Introduction
In 2020, breast cancer in females emerged as the
global cancer burden, accounting for ~2.3 million newly diagnosed
cases, which constituted 11.7% of all reported cancer cases.
Worldwide, there were 685,000 deaths recorded due to breast cancer,
ranking breast cancer 5th as regards cancer-related mortality
globally (1). In 2020, in India,
there were a total of 179,790 reported cases of breast cancer and
this constituted 10% of all cancer cases in that year. In urban
India, breast cancer among females is the most common malignancy.
The National Program of Cancer Registries (NPCR) reported an annual
percentage change (APC) of 0.68% for cancers at all sites and an
APC of 2% for breast cancer during the 3-year period from
2011-2014(2).
The risk factors for breast cancer are old age, late
menopause, early menarche, obesity, smoking, alcohol, a family
history of the disease, hormone-replacement-therapy, the use of
oral contraceptives and nulliparity. A maternal age of >30 years
at the first live birth is associated with a higher risk and <20
years with a lower risk of developing the disease. The most common
genes involved are p53 and BRCA1; 85% of breast cancers are
sporadic, while 10-15% are hereditary. Luminal-A breast cancer
(estrogen receptor-positive, and progesterone receptor-positive and
Her2neu-negative) is most common type and is associated with the
optimal prognosis. Basal-triple-negative breast cancer (TNBC) is
associated with outcomes and high chances of visceral metastasis
(3). Mutations of BRCA predispose
to hereditary breast and ovarian cancer syndrome (HBOC). HBOC
syndrome requires that all primary relatives are counseled and
tested for BRCA gene mutations (4).
The early diagnosis and prompt management of breast
cancer will eventually greatly reduce the morbidity and mortality.
Currently, the majority of cases are diagnosed using the following
techniques: Biopsy, mammography, magnetic resonance imaging,
ultrasonography, computerized tomography and positron emission
tomography (5).
Tissue gene biomarkers improve the diagnosis of
breast cancer to a great extent; however, their applications are
limited as sample collection is invasive, and there is a risk of
hemorrhage and the unpleasant nature of diagnostic procedures.
Thus, there is need for non-invasive marker or minimally-invasive
markers which are highly sensitive and specific, and which can be
used to detect early-stage breast cancer during screening, thus
aiding in diagnosis and prognosis (6).
MicroRNAs (miRNAs/miRs) are short oligonucleotides
(20-25 nucleotides in length), non-coding sequences. They regulate
the expression of post-transcriptional target genes negatively by
silencing mRNA translation and inducing mRNA degradation. miRNAs
contain complementary sequences with their respective target gene
promoter region and complementary sequences with mRNA transcript
coding sequences. The RNA-induced silencing complex (RISC) is
present in mRNA transcripts both on the 3' and 5'untranslated
regions. miRNAs recruit RISC to complementary sites and regulates
the target gene expression (7). On
the target genes, the miRNAs have an increased control on
amplification and decreased control on its deletion. Oncosuppressor
miRNAs inhibit the genes which promote cell growth and onco-miRNAs
increase cell proliferation by downregulating tumor suppressor
genes and promoting apoptosis (8).
Of note, >1,200 miRNAs have been identified to be
strongly associated with breast cancer; however, not one has been
translated for use in clinical diagnostics due to a lack of
validation and quality assessment (9). Several studies have been performed to
assess the specificity and sensitivity of miRNAs as screening tools
for the detection of early-stage breast cancer (10,11).
miR-27a has been shown to be associated with tumor proliferation
and metastasis, and its expression is elevated in the circulation
in the presence of breast cancer, suggesting that it may serve as
an early detection marker (12,13).
miRNA-122 facilitates breast cancer metastasis by preparing fuel
utilization in the premetastatic niche and has been tested as a
screening biomarker for early-stage BC (14-16).
Higher circulating levels of miR-155 in patients with breast cancer
are attributed to its dysregulation of tumor suppressors and its
involvement in the crosstalk of genetic and epigenetic messages for
malignant transformations in BC (17,18).
With the research performed over the past two
decades on breast cancer, circulating miRNA levels in blood serve
as minimally invasive or non-invasive biomarkers (6). Recently, reverse
transcription-quantitative PCR (RT-qPCR) was widely used in
detecting circulating miRNAs. It is a highly sensitive and requires
a minimal amount of RNA input (19). The present study aimed to
investigate the diagnostic role of circulating miRNAs (miR-27a,
-122 and 155) in detecting early-stage breast cancer and to compare
the assayed levels with those found in women with benign breast
tumors and healthy controls.
Materials and methods
Ethics approval and study
participants
Ethical clearance obtained from the Institutional
Ethics Committee (IEC) of All India Institute of Medical Sciences
(AIIMS), Bhubaneswar, India (IEC/AIIMS BBSR/PG Thesis/2021-22/32).
After obtaining written informed consent, blood samples were
collected from the study participants (from August, 2021 to March,
2023). The study was conducted on individuals visiting the Surgery
or Onco-Surgery Outpatient Department in our tertiary care
hospital, All India Institute of Medical Sciences, Bhubaneswar,
India. All patients resided in Eastern India, namely from the
states of Odisha and West Bengal. They were all of South Asian
Pacific ethnicity. The participants were classified into three
groups as follows: Group 1, included cases with early-stage breast
cancer; group 2, included cases with benign breast tumors; and
group 3, included healthy age-matched females with no breast lumps
or any other tumor or comorbidities.
The study sample size was calculated on the basis of
the results of the study by Swellam et al (13), in which the presence of the miRNA
marker was in 90% of the cases and in 10% of the controls. With an
α error of 0.05 and 80%, the sample size calculated was 56 in each
group, namely 56 healthy controls, 56 benign breast tumor cases and
56 early-stage breast cancer cases that had stage I, stage IIa and
stage IIb disease.
According to the study design, the inclusion
criteria were women within the age group of 20-60 years, women who
had early-stage breast-cancer or benign breast tumors, confirmed by
a FNAC/biopsy report, who had not received any intervention and had
not reported any other malignancies. Blood samples were collected
from all participants in the study from August, 2021 to March, 2023
and serum was separated. A total of 1 ml serum was used for
biochemical analyses (liver and kidney function tests) on the AU
5800 auto analyzer (Beckman Coulter, Inc.), using reagents provided
by the same vendor. Another 1 ml serum was stored in RNase free
aliquots at -80˚C till the extraction of RNA was performed.
miRNA extraction
miRNA was extracted from the serum samples using the
miRNeasy Serum/Plasma kit (cat. no. 217184, Qiagen, Inc.),
according to manufacturer's instructions; all procedures were
performed in RNase free conditions. Finally, elution with 14 µl
RNase-free water resulted into a 12-µl eluate. The purity of the
extracted miRNAs was detected using a Nanodrop spectrophotometer
(Nano Bio-analytical Technologies Limited).
First-strand cDNA synthesis
This protocol involves the conversion of total miRNA
into cDNA by reverse transcription. First-strand cDNA synthesis
reactions were carried out using the miRCURY LNA miRNA PCR Starter
kit (cat. no. 339320, Qiagen, Inc.) and miRCURY LNA RT kit (cat.
no. 339340, Qiagen, Inc.). First the volume of template RNA was
calculated using the following formula: [Template RNA (µl)=elution
volume (µl)/original sample volume (µl)x16 (µl)]. Thus, for a 12-µl
elution volume and a 100-µl original sample volume, the template
RNA volume was 1.92 µl. As recommended in the manufacturer's
instructions, the total volume of 10 µl reverse transcription
reaction components were as follows: 2 µl 5X miRCURY
SYBR®-Green RT Reaction Buffer, 1 µl 10X miRCURY RT
Enzyme Mix, 0.5 µl UniSp6 RNA spike-in, 1.92 µl template miRNA and
4.58 µl RNase-free water. PCR tubes with the 10-µl total reaction
volume were placed in thermal cycler (Eppendorf) and incubated for
60 min at 42˚C for the reverse transcription step, for 5 min at
95˚C to inactivate reverse transcriptase and finally cooled to 4˚C
immediately. PCR tubes were stored at 4˚C until quantitative PCR
was performed.
Quantitative PCR
Quantitative PCR was carried using the miRCURY LNA
miRNA PCR Starter kit (cat. no. 339320, Qiagen, Inc.) and the
miRCURY LNA SYBR®-Green PCR kit (cat. no, 339346,
Qiagen, Inc.) on a real-time thermal cycler qTOWER³ (Analytik Jena
India Private Limited). First, the cDNA volume (10 µl total RT
reaction volume) was diluted to 1:30 by the addition of 290 µl
RNase-free water to a 10-µl RT reaction prior to use. The primers
of the miRCURY LNA miRNA PCR assays used were for miR-16, miR-27a,
miR-122 and miR-155. All reactions were carried out in duplicate.
The reaction mixture and real-time cycler program were set up as
per the instructions of the manufacturer. The florescence was
detected and cycle-threshold (Cq) values were obtained. In the
present study, miR-16 was used as an endogenous control to
normalize the expression levels of miR-27a, miR-122 and
miR155(20). Data were analyzed
using the comparative ∆∆Cq method (2-∆∆Cq) (21). The data of miR-27a, miR-122 and
miR155 expression levels were calculated as follows: ∆∆Cq=Cq
(target miR)-[Cq (target miR)-Cq (miR-16)] mean of controls. The
relative quantification was calculated using the 2-∆∆Cq
method for all miRNAs (miR-27a, miR-122 and miR-155) (Fig. 1).
Statistical analysis
Data analysis was performed using SPSS software
(SPSS software version 29.0.1.0; IBM Corp.). The general
characteristics of the cases and controls were compared using the
t-test; the t-test was used for the comparison of continuous
variables and the Chi-squared was used for categorical data where
the data could be placed in a 2x2 table representation. The
Kruskal-Wallis-H test was used to compare differences in the serum
levels of miRNAs among the three groups (patients with early-stage
breast cancer, benign breast tumors and healthy controls). Dunn's
post hoc test was used following the Kruskal- Wallis test. The
Mann-Whitney test was used for those continuous variables which did
not have normal distribution of data as demonstrated by the
Shapiro-Wilk test. In all analyses, a value of P<0.05 was
considered to indicate a statistically significant difference. For
assessment of the diagnostic potential of miRNAs, receiver
operating characteristic (ROC) curve analysis was performed and the
area under the ROC curve (AUC) was calculated. The sensitivity,
specificity, negative predictive value (NPV), positive predictive
value (PPV) and likelihood ratios (LRs) for differentiating between
early-stage breast cancer, benign breast tumors and healthy control
were also calculated.
Results
Expression levels of miRNA-27a, -122
and -155 in the study samples
The present study included 112 cases (56 patients
with early-stage breast cancer and 56 patients with benign breast
tumors) and 56 healthy controls to determine the circulating levels
of miRNA-27a, -122 and -155.
The comparison of the age, parity, menstrual status
and body mass index revealed that there were no statistically
significant differences between the cases and healthy controls
(P-value >0.05); however, statistically significant differences
were found in family history and personal habits (tobacco intake)
between the cases and healthy controls (P-value <0.05).
Biochemical parameters, such as alanine transaminase (ALT),
aspartate aminotransferase (AST), alkaline phosphatase (ALP), urea,
creatinine, hemoglobin (Hb), total white blood cell (WBC) and
platelet count exhibited no statistically significant differences
between the cases and healthy controls (P-value >0.05) (Table I).
 | Table IGeneral examination parameters and
biochemical parameters. |
Table I
General examination parameters and
biochemical parameters.
| Parameter | Cases (n=112) | Controls (n=56) | P-value |
|---|
| Age group | | | P=0.860c |
|
<46
years | 78 (69.6%) | 38 (67.9%) | |
|
≥46
years | 34 (30.4%) | 18 (32.1%) | |
| Age (years) | 37.43±12.51 | 40.5±10.6 | P=0.099
c |
| Parity | | | P=0.589 |
|
Nullipara | 33 (29.5%) | 14 (25%) | |
|
Para | 79 (70.5%) | 42 (75%) | |
| Menstrual
status | | | P=0.721 |
|
Premenopausal | 32 (28.6%) | 18 (32.1%) | |
|
Postmenopausal | 80 (71.4%) | 38 (67.9%) | |
| BMI | 23.28±4.15 | 22.83±2.91 | P=0.416
c |
| Family history | | | P=0.032
c |
|
Negative | 102 (91%) | 56 (100%) | |
|
Positive | 10 (9%) | 0 | |
| Personal habits:
Tobacco intake | | | P=0.03
c |
|
No | 103 (92%) | 56 (100%) | |
|
Yes | 9 (8%) | 0 | |
| ALT (U/l) mean ±
2SD | 26±11.25 | 27.1±8.5 |
P=0.514t |
| AST (U/l) mean ±
2SD | 23.82±14.68 | 27.29±10.39 | P=0.08
t |
| ALP (U/l) mean ±
2SD | 83.7±19.32 | 80.8±18.13 | P=0.342
t |
| Urea(mg/dl) mean ±
2SD | 20.19±5.98 | 20.77±5.95 | P=0.557
t |
| Creatinine (mg/dl)
mean ± 2SD | 0.79±0.13 | 0.83±0.114 | P=0.108
t |
| Hb (g/dl) mean ±
2SD | 11.74±14.68 | 11.79±14.68 | P=0.735
t |
| WBC
(x103/µl) mean ± 2SD | 8.14±2.26 | 7.93±1.91 | P=0.548
t |
| Platelets
(x103/µl) mean ± 2SD | 268.38±79.5 | 262.29±66.3 | P=0.601
t |
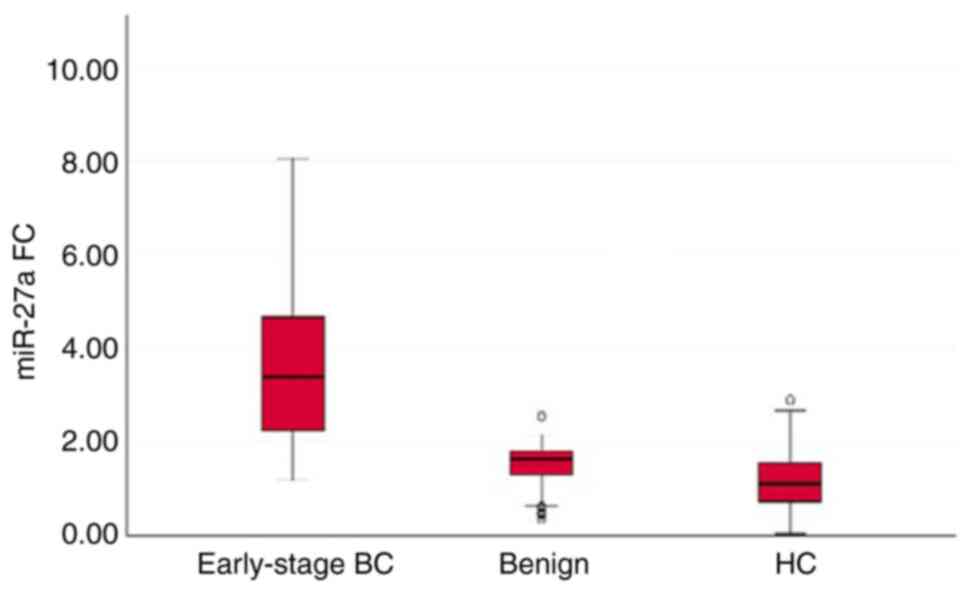
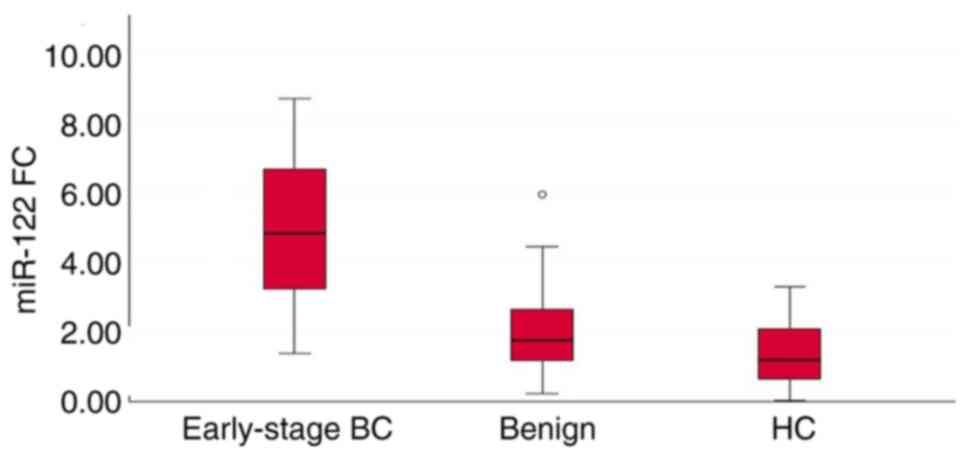
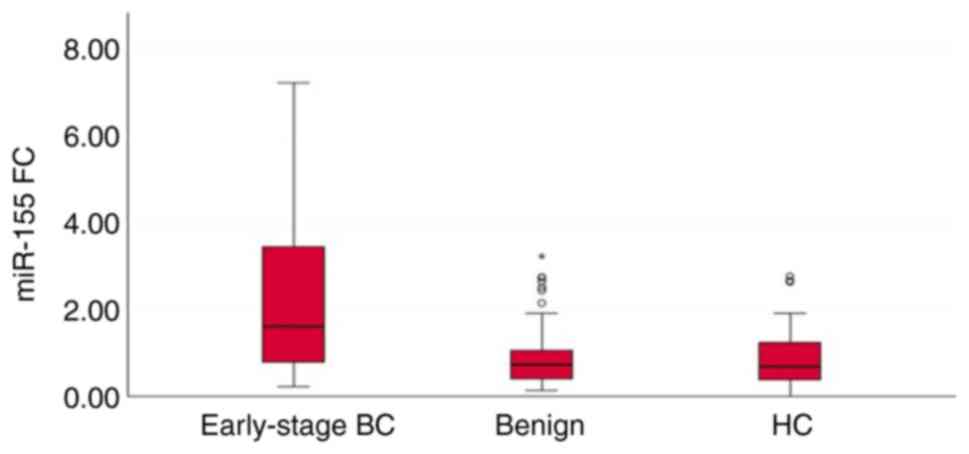
The results revealed that the expression levels of
the three miRNAs (miR-27a, miR-122 and miR-155) in serum were
significantly increased in patients with early-stage breast cancer
when compared to those of patients with benign breast tumors and
healthy controls (P<0.001). There was a significant difference
in the serum levels of miR-27a (P=0.015) and miR-122 (P=0.028),
between the patients with benign breast tumors and healthy
controls; the serum levels of these two miRNAs were upregulated in
patients with benign breast tumors when compared to those of the
healthy controls. However, there was no significant difference in
the serum levels of miR-155 (P=0.935) between the healthy controls
and patients with benign breast tumors (Table II and Figs. 2, 3
and 4). It was also found that the
miRNA-16 expression levels were relatively constant (mean, 1.4586;
SD, 0.08; data not shown).
 | Table IISerum levels of miR-27a, -122 and
-155 in patients with early-stage breast cancer, patients with
benign breast tumors and healthy controls. |
Table II
Serum levels of miR-27a, -122 and
-155 in patients with early-stage breast cancer, patients with
benign breast tumors and healthy controls.
| A, Based on the
total study population (n=168) |
|---|
| Variables | miR-27a | P-value | miR-122 | P-value | miR-155 | P-value |
|---|
| Early-stage breast
cancer (n=56) | 3.75±1.98, 3.3995
(1.19-8.07) |
<0.001a,b,c | 4.94±2.07, 4.8356
(1.37-8.71) |
<0.001a,b,c | 2.22±1.77, 1.6279
(0.22-7.23) |
<0.001a,b |
| Benign breast
tumors (n=56) | 1.52±0.49, 1.6566
(0.38-2.56) | | 1.97±1.19, 1.7683
(2.23-5.95) | | 0.95±0.77, 0.7364
(0.15-3.24) | |
| Healthy controls
(n=56) | 1.22±0.69, 1.1175
(0.08-2.92) | | 1.36±0.87, 1.2031
(0.05-3.29) | | 0.91±0.67, 0.6949
(0.02-2.78) | |
| B, Based on age
status (years) |
| Variables | miR-27a | P-value | miR-122 | P-value | miR-155 | P-value |
| <46 years | | | | | | |
|
Early-stage
breast cancer (n=32) | 3.10±1.92, 2.5137
(1.19-8.00) |
<0.001a,b | 4.26±1.95, 4.0168
(1.37-8.71) |
<0.001a,b,c | 1.43±1.21, 1.1016
(0.22-4.87) | 0.172 |
|
Benign
breast tumors (n=46) | 1.53±0.49, 1.6567
(0.38-2.56) | | 1.93±1.19, 1.6457
(0.23-5.95) | | 1.02±0.82, 0.7364
(0.15-3.24) | |
|
Healthy
controls (n=56) | 1.32±0.69, 1.2012
(0.14-2.92) | | 1.52±0.73, 1.3465
(0.32-2.94) | | 1.07±0.72, 0.7789
(0.21-2.78) | |
| ≥46 | | | | | | |
|
Early-stage
breast cancer (n=24) | 4.62±1.75, 3.9969
(1.54-8.07) |
<0.001a,b,c | 5.85±1.89, 6.5157
(1.39-8.15) |
<0.001a,b,c | 3.26±1.86, 2.6926
(0.55-7.24) |
<0.001a,b,c |
|
Benign
breast tumors (n=10) | 1.47±0.55, 1.7148
(0.51-1.91) | | 2.19±2.21, 2.0998
(0.36-4.09) | | 0.63±0.35, 0.6560
(0.22-1.38) | |
|
Healthy
controls (n=18) | 1.00±0.64, 1.0235
(0.08-2.56) | | 1.01±1.05, 0.5133
(0.05-3.29) | | 0.58±0.42, 0.5117
(0.02-1.39) | |
| C, Based on
menstrual status |
| Variables | miR-27a | P-value | miR-122 | P-value | miR-155 | P-value |
| Premenopausal | | | | | | |
|
Early-stage
breast cancer (n=29) | 2.74±1.51, 2.3347
(1.19-6.99) |
<0.001a,b,c | 3.99±1.82,3 .5368
(1.37-8.71) |
<0.001a,b,c | 1.16±0.89, 0.8317
(0.22-4.82) | <0.412 |
|
Benign
breast tumors (n=51) | 1.54±0.48, 1.6662
(0.38-2.56) | | 2.01±1.19, 1.8169
(0.23-5.95) | | 0.98±0.71, 0.7406
(0.15-3.24) | |
|
Healthy
controls (n=38) | 1.35±0.67, 1.2012
(0.41-2.92) | | 1.51±0.71, 1.3465
(0.32-2.94) | | 1.07±0.70, 0.8378
(0.21-2.78) | |
|
Early-stage
breast cancer (n=27) | 4.84±1.87, 4.0706
(1.54-8.07) |
<0.001a,b,c | 5.95±1.84, 6.5415
(1.39-8.25) |
<0.001a,b,c | 3.36±1.77, 3.0635
(1.24-7.24) |
<0.001a,b |
|
Benign
breast tumors (n=5) | 1.30±0.66, 1.6375
(0.51-1.90) | | 1.62±1.29, 1.7196
(0.36-3.48) | | 0.64±0.47, 0.5599
(0.22-1.38) | |
|
Healthy
controls (n=18) | 0.95±0.67, 0.8877
(0.08-2.56) | | 1.03±1.08, 0.5133
(0.05-3.29) | | 0.58±0.46, 0.4451
(0.02-1.75) | |
To examine the effect of aging and menstrual status
on the serum levels of miR-27a, miR-122 and miR-155, the samples
were classified into different groups. In the group aged <46
years and in the premenopausal group, the serum levels of miR-27a
and miR-122 were significantly increased in the patients with
early-stage breast cancer when compared to those with benign breast
tumors and the healthy controls (P<0.01); however, there was no
significant difference between the patients with benign breast
tumors and the healthy controls (miR-27a, P>0.05; and miR-122,
P>0.05). The serum levels of miR-155 exhibited no significance
difference among all three groups (P>0.05). In the group aged
≥46 years and in the postmenopausal group, the serum levels of all
three miRNAs were significantly increased in the patients with
early-stage breast cancer when compared to those with benign breast
tumors and the healthy controls (P<0.01); however, there was a
no significant difference between the patients with benign breast
tumors and the healthy controls (P>0.05) (Table II).
The serum levels of miR-27a, miR-122 and miR-155
exhibited a statistically significant difference between the groups
as regards clinicopathological features, such as Breast Imaging
Reporting and Data System (BI-RADS) scoring and pathological types;
however, there were no statistically significant differences
(P>0.05) among the groups in parameters, such as hormone
receptors (molecular subtypes: Luminal-A, Luminal-B, Her2-enriched
and TNBC), Ki67 proliferation-index (<14 group and ≥14 group)
and the staging of early-stage breast cancer (stage I, IIA and IIB)
(Table III).
 | Table IIIComparison of clinicopathological
characteristics of the patients and the serum levels of the three
miRNAs. |
Table III
Comparison of clinicopathological
characteristics of the patients and the serum levels of the three
miRNAs.
| | | miR-27a | miR-122 | miR-155 |
|---|
| Variables | No. of cases | Mean ± 2SD, median
(min-max) | P-value | Mean ± 2SD, median
(min-max) | P-value | Mean ± 2SD, median
(min-max) | P-value |
|---|
| BI-RADS
scoring | | | | | | | |
|
Group 1: 0-2
(0,1,2) | 27 | 1.67±0.37, 1.7119
(0.49-2.56) |
<0.001b,c | 1.94±1.25, 1.6945
(0.45-5.95) |
<0.001a,b,c | 0.77±0.63, 0.5760
(0.15-2.75) |
<0.001b,c |
|
Group 2: 3-5
(3,4,5) | 29 | 1.39±0.56, 1.5369
(0.38-2.17) | | 2.00±1.16, 1.9790
(0.23-4.45) | | 1.11±0.85, 0.8201
(0.15-3.24) | |
|
Group 3:
6 | 56 | 3.75±1.98, 3.3995
(1.19-8.07) | | 4.94±2.07, 4.8356
(1.37-8.71) | | 2.22±1.77, 1.6279
(0.22-7.24) | |
| Pathological
types | | | | | | | |
|
Fibroadenoma
(benign breast tumor) | 56 | 1.52±0.49, 1.6567
(0.38-2.56) |
<0.001a,b | 1.97±1.19, 1.7683
(0.23-5.95) |
<0.001a,b,c | 0.95±0.77, 0.7364
(0.15-3.24) |
<0.001a,b,c |
|
Group 1:
IDC | 49 | 3.73±2.02, 3.2804
(1.25-8.07) | | 4.85±2.13, 4.6807
(1.37-8.71) | | 2.19±1.77, 1.6275
(0.22-7.24) | |
|
Group 2:
ILC | 7 | 3.91±1.80, 1.0706
(1.19-7.05) | | 5.54±1.51, 5.4832
(3.52-7.51) | | 2.44±1.86, 1.6283
(0.71-5.65) | |
| Hormone
receptors | | | | | | | |
|
Group 1:
Luminal A | 26 | 3.23±1.78, 2.9518
(1.19-7.02) | 0.119 | 4.59±2.05, 4.3141
(1.37-7.93) | 0.139 | 2.19±1.85, 1.4834
(0.41-6.67) | 0.931 |
|
Group 2:
Luminal B | 13 | 3.84±1.64, 3.9466
(1.34-7.05) | | 5.86±1.79, 6.0528
(2.79-8.71) | | 2.07±1.35, 1.8185
(0.51-4.96) | |
|
Group 2:
Her2-enriched | 9 | 4.14±2.56, 3.2271
(1.31-8.07) | | 4.14±2.01, 4.2586
(1.46-7.34) | | 2.54±2.33, 1.5536
(0.22-7.24) | |
|
Group 4:
TNBC | 8 | 4.89±2.18, 4.0526
(1.96-8.00) | | 5.46±2.30, 5.9865
(1.89-8.25) | | 2.19±1.67, 1.5673
(0.59-4.87) | |
| Ki67 | | | | | | | |
|
Group 1:
<14 | 28 | 3.54±1.93, 3.4505
(1.19-8.07) | 0.523 | 5.04±1.88, 4.8381
(1.92-8.71) | 0.819 | 2.12±1.65, 1.7569
(0.51-6.67) | 0.819 |
|
Group 2:
≥14 | 28 | 3.96±2.05, 3.3030
(1.30-8.00) | | 4.83±2.27, 4.8355
(1.37-8.25) | | 2.32±1.90, 1.5673
(0.22-7.24) | |
| Tumor staging | | | | | | | |
|
Group 1:
I | 20 | 3.70±1.91, 3.4505
(1.25-8.07) | 0.997 | 4.49±2.01, 4.2114
(1.39-7.51) | 0.104 | 2.53±1.97, 1.8038
(0.22-7.24) | 0.663 |
|
Group 2:
IIA | 18 | 3.83±2.20, 3.2149
(1.19-8.00) | | 4.58±2.21, 4.6483
(1.37-8.25) | | 2.09±1.83, 1.3606
(0.41-6.67) | |
|
Group 3:
IIB | 18 | 3.73±1.94, 3.5885
(1.31-7.23) | | 5.79±1.81, 5.9668
(1.77-8.71) | | 1.99±1.48, 1.5604
(0.49-4.82) | |
Diagnostic efficacy of miR-27a,
miR-122 and miR-155 evaluated by ROC curve analysis
The diagnostic efficacy of the three miRNAs
(miR-27a, miR-122 and miR-155) in differentiating between patients
with early-stage breast cancer and between patients with benign
breast tumors and healthy controls was then determined. Cut-off
fold change values, P-values, PPV and NPV (Table IV) were obtained. The results
revealed that miR-27a and miR-122 exhibited significant efficacy in
differentiating all groups; however, miR-155 did not exhibit
significant efficacy in differentiating between benign tumors and
healthy controls. The corresponding AUC (95% CI), sensitivity and
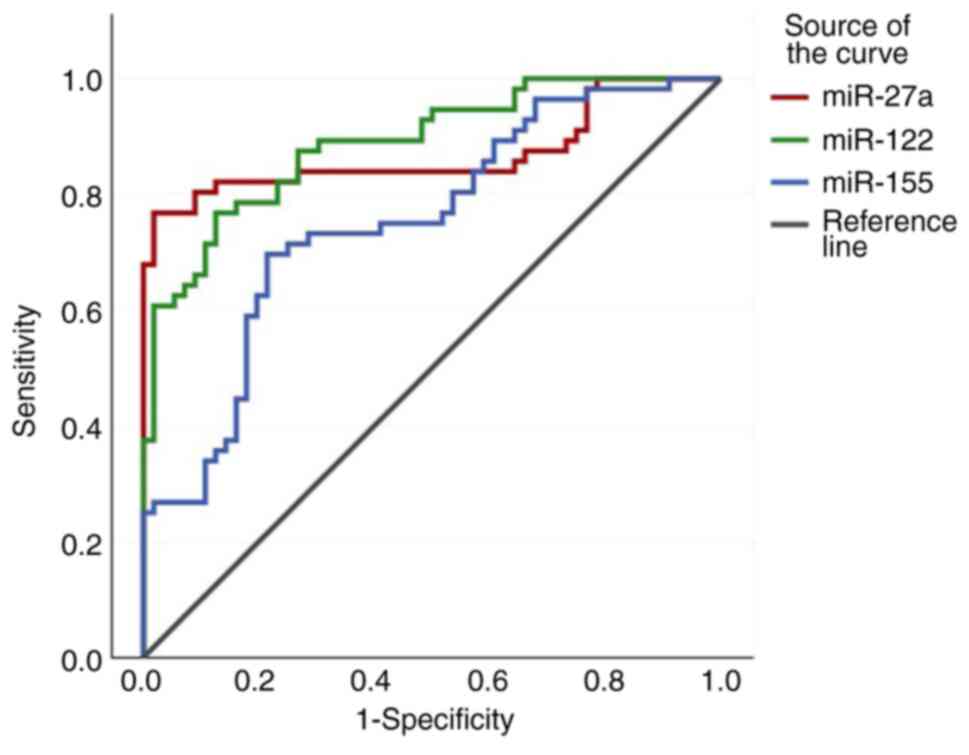
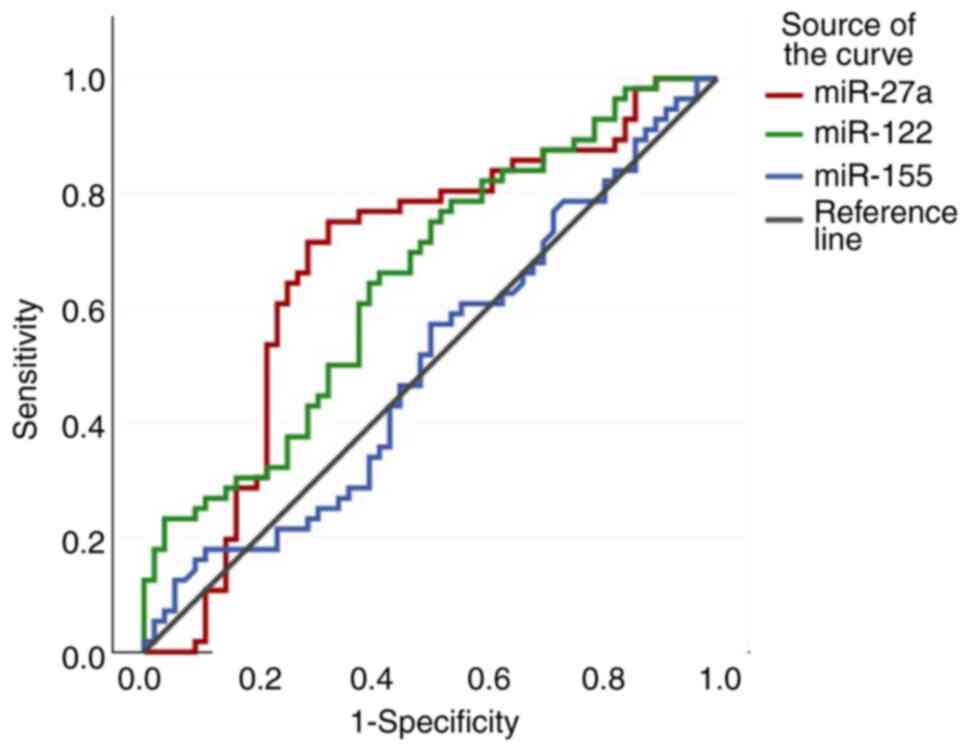
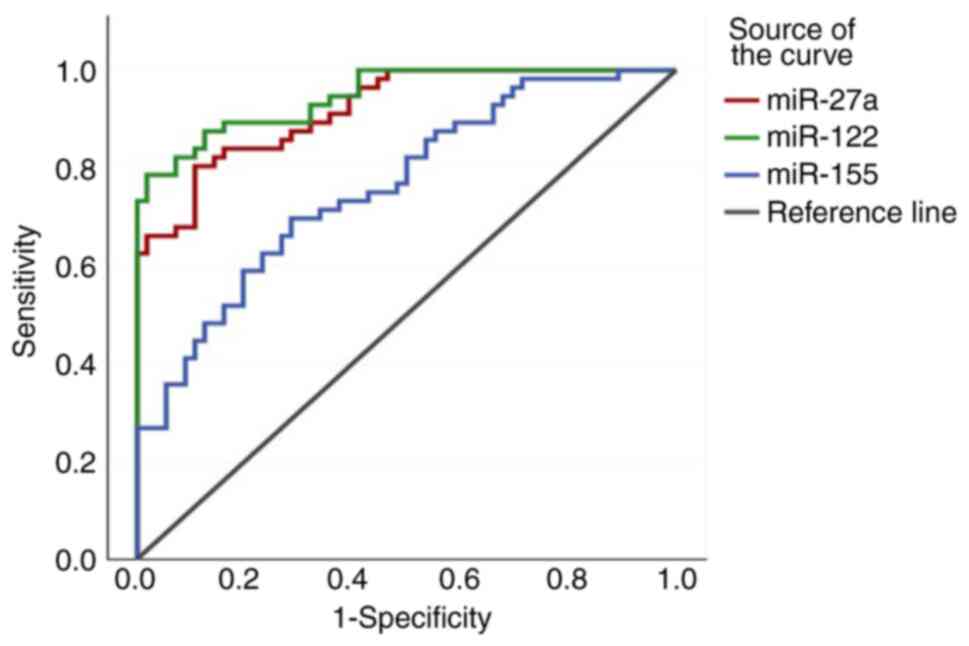
specificity of the ROC curves are depicted in Figs. 5, 6
and 7 and Tables V, VI and VII.
 | Table IVSensitivity, specificity, NPV, PPV
and LRs of the three miRNAs. |
Table IV
Sensitivity, specificity, NPV, PPV
and LRs of the three miRNAs.
| Compared
Groups | miRNAs | P-value | Cut-off FC
value | AUC (95% CI) | Sensitivity | Specificity | PPV | NPV | LR+ | LR- |
|---|
| Early stage breast
cancer (BC)-healthy controls | miR-27a | <0.001 | 2.0651 | 0.920 | 80.4 | 89.3 | 88.2 | 81.9 | 7.51 | 0.22 |
| | miR-122 | <0.001 | 3.0163 | 0.947 | 78.6 | 98.2 | 97.7 | 82 | 43.6 | 0.22 |
| | miR-155 | <0.001 | 1.1272 | 0.762 | 69.6 | 71.4 | 70.9 | 29.8 | 2.43 | 0.42 |
| Early stage breast
cancer-benign tumour | miR-27a | <0.001 | 2.2016 | 0.870 | 76.8 | 98.2 | 97.7 | 80.8 | 42.66 | 0.24 |
| | miR-122 | <0.001 | 3.1888 | 0.888 | 78.6 | 87.5 | 86 | 79 | 6.14 | 0.26 |
| | miR-155 | <0.001 | 1.1336 | 0.753 | 69.6 | 78.6 | 76.4 | 72.1 | 3.25 | 0.39 |
| Benign
tumour-healthy controls | miR-27a | 0.015 | 1.3492 | 0.675 | 75 | 67.9 | 70 | 73 | 2.33 | 0.37 |
| | miR-122 | 0.028 | 0.9302 | 0.651 | 82.1 | 58.9 | 58.2 | 69.6 | 1.39 | 0.43 |
| | miR-155 | 0.935 | 0.6101 | 0.502 | 60.7 | 44.6 | 52 | 53.1 | 1.09 | 0.88 |
 | Table VROC curve analysis of the three
miRNAs for differentiating between patients with early-stage breast
cancer and healthy controls (corresponding to Fig. 5). |
Table V
ROC curve analysis of the three
miRNAs for differentiating between patients with early-stage breast
cancer and healthy controls (corresponding to Fig. 5).
| miRNA | Test | Early-stage breast
cancer | Healthy
controls | Total | P-value from
χ2 test |
|---|
| miR-27a cut-off
fold | Test+ | 45 | 6 | 51 | 0.001 |
| change=2.06519 | Test- | 11 | 50 | 61 | |
| Total | | 56 | 56 | 112 | |
| miR-122 cut-off
fold | Test+ | 44 | 1 | 45 | 0.001 |
|
change=3.016371 | Test- | 12 | 55 | 67 | |
| Total | | 56 | 56 | 112 | |
| miR-155 cut-off
fold | Test+ | 39 | 16 | 55 | 0.001 |
| change=1.12727 | Test- | 17 | 40 | 57 | |
| Total | | 56 | 56 | 112 | |
 | Table VIROC curve analysis of three miRNAs
for differentiating between patients with early-stage breast cancer
and patients with benign breast tumors (corresponding to Fig. 6). |
Table VI
ROC curve analysis of three miRNAs
for differentiating between patients with early-stage breast cancer
and patients with benign breast tumors (corresponding to Fig. 6).
| miRNA | Test | Early-stage breast
cancer | Healthy
controls | Total | P-value from
χ2 test |
|---|
| miR-27a cut-off
fold | Test+ | 43 | 1 | 44 | 0.001 |
| change=2.20164 | Test- | 13 | 55 | 68 | |
| Total | | 56 | 56 | 112 | |
| miR-122 cut-off
fold | Test+ | 43 | 7 | 60 | 0.001 |
| change=3.18884 | Test- | 13 | 49 | 62 | |
| Total | | 56 | 56 | 112 | |
| miR-155 cut-off
fold | Test+ | 39 | 12 | 51 | 0.001 |
| change=1.13369 | Test- | 17 | 44 | 61 | |
| Total | | 56 | 56 | 112 | |
 | Table VIIROC curve analysis of the three
miRNAs for differentiating between patients with benign breast
tumors and healthy controls (corresponding to Fig. 7). |
Table VII
ROC curve analysis of the three
miRNAs for differentiating between patients with benign breast
tumors and healthy controls (corresponding to Fig. 7).
| miRNA | Test | Early-stage breast
cancer | Healthy
controls | Total | P-value from
χ2 test |
|---|
| miR-27a cut-off
fold | Test+ | 42 | 18 | 60 | 0.001 |
| change=1.34919 | Test- | 14 | 38 | 52 | |
| Total | | 56 | 56 | 112 | |
| miR-122 cut-off
fold | Test+ | 46 | 33 | 79 | 0.001 |
| change=0.93025 | Test- | 10 | 23 | 33 | |
| Total | | 56 | 56 | 112 | |
| miR-155 cut-off
fold | Test+ | 34 | 31 | 65 | 0.001 |
| change=0.61011 | Test- | 22 | 25 | 47 | |
| Total | | 56 | 56 | 112 | |
Discussion
Tissue gene biomarkers have improved the diagnosis
of breast cancer to a great extent; however, there are limitations
to their applications due to invasiveness, risk of hemorrhage and
the unpleasant nature of the diagnostic procedures. The estimation
of non-invasive or minimally invasive biomarkers, such as miRNA
expression levels in blood and serum, is a highly sensitive and
specific method which can benefit both clinicians and patients in
the screening, diagnosis and prognosis of patients with early-stage
breast cancer (6). Although there
has been substantial success in the discovery of serum biomarkers,
it is imperative to identify biomarkers that are useful for the
early diagnosis of affected patients.
Of note, in the present study, immunohistochemistry
was performed as routine diagnostic workup for sub-classifying the
patients with breast cancer. In the present study, the expression
levels of the three miRNAs (miR-27a, miR-122 and miR-155) were
significantly upregulated in the serum of patients with early-stage
breast cancer compared to those in patients with benign breast
tumors and healthy controls. These findings indicate that these
three miRNA are more likely related to early-stage breast cancer.
These findings also indicate these three miRNAs may be beneficial
biomarkers for the diagnosis of early-stage breast cancer.
In their study, Swellam et al (13) examined the expression levels of
miR-27a and tumor markers (CEA and CA15.3) among three investigated
groups; patients with primary breast cancer, patients with benign
breast tumors and healthy controls; they demonstrated that
miRNA-27a was superior to the two tumor markers in specificity and
accuracy for the detection of early-stage breast cancer. The
expression levels of miR-27a in serum were significantly increased
in patients with primary breast cancer when compared to those in
patients with benign breast tumors followed by the healthy controls
who had the lowest serum level (13). The results of their study are
comparable to those of the present study as regards the serum
levels of miR-27a. Previous studies performed by Wu et al
(22) and Ding et al
(23) reported the oncogenic role
of miR-27a among several other types of cancer. It was observed
that, the upregulation of miR-27a targeted the FOXO1 gene, which
was responsible for cell cycle regulation, thus promoting evasion
from apoptosis and cell proliferation in breast cancer (24).
Saleh et al (25) demonstrated that miR-122 exhibited an
improved specificity and sensitivity than tumor markers (CEA and
CA15-3) in the diagnosis of breast cancer and in predicting breast
cancer metastasis. In a cohort study, Wu et al (26) demonstrated that circulating levels
of miRNA-122 had a good specificity and sensitivity in predicting
metastasis. It was also found that the miR-122 level was increased
in patients who had relapsed after recovery and was able to
determine relapse in patients with stage IIa, IIb and III breast
cancer (26). On the other hand,
Wang et al (27)
demonstrated that miR-122 functioned as a tumor-suppressor in
breast cancer cells by suppressing the IGF1R-mediated downstream
Akt/mTOR/p70S6K signaling pathway and inhibiting breast cancer cell
growth. They demonstrated that the miR-122 expression levels
decreased in breast cancer cells when compared to normal breast
tissue (27). This discrepancy in
the results of miR-122 levels, between studies performed on breast
tissue samples and blood samples can be explained in the study by
Fong et al (28). Fong et
al (28) demonstrated that
breast cancer cells secrete large amounts of miR-122-secreting
vesicles, which leads to a decrease in miR-122 levels
intracellularly. This secreted miR-122 affects non-cancer cells and
decreases their uptake of glucose by downregulating glycolytic
enzyme pyruvate kinase. In this manner, miR-122 supports cancer
cell viability and metastasis by increasing nutrient supply
(28). Thus, the majority of breast
cancer cells have decreased levels of miR-122. In the present
study, miR-122 expression levels in serum were increased in
patients with early-stage breast cancer compared to those with
benign breast tumors and healthy controls. miR-122 also exhibited
improved sensitivity and specificity in detecting early-stage
breast cancer.
In the study by Hosseini Mojahed et al
(29), it was demonstrated that the
miR-155 expression levels in serum were significantly higher in
patients with breast cancer when compared with healthy controls.
The combination of three miRNAs, miR 17-5p, miR-155 and miR-222
levels, when compared with tumour markers (CEA and CA15.3) for
breast cancer screening, has revealed that the three miRNAs were
more effective than tumour markers for the early diagnosis of
breast cancer, particularly in high-risk groups (13,30).
In another study by Bašová et al (31), the serum levels of miR-155 were
highly predictive in determining relapse in early-stage breast
cancer. Another study demonstrated that the upregulation of miR-155
inhibited the translation of mRNAs, such as RhoA, FOXO3A, and
SOCS1. which led to the evasion of apoptosis and increased cell
proliferation (32). The decreased
levels of BRCA1 due to mutation upregulate the levels of miR-155 in
serum. This occurs since BRCA1 regulates miR-155 by binding to the
miR-155 promotor and recruiting histone deacetylases (33). In the present study, the levels of
miR-155 were significantly increased in patients with early-stage
breast cancer, when compared to healthy controls and have good
sensitivity and specificity were observed.
In the present study, ROC analysis revealed that the
serum levels of miR-27a, miR-122 and miR-155 exhibited good
accuracy with AUC values of 0.920, 0.947 and 0.762, respectively
for differentiating between early-stage breast cancer and healthy
controls. These three miRNAs also exhibited fair accuracy with AUC
values of 0.870, 0.888 and 0.753, respectively for differentiating
between early-stage breast cancer and benign breast tumors.
In conclusion, the results of the present study
indicate that the levels of miR-27a, miR-122 and miR-155 are
increased in patients with breast cancer. Their serum levels have
reliable sensitivity and specificity in detecting early-stage
breast cancer. These three miRNAs may thus potential for use as
non-invasive biomarkers in the diagnosis and screening of breast
cancer. However, their clinical applicability needs to be evaluated
in multiple centers in order to confirm their diagnostic
efficacy.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by a ICMR-MD/MS thesis
grant (registration no. MD21DEC-0045,
No.3/2/December-2021/PG-Thesis-HRD(14) dated May 30, 2022.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SSr and SSa wrote the manuscript and performed the
experiments and generated the figures. DKM and TSM collected the
clinical data. PM and SM provided the data of the recruited
patients from the Departments of Pathology and Radiodiagnosis, All
India Institute of Medical Sciences (Bhubaneswar, India),
respectively. All authors made critical revisions, and all authors
have read and approved the final manuscript. All authors (SSr, SSa,
DKM, TSM, PM and SM) confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Ethical clearance obtained from the Institutional
Ethics Committee (IEC) of All India Institute of Medical Sciences
(AIIMS), Bhubaneswar, India (IEC/AIIMS BBSR/PG Thesis/2021-22/32).
Each patient was explained the nature of study and after obtaining
their written consent, they were inducted into the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Malvia S, Bagadi SA, Dubey US and Saxena
S: Epidemiology of breast cancer in Indian women. Asia Pac J Clin
Oncol. 13:289–295. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Momenimovahed Z and Salehiniya H:
Epidemiological characteristics of and risk factors for breast
cancer in the world. Breast Cancer (Dove Med Press). 11:151–164.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Practice bulletin no 182: Hereditary
breast and ovarian cancer syndrome. Obstet Gynecol. 130:e110–e126.
2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Jafari SH, Saadatpour Z, Salmaninejad A,
Momeni F, Mokhtari M, Nahand JS, Rahmati M, Mirzaei H and Kianmehr
M: Breast cancer diagnosis: Imaging techniques and biochemical
markers. J Cell Physiol. 233:5200–5213. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang L: Early diagnosis of breast cancer.
Sensors (Basel). 17(1572)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hussen BM, Hidayat HJ, Salihi A, Sabir DK,
Taheri M and Ghafouri-Fard S: MicroRNA: A signature for cancer
progression. Biomed Pharmacother. 138(111528)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Heneghan HM, Miller N and Kerin MJ:
Circulating microRNAs: Promising breast cancer biomarkers. Breast
Cancer Res. 13(402)2011.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma
ES, Pang R, Chua D, Chu KM, Law WL, et al: Circulating microRNAs as
specific biomarkers for breast cancer detection. PLoS One.
8(e53141)2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Giordano C, Accattatis FM, Gelsomino L,
Del Console P, Győrffy B, Giuliano M, Veneziani BM, Arpino G, De
Angelis C, De Placido P, et al: miRNAs in the box: Potential
diagnostic role for extracellular vesicle-packaged miRNA-27a and
miRNA-128 in breast cancer. Int J Mol Sci. 24(15695)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ljepoja B, García-Roman J, Sommer AK,
Wagner E and Roidl A: MiRNA-27a sensitizes breast cancer cells to
treatment with selective estrogen receptor modulators. Breast Edinb
Scotl. 43:31–38. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Swellam M, Zahran RFK, Ghonem SA and
Abdel-Malak C: Serum MiRNA-27a as potential diagnostic nucleic
marker for breast cancer. Arch Physiol Biochem. 127:90–96.
2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Elghoroury EA, Abdelghafar EE, Kamel S,
Awadallah E, Shalaby A, El-Saeed GSM, Mahmoud E, Kamel MM, Abobakr
A and Yousef RN: Dysregulation of miR-122, miR-574 and miR-375 in
Egyptian patients with breast cancer. PLoS One.
19(e0298536)2024.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ranjbari S, Rezayi M, Arefinia R,
Aghaee-Bakhtiari SH, Hatamluyi B and Pasdar A: A novel
electrochemical biosensor based on signal amplification of Au
HFGNs/PnBA-MXene nanocomposite for the detection of miRNA-122 as a
biomarker of breast cancer. Talanta. 255(124247)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li M, Zou X, Xia T, Wang T, Liu P, Zhou X,
Wang S and Zhu W: A five-miRNA panel in plasma was identified for
breast cancer diagnosis. Cancer Med. 8:7006–7017. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ali SA, Abdulrahman ZFA and Faraidun HN:
Circulatory miRNA-155, miRNA-21 target PTEN expression and activity
as a factor in breast cancer development. Cell Mol Biol
(Noisy-le-grand). 66:44–50. 2020.PubMed/NCBI
|
|
18
|
Dziechciowska I, Dąbrowska M, Mizielska A,
Pyra N, Lisiak N, Kopczyński P, Jankowska-Wajda M and Rubiś B:
miRNA expression profiling in human breast cancer diagnostics and
therapy. Curr Issues Mol Biol. 45:9500–9525. 2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods San
Diego Calif. 50:298–301. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Roth C, Rack B, Müller V, Janni W, Pantel
K and Schwarzenbach H: Circulating microRNAs as blood-based markers
for patients with primary and metastatic breast cancer. Breast
Cancer Res. 12(R90)2010.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu XJ, Li Y, Liu D, Zhao LD, Bai B and Xue
MH: miR-27a as an oncogenic microRNA of hepatitis B virus- related
hepatocellular carcinoma. Asian Pac J Cancer Prev. 14:885–889.
2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ding L, Zhang S, Xu M, Zhang R, Sui P and
Yang Q: MicroRNA-27a contributes to the malignant behavior of
gastric cancer cells by directly targeting PH domain and
leucine-rich repeat protein phosphatase 2. J Exp Clin Cancer Res.
36(45)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Saleh AA, Soliman SE, Habib MSE, Gohar SF
and Abo-Zeid GS: Potential value of circulatory microRNA122 gene
expression as a prognostic and metastatic prediction marker for
breast cancer. Mol Biol Rep. 46:2809–2818. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu X, Somlo G, Yu Y, Palomares MR, Li AX,
Zhou W, Chow A, Yen Y, Rossi JJ, Gao H, et al: De novo sequencing
of circulating miRNAs identifies novel markers predicting clinical
outcome of locally advanced breast cancer. J Transl Med.
10(42)2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang B, Wang H and Yang Z: MiR-122
inhibits cell proliferation and tumorigenesis of breast cancer by
targeting IGF1R. PLoS One. 7(e47053)2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fong MY, Zhou W, Liu L, Alontaga AY,
Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, et al:
Breast-cancer-secreted miR-122 reprograms glucose metabolism in
premetastatic niche to promote metastasis. Nat Cell Biol.
17:183–194. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Hosseini Mojahed F, Aalami AH, Pouresmaeil
V, Amirabadi A, Qasemi Rad M and Sahebkar A: Clinical evaluation of
the diagnostic role of MicroRNA-155 in breast cancer. Int J
Genomics. 2020(9514831)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Swellam M, Zahran RFK, Abo El-Sadat Taha
H, El-Khazragy N and Abdel-Malak C: Role of some circulating MiRNAs
on breast cancer diagnosis. Arch Physiol Biochem. 125:456–464.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bašová P, Pešta M, Sochor M and Stopka T:
Prediction potential of serum miR-155 and miR-24 for relapsing
early breast cancer. Int J Mol Sci. 18(2116)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Mattiske S, Suetani RJ, Neilsen PM and
Callen DF: The oncogenic role of miR-155 in breast cancer. Cancer
Epidemiol Biomark Prev. 21:1236–1243. 2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chang S, Wang RH, Akagi K, Kim KA, Martin
BK and Cavallone L: Kathleen Cuningham Foundation Consortium for
Research into Familial Breast Cancer (kConFab). Haines DC, Basik M,
Mai P, et al: Tumor suppressor BRCA1 epigenetically controls
oncogenic microRNA-155. Nat Med. 17:1275–1282. 2011.PubMed/NCBI View
Article : Google Scholar
|