Introduction
Inhibitor of DNA binding (ID)-1 protein is a member
of the helix-loop-helix (HLH) protein family which is expressed in
actively proliferating cells. ID-1 regulates gene transcription by
heterodimerization with the basic HLH transcription factors, and
therefore inhibits them from DNA binding and transactivation of
their target genes (1). ID-1
functions mainly as a regulator in cellular differentiation of
muscle cells (1). ID-1 regulates
cellular senescence (2,3), induces cancer cell growth (4,5),
promotes cell survival as an oncogene (6) and invasion of cancer cells (7,8), and
up-regulates matrix metalloproteinase (MMP), which is necessary for
the breakdown of the basement membrane and extracellular matrix
that restrains tumor growth and metastasis. Expression of the
120-kDa MMP protein is strongly associated with the increase in
motility and invasiveness of mammary cells (7). Moreover, screening of breast cancer
cell lines revealed that expression of ID-1 as well as the 120-kDa
MMP protein was directly correlated with the invasiveness of these
cell lines (7), suggesting that
ID-1 regulates MMP protein as well as invasion. Furthermore,
down-regulating ID-1 expression through antisense or small
interfering RNA (siRNA) treatment suppressed the invasion and
metastatic ability of breast cancer cells (9,10).
ID-1 is frequently overexpressed in cancers of the breast (11), prostate (12), thyroid (13), pancreas (14) and esophagus (15). In addition, ID-1 is associated with
tumor stage and poor prognosis in prostate (16), breast (17), ovarian (18) and uterine cervical cancers
(19).
In a previous study, ID-1 overexpression was
associated with invasive behavior in uterine endometrial cancer
(20). However, the molecular
mechanism for this overexpression is not clear. This prompted us to
study the manner of expression of ID proteins in uterine
endometrial cancer according to clinical backgrounds with
angiogenic activity in tumors.
Materials and methods
Patients and tissues
Prior informed consent for the present study was
obtained from all patients and approval was given by the Research
Committee for Human Subjects, Gifu University School of Medicine.
Fifty patients ranging from 34 to 79 years of age with endometrioid
adenocarcinomas of the uterine endometrium [stage I, 20 cases;
stage II, 17 cases; stage III, 13 cases; well-differentiated
adenocarcinoma (G1), 22 cases; moderately differentiated
adenocarcinoma (G2), 11 cases; poorly differentiated adenocarcinoma
(G3), 17 cases] underwent hysterectomy at the Department of
Obstetrics and Gynecology, Gifu University School of Medicine,
between March 1996 and September 2004. Patient prognosis was
analyzed in relation to the 60-month survival rate. None of the
patients had received any pre-operative therapy before the uterine
endometrial cancer tissue was taken in surgery. A part of each
tissue of uterine endometrial cancers was snap-frozen in liquid
nitrogen and stored at −80°C to determine ID-1, ID-2 and ID-3 mRNA
levels, and those for immunohistochemistry were fixed with 10%
formalin and embedded in paraffin wax. The clinical stage of
uterine endometrial cancers was determined by the International
Federation of Obstetrics and Gynecology (FIGO) classification
(21).
Immunohistochemistry
Sections (4-μm) of formalin-fixed
paraffin-embedded tissue samples from uterine endometrial cancers
were cut with a microtome and dried overnight at 37°C on a
silanized slide (Dako, Carpinteria, CA, USA). The protocol of the
Universal Dako Labelled Streptavidin-Biotin kit (Dako, USA) was
followed for each sample. Samples were deparaffinized in xylene at
room temperature for 30 min, rehydrated with graded ethanol and
washed in phosphate-buffered saline (PBS). The samples were then
placed in 10 mM citrate buffer (pH 6.0) and boiled in a microwave
for 10 min for epitope retrieval. Endogenous peroxidase activity
was quenched by incubating tissue sections in 3%
H2O2 for 10 min. The primary antibodies,
rabbit anti-human ID-1 (SC-734; Santa Cruz Biotechnology Inc.,
Santa Cruz, CA, USA), mouse CD34 (Dako, Glostrup, Denmark) and
rabbit anti-factor VIII-related antigen (Zymed, San Francisco, CA,
USA) were used overnight at 4°C at dilutions of 1:50, 1:40 and 1:2,
respectively. The slides were washed, and biotinylated secondary
antibody (Dako, USA) was applied for 30 min after rinsing in PBS,
after which streptavidin-conjugated horseradish peroxidase (Dako,
USA) was added for 30 min. Slides were then washed and treated with
the chromogen 3,3′-diaminobenzidine (Dako, USA) for 5 min, rinsed
in PBS and counterstained with Mayer's haematoxylin, dehydrated in
graded ethanols, cleared in xylene and cover-slipped with a
mounting medium (Entellan New; Merck, Darmstadt, Germany). For
confirmation of the specificity for the ID-1 antigen, we also used
another ID-1 (SC-488) rabbit polyclonal antibody (Santa Cruz
Biotechnology Inc.), and we observed the exact identification of
staining for ID-1 expression in tumor cells as ID-1 (SC-734)
antibody. For the negative controls, the primary antibodies of
ID-1, CD34 and factor VIII-related antigen were omitted, and the
corresponding preimmune animal serums (rabbit, mouse and rabbit,
respectively) (Dako, USA) were used instead.
Assessment of histochemical score
(histoscore)
All sections for immunohistochemical staining for
ID-1 were evaluated in a semiquantitative fashion according to the
method described by McCarty et al (22), which considers both the intensity
and the percentage of cells stained in each of five intensity
categories. Intensities were classified as 0 (no staining), 1 (weak
staining), 2 (distinct staining), 3 (strong staining) and 4 (very
strong staining). For each stained section, a value-designated
histoscore was obtained by application of the following algorithm:
histoscore = ∑(i+1) × Pi, where i and Pi represent intensity and
percentage of cells that stain at each intensity, respectively, and
corresponding histoscores were calculated separately. Results were
assigned to four groups according to their overall scores: weak,
<160; distinct, 161–220; strong, 221–280; very strong,
>280.
Assessment of microvessel density
(MVD)
MVD was assessed with microvessel counts (MVCs) in
sequential tissue sections stained with mouse CD34 and rabbit
factor VIII-related antigen antibodies. Blood vessels with a
clearly defined lumen or a well-defined linear vessel shape, but
not single endothelial cells, were taken into account for
microvessel counting (23). Fives
areas of highest vascular density were chosen, and microvessel
counting was performed at ×200 magnification by two investigators.
MVCs were determined as the mean of the vessel counts obtained from
these fields (24).
Preparation of standard template for
real-time reverse transcription-polymerase chain reaction
(RT-PCR)
The internal standard template for real-time PCR was
produced by PCR amplification using the primers of the ID-1 gene,
418–782 in the cDNA (ID-1-TS, 5′-TTGGAGCTGAACTCGGAA-3′ and
ID-1-TAS, 5′-TCTCTGGTGACTAGTAGGT-3′); ID-2 gene, 907–1253 in the
cDNA (ID-2-TS, 5′-CTAAGCAGACTTT GCCTTT-3′ and ID-2-TAS,
5′-CTGAAATAAAGCAGGCA ATC-3′); ID-3 gene, 686–1009 in the cDNA
(ID-3-TS, 5′-GA ACTTGTCATCTCCAACGA-3′ and ID-3-TAS, 5′-CACGCT
CTGAAAAGACCT-3′). The DNA template was purified using a GeneClean
II kit (Qbiogene, Irvine, CA, USA). The copy numbers of the
standard template were determined to quantitate the ID-1, -2 and -3
mRNA level in samples for real-time RT-PCR.
Real-time RT-PCR to amplify ID-1, -2 and
-3 mRNAs
Total RNA was extracted with the acid guanidinium
thiocyanatephenol-chloroform method (25). Total RNA (3 μg) was reverse
transcribed using Moloney murine leukemia virus reverse
transcriptase (MMLV-RT, 200 U/μl; Invitrogen, Carlsbad, CA,
USA) and the following reagents: 250 mM Tris-HCl, pH 8.3, 375 mM
KCl, 15 mM MgCl2, 0.1 M dithiothreitol, 10 mM
deoxynucleotide (deoxyadenosine, deoxythymidine, deoxyguanosine and
deoxycystidine) tri-phosphates (dNTPs) mixture and random hexamers
(Invitrogen) at 37°C for 1 h. The reaction mixture was heated for 5
min at 94°C to inactivate MMLV-RTase.
The real-time PCR reaction was performed with a
Takara Premix Ex Taq (Perfect Real Time) R-PCR kit (Takara, Otsu,
Japan), using a smart cycler system (Cepheid, Sunnyvale, CA, USA).
The reaction solution (25 μl) contained Takara Premix Ex Taq
(2X), SYBR Green I (1:1000 dilution; CambrexBio Science, Rockland
Inc., Rockland, ME, USA) and 20 μM of the primers of the
ID-1 gene, 545–675 in the cDNA (ID-1-S, 5′-ACGATCGCATCTTGTGTC-3′
and ID-1-AS, 5′-CTTGTTC TCCCTCAGATCC-3′); ID-2 gene, 907-1026 in
the cDNA (ID-2-S, 5′-CTAAGCAGACTTTGCCTTT-3′ and ID-2-AS, 5′-C
ATTCAGTAGGCTTGTGTC-3′); ID-3 gene, 709–873 in the cDNA (ID-3-S,
5′-AAGGAGCTTTTGCCACTGA-3′ and ID-3-AS, 5′-CCAGGAAGGGATTTGGTGAA-3′)
with the transcribed total RNA from the tissue and a serially
diluted standard template. The real-time PCR reactions were
initially denatured by heating at 95°C for 30 sec, followed by 40
cycles consisting of denaturation at 94°C for 10 sec, annealing at
55°C for 5 sec and extension at 72°C for 20 sec. A strong linear
relationship between the threshold cycle and the log concentration
of the starting DNA copy number was always shown (correlation
coefficient >0.99). Quantitative analysis was performed to
determine the copy number of each sample.
Statistical analysis
ID-1, -2 and -3 mRNA levels were determined from
three parts taken from each tumor, and each sample was analyzed in
triplicate. ID-1 histoscores and mRNA levels were compared using
the Student's t-test. The 60-month survival rate was calculated
according to the Kaplan-Meier method and analyzed with the log-rank
test. The correlations between ID-1 histoscores and mRNA levels
with MVCs were performed with bivariate Pearson's correlation.
Differences were considered significant at p<0.05.
Results
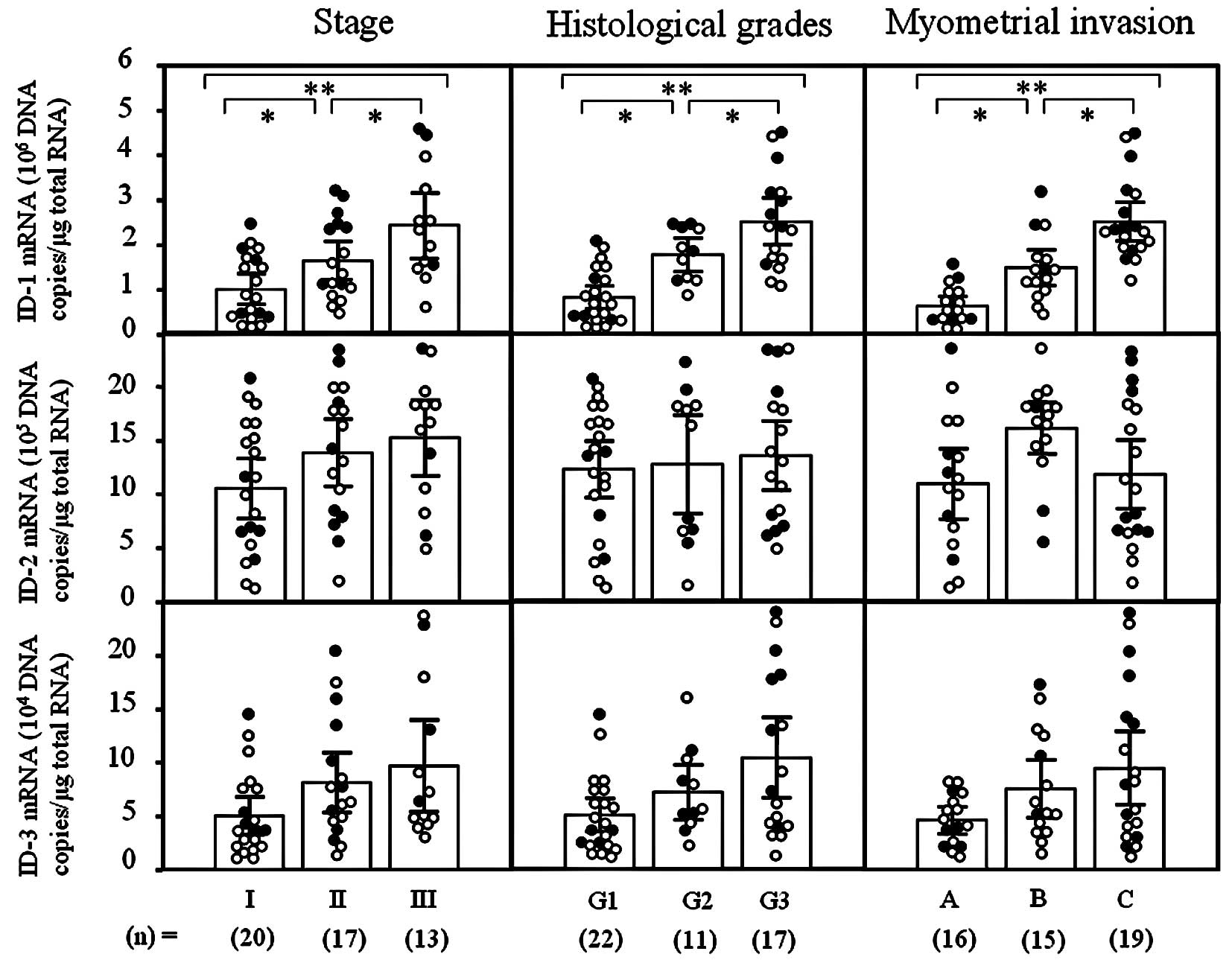
ID-1 mRNA levels significantly increased with
increasing clinical stages (I<II<III; p<0.05),
histological grades (G1<G2<G3; p<0.05) and depth of
myometrial invasion (A<B<C; p<0.05) of the uterine
endometrial cancers (Fig. 1).
However, there was no significant difference in ID-2 or -3 mRNA
levels according to clinical stage, histological grade or depth of
myometrial invasion in uterine endometrial cancers, as shown in
Fig. 1. These results prompted us
to concentrate our investigation on ID-1 in uterine endometrial
cancers.
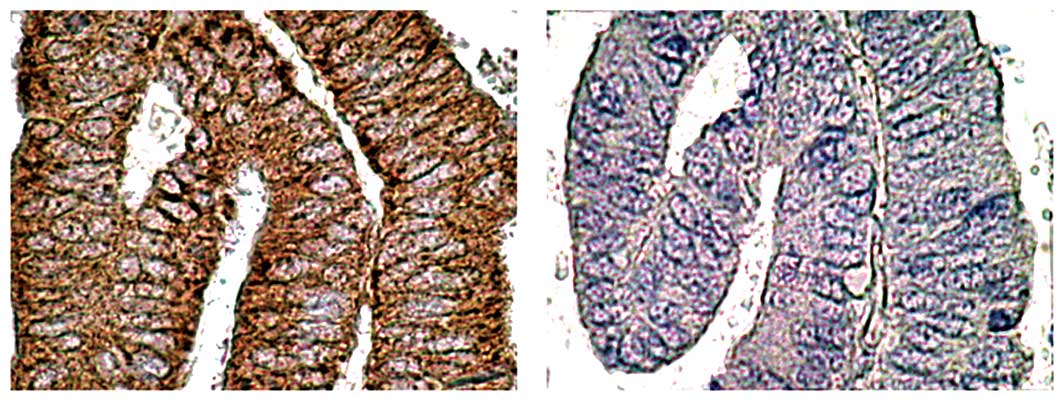
ID-1 staining was diffusely located in the cancer
cells (Fig. 2; a representative
case of well-differentiated endometrioid adenocarcinoma of the
uterine endometrium). Since ID-1 is not a transcription factor
per se, it lacks the nuclear localization signal found on
many basic HLH proteins but gives a cytoplasmic signal instead
(8,26). ID-1 diffuse cytoplasmic staining
from moderate to strong intensity was noted in most cases whereas
nuclear staining was observed only occasionally.
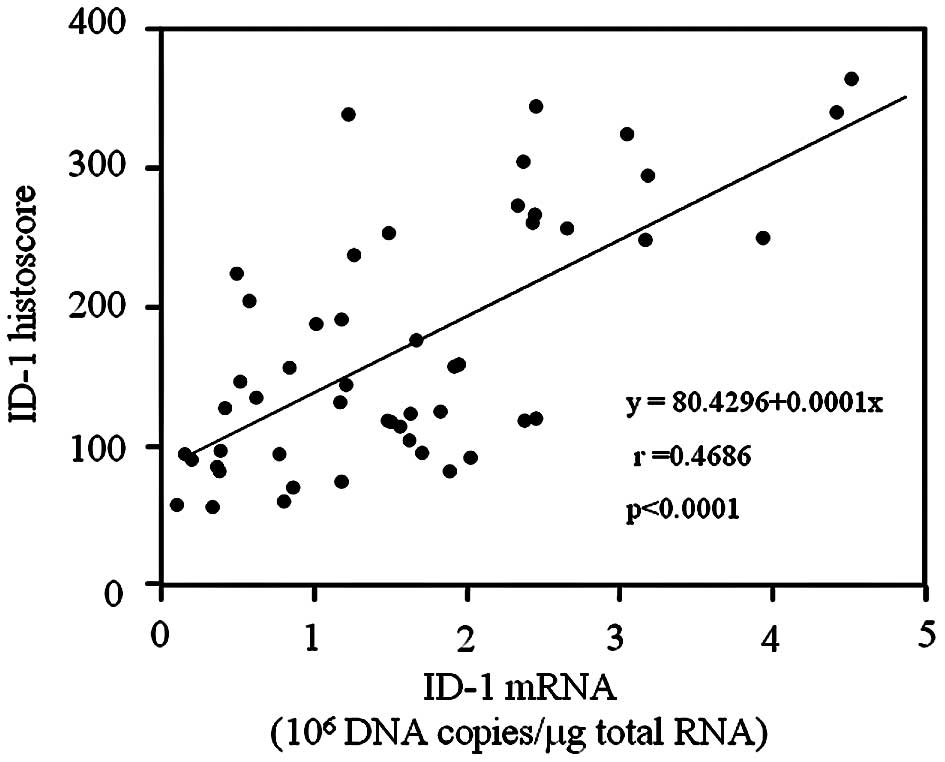
The ID-1 histoscore in cancer cells was
significantly (p<0.001) correlated with corresponding mRNA
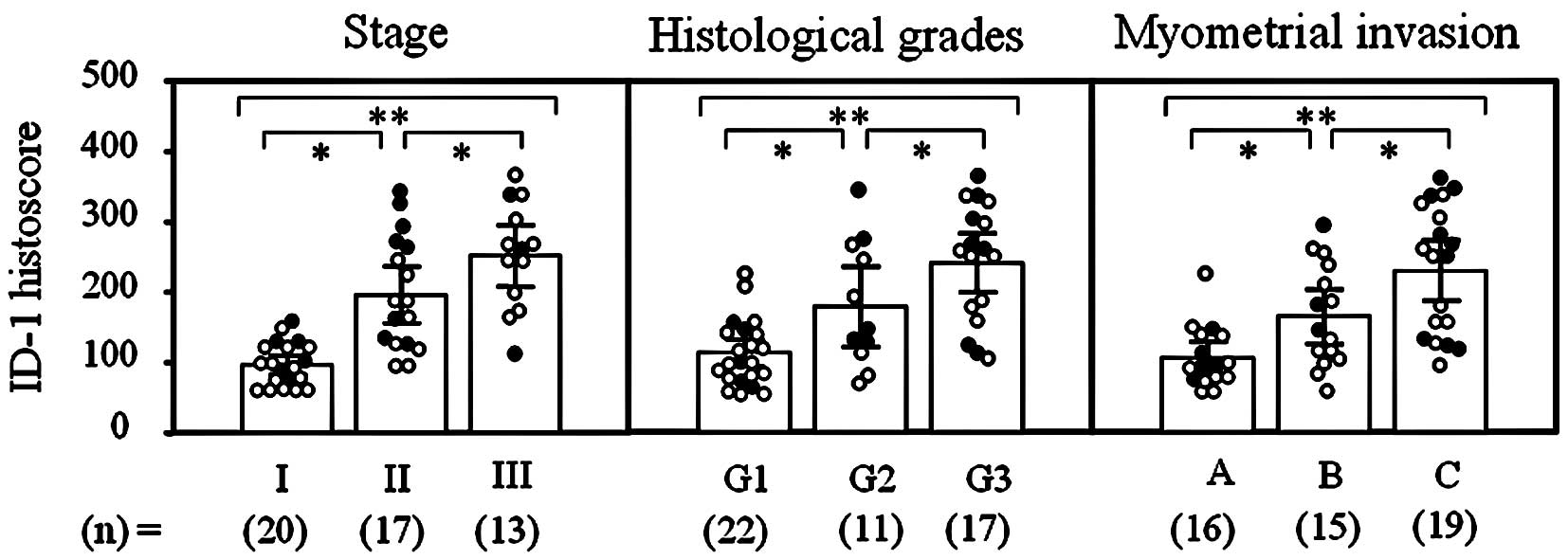
levels in each tissue, as shown in Fig. 3. Moreover, ID-1 histoscores
significantly (I<II<III; p<0.05) increased with increasing
clinical stages, histological grades (G1<G2<G3; p<0.05)
and depth of myometrial invasion (A<B<C; p<0.05) of the
uterine endometrial cancers (Fig.
4), as did ID-1 mRNA.
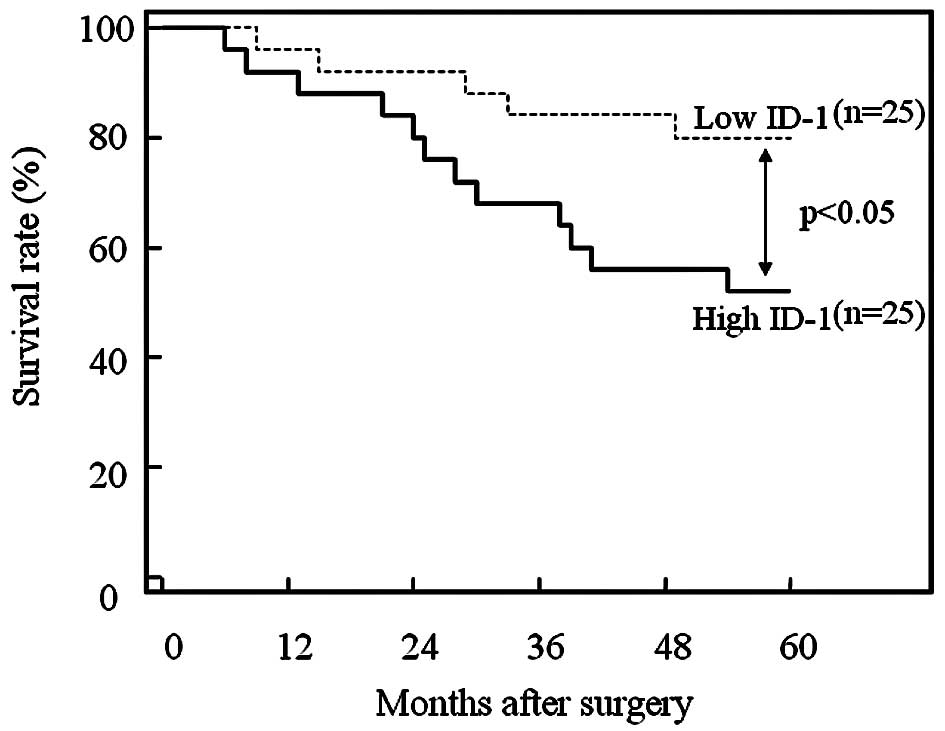
Furthermore, the 50 patients who underwent
hysterectomy were divided into two equal groups based on ID-1
histoscores and mRNA levels, with the midpoint being a histoscore
of 150 and mRNA of 1.5×106 copies/μg total RNA,
respectively. The two groups, determined independently by the ID-1
histoscores and mRNA levels, consisted of exactly the same
patients. The prognosis of the 25 patients with high ID-1 (>150
histoscore and >1.5×106 copies/μg total RNA)
in the uterine endometrial cancers was poor (52%), while the
60-month survival rate of the other 25 patients with low ID-1
(<150 histoscore and <1.5×106 copies/μg
total RNA) was higher (80%), as shown in Fig. 5.
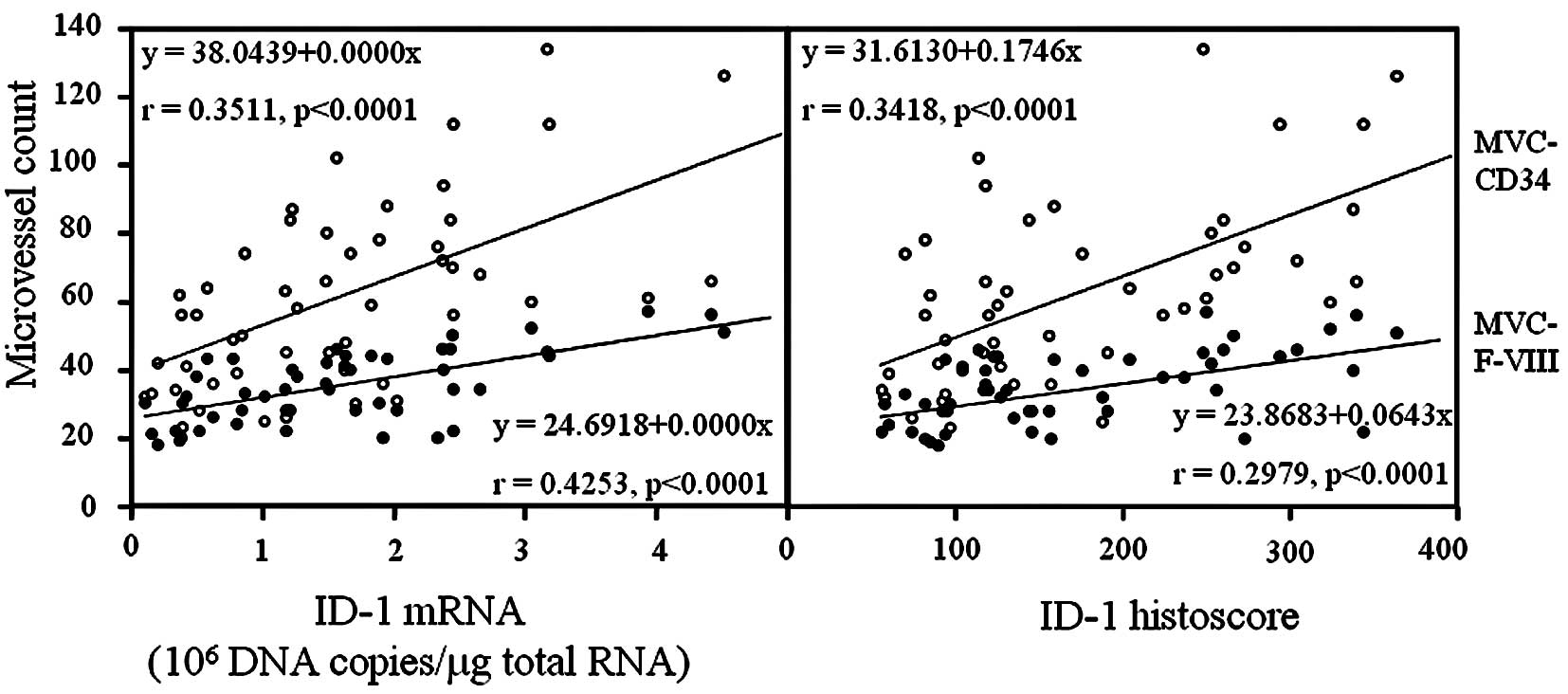
ID-1 expression was negative in endothelial cells,
although CD34 and factor VIII-related antigen expression was
strong. ID-1 histoscores significantly correlated with MVC-CD34
(MVCs determined by immunohistochemistry for CD34; r=0.3418,
p<0.0001) and MVC-F-VIII (MVCs determined by
immunohistochemistry for factor VIII-related antigen; r=0.2978,
p<0.0001); ID-1 mRNA levels were also correlated with MVC-CD34
(r=0.3511, p<0.0001) and MVC-F-VIII (r=0.4253, p<0.0001), as
shown in Fig. 6.
Discussion
In this study, ID-1 expression significantly
increased with more advanced clinical stage, histological grades
and depth of myometrial invasion. In addition, the patients with
high ID-1 expression had a lower survival rate compared with
patients with low ID-1 expression. Therefore, ID-1 contributes to
tumor progression and can be recognized as a novel indicator of
tumor advancement and patient prognosis in uterine endometrial
cancers. In a previous study, abundant ID-1 immunoreactivity was
found in endometrial carcinoma cells; ID-1 expression was
associated with histological grade and invasion to the myometrium
(20). However, no evidence is
available on the prognostic implications of ID-1 or the
relationship between ID-1 expression and angiogenesis in uterine
endometrial cancer.
Studies with ID-1 knock-out mice revealed that ID-1
expression is required, not only for normal angiogenesis, but is
also essential for tumor-associated angiogenesis during cancer
progression (27,28). Both VEGF and its receptor were
down-regulated in endothelial cells of ID-1/ID-3 double knock-out
mice (27), suggesting a possible
linkage between ID-1 and VEGF functioning. Accordingly, VEGF is a
downstream target of the ID-1 protein, and inactivation of ID-1 by
siRNA transfection, resulted in down-regulation of both VEGF gene
transactivation and protein secretion (29). As VEGF is one of the most potent
tumor angiogenic factors that activates, not only endothelial cell
proliferation, but also blood vessel formation, these findings
indicate a role of ID-1 in the induction of tumor angiogenesis. In
the present study, ID-1 expression correlated with microvessel
counts in endometrial cancers. Therefore, ID-1 might contribute to
angiogenesis related to the tumor progression of uterine
endometrial cancers.
In summary, this study demonstrates that ID-1
over-expression plays a role in tumor advancement via angiogenesis
and that ID-1 can be used as a prognostic indicator in uterine
endometrial cancers. Moreover, ID-1 might be a novel anticancer
target molecule for anti-angiogenic drug design in cancer
treatment.
References
|
1.
|
Benezra R, Davis RL, Lockshon D, Turner DL
and Weintraub H: The protein id: a negative regulator of
helix-loop-helix DNA binding proteins. Cell. 61:49–59. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Alani RM, Hasskarl J, Grace M, Hernandez
MC, Israel MA and Munger K: Immortalization of primary human
keratinocytes by the helix-loop-helix protein, id-1. Proc Natl Acad
Sci USA. 96:9637–9641. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Ohtani N, Zebedee Z, Huot TJ, Stinson JA,
Sugimoto M, Ohashi Y, Sharrocks AD, Peters G and Hara E: Opposing
effects of ets and id proteins on p16ink4a expression during
cellular senescence. Nature. 409:1067–1070. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ling MT, Wang X, Ouyang XS, Lee TK, Fan
TY, Xu K, Tsao SW and Wong YC: Activation of MAPK signaling pathway
is essential for id-1 induced serum independent prostate cancer
cell growth. Oncogene. 21:8498–8505. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Swarbrick A, Akerfeldt MC, Lee CS, Sergio
CM, Caldon CE, Hunter LJ, Sutherland RL and Musgrove EA: Regulation
of cyclin expression and cell cycle progression in breast
epithelial cells by the helix-loop-helix protein id1. Oncogene.
24:381–389. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Ling MT, Wang X, Ouyang XS, Xu K, Tsao SW
and Wong YC: Id-1 expression promotes cell survival through
activation of nf-kappab signalling pathway in prostate cancer
cells. Oncogene. 22:4498–4508. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Desprez PY, Lin CQ, Thomasset N, Sympson
CJ, Bissell MJ and Campisi J: A novel pathway for mammary
epithelial cell invasion induced by the helix-loop-helix protein
id-1. Mol Cell Biol. 18:4577–4588. 1998.PubMed/NCBI
|
|
8.
|
Lin CQ, Singh J, Murata K, Itahana Y,
Parrinello S, Liang SH, Gillett CE, Campisi J and Desprez PY: A
role for id-1 in the aggressive phenotype and steroid hormone
response of human breast cancer cells. Cancer Res. 60:1332–1340.
2000.PubMed/NCBI
|
|
9.
|
Fong S, Itahana Y, Sumida T, Singh J,
Coppe JP, Liu Y, Richards PC, Bennington JL, Lee NM, Debs RJ and
Desprez PY: Id-1 as a molecular target in therapy for breast cancer
cell invasion and metastasis. Proc Natl Acad Sci USA.
100:13543–13548. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu
W, Giri DD, Viale A, Olshen AB, Gerald WL and Massague J: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Jang TJ, Jung KH and Choi EA: Id-1 gene
downregulation by sulindac sulfide and its upregulation during
tumor development in gastric cancer. Int J Cancer. 118:1356–1363.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Coppe JP, Itahana Y, Moore DH, Bennington
JL and Desprez PY: Id-1 and id-2 proteins as molecular markers for
human prostate cancer progression. Clin Cancer Res. 10:2044–2051.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Kebebew E, Peng M, Treseler PA, Clark OH,
Duh QY, Ginzinger D and Miner R: Id1 gene expression is
up-regulated in hyperplastic and neoplastic thyroid tissue and
regulates growth and differentiation in thyroid cancer cells. J
Clin Endocrinol Metab. 89:6105–6111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Lee KT, Lee YW, Lee JK, Choi SH, Rhee JC,
Paik SS and Kong G: Overexpression of id-1 is significantly
associated with tumour angiogenesis in human pancreas cancers. Br J
Cancer. 90:1198–1203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Yuen HF, Chan YP, Chan KK, Chu YY, Wong
ML, Law SY, Srivastava G, Wong YC, Wang X and Chan KW: Id-1 and
id-2 are markers for metastasis and prognosis in oesophageal
squamous cell carcinoma. Br J Cancer. 97:1409–1415. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Ouyang XS, Wang X, Lee DT, Tsao SW and
Wong YC: Overexpression of id-1 in prostate cancer. J Urol.
167:2598–2602. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Jang KS, Han HX, Paik SS, Brown PH and
Kong G: Id-1 over-expression in invasive ductal carcinoma cells is
significantly associated with intratumoral microvessel density in
er-negative/node-positive breast cancer. Cancer Lett. 244:203–210.
2006. View Article : Google Scholar
|
|
18.
|
Schindl M, Schoppmann SF, Strobel T,
Heinzl H, Leisser C, Horvat R and Birner P: Level of id-1 protein
expression correlates with poor differentiation, enhanced malignant
potential and more aggressive clinical behavior of epithelial
ovarian tumors. Clin Cancer Res. 9:779–785. 2003.
|
|
19.
|
Maw MK, Fujimoto J and Tamaya T:
Expression of the inhibitor of DNA-binding (id)-1 protein as an
angiogenic mediator in tumour advancement of uterine cervical
cancers. Br J Cancer. 99:1557–1563. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Takai N, Miyazaki T, Fujisawa K, Nasu K
and Miyakawa I: Id1 expression is associated with histological
grade and invasive behavior in endometrial carcinoma. Cancer Lett.
165:185–193. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
International Federation of Obstetrics and
Gynecology (FIGO) News: Int J Gynecol Obstet, FIGO News.
28:189–193. 1989. View Article : Google Scholar
|
|
22.
|
McCarty KS Jr, Miller LS, Cox EB, Konrath
J and McCarty KS Sr: Estrogen receptor analyses. Correlation of
biochemical and immunohistochemical methods using monoclonal
antireceptor antibodies. Arch Pathol Lab Med. 109:716–721.
1985.
|
|
23.
|
Giatromanolaki A, Sivridis E, Brekken R,
Thorpe PE, Anastasiadis P, Gatter KC, Harris AL and Koukourakis MI:
The angiogenic ‘vascular endothelial growth factor/flk-1(kdr)
receptor’ pathway in patients with endometrial carcinoma:
prognostic and therapeutic implications. Cancer. 92:2569–2577.
2001.
|
|
24.
|
Maeda K, Chung YS, Ogawa Y, Takatsuka S,
Kang SM, Ogawa M, Sawada T, Onoda N, Kato Y and Sowa M: Thymidine
phosphorylase/platelet-derived endothelial cell growth factor
expression associated with hepatic metastasis in gastric carcinoma.
Br J Cancer. 73:884–888. 1996. View Article : Google Scholar
|
|
25.
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Maruyama H, Kleeff J, Wildi S, Friess H,
Buchler MW, Israel MA and Korc M: Id-1 and id-2 are overexpressed
in pancreatic cancer and in dysplastic lesions in chronic
pancreatitis. Am J Pathol. 155:815–822. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Lyden D, Young AZ, Zagzag D, Yan W, Gerald
W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K and Benezra
R: Id1 and id3 are required for neurogenesis, angiogenesis and
vascularization of tumour xenografts. Nature. 401:670–677. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Volpert OV, Pili R, Sikder HA, Nelius T,
Zaichuk T, Morris C, Shiflett CB, Devlin MK, Conant K and Alani RM:
Id1 regulates angiogenesis through transcriptional repression of
thrombospondin-1. Cancer Cell. 2:473–483. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Ling MT, Lau TC, Zhou C, Chua CW, Kwok WK,
Wang Q, Wang X and Wong YC: Overexpression of id-1 in prostate
cancer cells promotes angiogenesis through the activation of
vascular endothelial growth factor (VEGF). Carcinogenesis.
26:1668–1676. 2005. View Article : Google Scholar : PubMed/NCBI
|