature distal contractions with normal
EGJ relaxation. In HRM, premature contractions can also be observed
in type III achalasia, and some achalasia cases may present with
normal IRP values, making it easily confused with DES. Moreover,
EGJOO cannot be accurately diagnosed based solely on HRM findings;
additional evaluations, such as endoscopy, timed barium esophagram,
or FLIP, are required to confirm true obstruction. Therefore, we
considered DES and EGJOO to be inappropriate as the initial targets
for AI model development.
In this study, we developed and validated an
achalasia prediction and classification model using HRM with deep
learning. To the best of our knowledge, this is the first time AI
has demonstrated the potential to support medical practice by
visualizing AI prediction sites using Grad-CAM and reevaluating
them from a physician's perspective. Few reports are available on
the use of deep learning for HRM images. For example, models based
on VAE and DenseNet201(35) have
been shown to achieve high accuracy rates, reaching approximately
93% in classifying esophageal motility disorders (36). In addition, Kou et al
(37) reported that a Long
Short-Term Memory (LSTM) model achieved a correct classification
rate of 88% for the Chicago Classification using HRM images. We
employed a relatively generic convolutional neural network,
ResNet-34(38), with transfer
learning for HRM image classification, achieving a correct
classification rate of 95%, consistent with prior deep-learning
studies. To investigate the underlying cause, we conducted PCA and
3D-tSNE analyses, which indicated that the discriminative challenge
could be advantageous for AI in distinguishing between normal HRM
and achalasia cases, given the fundamental differences in the image
features. Although the internal validation yielded AUCs of 1.000
and very high sensitivity and specificity, these figures should be
regarded as optimistic estimates rather than evidence of perfect
clinical performance. The validation sets were small (22 cases for
the multiclass task and 30 cases for the binary task), and with
such sample sizes, an empirically perfect ROC curve can occur when
the model ranks all cases correctly in a single split. In addition,
we fine-tuned a pre-trained ResNet-34 model on data from a single
source, which increases the risk of overfitting to the local data
distribution. Therefore, the present results should be interpreted
as showing that the model can separate these particular data well,
and not that it will achieve 100% accuracy in broader practice.
Future studies should include repeated cross-validation and, more
importantly, external validation in a larger and more heterogeneous
cohort.
Previous studies have reported that AI prediction of
HRM images is helpful for the Chicago classification of achalasia,
achieving a high accuracy level of 81-93% (37,39).
In this study, our validation dataset yielded a classification
accuracy of 95% for the Chicago Classification groups. Furthermore,
we not only developed a classification model but also applied
Grad-CAM to visualize the regions contributing to the AI's
predictions and reviewed all images from the perspective of
esophageal specialists. The Grad-CAM results also focused on LES
pressure in the HRM image, demonstrating that the LES plays a
crucial role in representing pathological function, as mentioned
previously (40). These findings
are valuable for guiding treatment choices. Grad-CAM images can be
instrumental in determining whether to cut or preserve the LES
during procedures such as peroral endoscopic myotomy, diffuse
esophageal spasm, and hypercontractile esophagus, which are
collectively referred to as esophageal motility disorders (30).
In the future, we plan to introduce our developed
model in medical practice and investigate its potential not only
for the identification of achalasia but also as a tool for medical
education, supporting residents and other physicians through
AI-assisted diagnostics.
A limitation of HRM is that it is primarily used to
evaluate rare diseases, making validation with large datasets and
balanced case designs challenging. Therefore, validation on large
datasets and with a balanced case design is difficult. Although a
discriminator and classification model for achalasia was developed
in this study, further development and validation of the model were
constrained by the lack of sufficient training data for other rare
esophageal diseases. Future directions may include the
implementation of abnormality-detection models based on generative
adversarial networks, which hold promise for addressing these
limitations.
In conclusion, our AI-based HRM assistance system
not only enhances diagnostic accuracy but also offers clinicians
new insights and interpretative perspectives for HRM evaluation.
Further large-scale studies are required to validate its
utility.
Supplementary Material
Binary ROC curve for control vs.
achalasia (positive=achalasia). The solid line is the ROC curve;
the diagonal dotted line indicates the line of no discrimination.
AUC, 1.000 (bootstrap 95% CI, 1.000-1.000). The sensitivity,
specificity and accuracy at a fixed probability threshold of 0.5
are reported in Table SI. AUC,
area under the curve; ROC, receiver operating characteristic.
Multiclass ROC curves (one vs. rest)
for type 1, type 2 and type 3 achalasia, and hypercontractile
esophagus. Solid lines denote class-specific ROC curves; the dashed
line indicates the micro-average ROC. The diagonal dotted line
indicates the line of no discrimination. AUCs: Class-wise, 1.000
for all classes; micro-average, 0.999 (95% CI, 0.996-1.000);
macro-average, 1.000 (95% CI, 1.000-1.000). The macro-average AUC
value is reported; however, a macro-average ROC curve is not
displayed because the macro-average AUC represents an average of
the class-specific AUCs and does not correspond to a single ROC
curve. AUC, area under the curve; ROC, receiver operating
characteristic.
Examples of gradient class activation
map images. In achalasia type 2, a persistent increase in
panesophageal pressure was observed accompanied by lower esophageal
sphincter and UES contraction (upper panel; yellow arrow). By
contrast, patients with achalasia type 1 exhibit no concurrent
increase in esophageal pressure during UES contraction (lower
panel; yellow arrow). This observation may serve as an imaging
characteristic specific to achalasia type 2. The percentage
indicates the predicted probability for each class obtained from
the softmax output. UES, upper esophageal sphincter.
Comprehensive performance metrics for
binary tasks (control vs. achalasia; positive = achalasia).
Comprehensive performance metrics for
multiclass tasks (type 1, type 2 and type 3 achalasia, and
hypercontractile esophagus).
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study are not
publicly available because they contain information that could
compromise the privacy of research participants, but may be
requested from the corresponding author.
Authors' contributions
MT and YN conducted the experiments and wrote the
manuscript. HMin, NY, YA and HMiy made substantial contributions to
the conception and design of the study. MT, HI, JS, TA, KH, MK, KM
and HMin contributed to acquisition and analysis of data. NY, YA
and HMiy critically revised the manuscript for important
intellectual content. HMin, JS and YN confirm the authenticity of
all the raw data. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
The study protocol was reviewed and approved by the
Nagasaki University Hospital Research Ethics Committee (approval
no. 23041703-2; Nagasaki, Japan), and the need for informed consent
was waived. The hospital website ensures that patients have the
right to refuse to share information through an opt-out system.
Patient consent for publication
Patient consent for publication was obtained using
an opt-out approach, whereby the study information was publicly
disclosed, as approved by the Nagasaki University Hospital Research
Ethics Committee.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, artificial
intelligence tools were used to improve the readability and
language of the manuscript or to generate images, and subsequently,
the authors revised and edited the content produced by the
artificial intelligence tools as necessary, taking full
responsibility for the ultimate content of the present
manuscript.
References
|
1
|
Boeckxstaens GE, Zaninotto G and Richter
JE: Achalasia. Lancet. 383:83–93. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Vaezi MF, Pandolfino JE, Yadlapati RH,
Greer KB and Kavitt RT: ACG clinical guidelines: Diagnosis and
management of achalasia. Am J Gastroenterol. 115:1393–1411.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vaezi MF, Pandolfino JE and Vela MF: ACG
clinical guideline: Diagnosis and management of achalasia. Am J
Gastroenterol. 108:1238–1249, 1250. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Naik RD, Vaezi MF, Gershon AA,
Higginbotham T, Chen JJ, Flores E, Holzman M, Patel D and Gershon
MD: Association of achalasia with active varicella zoster virus
infection of the esophagus. Gastroenterology. 161:719–721.e2.
2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gaber CE, Cotton CC, Eluri S, Lund JL,
Farrell TM and Dellon ES: Autoimmune and viral risk factors are
associated with achalasia: A case-control study. Neurogastroenterol
Motil. 34(e14312)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yamasaki T, Tomita T, Mori S, Takimoto M,
Tamura A, Hara K, Kondo T, Kono T, Tozawa K, Ohda Y, et al:
Esophagography in patients with esophageal achalasia diagnosed with
high-resolution esophageal manometry. J Neurogastroenterol Motil.
24:403–409. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Kahrilas PJ, Bredenoord AJ, Fox M, Gyawali
CP, Roman S, Smout AJ and Pandolfino JE: International High
Resolution Manometry Working Group. The Chicago Classification of
esophageal motility disorders, v3.0. Neurogastroenterol Motil.
27:160–174. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
do Carmo GC, de Assis Mota G, da Silva
Castro Perdoná G and de Oliveira RB: Integrated relaxation pressure
and its diagnostic ability may vary according to the conditions
used for HREM recording. Dysphagia. 39:746–756. 2024.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Cha B and Jung KW: Diagnosis of dysphagia:
High resolution manometry & EndoFLIP. Korean J Gastroenterol.
77:64–70. 2021.PubMed/NCBI View Article : Google Scholar : (In Korean).
|
|
10
|
Czako Z, Surdea-Blaga T, Sebestyen G,
Hangan A, Dumitrascu DL, David L, Chiarioni G, Savarino E and Popa
SL: Integrated relaxation pressure classification and probe
positioning failure detection in high-resolution esophageal
manometry using machine learning. Sensors (Basel).
22(253)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gong EJ: Integrated relaxation pressure
during swallowing: An Ever-changing metric. J Neurogastroenterol
Motil. 27:151–152. 2021.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Kou W, Carlson DA, Baumann AJ, Donnan E,
Luo Y, Pandolfino JE and Etemadi M: A deep-learning-based
unsupervised model on esophageal manometry using variational
autoencoder. Artif Intell Med. 112(102006)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Surdea-Blaga T, Sebestyen G, Czako Z,
Hangan A, Dumitrascu DL, Ismaiel A, David L, Zsigmond I, Chiarioni
G, Savarino E, et al: Automated Chicago classification for
esophageal motility disorder diagnosis using machine learning.
Sensors (Basel). 22(5227)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Luus F, Khan N and Akhalwaya I: Active
learning with tensorboard projector. arXiv.org, 2019.
|
|
15
|
Van der Maaten L and Hinton G: Visualizing
data using t-SNE. J Mach Learn Res. 9:2579–2605. 2008.
|
|
16
|
Kanamori K, Koyanagi K, Ozawa S, Yamamoto
M, Ninomiya Y, Yatabe K, Higuchi T and Tajima K: Multimodal therapy
for esophageal squamous cell carcinoma according to TNM staging in
Japan-a narrative review of clinical trials conducted by Japan
clinical oncology group. Ann Esophagus. 6(32)2023.
|
|
17
|
Koyanagi K, Kanamori K, Ninomiya Y, Yatabe
K, Higuchi T, Yamamoto M, Tajima K and Ozawa S: Progress in
multimodal treatment for advanced esophageal squamous cell
carcinoma: Results of multi-institutional trials conducted in
Japan. Cancers (Basel). 13(51)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Higuchi T, Shoji Y, Koyanagi K, Tajima K,
Kanamori K, Ogimi M, Yatabe K, Ninomiya Y, Yamamoto M, Kazuno A, et
al: Multimodal treatment strategies to improve the prognosis of
locally advanced thoracic esophageal squamous cell carcinoma: A
narrative review. Cancers (Basel). 15(10)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bolger JC, Donohoe CL, Lowery M and
Reynolds JV: Advances in the curative management of oesophageal
cancer. Br J Cancer. 126:706–717. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yang H, Wang F, Hallemeier CL, Lerut T and
Fu J: Oesophageal cancer. Lancet. 404:1991–2005. 2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Nishizawa T and Suzuki H: Long-term
outcomes of endoscopic submucosal dissection for superficial
esophageal squamous cell carcinoma. Cancers (Basel).
12(2849)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Averbukh LD and Tadros M: The role of
automatically generated Chicago classification in delayed achalasia
diagnosis. ACG Case Rep J. 7(e00345)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Müller M, Förschler S, Wehrmann T, Marini
F, Gockel I and Eckardt AJ: Atypical presentations and pitfalls of
achalasia. Dis Esophagus. 36(doad029)2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Richter JE: High-resolution manometry in
diagnosis and treatment of achalasia: Help or hype. Curr
Gastroenterol Rep. 16(420)2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Inoue H, Minami H, Kobayashi Y, Sato Y,
Kaga M, Suzuki M, Satodate H, Odaka N, Itoh H and Kudo S: Peroral
endoscopic myotomy (POEM) for esophageal achalasia. Endoscopy.
42:265–271. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Schlottmann F and Patti MG: Esophageal
achalasia: Current diagnosis and treatment. Expert Rev
Gastroenterol Hepatol. 12:711–721. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sato H, Takahashi K, Mizuno KI, Hashimoto
S, Yokoyama J and Terai S: A clinical study of peroral endoscopic
myotomy reveals that impaired lower esophageal sphincter relaxation
in achalasia is not only defined by high-resolution manometry. PLoS
One. 13(e0195423)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kim E, Yoo IK, Yon DK, Cho JY and Hong SP:
Characteristics of a subset of achalasia with normal integrated
relaxation pressure. J Neurogastroenterol Motil. 26:274–280.
2020.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Kawami N, Hoshino S, Hoshikawa Y,
Takenouchi N, Hanada Y, Tanabe T, Goto O, Kaise M and Iwakiri K:
Validity of the cutoff value for integrated relaxation pressure
used in the Starlet high-resolution manometry system. J Nippon Med
Sch. 86:322–326. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Morley TJ, Mikulski MF, Rade M, Chalhoub
J, Desilets DJ and Romanelli JR: Per-oral endoscopic myotomy for
the treatment of non-achalasia esophageal dysmotility disorders:
Experience from a single high-volume center. Surg Endosc.
37:1013–1020. 2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Rogers AB, Rogers BD and Gyawali CP:
Pathophysiology of achalasia. Ann Esophagus. 3(27)2020.
|
|
32
|
Rohof WOA and Bredenoord AJ: Chicago
classification of esophageal motility disorders: Lessons learned.
Curr Gastroenterol Rep. 19(37)2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yadlapati R, Kahrilas PJ, Fox MR,
Bredenoord AJ, Prakash Gyawali C, Roman S, Babaei A, Mittal RK,
Rommel N, Savarino E, et al: Esophageal motility disorders on
high-resolution manometry: Chicago classification version
4.0©. Neurogastroenterol Motil.
33(e14058)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fox MR, Sweis R, Yadlapati R, Pandolfino
J, Hani A, Defilippi C, Jan T and Rommel N: Chicago classification
version 4.0© technical review: Update on standard
high-resolution manometry protocol for the assessment of esophageal
motility. Neurogastroenterol Motil. 33(e14120)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Huang G, Liu Z, Pleiss G, van der Maaten L
and Weinberger KQ: Convolutional networks with dense connectivity.
IEEE Trans Pattern Anal Mach Intell. 44:8704–8716. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Popa SL, Surdea-Blaga T, Dumitrascu DL,
Chiarioni G, Savarino E, David L, Ismaiel A, Leucuta DC, Zsigmond
I, Sebestyen G, et al: Automatic diagnosis of high-resolution
esophageal manometry using artificial intelligence. J
Gastrointestin Liver Dis. 31:383–389. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kou W, Galal GO, Klug MW, Mukhin V,
Carlson DA, Etemadi M, Kahrilas PJ and Pandolfino JE: Deep
learning-based artificial intelligence model for identifying
swallow types in esophageal high-resolution manometry.
Neurogastroenterol Motil. 34(e14290)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
He K, Zhang X, Ren S and Sun J: Deep
residual learning for image recognition. arXiv. [csCV]:770–778.
2015.
|
|
39
|
Kou W, Carlson DA, Baumann AJ, Donnan EN,
Schauer JM, Etemadi M and Pandolfino JE: A multi-stage machine
learning model for diagnosis of esophageal manometry. Artif Intell
Med. 124(102233)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cohen S, Lipshutz W and Hughes W: Role of
gastrin supersensitivity in the pathogenesis of lower esophageal
sphincter hypertension in achalasia. J Clin Invest. 50:1241–1247.
1971.PubMed/NCBI View Article : Google Scholar
|