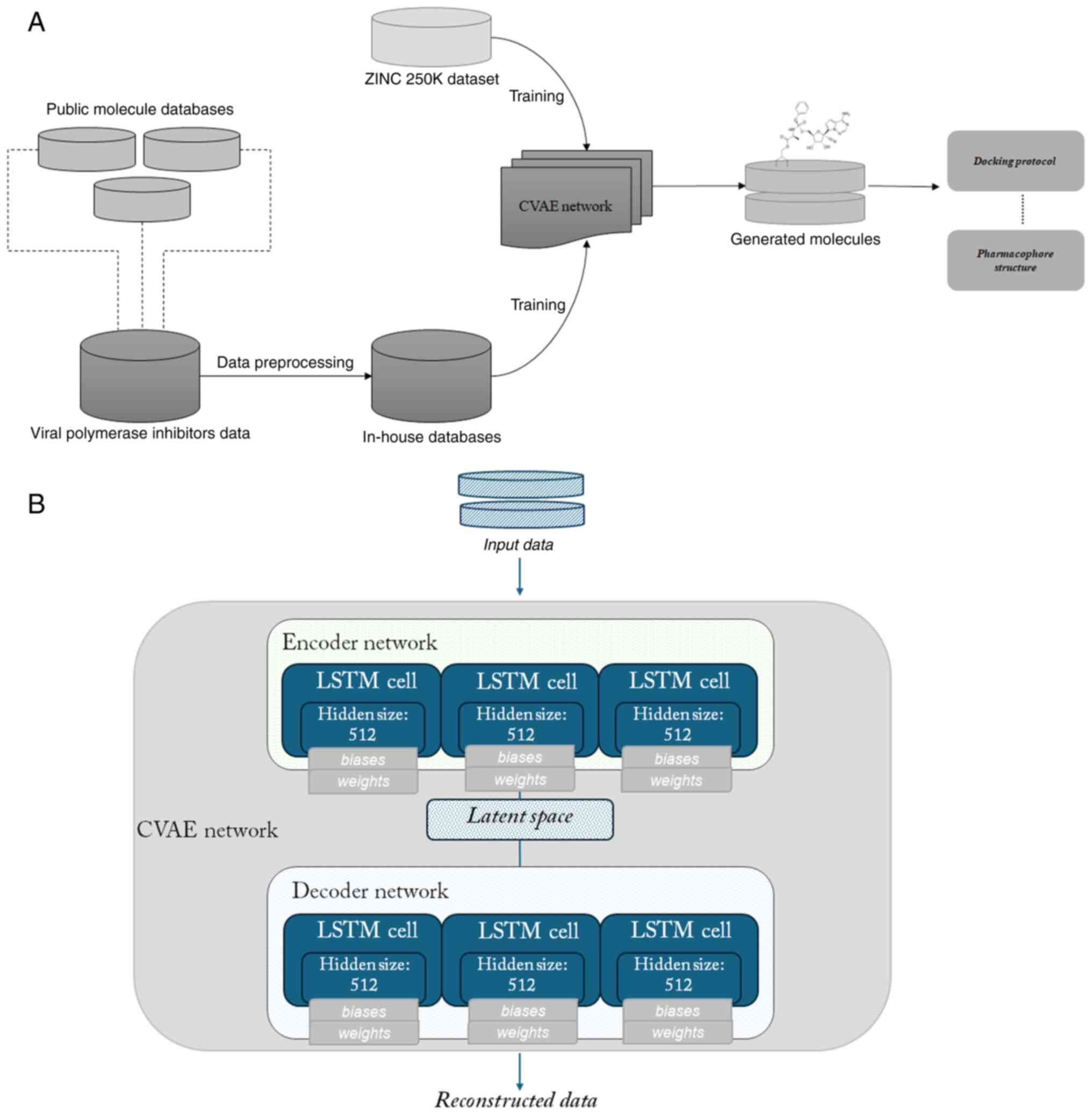
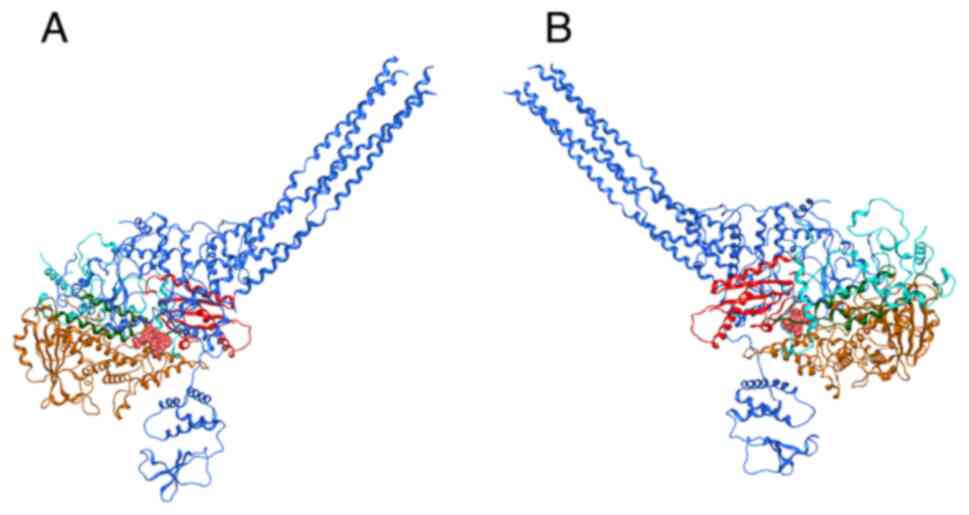
|
1
|
Clegg J and Lloyd G: Ebola haemorrhagic
fever in Zaire, 1995. Curr Opin Infect Dis. 8:225–228. 1995.
|
|
2
|
Muyembe-Tamfum JJ, Mulangu S, Masumu J,
Kayembe JM, Kemp A and Paweska JT: Ebola virus outbreaks in Africa:
Past and present. Onderstepoort J Vet Res. 79(451)2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zheng H, Yin C, Hoang T, He RL, Yang J and
Yau SS: Ebolavirus classification based on natural vectors. DNA
Cell Biol. 34:418–428. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ajelli M, Merler S, Fumanelli L, Pastore
Y, Piontti A, Dean NE, Longini IM Jr, Halloran ME and Vespignani A:
Spatiotemporal dynamics of the Ebola epidemic in Guinea and
implications for vaccination and disease elimination: A
computational modeling analysis. BMC Med. 14(130)2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Branda F, Mahal A, Maruotti A, Pierini M
and Mazzoli S: The challenges of open data for future epidemic
preparedness: The experience of the 2022 Ebolavirus outbreak in
Uganda. Front Pharmacol. 14(1101894)2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kucharski AJ and Edmunds WJ: Case fatality
rate for Ebola virus disease in west Africa. Lancet.
384(1260)2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Gostin LO and Friedman EA: A retrospective
and prospective analysis of the west African Ebola virus disease
epidemic: Robust national health systems at the foundation and an
empowered WHO at the apex. Lancet. 385:1902–1909. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Brolin Ribacke KJ, Saulnier DD, Eriksson A
and von Schreeb J: Effects of the West Africa ebola virus disease
on health-care utilization - A systematic review. Front Public
Health. 4(422)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Marí Saéz A, Weiss S, Nowak K, Lapeyre V,
Zimmermann F, Düx A, Kühl HS, Kaba M, Regnaut S, Merkel K, et al:
Investigating the zoonotic origin of the West African Ebola
epidemic. EMBO Mol Med. 7:17–23. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Plowright RK, Parrish CR, McCallum H,
Hudson PJ, Ko AI, Graham AL and Lloyd-Smith JO: Pathways to
zoonotic spillover. Nat Rev Microbiol. 15:502–510. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Koch LK, Cunze S, Kochmann J and Klimpel
S: Bats as putative Zaire ebolavirus reservoir hosts and their
habitat suitability in Africa. Sci Rep. 10(14268)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Osterholm MT, Moore KA, Kelley NS,
Brosseau LM, Wong G, Murphy FA, Peters CJ, LeDuc JW, Russell PK,
Van Herp M, et al: Transmission of ebola viruses: What We know and
what we do not know. mBio. 6(e00137)2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Velásquez GE, Aibana O, Ling EJ, Diakite
I, Mooring EQ and Murray MB: Time from infection to disease and
infectiousness for ebola virus disease, a systematic review. Clin
Infect Dis. 61:1135–1140. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bettini A, Lapa D and Garbuglia AR:
Diagnostics of ebola virus. Front Public Health.
11(1123024)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Verbeek JH, Rajamaki B, Ijaz S, Sauni R,
Toomey E, Blackwood B, Tikka C, Ruotsalainen JH and Kilinc Balci
FS: Personal protective equipment for preventing highly infectious
diseases due to exposure to contaminated body fluids in healthcare
staff. Cochrane Database Syst Rev. 4(CD011621)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ghosh S, Saha A, Samanta S and Saha RP:
Genome structure and genetic diversity in the Ebola virus. Curr
Opin Pharmacol. 60:83–90. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Judson S, Prescott J and Munster V:
Understanding ebola virus transmission. Viruses. 7:511–521.
2015.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Nanbo A, Imai M, Watanabe S, Noda T,
Takahashi K, Neumann G, Halfmann P and Kawaoka Y: Ebolavirus is
internalized into host cells via macropinocytosis in a viral
glycoprotein-dependent manner. PLoS Pathog.
6(e1001121)2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wolf J, Jannat R, Dubey S, Troth S,
Onorato MT, Coller BA, Hanson ME and Simon JK: Development of
pandemic vaccines: ERVEBO case study. Vaccines (Basel).
9(190)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Dolzhikova IV, Zubkova OV, Tukhvatulin AI,
Dzharullaeva AS, Tukhvatulina NM, Shcheblyakov DV, Shmarov MM,
Tokarskaya EA, Simakova YV, Egorova DA, et al: Safety and
immunogenicity of GamEvac-Combi, a heterologous VSV- and
Ad5-vectored Ebola vaccine: An open phase I/II trial in healthy
adults in Russia. Hum Vaccin Immunother. 13:613–620.
2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li Y, Wang L, Zhu T, Wu S, Feng L, Cheng
P, Liu J and Wang J: Establishing China's national standard for the
recombinant adenovirus type 5 vector-based ebola vaccine (Ad5-EBOV)
virus titer. Hum Gene Ther Clin Dev. 29:226–232. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Malik S and Waheed Y: Tracing down the
updates on Ebola virus surges: An update on anti-ebola therapeutic
strategies. J Transl Int Med. 11:216–225. 2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lee JS, Adhikari NKJ, Kwon HY, Teo K,
Siemieniuk R, Lamontagne F, Chan A, Mishra S, Murthy S, Kiiza P, et
al: Anti-Ebola therapy for patients with Ebola virus disease: A
systematic review. BMC Infect Dis. 19(376)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Taki E, Ghanavati R, Navidifar T, Dashtbin
S, Heidary M and Moghadamnia M: Ebanga™: The most recent
FDA-approved drug for treating Ebola. Front Pharmacol.
14(1083429)2023.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Saxena D, Kaul G, Dasgupta A and Chopra S:
Atoltivimab/maftivimab/odesivimab (Inmazeb) combination to treat
infection caused by Zaire ebolavirus. Drugs Today (Barc).
57:483–490. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tchesnokov EP, Feng JY, Porter DP and
Götte M: Mechanism of inhibition of Ebola virus RNA-Dependent RNA
polymerase by remdesivir. Viruses. 11(326)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sun D, Gao W, Hu H and Zhou S: Why 90% of
clinical drug development fails and how to improve it? Acta Pharm
Sin B. 12:3049–3062. 2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Vlachakis D: Genetic and structural
analyses of ssRNA viruses pave the way for the discovery of novel
antiviral pharmacological targets. Mol Omics. 17:357–364.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Papageorgiou L, Loukatou S, Sofia K,
Maroulis D and Vlachakis D: An updated evolutionary study of
Flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase
reveals novel invariable motifs as potential pharmacological
targets. Mol Biosyst. 12:2080–2093. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dara S, Dhamercherla S, Jadav SS, Babu CM
and Ahsan MJ: Machine learning in drug discovery: A review. Artif
Intell Rev. 55:1947–1999. 2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li W, Zhou Q, Liu W, Xu C, Tang ZR, Dong
S, Wang H, Li W, Zhang K, Li R, et al: A machine learning-based
predictive model for predicting lymph node metastasis in patients
with ewing's sarcoma. Front Med (Lausanne).
9(832108)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Dong S, Yang H, Tang ZR, Ke Y, Wang H, Li
W and Tian K: Development and Validation of a predictive model to
evaluate the risk of bone metastasis in kidney cancer. Front Oncol.
11(731905)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Davies M, Nowotka M, Papadatos G, Dedman
N, Gaulton A, Atkinson F, Bellis L and Overington JP: ChEMBL web
services: Streamlining access to drug discovery data and utilities.
Nucleic Acids Res. 43:W612–W620. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Papadatos G, Davies M, Dedman N, Chambers
J, Gaulton A, Siddle J, Koks R, Irvine SA, Pettersson J, Goncharoff
N, et al: SureChEMBL: A large-scale, chemically annotated patent
document database. Nucleic Acids Res. 44:D1220–D1228.
2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pickett BE, Sadat EL, Zhang Y, Noronha JM,
Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, et al: ViPR:
An open bioinformatics database and analysis resource for virology
research. Nucleic Acids Res. 40:D593–D598. 2012.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kim S, Chen J, Cheng T, Gindulyte A, He J,
He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, et al: PubChem in
2021: New data content and improved web interfaces. Nucleic Acids
Res. 49:D1388–D1395. 2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Polanski J and Gasteiger J: Computer
Representation of Chemical Compounds. Springer International
Publishing, pp1997-2039, 2017.
|
|
39
|
Silva Y, Almeida I and Queiroz M: SQL:
From Traditional Databases to Big Data. SIGCSE. 413–418. 2016.
|
|
40
|
Landrum G: RDKit. Open-Source
Cheminformatics Software. 2010. Available from: https://www.rdkit.org.
|
|
41
|
Goodfellow I, Pouget-Abadie J, Mirza M, Xu
B, Warde-Farley D, Ozair S, Courville A and Bengio Y: Generative
Adversarial Networks. Vol 27. Advances in Neural Information
Processing Systems, 2014.
|
|
42
|
Kingma DP and Welling M: An introduction
to variational autoencoders. ArXiv: Dec 11, 2019 (Epub ahead of
print).
|
|
43
|
Abadi M, Barham P, Chen J, Chen Z, Davis
A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al:
TensorFlow: A system for large-scale machine learning. ArXiv: May
31, 2016 (Epub ahead of print).
|
|
44
|
Hodson TO: Root-Mean-Square Error (RMSE)
or Mean Absolute Error (MAE): When to use them or not.
Geoscientific Model Development. Vol 15. European Geosciences
Union, pp5481-5487, 2022.
|
|
45
|
DiPietro R and Hager G: Deep learning:
RNNs and LSTM. pp503-519, 2020.
|
|
46
|
Akhmetshin T, Lin A, Mazitov D, Ziaikin E,
Madzhidov T and Varnek A: HyFactor: Hydrogen-count labelled
graph-based defactorization Autoencoder. Comput Theor Chem: Dec 6,
2021 (Epub ahead of print).
|
|
47
|
Chemical Computing Group: Computer-Aided
Molecular Design. Molecular Operating Environment (MOE). 2024.
Available from: https://www.chemcomp.com/en/Products.htm.
|
|
48
|
Szymański P, Markowicz M and
Mikiciuk-Olasik E: Adaptation of high-throughput screening in drug
discovery-toxicological screening tests. Int J Mol Sci. 13:427–452.
2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Giordano D, Biancaniello C, Argenio MA and
Facchiano A: Drug design by pharmacophore and virtual screening
approach. Pharmaceuticals (Basel). 15(646)2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Prasanna S and Doerksen RJ: Topological
polar surface area: A useful descriptor in 2D-QSAR. Curr Med Chem.
16:21–41. 2009.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Leung DW, Prins KC, Basler CF and
Amarasinghe GK: Ebolavirus VP35 is a multifunctional virulence
factor. Virulence. 1:526–531. 2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yuan B, Peng Q, Cheng J, Wang M, Zhong J,
Qi J, Gao GF and Shi Y: Structure of the Ebola virus polymerase
complex. Nature. 610:394–401. 2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kalinowsky L, Weber J, Balasupramaniam S,
Baumann K and Proschak E: A diverse benchmark based on 3D matched
molecular pairs for validating scoring functions. ACS Omega.
3:5704–5714. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Leon M, Perezhohin Y, Peres F, Popovič A
and Castelli M: Comparing SMILES and SELFIES tokenization for
enhanced chemical language modeling. Sci Rep.
14(25016)2024.PubMed/NCBI View Article : Google Scholar
|
|
55
|
van Tilborg D and Grisoni F: Traversing
chemical space with active deep learning for low-data drug
discovery. Nat Comput Sci. 4:786–796. 2024.PubMed/NCBI View Article : Google Scholar
|