Introduction
One of the most lethal features of pancreatic cancer
is its apparent capacity for early invasion and metastasis to the
liver and other organs. Apart from surgery, there is no effective
therapy and even resected patients usually die within one year
postoperatively. Reasons for the poor prognosis include the
occurrence of local recurrences and/or distant metastasis after
surgery. However, to date, the cellular and molecular mechanisms of
the invasion-metastasis of pancreatic cancer remain unclear.
Detection of the factors related to the differences in potential
for invasion and metastasis of cancer cells could provide useful
information for the development of new therapeutic methods to
prevent the invasion and metastasis of pancreatic cancer.
To investigate the mechanisms of invasion-metastasis
of pancreatic cancer, two hamster pancreatic cancer cell lines with
a different potential for invasion and metastasis, PC-1 with a low
potential and PC-1.0 with a high potential after intrapancreatic
transplantation, were established from a pancreatic ductal
carcinoma induced by N-nitrosobis (2-oxopropyl) amine (BOP) in a
Syrian golden hamster in our previous investigation (1,2).
cDNA microarray is a new emerging technique in the
post genomic era. Large-scale analysis of gene expression with cDNA
microarray allows us to evaluate the gene expression profiles of
hundreds to tens of thousands of genes in a single experiment
(3). Therefore, the cDNA
microarray is a promising tool to provide new insight into the
mechanisms of cancer invasion and metastasis.
In the present study, we analyzed alteration in the
invasion-metastasis-related gene expression patterns of 27,000
genes in highly invasive and metastatic pancreatic cancer cells
(PC-1.0) in comparison to weakly invasive and metastatic pancreatic
cancer cells (PC-1) utilizing powerful cDNA microarray
technology.
Materials and methods
Cell lines and cell culture
Two hamster pancreatic cancer cell lines, weakly
invasive and metastatic cells (PC-1) and highly invasive and
metastatic cells (PC-1.0) were used. The PC-1 cell line was
established from pancreatic ductal/ductular adenocarcinomas induced
by BOP in a Syrian golden hamster (1). The PC-1.0 cell line was established
from a subcutaneous tumor produced after inoculation of PC-1 cells
(2). In vitro, PC-1 cells
grow mainly as island-like cell colonies, whereas PC-1.0 cells
exhibit the growth pattern of single cells. In vivo, local
expansion of PC-1 cells and local invasion of PC-1.0 cells are
observed (1,2).
The PC-1 and PC-1.0 cells were incubated in
RPMI-1640 (Gibco-BRL, Grand Island, NY, USA), supplemented with 10%
fetal bovine serum (Bioserum, Victoria, Australia), 100 U/ml
penicillin G and 100 μg/ml streptomycin at 37°C in a
humidified atmosphere of 5% CO2/95% air.
Preparation of total RNA
Total RNA of the PC-1.0 and PC-1 cells was extracted
using the TRIzol reagent according to the manufacturer's
instructions (Invitrogen). After TRIzol purification, RNA was
further purified with RNeasy mini spin column kit (Qiagen,
Valencia, CA, USA). The concentration and qualify of the RNA were
assessed via spectrophotometry and agarose gel electrophoresis.
cDNA microarray and statistical analysis
of data
Preparation of fluorescent dye-labeled DNA and
hybridizations was performed according to the protocol of the
reagent/kit manufacturers and previously reported methods (4). Briefly, RNA was reverse-transcribed
into cDNA with Oligo(dT)15 (Promega) as primer and Superscript II
choice for cDNA synthesis (Invitrogen) and subsequently labeled in
red (Cy5) or in green (Cy3) (Amersham Pharmacia Biotech). Cy5- and
Cy3-labeled cDNA was purified with a PCR purification kit (Qiagen).
DNA was mixed with 30 μl hybridization solution prior to
loading onto a rat gene microarray (Capitalbio Inc., Beijing, P.R.
China) which included 27,000 transcripts (Oligo library, Rat Genome
version 3.0.5; Qiagen). Arrays were hybridized at 42°C overnight.
The experiments were performed twice with reverse dye-labeled
cDNA.
The microarray plates were scanned by LuxScan 10KA
dual pathways laser scanner (Capitalbio), and images were analyzed
through GenePix Pro 4.0 image analysis software (Axon Instruments
Co.). Genes were considered to be differentially expressed,
integrated ratio of two experiments, at a change in increase
(>3.00) or decrease (<0.33) in the ratio of expression levels
between PC-1.0 and PC-1 cells.
Statistical analysis was carried out with the
t-test, and the expression of a given gene was considered changed
when the difference between means was significant (P<0.01).
Reverse transcriptase-polymerase chain
reaction (RT-PCR)
Total RNA was isolated from PC-1.0 and PC-1 cells,
and an aliquot of 1 μg of total RNA from each sample was
reverse-transcribed to cDNA using the SuperScript II kit (Life
Technologies, Inc.) as previously described (5). The primers used for PCR amplification
in this study are listed in Table
I. Amplification was run for 30 cycles at 95°C for 5 min, 95°C
for 40 sec, 55°C for 30 sec, 72°C for 1 min and finally extended at
72°C for 7 min.
 | Table I.Primers used for the RT-PCR of PC-1.0
and PC-1 cells. |
Table I.
Primers used for the RT-PCR of PC-1.0
and PC-1 cells.
| Gene name | Primer sequence | Product size
(bp) |
|---|
| Actin | F:
GTGGGGCGCCCCAGGCACCA
R: CTCCTTAAGTCACGCACGATTCC | 664 |
| nup107 | F:
GACAGAAGAGGCACAACGAC
R: ACCAGACTGTCCACCATCAC | 309 |
| tjp2 | F:
GCAGAGCGAACGAAGAGTATGG 245
R: TGACGGGATGTTGATGAGGGT | |
| MMP-13 | F:
CAGTCTTTCTTCGGCTTAG
R: CAGGGTCCTTGGAGTGGTC | 496 |
| Spc21 | F:
GTGGTGCTGAGTGGCAGTAT
R: CCAGTTCTGGCCTTCTTTGT | 246 |
| plau | F:
AGAATTCACCACCATCGAGA
R: ATCAGCTTCACAACAGTCAT | 474 |
| CD44 | F:
AAGGTGGAGCAAACACAACC
R: AACTGCAATGCAAACTGCAAG | 115 |
Gene Ontology and Pathway analysis of
differentially expressed genes
Using the Gene Ontology tool from http://www.pantherdb.org, the differentially expressed
genes were automatically assembled to categories of Biological
process, Molecular function and Cellular component. Biologically
related networks were automatically assembled from identified genes
on microarrays by the BioRag (http://www.biorag.org), which enables the analysis of
pathways among interested genes according to Kegg (http://www.genome.ad.jp/kegg) or GenMAPP (http://www.genmapp.org). The Fisher's exact test was
performed to detect the significantly regulated gene and pathway, A
P-value <0.01 was considered significantly overrepresented.
Results
Differentially expressed genes identified
by cDNA microarray in the highly (PC-1.0) and weakly (PC-1)
invasive and metastatic pancreatic cancer cells
To clarify the differentially expressed genes
between highly (PC-1.0) and weakly (PC-1) invasive and metastatic
cells, the expression level for each gene in the two pancreatic
cancer cell lines was compared. Of the 27,000 genes analyzed
through microarray experiments, a total of 141 genes revealed
differential expression using a fold ratio >3 as the criteria
for cut-off. Of the 141 genes, the expression of 46 genes (32.6%)
was markedly increased in the highly invasive and metastatic cells
(PC-1.0) as compared with the weakly invasive and metastatic cells
(PC-1) (Table II). On the other
hand, the expression of 95 genes (67.4%) was significantly
decreased in the highly invasive and metastatic cells (PC-1.0) as
compared with the weakly invasive and metastatic cells (PC-1)
(Table III). The ratio represented
the expression value in PC-1.0 cells compared with the expression
level in PC-1 cells.
 | Table II.Genes up-regulated in highly invasive
and metastatic cells (PC-1.0) compared with weakly invasive and
metastatic cells (PC-1). |
Table II.
Genes up-regulated in highly invasive
and metastatic cells (PC-1.0) compared with weakly invasive and
metastatic cells (PC-1).
| Gene name | Gene ID | Gene symbol | Description | Ratio |
|---|
| Mlp |
ENSRNOG00000009113 | NM_030862 | MARCKS-like
protein | 71.9931 |
| Aldr1 |
ENSRNOG00000009513 | ALDR_RAT | Aldehyde reductase
1 | 33.6872 |
| MMP-13 |
ENSRNOG00000008478 | MM13_RAT | Matrix
metallopeptidase 13 | 30.0071 |
| MMP-12 |
ENSRNOG00000008993 | MM03_RAT | Matrix
metallopeptidase 12 | 26.2124 |
| Col5a2 |
ENSRNOG00000003736 | O70598 | Collagen, type V,
α2 | 20.8255 |
| Tnni2 |
ENSRNOG00000020276 | TRIF_RAT | Troponin 1, type
2 | 20.5716 |
| Tjp2 |
ENSRNOG00000015030 | P70625 | Tjp2 protein | 20.5690 |
| MMP-3 |
ENSRNOG00000008993 | MM03_RAT | Matrix
metallopeptidase 3 | 20.5668 |
| Snrpn |
ENSRNOG00000022595 | NM_130738 | Small nuclear
ribonucleoprotein N | 17.1193 |
| Syt8 |
ENSRNOG00000020245 | NM_053325 | Synaptotagmin 8 | 15.8243 |
| S100a5 |
ENSRNOG00000011748 | S105_MOUSE | S100 calcium binding
protein A5 | 11.6390 |
| Ndrg2 |
ENSRNOG00000010389 | NM_133583 | N-myc downstream
regulated gene 2 | 11.4480 |
| MMP-10 |
ENSRNOG00000008993 | MM03_RAT | Matrix
metallopeptidase 10 | 10.7218 |
| Tf |
ENSRNOG00000009434 | TRFE_RAT | Transferrin | 8.7425 |
| Anxa6 |
ENSRNOG00000010668 | ANX6_RAT | Annexin A6 | 6.5978 |
| Nup107 |
ENSRNOG00000006541 | N107_RAT | Nucleoporin 107 | 6.5895 |
| Spnb3 |
ENSRNOG00000019564 | SPCP_RAT | β-spectrin 3 | 6.3763 |
| Cdk4 |
ENSRNOG00000025602 | CDK4_RAT | Cyclin-dependent
kinase 4 | 6.2099 |
| Fap |
ENSRNOG00000005679 | NM_138850 | Fibroblast activation
protein | 5.7157 |
| Eno3 |
ENSRNOG00000004078 | ENOB_RAT | Enolase 3,β | 5.6429 |
 | Table III.Genes down-regulated in highly
invasive and metastatic cells (PC-1.0) compared with weakly
invasive and metastatic cells (PC-1). |
Table III.
Genes down-regulated in highly
invasive and metastatic cells (PC-1.0) compared with weakly
invasive and metastatic cells (PC-1).
| Gene name | Gene ID | Gene symbol | Description | Ratio |
|---|
| App |
ENSRNOG00000001546 | A4_RAT | Amyloid β (A4)
precursor protein | 0.1286 |
| Col9a1 |
ENSRNOG00000012920 | CA19_RAT | Procollagen, type IX,
α 1 | 0.1269 |
| CD44 |
ENSRNOG00000013562 | CD44_RAT | CD44 antigen | 0.1205 |
| Serpinh1 |
ENSRNOG00000016831 | HS47_RAT | Serine proteinase
inhibitor 1, clade H | 0.1190 |
| Tcf4 |
ENSRNOG00000012405 | ITF2_RAT | Transcription factor
4 | 0.1039 |
| Chn2 |
ENSRNOG00000009411 | CHIO_RAT | Chimerin (chimaerin)
2 | 0.0913 |
| Plau |
ENSRNOG00000010516 | UROK_RAT | Plasminogen
activator, urokinase | 0.0895 |
| Sphk1 |
ENSRNOG00000010626 | NM_133386 | Sphingosine kinase
1 | 0.0882 |
| Apom |
ENSRNOG00000000850 | APOM_RAT | Apolipoprotein M | 0.0850 |
| Psmb8 |
ENSRNOG00000000456 | PSB8_RAT | Proteosome subunit, β
type 8 | 0.0844 |
| Ldhb |
ENSRNOG00000013000 | LDHB_RAT | Lactate dehydrogenase
B | 0.0737 |
| Spc21 |
ENSRNOG00000017036 | SPC3_RAT | Microsomal signal
peptidase 21 kDa subunit | 0.0712 |
| Klf4 |
ENSRNOG00000016299 | NM_053713 | Kruppel-like factor
4 | 0.0669 |
| Cntn4 |
ENSRNOG00000005652 | NM_053746 | Contactin 4 | 0.0508 |
| Ephx1 |
ENSRNOG00000003515 | HYEP_RAT | Epoxide hydrolase
1 | 0.0493 |
| Serpinb2 |
ENSRNOG00000002460 | PAI2_RAT | Plasminogen
activator inhibitor 2 | 0.0465 |
| Pmp22 |
ENSRNOG00000003338 | PM22_RAT | Peripheral myelin
protein 22 | 0.0337 |
| Pde1c |
ENSRNOG00000012337 | CN1C_RAT | Phosphodiesterase
1C | 0.0322 |
| Ngfrap1 |
ENSRNOG00000012646 | NM_053401 | Nerve growth factor
receptor associated protein 1 | 0.0218 |
| Hspb1 |
ENSRNOG00000023546 | HS27_RAT | Heat shock 27 kDa
protein 1 | 0.0177 |
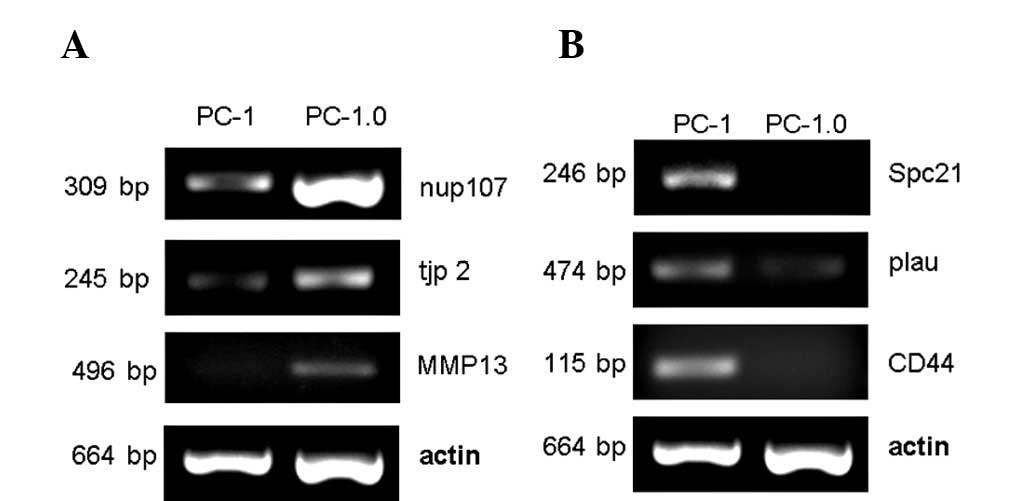
Validation of selected genes with
RT-PCR
To verify the reliability of the microarray data, we
selected three up-regulated genes (nup107, tjp2 and MMP13) and
three down-regulated genes (Spc21, plau and CD44) to measure their
expression levels by RT-PCR. The results were very similar to the
cDNA microarray data on these genes and supported the reliability
of our expression data (Fig.
1).
Gene Ontology and Pathway analysis of
differentially expressed genes
Gene Ontology (GO) and Pathway analysis was applied
in order to generate groups of genes that belong to similar
biological processes correlated with invasion and metastasis of
pancreatic cancer cells.
The differentially expressed genes between highly
(PC-1.0) and weakly (PC-1) invasive and metastatic cells were
summarized in Molecular function, Biological process and Cellular
component, respectively. These are the three types of categories of
GO analysis. The ten most correlated (the lowest P-value) GO
categories of Molecular function, Biological Process and Cellular
Component are presented in Tables
IV, V and VI, respectively.
 | Table IV.Gene Ontology analysis – Molecular
function. |
Table IV.
Gene Ontology analysis – Molecular
function.
| GO Term | Total | P-value | Gene | Input symbol |
|---|
| GO:0004852
uroporphyrinogen-III synthase activity | 1 | 0.0043 | Uros | Rn30016380 |
| GO:0000900
translation repressor activity | 1 | 0.0043 | Purb | Rn30006362 |
| GO:0005131 growth
hormone receptor binding | 1 | 0.0086 | Socs2 | R002975_01 |
| GO:0030161 calpain
inhibitor activity | 1 | 0.0086 | Cast | R001975_01 |
| GO:0046980 tapasin
binding | 1 | 0.0086 | Tap2 | Rn30000347 |
| GO:0004308
exo-α-sialidase activity | 1 | 0.0129 | Neu1 | R003273_01 |
| GO:0005518 collagen
binding | 1 | 0.0172 | Serpinh1 | R003232_01 |
| GO:0008538
proteasome activator activity | 1 | 0.0172 | Psme1 | Rn30017518 |
| GO:0008243
plasminogen activator activity | 1 | 0.0214 | Plau | Rn30009672 |
| GO:0019838 growth
factor binding | 1 | 0.1738 | axl | Rn30019093 |
 | Table V.Gene Ontology analysis – Biological
process. |
Table V.
Gene Ontology analysis – Biological
process.
| GO Term | Total | P-value | Gene | Input symbol |
|---|
| GO:0015914
phospholipid transport | 1 | 1.10E-4 | Plscr1 | Rn30007316 |
| GO:0006983 ER
overload response | 1 | 1.83E-4 | Ddit3 | Rn30006089 |
| GO:0006955 immune
response | 4 | 0.0068 | Tap2 | Rn30000347 |
| | | Ada | R004405_01 |
| | | Psme1 | Rn30017518 |
| | | Plscr1 | Rn30007316 |
| GO:0007034 vacuolar
transport | 1 | 0.0086 | Vps26a | Rn30000282 |
| GO:0031100 organ
regeneration | 1 | 0.0172 | axl | Rn30019093 |
| GO:0007520 myoblast
fusion | 1 | 0.0172 | Cast | R001975_01 |
| GO:0009968 negative
regulation of signal transduction | 2 | 0.0183 | Socs2 | R002975_01 |
| | | Rgs10 | Rn30018565 |
| GO:0050892
intestinal absorption | 1 | 0.0257 | Vdr | Rn30007787 |
| GO:0001558
regulation of cell growth | 1 | 0.0331 | Igfbp6 | Rn30010107 |
| GO:0035023
regulation of Rho protein signal transduction | 1 | 0.0382 | Net1 | Rn30016337 |
 | Table VI.Gene Ontology analysis – Cellular
component. |
Table VI.
Gene Ontology analysis – Cellular
component.
| Go Term | Total | P-value | Gene | Input symbol |
|---|
| GO:0005923 tight
junction | 1 | 0.0156 | Tjp2 | Rn30013789 |
| GO:0005788
endoplasmic reticulum lumen | 1 | 0.0340 | Tap2 | Rn30000347 |
| GO:0030904 retromer
complex | 1 | 0.0043 | Vps26a | Rn30000282 |
| GO:0008537
proteasome activator complex | 1 | 0.0172 | Psme1 | Rn30017518 |
| GO:0005662 DNA
replication factor A complex | 1 | 0.0040 | Purb | Rn30006362 |
| GO:0042589 zymogen
granule membrane | 1 | 0.0214 | Scamp1 | R004473_01 |
| GO:0005793 ER-Golgi
intermediate compartment | 1 | 0.0506 | Serpinh1 | R003232_01 |
| GO:0005905 coated
pit | 1 | 0.0949 | Vldlr | Rn30025704 |
| GO:0019717
synaptosome | 1 | 0.0382 | Vamp3 | Rn30017017 |
| GO:0016020
membrane | 1 | 0.6022 | axl | Rn30019093 |
In addition, Pathway analysis of differentially
expressed genes was also applied using the public database (Kegg
and GenMAPP). The ten most correlated pathways obtained from the
Kegg and GenMAPP are listed in Tables
VII and VIII,
respectively.
 | Table VII.Pathway analysis – Kegg. |
Table VII.
Pathway analysis – Kegg.
| Pathway name | Total | P-value |
|---|
| Pentose and
glucuronate interconversions | 4 | 0.0000 |
| Antigen processing
and presentation | 8 | 2.7E-5 |
| Starch and sucrose
metabolism | 3 | 3.15E-4 |
| Porphyrin and
chlorophyll metabolism | 2 | 3.65E-4 |
| Sphingolipid
metabolism | 5 | 3.65E-4 |
| Fructose and
mannose metabolism | 5 | 9.51E-4 |
| Phenylalanine,
tyrosine and tryptophan biosynthesis | 3 | 0.0011 |
| Type I diabetes
mellitus | 4 | 0.0028 |
| Metabolism of
xenobiotics P450 by cytochrome | 3 | 0.0028 |
| SNARE interactions
in vesicular transport | 4 | 0.0035 |
 | Table VIII.Pathway analysis – GenMAPP. |
Table VIII.
Pathway analysis – GenMAPP.
| Pathway name | Total | P-value |
|---|
| GTP binding | 23 | 2.0E-6 |
| Guanyl nucleotide
binding | 23 | 3.0E-6 |
| Cytosol | 15 | 4.0E-6 |
| Endoplasmic
reticulum | 23 | 7.0E-6 |
| Binding | 21 | 8.0E-6 |
| Cytoplasm | 21 | 7.30E-5 |
| Electron
transport | 20 | 2.11E-4 |
| Magnesium ion
binding | 11 | 2.74E-4 |
| Protein
folding | 14 | 3.52E-4 |
| RNA binding | 20 | 3.69E-4 |
| Metabolism | 21 | 3.73E-4 |
The complete data of the GO and Pathway analysis is
available upon request.
Discussion
To date, there have been some reports regarding the
molecular mechanisms involved in the development of pancreatic
cancer, including some reports utilizing cDNA microarray (6,7).
However, thus far, most of these cDNA microarray studies have
focused on the differences between pancreatic cancer tissue and
normal tissue (8); few studies
have investigated the mechanism of invasion and metastasis in
pancreatic cancer cells using highly and weakly invasive and
metastatic pancreatic cancer cell lines. Yet, these tissue samples
have considerable disadvantages. They are highly complex and are
usually composed of several different cell types and extracellular
matrices; for example, non-neoplastic pancreatic tissue includes
ductal and acinar cells, various neuroendocrine cells and
mesenchymal cells. Thus, one has to be aware that using samples of
tissue homogenates does not simply mean a comparison of neoplastic
vs. non-neoplastic epithelial cells, but a complex mixture of genes
of diverse origin, some of them deriving from epithelial cells. In
contrast, one advantage of using cancer cell lines is that pure
tumor cells are tested without any contamination from surrounding
stromal elements.
In particular, the highly (PC-1.0) and weakly (PC-1)
invasive and metastatic pancreatic cancer cell lines, which are
established from the experimental pancreatic cancer model in our
previous study (1,2), show an obviously different potential
for invasion and metastasis (9,10).
Therefore, this cell line model is suitable for the investigation
of invasion-metastasis-related specific factors in pancreatic
cancer.
In the present study, using cDNA microarray
analysis, we found that a total of 141 genes were differentially
expressed between the PC-1.0 and PC-1 cells, including 46
up-regulated genes and 95 down-regulated genes. We selected several
differentially expressed genes (nup107, tjp-2, MMP-13, Spc21, plau
and CD44) for validation by RT-PCR. The results of RT-PCR were in
accordance with those of the cDNA microarray analysis. In addition,
several of the identified genes (i.e., MMP-13, plau and CD44) have
been previously reported to be correlated with invasion and
metastasis (11–13), and the other differentially
expressed genes (i.e., nup107, tjp-2 and Spc21) have not been
reported to be associated with the invasion-metastasis of
pancreatic cancer.
Of the identified genes not previously reported to
be associated with the invasion-metastasis of pancreatic cancer,
Nup107 is a critical component of the nucleoporin 107–160
subcomplex, which is the key building block of the nuclearpore
complex (NPC). From yeast to humans, the function of NPC is the
regulation of nuclear import and export (14). The Nup107–160 complex thus
additionally offers an attractive point for regulation of nuclear
pore complex assembly (15).
Although nup107 has been identified from the comparison of gene
expression in highly and weakly invasive and metastatic pancreatic
cancer cells in the present study, the molecular mechanism of
involvement of nup107 in the invasion-metastasis of pancreatic
cancer needs to be further tested and assessed.
Several studies have demonstrated that tight
junction proteins (TJPs) associate with each other and directly
and/or indirectly to actin filaments (16) and also recruit factors involved in
signal transduction and the regulation of proliferation and
differentiation (17). The zonula
occludens (ZO) protein is one of the tight junction proteins and
belongs to the membrane associated guanylate kinase-like (MAGUK)
protein family. It includes three members, TJP1/ZO-1, TJP2/ZO-2 and
TJP3/ZO-3 (18). mRNA levels of
ZO-2 were found to be elevated in tumor tissues compared with
controls using quantitative PCR. Moreover, ZO-2 exhibits a 23-amino
acid truncation at the N-terminus, which may play a role in
limiting tumor development in pancreatic cells. In another
investigation, ZO-2 was found to be associated with the progression
of breast cancer (19).
Moreover, Spc21 was identified as a down-regulated
gene in this study, suggesting that dysregulation of this gene is
likely to be associated with the invasion and metastasis of
pancreatic cancer cells. Fish and ISH analysis for this gene
demonstrated a significant correlation between genetic deletion and
corresponding mRNA down-regulation, raising the possibility that
the Spc21 gene may play a putative role as a tumor suppressor
(20). However, little is known
about the biological role of this gene, although it belongs to the
peptidase S26B family and functions as part of the signal peptidase
complex (20).
In conclusion, our results suggest that a highly
organized and structured process of invasion and metastasis exists
in the pancreas. Analysis of gene expression profiles by cDNA
microarray can provide useful information for clarifying the
mechanism underlying the invasion and metastasis of pancreatic
cancer cells. Furthermore, the identification of
invasion-metastasis-specific genes may allow us to develop new
therapeutic and diagnostic targets for the invasion-metastasis of
pancreatic cancer.
Acknowledgements
This study was supported by a
grant-in-aid from the China Postdoctoral Science Foundation (no.
20060390302). We thank Professor Hideo Baba for the kind gift of
PC-1 and PC-1.0 cell lines.
References
|
1.
|
Egami H, Takiyama Y, Cano M, Houser WH and
Pour PM: Establishment of hamster pancreatic ductal carcinoma cell
line (PC-1) producing blood group-related antigens. Carcinogenesis.
10:861–869. 1989.
|
|
2.
|
Egami H, Tomioka T, Tempero M, Kay D and
Pour PM: Development of intrapancreatic transplantable model of
pancreatic duct adenocarcinoma in Syrian golden hamsters. Am J
Pathol. 138:557–561. 1991.
|
|
3.
|
Goggins M: Identifying molecular markers
for the early detection of pancreatic neoplasia. Semin Oncol.
34:303–310. 2007.
|
|
4.
|
Shi YH, Zhu SW, Mao XZ, et al:
Transcriptome profiling, molecular biological and physiological
studies reveal a major role for ethylene in cotton fiber cell
elongation. Plant Cell. 18:651–664. 2006.
|
|
5.
|
Tan X, Egami H, Kamohara H, et al:
Involvement of the mitogen-activated protein kinase kinase 2 in the
induction of cell dissociation in pancreatic cancer. Int J Oncol.
24:65–73. 2004.
|
|
6.
|
Duerr EM, Mizukami Y, Ng A, et al:
Defining molecular classifications and targets in
gastroenteropancreatic neuroendocrine tumors through DNA microarray
analysis. Endocr Relat Cancer. 15:243–256. 2008.
|
|
7.
|
Jones S, Zhang X, Parsons DW, et al: Core
signaling pathways in human pancreatic cancers revealed by global
genomic analysis. Science. 321:1801–1806. 2008.
|
|
8.
|
Capurso G, Lattimore S, Crnogorac-Jurcevic
T, et al: Gene expression profiles of progressive pancreatic
endocrine tumours and their liver metastases reveal potential novel
markers and therapeutic targets. Endocr Relat Cancer. 13:41–58.
2006.
|
|
9.
|
Pour PM, Egami H and Takiyama Y: Patterns
of growth and metastases of induced pancreatic cancer in relation
to the prognosis and its clinical implication. Gastroenterology.
100:529–536. 1991.
|
|
10.
|
Kurizaki T, Egami H, Hirota M, et al:
Characterization of cancer cell dissociation factor in a highly
invasive pancreatic cancer cell line. Cancer. 75:1554–1561.
1995.
|
|
11.
|
Morgia G, Falsaperla M, Malaponte G, et
al: Matrix metalloproteinases as diagnostic (MMP-13) and prognostic
(MMP-2, MMP-9) markers of prostate cancer. Urol Res. 33:44–50.
2004.
|
|
12.
|
Iiizumi M, Liu W, Pai SK, Furuta E and
Watabe K: Drug development against metastasis-related genes and
their pathways: a rationale for cancer therapy. Biochim Biophys
Acta. 1786:87–104. 2008.
|
|
13.
|
Klingbeil P and Marhaba R: CD44 variant
isoforms promote metastasis formation by a tumor cell-matrix
cross-talk that supports adhesion and apoptosis resistance. Mol
Cancer Res. 7:168–179. 2009.
|
|
14.
|
Schuldt A: Nuclear pore assembly: locating
the linchpin. Nat Cell Biol. 5:4972003.
|
|
15.
|
Harel A, Orjalo AV, Vincent T, et al:
Removal of a single pore subcomplex results in vertebrate nuclei
devoid of nuclear pores. Mol Cell. 11:853–864. 2003.
|
|
16.
|
Utepbergenov DI, Fanning AS and Anderson
JM: Dimerization of the scaffolding protein ZO-1 through the second
PDZ domain. J Biol Chem. 281:24671–24677. 2006.
|
|
17.
|
Matter K and Balda MS: Functional analysis
of tight junctions. Methods. 30:228–234. 2003.
|
|
18.
|
Sato N, Fukushima N, Maitra A, et al:
Discovery of novel targets for aberrant methylation in pancreatic
carcinoma using high-throughput microarrays. Cancer Res.
63:3735–3742. 2003.
|
|
19.
|
Martin TA, Watkins G, Mansel RE and Jiang
WG: Loss of tight junction plaque molecules in breast cancer
tissues is associated with a poor prognosis in patients with breast
cancer. Eur J Cancer. 40:2717–2725. 2004.
|
|
20.
|
Harada T, Baril P, Gangeswaran R, et al:
Identification of genetic alterations in pancreatic cancer by the
combined use of tissue microdissection and array-based comparative
genomic hybridization. Br J Cancer. 96:373–382. 2007.
|