Introduction
Breast cancer is among the most prevalent
malignancies in women worldwide (1). Triple-negative breast cancer (TNBC)
is an aggressive breast cancer subtype characterized by the absence
of estrogen receptors (ERs), progesterone receptors and human
epidermal growth factor receptor-2 (HER-2). Therefore, it does not
respond to typical endocrine and anti-HER-2 therapies.
Consequently, TNBC is associated with higher recurrence and
metastasis rates compared with other breast cancer subtypes.
Current treatment strategies primarily rely on chemotherapy,
however the outcomes remain suboptimal (2), necessitating the identification of
novel therapeutic targets.
ERβ [also known as estrogen receptor 2 (ESR2)] is
highly expressed in normal breast tissues, however expression is
markedly decreased during breast carcinogenesis (3-6).
ERβ is detected in all breast cancer subtypes, especially in TNBC,
with ~30% of TNBC tumors expressing ERβ and elevated ERβ levels
being associated with its favorable prognosis (7,8).
Preclinical studies have demonstrated the tumor-suppressive role of
ERβ (9-11),
highlighting it as a promising therapeutic target for TNBC.
However, the mechanisms driving this downregulation remain
unclear.
Tumor-associated macrophages (TAMs) are key
components of the breast cancer microenvironment. High TAM
infiltration is associated with poor prognosis (12-14).
TAMs regulate ERα expression in cancer cells (15,16).
However, the specific association between TAMs and ERβ expression
and the mechanisms by which TAMs regulate ERβ expression remains
unknown. Therefore, in the present study, the association between
TAMs and ERβ expression in TNBC cells was investigated and the
molecular mechanisms underlying TAM-mediated ERβ suppression was
explored.
Materials and methods
Mice and cell lines
Female BALB/c mice (6-8 weeks old; 20-25 g) were
housed in a pathogen-free facility at the Experimental Animal
Center of Tongji Medical College, Huazhong University of Science
and Technology (Wuhan, China). The animals were maintained at a
constant temperature of 22±2˚C and a relative humidity of 50±10%
under a 12-h light/dark cycle, with free access to food and water.
All procedures were approved by the Institutional Animal Care and
Use Committee of Huazhong University of Science and Technology.
Following tumor cell implantation, mice (n=12) were monitored every
2 days for body weight, behavior, tumor size and vital signs, with
food, water and bedding refreshed as needed. Humane endpoints
included >20% body weight loss, dyspnea, tumor
ulceration/necrosis or inability to access food and water. Based on
prior reports and preliminary studies, mice were sacrificed at week
5 to allow assessment of pulmonary metastases while minimizing
mortality. Sacrifice was performed with intraperitoneal sodium
pentobarbital (50 mg/kg) followed by cervical dislocation and
mortality was verified by absence of heartbeat, respiration and
corneal reflexes. The murine 4T1 cell line was purchased from the
Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and
incubated in humidified conditions at 37˚C. Bone marrow-derived
macrophages with M2 polarization and conditioned medium (CM) from
M2 macrophages were obtained as previously described (17).
Data acquisition
Expression profiles and related clinical data of
patients with TNBC were obtained from The Cancer Genome Atlas
(TCGA) datasets (https://portal.gdc.cancer.gov/). Single-cell RNA
sequencing datasets were downloaded from the Gene Expression
Omnibus database [GEO; https://www.ncbi.nlm.nih.gov/geo/; GSE202624(18)]. UCALAN (http://ualcan.path.uab.edu/) was used to compare the
ESR2 mRNA levels among breast cancer gene (BRCA) subtypes and Human
Protein Atlas (HPA; https://www.proteinatlas.org/) datasets were used to
compare the ERβ protein levels between breast cancer and normal
tissues. AnimalTFDB (version 4.0; https://guolab.wchscu.cn/AnimalTFDB4) was used to
predict transcription factor (TF) binding sites.
Bioinformatics analysis
Bioinformatics analyses of the data of patients with
TNBC obtained from TCGA and single-cell RNA sequencing datasets
(GEO: GSE202624) were performed using R (version 4.0.2; http://www.R-project.org). Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analysis and gene set enrichment
analysis (GSEA) of differentially expressed genes were performed
using the ‘ClusterProfiler’ R package (version 3.8; https://bioconductor.org/packages/clusterProfiler/).
Survival statistics were plotted using the K-M Plotter database
(https://kmplot.com/). Immune infiltration and
mRNA expression data from Gene Set Cancer Analysis (GSCA;
https://guolab.wchscu.cn/GSCA) were used
to investigate the association between mRNA expression and immune
cell infiltration through Spearman's rank correlation analysis.
Additionally, the Tumor Immune Estimation Resource 2.0 database
(TIMER2.0; http://timer.cistrome.org/) was used
to explore the association between macrophage scavenger receptor 1
(MSR1) and a number of epithelial-mesenchymal transition (EMT)
markers in TNBC.
Reverse transcription quantitative PCR
(RT-qPCR)
RNAiso Plus (Takara Bio, Inc.) was used to extract
the total RNA of 4T1 cells, which was reverse transcribed into cDNA
using the ReverTra Ace™ qPCR RT Kit (Toyobo Co., Ltd.)
according to the manufacturer's instructions. qPCR analyses with
SYBR® Green Real Time PCR Master Mix (Toyobo Co., Ltd.)
and specific primers were performed to measure the target gene mRNA
transcript levels relative to the β-actin levels. The thermocycling
conditions were as follows: Initial denaturation at 95˚C for 1 min;
40 cycles of 95˚C for 5 sec (denaturation), 60˚C for 15 sec
(annealing) and 72˚C for 45 sec (extension), with fluorescence data
collection. A melting-curve analysis was performed at the end of
each run to confirm the specificity of amplification. All reactions
were carried out according to the manufacturers' protocols. Data
were normalized to the β-actin levels and analyzed through the
2-ΔΔCq method (19).
All primer sequences are listed in the Table SI.
Mouse model of breast cancer and
macrophage depletion
4T1 cell lines (1x106) were injected into
the mammary fat pads of BALB/c mice. Clodronate liposomes obtained
from Vrije Universiteit (Amsterdam) were used to deplete the
macrophages, as previously described (17). LY294002 (MedChemExpress), a
selective PI3K inhibitor, was dissolved at 5 mg/ml in 10% DMSO and
intraperitoneally injected at 50 mg/kg (200 µl/injection), twice
weekly for five weeks (20,21).
ERB041 (Tocris Bioscience), an ERβ-selective agonist, was dissolved
at 2 mg/ml in 10% DMSO and administered subcutaneously at 5 mg/kg
(50 µl/injection) once daily for five weeks, as previously
described (22). For vehicle
controls, mice received injections of 10% DMSO in 90% corn oil.
Tumor volume was calculated as follows: Volume=(length x width x
width)/2.
Immunofluorescence assay and
immunohistochemistry
Mouse breast tumor paraffin-embedded sections (4 µm)
were stained with specific primary antibodies overnight at 4˚C. The
bound antibodies were detected using CoraLite488-goat anti-rabbit
IgG (1:400; cat. no. SA00013-2; Proteintech Group, Inc.) and
Coralite594-goat anti-rabbit IgG (1:400; cat. no. SA00013-4;
Proteintech Group, Inc.) at room temperature for 30 min. DAPI
(Wuhan Servicebio Technology Co., Ltd.) was used to stain the
nuclei at room temperature for 10 min in the dark. Finally, the
cells were observed under a laser confocal microscope (Olympus
Corporation). All antibodies used in the present study are listed
in the Table SII.
TNBC tissues from two female patients (age, 45 and
63 years) were collected at The Central Hospital of Wuhan, Tongji
Medical College, Huazhong University of Science and Technology
between January 2022 and December 2023. The present study was
approved by the Ethics Committee of The Central Hospital of Wuhan
(China; approval no. WHZXKYL2022-033). Immunohistochemistry
staining was performed by Wuhan Servicebio Technology Co., Ltd., on
formalin-fixed, paraffin-embedded tissue sections. The procedures
followed standard immunohistochemistry protocols, including
deparaffinization, antigen retrieval, blocking, incubation with
primary and secondary antibodies, and chromogenic detection. Slides
were counterstained, dehydrated and mounted according to routine
laboratory practices. All steps were carried out following the
company's standard operating procedures.
Flow cytometry
4T1 cells were cultured with M2 macrophages at 37˚C
in a humidified atmosphere containing 5% CO2 in the same
dish for 24 h, washed with cold PBS and stained with
anti-F4/80-BV421 (BD Pharmingen; BD Biosciences) and BV421 rat IgG
isotype antibody (BioLegend, Inc.) for 30 min at 4˚C. Subsequently,
the cells were fixed, made permeable (Fixation/Permeabilization
Kit; cat. no. 554714; BD Pharmingen; BD Biosciences) according to
the manufacturer's instructions, and intracellularly stained with
anti-ERβ-phycoerythrin antibodies (Novus Biologicals) for 60 min at
4˚C. Flow cytometry (BD FACSAria™ II) was used to
characterize the cells and FlowJo™ v10 software (BD
Biosciences) was used for data analysis. Cells were first gated
based on forward scatter (FSC) and side scatter (SSC) to select the
live single-cell population, excluding debris and aggregates.
Macrophages were gated for F4/80+ and mean fluorescence
intensity of ERβ in 4T1 cells (F4/80-) was analyzed. All
antibodies used in the present study are listed in Table SII.
Transwell assay and wound healing
assay
After digestion with trypsin for 3 min at 37˚C, 4T1
cells were resuspended in a serum-free medium. Then, 200 µl cell
suspension with 2x104 cells was added to the upper
chamber and 500 µl culture medium containing 10% fetal bovine serum
(Wuhan Servicebio Technology Co., Ltd.) was added to the lower
chamber of the Transwell system (Corning, Inc.) at 37˚C. After 24
h, the migrated cells were fixed with 4% paraformaldehyde and
stained with crystal violet for 15 min at room temperature, images
were acquired using an inverted light microscope (IX81; Olympus
Corporation) and cells were counted using ImageJ software (version
1.8.0; National Institute of Health). Briefly, images were
converted to 8-bit grayscale, the background was subtracted using
the ‘Subtract Background’ function and cells were distinguished
from the background by applying a threshold. The ‘Analyze
Particles’ function was used to count cells, with size and
circularity parameters adjusted to exclude debris. For each well,
three random fields were analyzed, and the mean cell number was
calculated. A total of 1x105 cells were seeded in a
six-well plate and cultured until they reached 90% confluence. A
scratch was then created using a 20 µl pipette tip, then washed
three times with PBS. Images were captured at 0 and 24 h to
evaluate cell migration.
Chromatin immunoprecipitation (ChIP)
assay
A total of 1x107 4T1 cells were used for
each experimental condition. Briefly, 4T1 cells were fixed with 1%
formaldehyde for 10 min at room temperature, and cross-linked
protein-DNA complexes were extracted using the Magna
ChIP® A/G Chromatin Immunoprecipitation Kit (cat. no.
17-10085; MilliporeSigma) according to the manufacturer's
instructions. Immunoprecipitation was performed by overnight
incubation at 4˚C with a primary antibody against FOXO3a or rabbit
IgG under gentle rotation (Table
SII), followed by incubation with magnetic beads at 4˚C for 2
h. DNA was purified from the complexes and analyzed through qPCR
using primers targeting the FOXO3a binding site within the mouse
ESR2 promoter (forward: 5'-GTGGGATAAGGGATTGTGAA-3'; reverse:
5'-AACGCAGGAGCAGAAGATAG-3'). ChIP signal enrichment was quantified
by RT-qPCR and expressed as the signal-to-input ratio (fold
enrichment=100x2ΔCq). Amplified products were resolved
by 1.5% agarose gel electrophoresis in 1X TAE buffer, stained with
ethidium bromide and detected under a UV transilluminator.
Statistical analysis
Statistical analyses were conducted using the
GraphPad Prism (version 8.0; GraphPad; Dotmatics) software.
Differences between two groups were evaluated using an unpaired
Student's t-test, while comparisons among three or more groups were
analyzed using one-way ANOVA followed by Tukey's post-hoc test.
Results are represented as the mean ± standard deviation. P<0.05
was considered to indicate a statistically significant difference.
*P<0.05, **P<0.01 and
***P<0.001.
Results
Clinical value and expression of ERβ
in TNBC
Survival analysis using GSCA data revealed that high
ESR2 levels in patients with breast cancer were associated with
improved progression-free survival (Fig. 1A) and overall survival (Fig. 1B). ESR2 levels were also found to
differ among breast cancer subtypes (Fig. 1C), with TNBC showing the highest
ESR2 levels (23). Further
validation using the Cancer Cell Line Encyclopedia (24) demonstrated that TNBC cell lines
expressed high ESR2 levels (P<0.05; Fig. 1D). Transwell assay results revealed
that the pharmacological activation of ERβ effectively suppressed
4T1 cell migration (P=0.003; Fig.
1E), suggesting ERβ as a potential therapeutic target for TNBC.
Furthermore, ESR2 expression levels were compared between normal
and tumor tissues using TCGA data. Findings demonstrated that ESR2
levels were significantly downregulated in TNBC (P<0.001;
Fig. 1F). Analysis of the HPA data
(25) revealed that protein ERβ
levels were lower in BRCA tissues compared with normal tissues
(Fig. 1G). Results consistently
showed lower ERβ levels in cancerous tissues compared with adjacent
non-malignant tissues (Fig. 1H).
Collectively, TNBC showed higher ERβ expression compared with other
breast cancer subtypes, indicating its therapeutic potential.
However, the levels remained markedly lower compared with those in
normal breast tissues. The underlying mechanisms of ERβ
downregulation remain to be elucidated and warrant further
investigation.
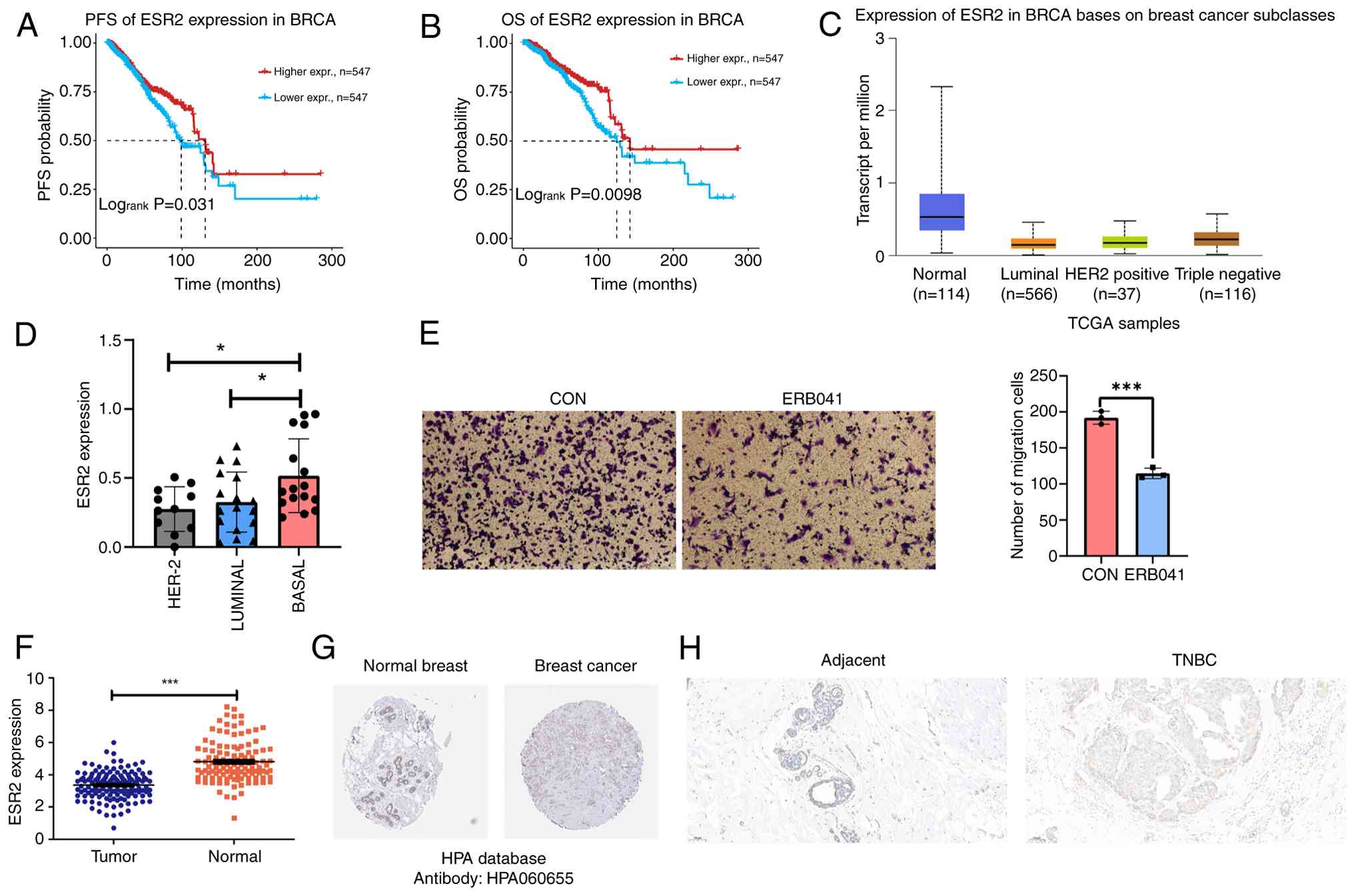 | Figure 1Clinical value and expression of ERβ
(also known as ESR2) in TNBC. (A) PFS analysis of ESR2 levels in
BRCA. (B) OS analysis of ESR2 levels in BRCA. (C) ESR2 mRNA levels
in different BRCA subtypes. (D) ESR2 mRNA levels in different
breast cancer subtype cell lines. (E) Migratory capacity of 4T1
cells treated with vehicle (ethanol) or ERβ agonist, ERB041 (100
nM), was assessed through Transwell assays. (F) Scatter plot
showing the differentially expressed ESR2 between TNBC tumor and
normal tissues. (G) Immunohistochemical staining from the Human
Protein Atlas database revealed ERβ protein expression in both
normal breast and BRCA tissues. (H) Immunohistochemistry was used
to evaluate the ERβ expression levels in paracancerous and breast
cancer tissues. Scale bar, 100 µm. Magnification, x100.
*P<0.05 and ***P<0.001. Erβ, estrogen
receptor-β; TNBC, triple negative breast cancer; PFS,
progression-free survival; OS, overall survival; BRCA; breast
cancer; HER-2, human epidermal growth factor receptor-2; CON,
control; ns, not significant; TCGA, The Cancer Genome Atlas. |
TAMs enhance tumor metastasis in
TNBC
TAM infiltration is a key feature of the breast
cancer microenvironment and is associated with a poor prognosis. To
assess the roles of TAMs in breast cancer, the expression levels of
TAM marker MSR1 were examined using TCGA data. MSR1 levels were
notably upregulated in the breast cancer tissues, including TNBC,
compared with those in the adjacent normal tissues (Fig. 2A). Survival analysis revealed that
high MSR1 expression was associated with poor overall survival in
TNBC (logrank P=0.015; Fig.
2B). Additionally, analysis of TIMER2.0 data (26) revealed positive correlations
between EMT-related genes [such as matrix metalloproteinase (MMP)-9
and Twist2] and MSR1 levels in TNBC (Fig. 2C). GSEA was performed on
single-cell RNA sequencing data from TAM-enriched and TAM-depleted
mouse breast tumors (GEO: GSE202624), obtained from Seok et
al (18). Findings revealed
that EMT-associated genes in breast cancer were downregulated in
the TAM-depleted group (Fig. 2D
and E). These findings suggested
that TAMs notably enhance TNBC metastasis.
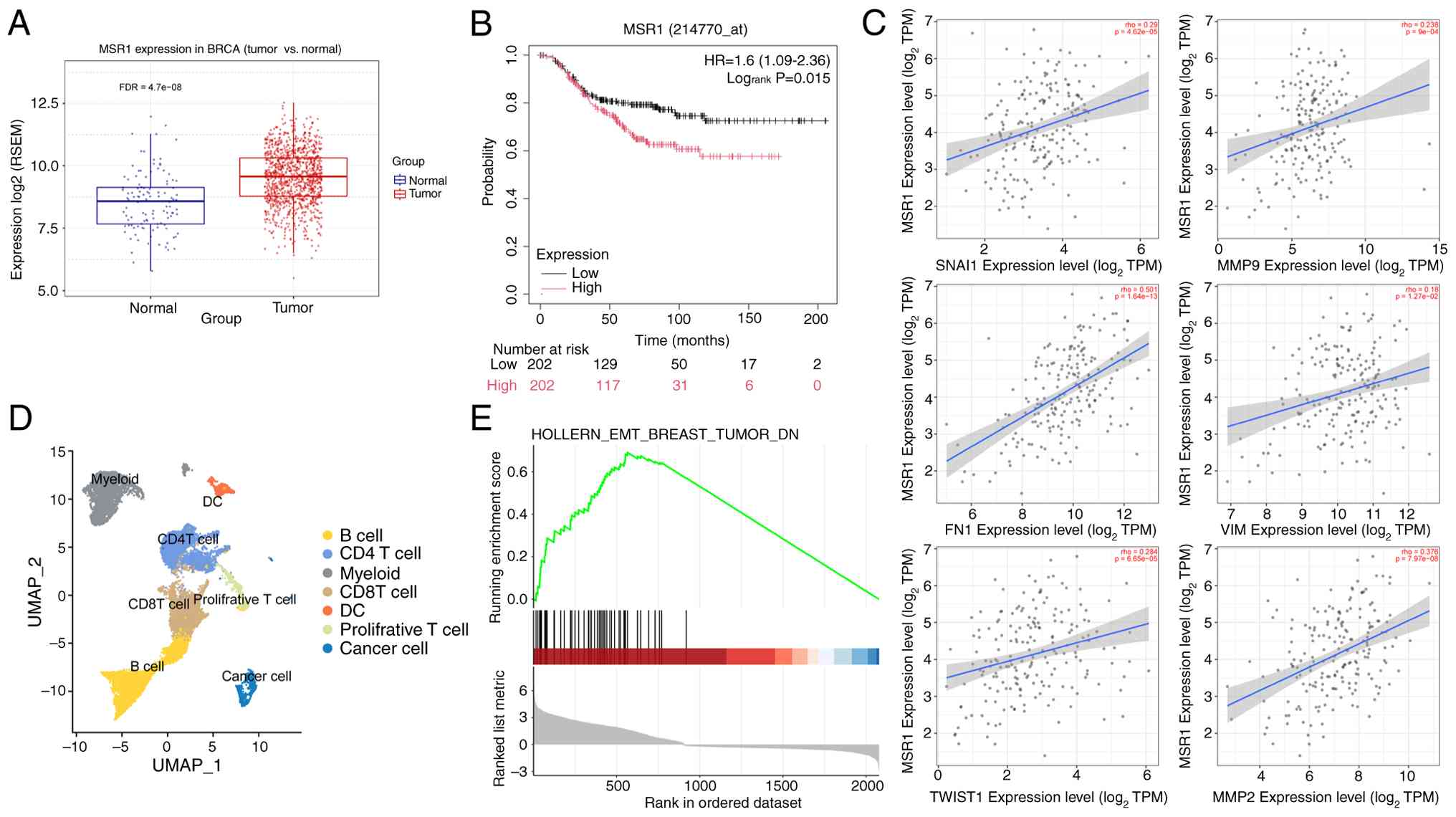 | Figure 2Tumor-associated macrophages enhance
tumor metastasis in TNBC. (A) Boxplots demonstrating the
differential expression of MSR1 between BRCA tumor tissue and
normal tissue. (B) Overall survival analysis of MSR1 levels in
TNBC. (C) Correlation analysis of MSR1 levels with Vim, Mmp-9,
Snai1, Twist1, Fn1 and Mmp2 levels in TNBC. (D) UMAP analysis of
total cells in mouse breast tumors. (E) Gene set enrichment
analysis of the epithelial-mesenchymal transition marker levels in
breast tumor. MSR1, macrophage scavenger receptor ; RSEM,
RNA-sequencing by expectation maximization; BRCA, breast cancer;
TPM, transcripts per million; Mmp-9, matrix metalloproteinase-9;
Vim, vimentin; Fn1, fibronectin 1; MMP2, matrix
metalloproteinase-2; DC, dendritic cells; FDR, false discovery
rate; TNBC, triple negative breast cancer; UMAP, uniform manifold
approximation and projection. |
TAMs suppress ERβ expression in
TNBC
Based on the aforementioned results, the correlation
between ESR2 levels and TAMs in TNBC was explored. Correlation
analysis using TCGA data revealed that TAM infiltration was
negatively correlated with ERβ expression in BRCA (Fig. 3A and B). In breast cancer, TAMs mainly exhibit
the M2 phenotype (17). To mimic
the TNBC tumor microenvironment, M2-polarized macrophages and 4T1
cells were added to the upper and lower chambers of the Transwell
system, respectively. ESR2 mRNA levels in 4T1 cells were markedly
reduced in the M2 co-culture group (P<0.05; Fig. 3C). To further assess whether TAMs
suppress ERβ expression, flow cytometry was used to analyze ERβ
level after treatment with varying proportions of M2 macrophages.
Notably, co-culture with M2 macrophages markedly reduced ERβ
expression, as evidenced by the decreasing mean fluorescence
intensity of ERβ in 4T1 cells with increasing macrophage
proportions (P<0.001; Fig. 3D).
Cellular immunofluorescence staining revealed that TAM-CM notably
suppressed ERβ expression (Fig.
3E). Subsequently, clodronate liposomes were used to
selectively deplete the macrophages in 4T1 mice model to validate
findings in vivo. Immunofluorescence staining was used to
analyze the tumor tissue sections. Compared with the control group,
clodronate liposome-treated group showed markedly reduced
CD204+ TAM levels, associated with increased
ERβ+ tumor cell levels (Fig. 3F). These results collectively
suggested TAMs inhibit ERβ expression in TNBC.
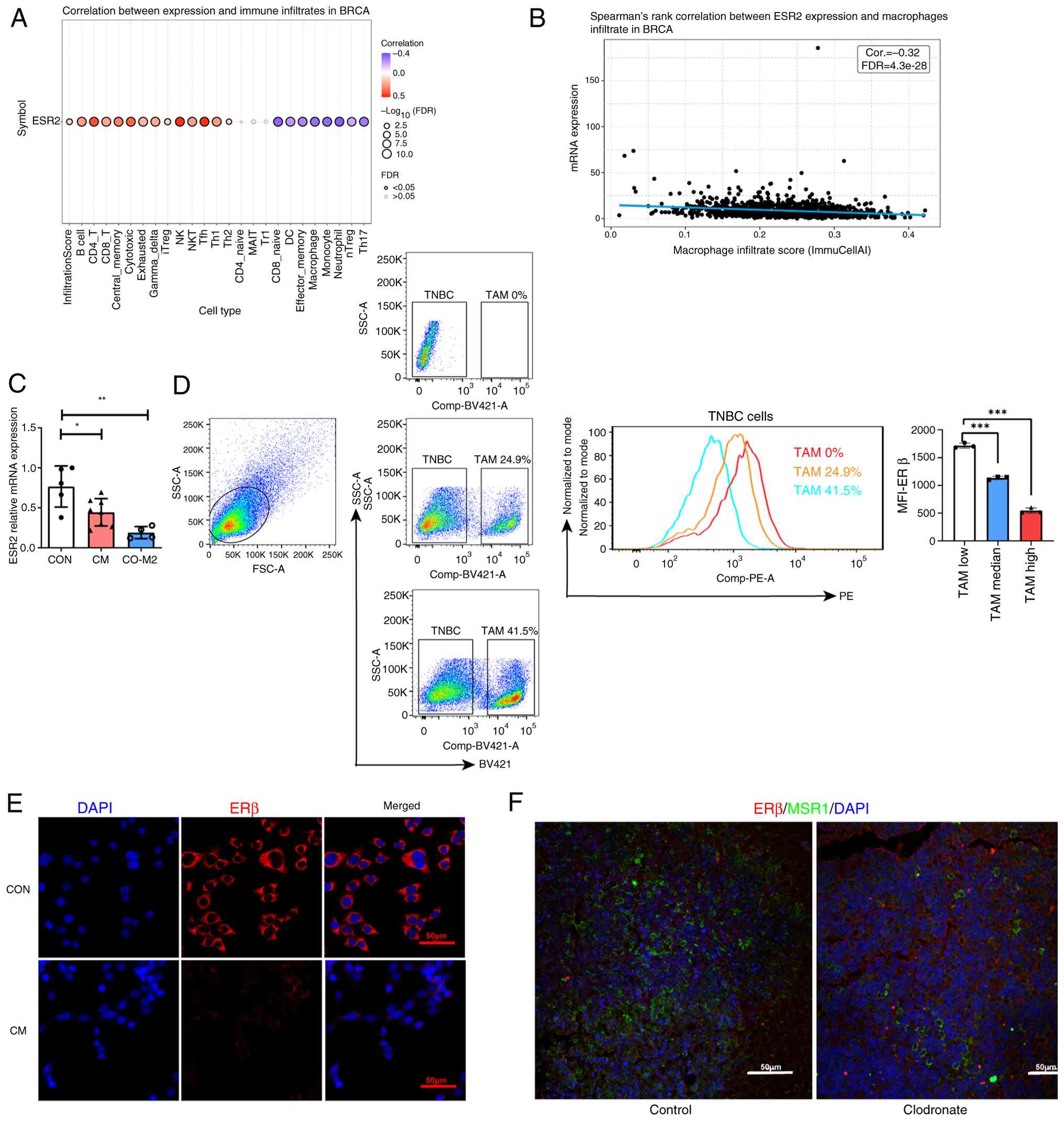 | Figure 3TAMs suppress ERβ expression in TNBC.
(A) Bubble plot demonstrating the correlation between ESR2 mRNA
levels and 24 immune cell type infiltrates in BRCA. Bubble size
correlates with FDR. Black outline border indicates FDR ≤0.05. (B)
Correlation between macrophages infiltration and ESR2 expression.
(C) Relative mRNA levels of ESR2 in different groups. (D)
Representative flow cytometric analysis of ERβ expression levels in
4T1 cells co-cultured with different proportion of TAMs (low,
median and high) for 24 h. Mean fluorescence intensity of ERβ in
the 4T1 cells of each group shown on the right (n=3). (E)
Representative cellular immunofluorescence images of ERβ (red) and
nuclei stained with DAPI (blue). (F) Immunofluorescence staining
for MSR1 (green) and ERβ (red) in the tumor tissues of the control
(left) and clodronate liposome-treated (right) groups. Original
magnification, x200. Scale bars, 50 µm. *P<0.05,
**P<0.01 and ***P<0.001. Comp-BV421-A
represents F4/80 staining, and Comp-PE-A represents ERβ staining.
ns, no significant; ERβ, estrogen receptor-β; TNBC, triple-negative
breast cancer; BRCA, breast cancer; FDR, false discovery rate; TAM,
tumor-associated macrophage; Cor., correlation; CON, control; CM,
conditioned medium; SSC-A, side scatter-area; FSC-A, forward
scatter-area; MFI, mean fluorescence intensity. |
TAM-mediated ERβ suppression
mechanisms
To elucidate TAM-mediated ERβ suppression
mechanisms, TNBC samples from TCGA database were stratified into
high and low TAM expression groups based on MSR1 levels. KEGG
pathway analysis revealed significant enrichment of the ‘PI3K-AKT
signaling pathway’ (Fig. 4A).
Further validation using the GSCA database corroborated the
differential activity of the PI3K/AKT pathway between the high and
low MSR1 expression groups (Fig.
4B). TAMs are reported to activate tumor-intrinsic PI3K/AKT
signaling across cancer types, promoting tumor invasion and therapy
resistance in digestive tract tumors and ER+ breast
cancer (27-29).
The role of TAMs in modulating the PI3K/AKT pathway was further
investigated using a 4T1 mouse model. Immunohistochemical analysis
of tumor tissues revealed that clodronate liposome-mediated
depletion of TAMs significantly reduced phosphorylated (p)-AKT
expression (Fig. 4C). To assess
the specific role of PI3K/AKT signaling in TAM-mediated ERβ
suppression, 4T1 cells were treated with the PI3K inhibitor,
LY294002, in a TAM-CM. RT-qPCR analysis revealed that PI3K/AKT
pathway inhibition reversed the TAM-induced suppression of ESR2 in
a dose-dependent manner (P<0.01; Fig. 4D). These findings suggested that
TAM-mediated PI3K/AKT activation inhibited ESR2 transcription.
Immunofluorescence assay results further corroborated these
findings, showing that p-AKT and ERβ co-localize in 4T1 cells,
appearing yellow in merged images, with ERβ levels increased in
CM-treated 4T1 cells following PI3K/AKT inhibition, supporting an
association between PI3K/AKT activation and ERβ expression
(Fig. 4E).
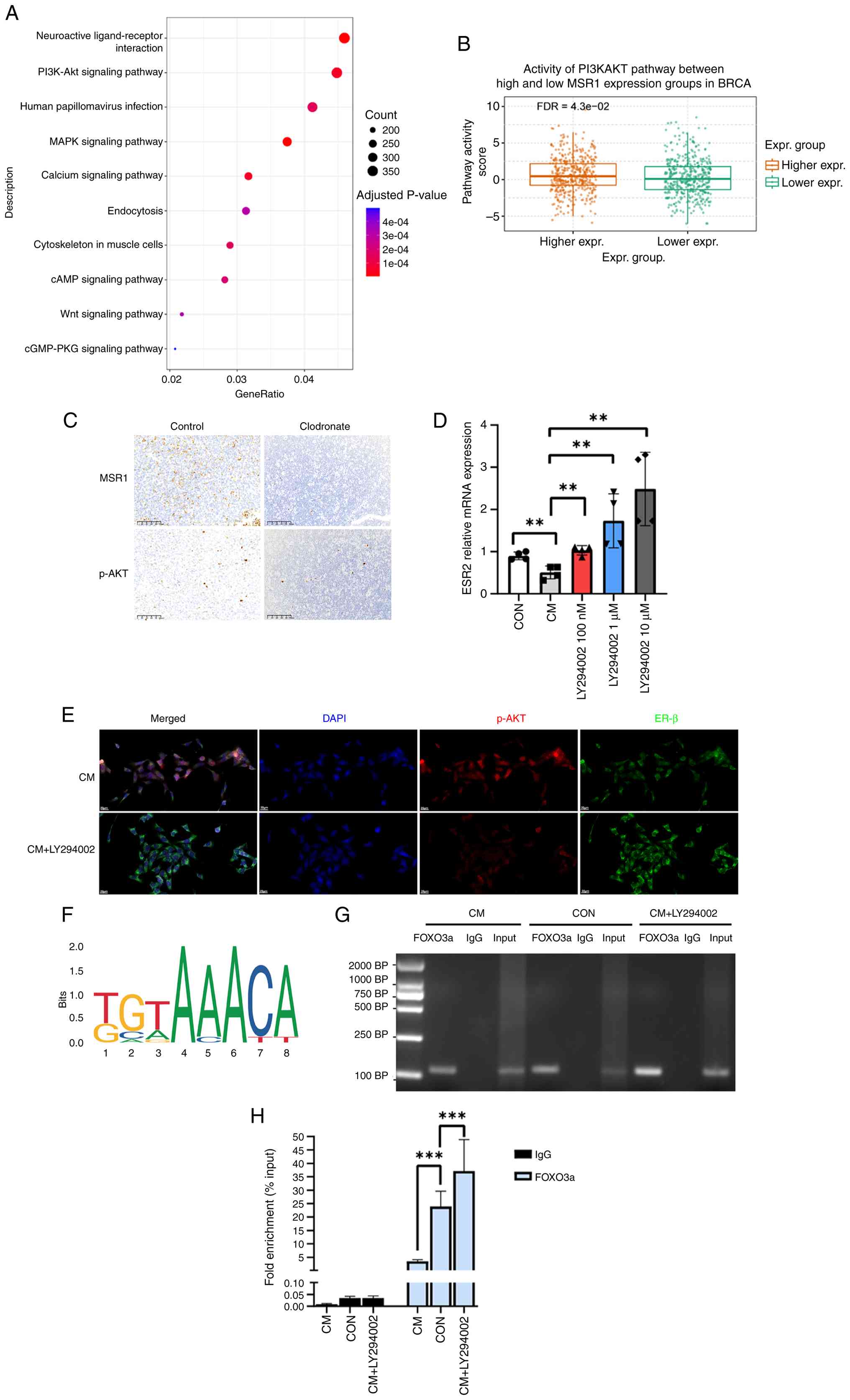 | Figure 4Tumor-associated macrophages suppress
ERβ expression through the PI3K/AKT pathway. (A) Top 10 Kyoto
Encyclopedia of Genes and Genomes enrichment pathways. (B) PI3K/AKT
pathway activity in high and low breast cancer MSR1 expression
groups. (C) Immunohistochemistry for MSR1 and p-AKT levels in the
control and clodronate-treated groups. Original magnification,
x200, scale bars, 100 µm. (D) 4T1 cells cultured in CM then treated
DMSO or 100 nM, 1 µM or 10 µM of LY294002 for 24 h. Relative mRNA
levels of ESR2 in different groups (n=4). (E) Co-localization of
p-AKT and ER β in 4T1 cells. Representative cellular
immunofluorescence images of 4T1 cells treated with CM or CM +
LY294002 (1 µM), ERβ (green), p-AKT (red) and DAPI (blue). Original
magnification, x400, scale bar, 20 µm. (F) Binding motif of FOXO3a
(from JASPAR). (G) ChIP-PCR shows the binding of FOXO3a on the
promoter of ESR2. Agarose gel electrophoresis of ChIP-PCR products.
(H) ChIP analysis for binding of FOXO3a to the ESR2 promoter in 4T1
cells upon CM, CON or CM + LY294002 treatment for 24 h. Data are
expressed as enrichment relative to the input.
**P<0.01 and ***P<0.001; ns, no
significant; CM, conditioned medium; ChiP; chromatin
immunoprecipitation; Erβ, estrogen receptor-β; CON, control; p-AKT;
phosphorylated AKT; MSR1, macrophage scavenger receptor 1; Expr.,
expression; FDR, false discovery rate. |
In a previous study, whole-transcriptome profiling
of 4T1 cells co-cultured with vehicle or M2 macrophages in
Transwell systems revealed marked enrichment of the FOXO signaling
pathway (17).
FOXO transcription factors govern diverse biological
programs. Activation of the PI3K/AKT pathway results in FOXO
phosphorylation, thereby influencing their localization and
transcriptional regulation (30).
Moreover, FOXO3a has been shown to directly drive ESR1
transcription in breast cancer (31). Therefore, it was hypothesized that
TAMs suppress ERβ expression through the PI3K/AKT pathway by
modulating the activity of the FOXO3a transcription factor. JASPAR
(https://jaspar.elixir.no) was used to analyze the
FOXO3a binding motif (Fig. 4F).
The sequence ‘CCTGTTTCCA’ was predicted as a potential FOXO3a
binding site within the mouse ESR2 promoter. ChIP assays were
performed to assess FOXO3a binding to the ESR2 promoter and to
determine whether activation of the PI3K/AKT pathway reduced this
binding, thereby suppressing ESR2 transcription. ChIP-PCR analysis
demonstrated that ESR2 was enriched by the anti-FOXO3a antibody,
supporting a potential role for FOXO3a as a transcriptional
regulator of ESR2 (Fig. 4G).
Compared with the control group, CM treatment markedly reduced
FOXO3a binding to the ESR2 promoter, whereas LY294002 treatment
restored and perhaps enhanced this binding (P<0.001, Fig. 4G and H). These findings established the
PI3K/AKT/FOXO3a axis as an important mechanism by which TAMs
suppress ERβ expression in TNBC.
PI3K/AKT inhibition and ERβ activation
inhibit tumor metastasis
Considering the invasive and migratory properties of
TNBC, the effects of PI3K/AKT inhibition and ERβ activation on
these properties were evaluated. Transwell assay and wound healing
assays demonstrated that LY294002 treatment suppressed 4T1 cell
migration. This effect was amplified when combined with the ERβ
agonist, ERB041 and involved LY294002-induced ERβ upregulation
(P<0.01, Fig. 5A-C).
Furthermore, 4T1 cells were orthotopically implanted into the
mammary fat pads of BALB/c mice, followed by their treatment with
DMSO, LY294002 and LY294002 + ERB041. Metastatic lesions in the
lungs were quantified and representative images of the tumor
nodules in each group taken. Notably, LY294002 potently suppressed
lung metastasis compared with that in the control group. Moreover,
LY294002 + ERB041 further decreased the metastatic burden
(P<0.05, Fig. 5D and E). It was also observed that LY294002
significantly suppressed tumor growth (P<0.01, Figs. 5F and S1). Consistent with the in vitro
results, PI3K/AKT activation was diminished, however ERβ expression
was restored in the primary tumor of the LY294002-treated group
(Fig. 5G). Body weights remained
stable across all groups during treatment (Fig. S2), demonstrating no evident
systemic toxicity. The present data indicated that PI3K/AKT pathway
inhibition and ERβ activation suppressed metastasis in TNBC mice
model.
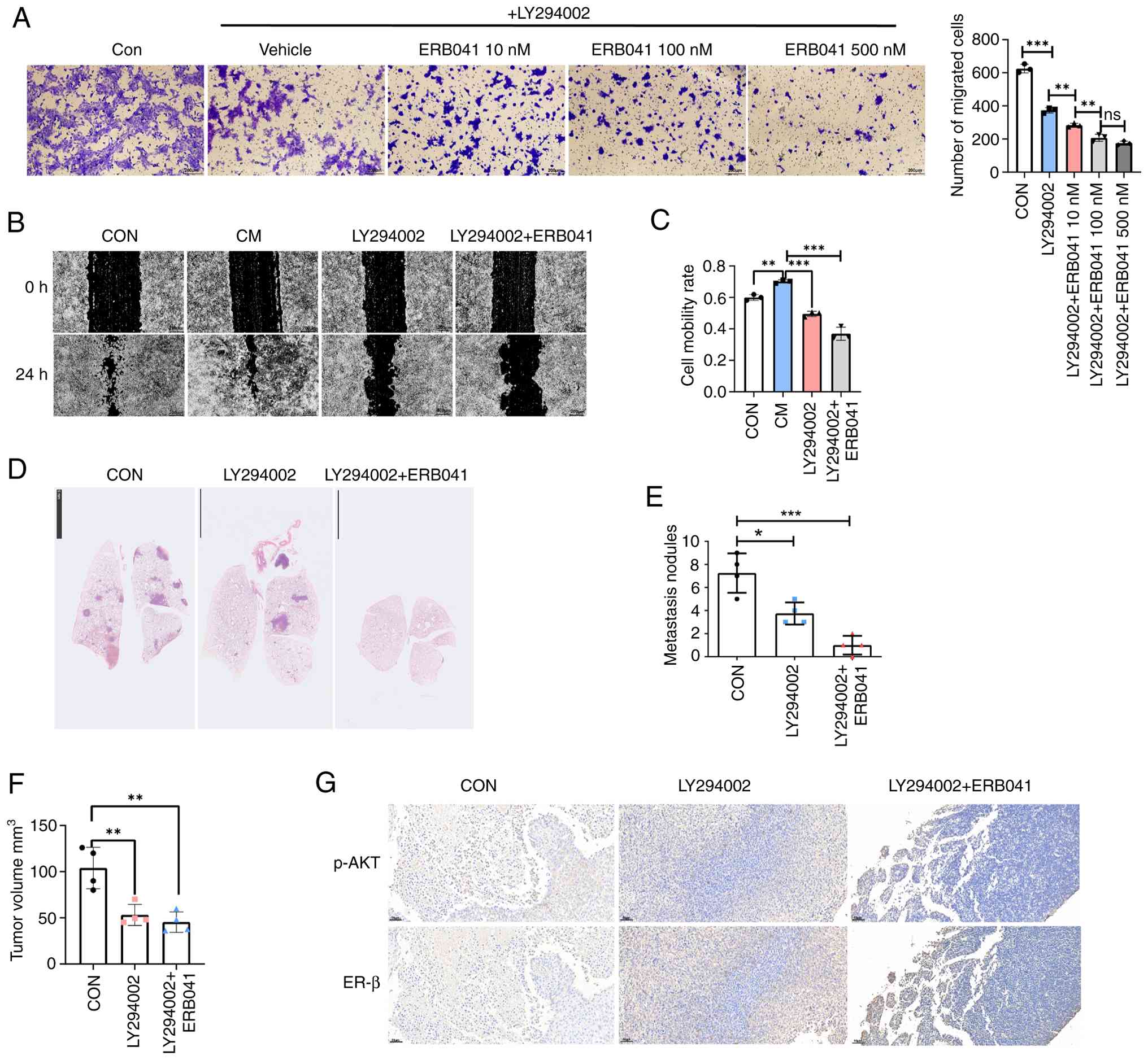 | Figure 5PI3K/AKT inhibition and ERβ
activation inhibit tumor metastasis. (A) Representative images of
Transwell migration assays after treatment with LY294002 (1 µM) or
a combination of LY294002 and ERB041 (10-500 nM). Graph of
Transwell assays (n=3). (B) Representative images of wound healing
assay at 0 and 24 h after treatment with CM, LY294002 (1 µM) or a
combination of LY294002 and ERB041 (100 nM). (C) Graph of wound
healing assay (n=3). (D) 4T1 cells (1x106) were injected
into the fat pads of BALB/c mice, then treated with DMSO, LY294002
(50 mg/kg intraperitoneally twice a week for five weeks) or
LY294002 combined with ERB041 (5 mg/kg subcutaneously daily for
five weeks). Representative images showing the H&E staining of
lung metastatic nodules in 4T1 mice model. Scale bar, 5 mm (E) Lung
metastatic nodule counts in different groups (n=4). (F) The volume
of the 4T1 tumor across all groups (n=4). (G) Representative
immunohistochemical staining images of p-AKT and ERβ in the breast
tumors of different treatment groups. Scale bar, 50 µm.
Magnification, x200. *P<0.05, **P<0.01
and ***P<0.001; ns, not significant. Erβ, estrogen
receptor-β; CM, conditioned medium; p-AKT, phosphorylated AKT; Con,
control. |
Discussion
In the present study, findings revealed that TAMs
suppressed ERβ expression in TNBC through the PI3K/AKT signaling
pathway, thereby promoting tumor progression and metastasis. This
finding highlights the key role of the tumor microenvironment in
modulating therapeutic targets, outlining a potential approach to
enhance ERβ-based treatment efficacy for TNBC.
TNBC is challenging to treat owing to its lack of
effective therapeutic targets and aggressive and metastatic nature
(32). Although ERβ exerts
anti-tumor effects against TNBC (9,11,33-35),
its clinical efficacy is limited due to reduced expression in tumor
tissues (36). ERβ expression is
gradually reduced during mammary tumorigenesis (4). Consistent with a previous report
(37), the present study observed
significantly downregulated ERβ levels in TNBC tumors. Therefore,
the findings suggested that ERβ downregulation compromises the
efficacy of ERβ agonists, marking a key event in TNBC progression.
Elucidation of the mechanisms underlying such key events will
facilitate the development of novel TNBC treatment strategies.
The present study focused on TAMs, the most abundant
infiltrating immune cells promoting metastasis through multiple
mechanisms in breast cancer (38,39).
Findings revealed that the proportion of TAMs, marked by MSR1
expression, were significantly higher in TNBC tumors compared with
in adjacent tissues and associated with poor overall survival. GSEA
analysis revealed strong associations between TAM proportions and
metastasis-related phenotypes in TNBC. Importantly, a negative
correlation between TAM infiltration and ERβ expression in breast
cancer tissues was identified and subsequently validated using
co-culture experiments and TAM-CM. Furthermore, in a murine model,
depletion of macrophages by clodronate liposomes restored the ERβ
expression in tumor tissues, further demonstrating the role of TAMs
as key mediators of ERβ suppression in TNBC.
To elucidate the underlying signaling mechanisms,
analysis of the pathways influenced by TAMs in TNBC samples was
conducted and the significant enrichment of the PI3K/AKT pathway
was observed. The PI3K/AKT pathway is a well-established oncogenic
driver of TNBC (40). Consistent
with a previous report of a negative correlation between PI3K/AKT
activation and ERβ expression in TNBC (8), RT-qPCR analyses revealed that
pharmacological blockade of the PI3K/AKT pathway reversed
TAM-induced ERβ downregulation in the present study. These results
suggest that TAMs suppress ERβ transcription through the PI3K/AKT
pathway.
Previous work revealed marked enrichment of the FOXO
signaling pathway in 4T1 cells co-cultured with M2 macrophages
(17). The PI3K/AKT pathway is
known to phosphorylate FOXO transcription factors, thereby altering
their subcellular localization and transcriptional activity
(30,31). Based on these findings, it was
hypothesized that TAMs suppress ERβ expression through
PI3K/AKT-mediated modulation of FOXO3a transcriptional activity.
ChIP-PCR analysis identified ESR2 as a direct transcriptional
target of FOXO3a. Notably, CM treatment markedly reduced FOXO3a
occupancy at the ESR2 promoter, whereas PI3K/AKT blockade restored
and even enhanced this binding, supporting a model in which
TAM-driven PI3K/AKT activation suppresses ERβ transcription through
FOXO3a.
Considering its strong anti-breast cancer activity,
a number of ERβ agonists have been evaluated in clinical trials for
TNBC treatment (41). However,
they have demonstrated limited efficacy and ERβ expression serves
as a key determinant of their efficacy in clinical trials (36).
Previous clinical trials suggest that PI3K/AKT
inhibitors improve the progression-free survival of patients with
metastatic TNBC (42,43). The findings of the present study
revealed that blocking the PI3K/AKT pathway induced ERβ expression,
warranting further investigation of the roles of ERβ-specific
agonist and PI3K/AKT inhibitor combinations. In the present study,
LY294002 suppressed TNBC cell migration and this effect was further
magnified by the ERβ-selective agonist, ERB041. In vitro
findings were further validated using a metastatic model, in which
the combination of LY294002 and ligand-induced ERβ activation
suppressed TNBC lung metastasis more potently compared with either
agent alone.
In conclusion, the present study demonstrated that
TAMs promoted tumor metastasis by suppressing ERβ expression
through the PI3K/AKT pathway. Overall, these findings provide a
mechanistic basis for the evaluation of PI3K/AKT inhibitor and
ERβ-selective agonist combinations as novel therapeutic agents for
TNBC.
Supplementary Material
Representative image of excised tumors
from mice in each treatment group. Tumors were excised from mice in
the control, LY294002 and LY294002 + ERB041 groups at the end of
the treatment period. Tumors are shown alongside a ruler to
indicate size. CON, control.
Body weight monitoring of mice during
treatment. Body weights of mice in the control, LY294002 and
LY294002 + ERB041 groups were recorded every 2 days during the
treatment period (5 weeks). Data are presented as mean ± SEM. ns,
not significant.
Primer Sequences.
Antibody List
Acknowledgements
The authors appreciate the assistance of Dr Guan Tan
(Huazhong University of Science and Technology) in conducting the
bioinformatics analysis.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Hubei Province (grant no. 2022CFB772).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
QZ designed the study and drafted the manuscript.
DG, GX and RX performed the experiments and acquired the data. QZ
and DG confirm the authenticity of all the raw data. YD analyzed
the data. JW and PF contributed to the conception and design of the
study, and provided revisions of the manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
All animal experiments were reviewed and approved by
the Institutional Animal Care and Use Committee of Huazhong
University of Science and Technology and all procedures were
conducted in accordance with the institutional guidelines for the
care and use of laboratory animals. Breast cancer tissues from two
patients with TNBC were collected at The Central Hospital of Wuhan,
Tongji Medical College, Huazhong University of Science and
Technology between January 2022 and December 2023. The present
study was approved by the Ethics Committee of The Central Hospital
of Wuhan (approval no. WHZXKYL2022-033). Written informed consent
was obtained from all patients, who also agreed to donate their
surgical specimens to the Biobank of The Central Hospital of Wuhan
for future breast cancer-related research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Giaquinto AN and Jemal A:
Cancer statistics, 2024. CA Cancer J Clin. 74:12–49.
2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
von Minckwitz G, Untch M, Blohmer JU,
Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich
J, Huober J, et al: Definition and impact of pathologic complete
response on prognosis after neoadjuvant chemotherapy in various
intrinsic breast cancer subtypes. J Clin Oncol. 30:1796–1804.
2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hieken TJ, Carter JM, Hawse JR, Hoskin TL,
Bois M, Frost M, Hartmann LC, Radisky DC, Visscher DW and Degnim
AC: ERβ expression and breast cancer risk prediction for women with
atypias. Cancer Prev Res (Phila). 8:1084–1092. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Roger P, Sahla ME, Mäkelä S, Gustafsson
JA, Baldet P and Rochefort H: Decreased expression of estrogen
receptor beta protein in proliferative preinvasive mammary tumors.
Cancer Res. 61:2537–2541. 2001.PubMed/NCBI
|
|
5
|
Shaw JA, Udokang K, Mosquera JM, Chauhan
H, Jones JL and Walker RA: Oestrogen receptors alpha and beta
differ in normal human breast and breast carcinomas. J Pathol.
198:450–457. 2002.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Huang B, Omoto Y, Iwase H, Yamashita H,
Toyama T, Coombes RC, Filipovic A, Warner M and Gustafsson JÅ:
Differential expression of estrogen receptor α, β1, and β2 in
lobular and ductal breast cancer. Proc Natl Acad Sci USA.
111:1933–1938. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Reese JM, Suman VJ, Subramaniam M, Wu X,
Negron V, Gingery A, Pitel KS, Shah SS, Cunliffe HE, McCullough AE,
et al: ERβ1: Characterization, prognosis, and evaluation of
treatment strategies in ERα-positive and -negative breast cancer.
BMC Cancer. 14(749)2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang J, Zhang C, Chen K, Tang H, Tang J,
Song C and Xie X: ERβ1 inversely correlates with PTEN/PI3K/AKT
pathway and predicts a favorable prognosis in triple-negative
breast cancer. Breast Cancer Res Treat. 152:255–269.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Dey P, Wang A, Ziegler Y, Kumar S, Yan S,
Kim SH, Katzenellenbogen JA and Katzenellenbogen BS: Estrogen
receptor beta 1: A potential therapeutic target for female triple
negative breast cancer. Endocrinology. 163(bqac172)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhao L, Huang S, Mei S, Yang Z, Xu L, Zhou
N, Yang Q, Shen Q, Wang W, Le X, et al: Pharmacological activation
of estrogen receptor beta augments innate immunity to suppress
cancer metastasis. Proc Natl Acad Sci USA. 115:E3673–E3681.
2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Reese JM, Bruinsma ES, Nelson AW,
Chernukhin I, Carroll JS, Li Y, Subramaniam M, Suman VJ, Negron V,
Monroe DG, et al: ERβ-mediated induction of cystatins results in
suppression of TGFβ signaling and inhibition of triple-negative
breast cancer metastasis. Proc Natl Acad Sci USA. 115:E9580–E9589.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mehraj U, Qayoom H and Mir MA: Prognostic
significance and targeting tumor-associated macrophages in cancer:
New insights and future perspectives. Breast Cancer. 28:539–555.
2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Williams CB, Yeh ES and Soloff AC:
Tumor-associated macrophages: Unwitting accomplices in breast
cancer malignancy. NPJ Breast Cancer. 2(15025)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Miyasato Y, Shiota T, Ohnishi K, Pan C,
Yano H, Horlad H, Yamamoto Y, Yamamoto-Ibusuki M, Iwase H, Takeya M
and Komohara Y: High density of CD204-positive macrophages predicts
worse clinical prognosis in patients with breast cancer. Cancer
Sci. 108:1693–1700. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ning C, Xie B, Zhang L, Li C, Shan W, Yang
B, Luo X, Gu C, He Q, Jin H, et al: Infiltrating macrophages induce
ERα expression through an IL17A-mediated epigenetic mechanism to
sensitize endometrial cancer cells to estrogen. Cancer Res.
76:1354–1366. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Stossi F, Madak-Erdoğan Z and
Katzenellenbogen BS: Macrophage-elicited loss of estrogen
receptor-α in breast cancer cells via involvement of MAPK and c-Jun
at the ESR1 genomic locus. Oncogene. 31:1825–1834. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang Q, Le K, Xu M, Zhou J, Xiao Y, Yang
W, Jiang Y, Xi Z and Huang T: Combined MEK inhibition and
tumor-associated macrophages depletion suppresses tumor growth in a
triple-negative breast cancer mouse model. Int Immunopharmacol.
76(105864)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chung H, Gyu-Mi P, Na YR, Lee YS, Choi H
and Seok SH: Comprehensive characterization of early-programmed
tumor microenvironment by tumor-associated macrophages reveals
galectin-1 as an immune modulatory target in breast cancer.
Theranostics. 14:843–860. 2024.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jiang H, Fan D, Zhou G, Li X and Deng H:
Phosphatidylinositol 3-kinase inhibitor(LY294002) induces apoptosis
of human nasopharyngeal carcinoma in vitro and in vivo. J Exp Clin
Cancer Res. 29(34)2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Fujiwara M, Izuishi K, Sano T, Hossain MA,
Kimura S, Masaki T and Suzuki Y: Modulating effect of the
PI3-kinase inhibitor LY294002 on cisplatin in human pancreatic
cancer cells. J Exp Clin Cancer Res. 27(76)2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Guo D, Liu X, Zeng C, Cheng L, Song G, Hou
X, Zhu L and Zou K: Estrogen receptor β activation ameliorates
DSS-induced chronic colitis by inhibiting inflammation and
promoting Treg differentiation. Int Immunopharmacol.
77(105971)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ghandi M, Huang FW, Jané-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357(eaan2507)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48 (W1):W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang B, Guo X, Huang L, Zhang Y, Li Z, Su
D, Lin L, Zhou P, Ye H, Lu Y and Zhou Q: Tumour-associated
macrophages and Schwann cells promote perineural invasion via
paracrine loop in pancreatic ductal adenocarcinoma. Br J Cancer.
130:542–554. 2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhang L, Lu X, Xu Y, La X, Tian J, Li A,
Li H, Wu C, Xi Y, Song G, et al: Tumor-associated macrophages
confer colorectal cancer 5-fluorouracil resistance by promoting
MRP1 membrane translocation via an intercellular
CXCL17/CXCL22-CCR4-ATF6-GRP78 axis. Cell Death Dis.
14(582)2023.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Qin Q, Ji H, Li D, Zhang H, Zhang Z and
Zhang Q: Tumor-associated macrophages increase COX-2 expression
promoting endocrine resistance in breast cancer via the
PI3K/Akt/mTOR pathway. Neoplasma. 68:938–946. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yu X, Wang R, Zhang Y, Zhou L, Wang W, Liu
H and Li W: Skp2-mediated ubiquitination and mitochondrial
localization of Akt drive tumor growth and chemoresistance to
cisplatin. Oncogene. 38:7457–7472. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jia X, Li C, Li L, Liu X, Zhou L, Zhang W,
Ni S, Lu Y, Chen L, Jeong LS, et al: Neddylation inactivation
facilitates FOXO3a nuclear export to suppress estrogen receptor
transcription and improve fulvestrant sensitivity. Clin Cancer Res.
25:3658–3672. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hammershoi Madsen AM, Lovendahl Eefsen RH,
Nielsen D and Kumler I: Targeted treatment of metastatic
triple-negative breast cancer: A systematic review. Breast J.
2024(9083055)2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hwang NM and Stabile LP: Estrogen receptor
ß in cancer: To ß(e) or not to ß(e)? Endocrinology.
162(bqab162)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Shanle EK, Zhao Z, Hawse J, Wisinski K,
Keles S, Yuan M and Xu W: Research resource: global identification
of estrogen receptor β target genes in triple negative breast
cancer cells. Mol Endocrinol. 27:1762–1775. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hinsche O, Girgert R, Emons G and Gründker
C: Estrogen receptor β selective agonists reduce invasiveness of
triple-negative breast cancer cells. Int J Oncol. 46:878–884.
2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wisinski KB, Xu W, Tevaarwerk AJ, Saha S,
Kim K, Traynor A, Dietrich L, Hegeman R, Patel D, Blank J, et al:
Targeting estrogen receptor beta in a phase 2 study of high-dose
estradiol in metastatic triple-negative breast cancer: A wisconsin
oncology network study. Clin Breast Cancer. 16:256–261.
2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Gao L, Qi X, Hu K, Zhu R, Xu W, Sun S,
Zhang L, Yang X, Hua B and Liu G: Estrogen receptor β promoter
methylation: A potential indicator of malignant changes in breast
cancer. Arch Med Sci. 12:129–136. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Ireland L, Santos A, Campbell F,
Figueiredo C, Hammond D, Ellies LG, Weyer-Czernilofsky U,
Bogenrieder T, Schmid M and Mielgo A: Blockade of insulin-like
growth factors increases efficacy of paclitaxel in metastatic
breast cancer. Oncogene. 37:2022–2036. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Fang C, Cheung MY, Chan RC, Poon IK, Lee
C, To CC, Tsang JY, Li J and Tse GM: Prognostic significance of
CD163+ and/or CD206+ tumor-associated macrophages is linked to
their spatial distribution and tumor-infiltrating lymphocytes in
breast cancer. Cancers (Basel). 16(2147)2024.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Pascual J and Turner NC: Targeting the
PI3-kinase pathway in triple-negative breast cancer. Ann Oncol.
30:1051–1060. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Shen K, Yu H, Xie B, Meng Q, Dong C, Shen
K and Zhou HB: Anticancer or carcinogenic? The role of estrogen
receptor β in breast cancer progression. Pharmacol Ther.
242(108350)2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Schmid P, Abraham J, Chan S, Wheatley D,
Brunt AM, Nemsadze G, Baird RD, Park YH, Hall PS, Perren T, et al:
Capivasertib plus paclitaxel versus placebo plus paclitaxel as
first-line therapy for metastatic triple-negative breast cancer:
The PAKT trial. J Clin Oncol. 38:423–433. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kim SB, Dent R, Im SA, Espié M, Blau S,
Tan AR, Isakoff SJ, Oliveira M, Saura C, Wongchenko MJ, et al:
Ipatasertib plus paclitaxel versus placebo plus paclitaxel as
first-line therapy for metastatic triple-negative breast cancer
(LOTUS): A multicentre, randomised, double-blind,
placebo-controlled, phase 2 trial. Lancet Oncol. 18:1360–1372.
2017.PubMed/NCBI View Article : Google Scholar
|



















