Introduction
According to statistics, ovarian cancer accounts for
2.5% of all malignant tumors, and accounts for 5% of female
cancer-related deaths, mainly due to late diagnosis. Despite recent
improvements in the diagnosis, ~70% of ovarian cancers are
diagnosed at an advanced stage, and only 30% of patients with
advanced-stage ovarian cancer survive for >5 years. Ovarian
cancer is a heterogeneous group of malignant tumors that vary in
etiology and molecular biology. Although the incidence and
mortality rates have decreased in recent years, there is still an
urgent need to explore the molecular biology of ovarian cancer in
order to further identify early diagnostic and therapeutic targets
(1,2).
There are numerous post-transcriptional
modifications in organisms, among which
N6-methyladenosine (m6A) is the most abundant
internal modification in eukaryotes (3), which plays a key role in various
biological processes, such as stem cell self-renewal and
differentiation, DNA damage and heat shock. m6A can be
regulated by specific enzymes known as ‘writers’, ‘erasers’ and
‘readers’. The ‘writers’ are methyltransferases, including
methyltransferase-like (METTL)3, METTL14 and Wilms tumor
1-associated protein. The ‘erasers’ are demethyltransferases,
including fat mass and obesity-associated and AlkB homolog 5, RNA
demethylase. The ‘readers’ are RNA-binding proteins, including the
YTH family. m6A-related proteins play a role in
modification and the regulation of the pathogenesis of various
types of cancer, such as leukemia, brain tumors, breast cancer,
liver cancer, cervical cancer and lung cancer (4).
Single nucleotide polymorphisms (SNPs) are DNA
sequence polymorphisms caused by variations in a single nucleotide
at the genomic level, which is the most common form of genetic
variation in humans (5). Some
studies have found that m6A and its polymorphisms are
associated with susceptibility to bladder cancer, gastric cancer,
pancreatic cancer and hepatoblastoma (6-9).
Moreover, METTL3 polymorphisms have been reported to affect the
susceptibility to neuroblastoma, hepatoblastoma and nephroblastoma
(10-12).
The association between METTL3 polymorphisms and the
development of ovarian cancer has rarely been reported, at least to
the best of our knowledge. Given that m6A and its
polymorphisms are associated with tumor susceptibility, it was
hypothesized that METTL3 SNPs may be associated with the risk of
developing ovarian cancer. In order to verify this hypothesis, the
present multicenter large sample case-control study was conducted
to investigate the association between METTL3 polymorphisms and the
susceptibility to ovarian cancer.
Patients and methods
Study population
Tissue samples from 244 patients with ovarian cancer
diagnosed by pathological analysis and blood samples from 276
normal controls were collected from the First Affiliated Hospital
of Jinan University (Guangzhou, China), Guangzhou Women's and
Children's Medical Center (Guangzhou, China) and Shunde Hospital of
Southern Medical University (Foshan, China). The present study was
approved by the ethics committees of the above three hospitals [the
Ethics Committee of the First Affiliated Hospital of Jinan
University (KY-2022-233), the Ethics Committee of Guangzhou Medical
University Women and Children's Medical Center (117A01), and the
Ethics Committee of Shunde Hospital, Southern Medical University
(KYLS20220903)]. In addition, written informed consent was obtained
from the subjects. The clinical data of the subjects has been
permitted for public disclosure, and the personal information of
the subjects has been concealed in the study results. The clinical
and pathological information of all subjects in the ovarian cancer
group was collected from the databases of the aforementioned
hospitals, including name, age, pregnancy and delivery, tumor
stage, pathological type and immunohistochemistry results. The
relevant information was obtained by querying the clinical medical
record system (Table SI).
SNP selection and genotyping
Genomic DNA was extracted from peripheral blood and
paraffin samples using the DNA extraction kit (Tiangen Biotech Co.,
Ltd.) (DP304-03). SNPs with potential biological functions were
screened using the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/) and SNPinfo (http://snpinfo.niehs.nih.gov/) online software. Of
note, four SNPs (rs1263801 G>C, rs1139130 G>A, rs1061027
C>A and rs1061026 T>G) were selected for analysis. The
sequences for these SNPs were as follows: rs1263801 G>C,
CTGCCAAGAAATGACCACTACAAAA[C/G] and AGTCGTTATAACTGAGGGAACAAAG;
rs1139130 G>A, ACACAACCACTACTTACCCCCAGAG[A/G] and
TTTAGACATTCTCTCCCCAACTCCA; rs1061027 C>A,
TTCTGTCCTTAATCATAAATAATAG[A/C] and CCCTTGAGGACTAGCCTGTTCTCTG;
rs1061026 T>G, AAAACAATGTGAAGCTCTACTAAGT[G/T] and
CTGTCCTTAATCATAAATAATAGCC. Genotyping of the extracted genomic DNA
was performed using a TaqMan assay with the TIANtough Genotyping
qPCR PreMix (Probe) (TianGen, Guangzhou Z-ZHI Biotechnology). The
PCR protocol consisted of an initial denaturation at 95˚C for 10
min, followed by 45 cycles of 95˚C for 15 sec and 60˚C for 60
sec.
SNP-SNP interaction analysis
Interactions between SNP loci and their epistasis
were verified using the multifactor dimensionality reduction (MDR)
method using MDR software v3.0.2 (Laboratory of Computational
Genetics, University of Pennsylvania, Philadelphia, PA, USA;
available free of charge at http://www.epist asis.org). This method can identify
correlations in studies with a small sample size and low SNP
penetrance. Cross-validation consistency (CVC) and test accuracy
were used to determine the optimal interaction model. The optimal
model was the one with the highest CVC and test accuracy values.
Values of P<0.05 were considered to indicate statistically
significant differences.
Statistical analysis
The Chi-squared test was used to determine whether
there was a statistically significant difference in age between the
experimental and control groups. Logistic regression analysis was
used to calculate odds ratios (ORs) and 95% confidence intervals
(CIs) to assess the association between METTL3 polymorphisms and
susceptibility to ovarian cancer, and age was corrected to avoid
the influence of confounding factors. Stratified analyses were
performed according to age, clinical stage, pregnancy outcomes and
immunohistochemistry results to investigate the association between
genotypes and susceptibility to ovarian cancer in each sub-stratum.
Haplotype analysis was performed using logistic regression
analysis, which was used to comprehensively evaluate the effect of
selected SNPs of the gene on susceptibility to ovarian cancer. The
goodness-of-fit test was used to determine whether the frequency
distribution of the genotypes of each SNP in the control group
satisfied the Hardy-Weinberg equilibrium (HWE); a value of
P>0.05 was considered to indicate statistically significant
difference, which indicated that the SNP locus in the control group
complied with the HWE. The Gene-Tissue Expression (GTEx) portal
(https://www.gtexportal.org/home/) was
also used for expression quantitative trait loci (eQTL) analysis to
predict potential associations between SNPs and gene expression
levels. Statistical analysis was performed using SAS 9.4 software
(SAS Institute Inc.
Results
Characteristics of the study
participants
Detailed information on the demographic and clinical
characteristics of the patients with ovarian cancer (n=244) and the
controls (n=276) is presented in Table
SI. There was no statistically significant difference in age
between the ovarian cancer and control groups (P=0.47).
Association of METTL3 gene
polymorphisms with susceptibility to ovarian cancer
Genotyping was performed on 244 patients and 276
control subjects. The association between METTL3 polymorphisms and
susceptibility to ovarian cancer is presented in Table I. None of the selected SNPs were
statistically different in HWE (P>0.05). First, single locus
analysis was performed which yielded the following results:
rs1263801 CC vs. GG: Adjusted OR, 0.480; 95% CI, 0.238-0.968;
P=0.0402; CC vs. GG/GC: Adjusted OR, 0.48; 95% CI, 0.246-0.966;
P=0.0395; rs1061027 CA vs. CC: Adjusted OR, 0.500; 95% CI,
0.331-0.754; P=0.001; CA/AA vs. CC: Adjusted OR, 0.629; 95% CI,
0.428-0.925; P=0.0185; rs1061026 TG vs. TT: Adjusted OR, 0.580; 95%
CI, 0.359-0.937; P=0.0262; TG/GG vs. TT: Adjusted OR, 0.0366; 95%
CI, 0.386-0.977; P=0.0395. Allelic variants reduced the risk of
developing ovarian cancer. However, rs1139130 was not associated
with the risk of developing ovarian cancer (Table I).
 | Table ILogistic regression analysis of
associations between METTL3 polymorphisms and susceptibility to
ovarian cancer. |
Table I
Logistic regression analysis of
associations between METTL3 polymorphisms and susceptibility to
ovarian cancer.
| Genotype | Cases (n=244), n
(%) | Controls (n=276), n
(%) | P-valuea | Crude OR (95%
CI) | P-value | Adjusted OR (95%
CI) | P-valueb |
|---|
| rs1263801 G>C
(HWE=0.68) | | | | | | | |
|
GG | 124 (51.88) | 128 (47.58) | | 1 | | 1.00 | |
|
GC | 102 (42.68) | 113 (42.01) | | 0.945
(0.659-1.355) | 0.7568 | 0.944
(0.658-1.354) | 0.7552 |
|
CC | 13 (5.44) | 28 (10.41) | | 0.486
(0.241-0.979) | 0.0435 | 0.480
(0.238-0.968) | 0.0402 |
|
Additive | | | 0.1132 | 0.794
(0.602-1.047) | 0.1017 | 0.790
(0.599-1.042) | 0.0949 |
|
Dominant | 11 5(48.12) | 141 (52.42) | 0.3334 | 0.842
(0.594-1.193) | 0.3335 | 0.839
(0.592-1.189) | 0.3236 |
|
Recessive | 226 (94.56) | 241 (89.59) | 0.0401 | 0.495
(0.250-0.980) | 0.0435 | 0.488
(0.246-0.966) | 0.0395 |
| rs1139130 G>A
(HWE=0.55) | | | | | | | |
|
GG | 102 (43.22) | 206 (39.41) | | 1 | | 1 | |
|
GA | 101 (42.80) | 122 (45.35) | | 0.850
(0.586-1.234) | 0.3935 | 0.847
(0.583-1.229) | 0.3812 |
|
AA | 33 (13.98) | 41 (15.24) | | 0.827
(0.488-1.402) | 0.4805 | 0.819
(0.482-1.390) | 0.4592 |
|
Additive | | | 0.6817 | 0.901
(0.701-1.158) | 0.4157 | 0.895
(0.696-1.151) | 0.3875 |
|
Dominant | 134 (56.78) | 163 (60.59) | 0.3848 | 0.854
(0.599-1.219) | 0.3849 | 0.847
(0.594-1.210) | 0.3615 |
|
Recessive | 203 (86.02) | 228 (84.76) | 0.6899 | 0.904
(0.551-1.485) | 0.6908 | 0.895
(0.545-1.471) | 0.6622 |
| rs1061027 C>A
(HWE=0.45) | | | | | | | |
|
CC | 156 (71.89) | 167 (61.85) | | 1 | | 1 | |
|
CA | 47 (21.66) | 88 (32.59) | | 0.505
(0.335-0.761) | 0.0011 | 0.500
(0.331-0.754) | 0.001 |
|
AA | 14 (6.45) | 15 (5.56) | | 0.882
(0.414-1.882) | 0.7460 | 0.894
(0.419-1.090) | 0.7726 |
|
Additive | | | 0.0276 | 0.771
(0.569-1.046) | 0.0951 | 0.770
(0.567-1.045) | 0.0937 |
|
Dominant | 61 (28.11) | 103 (38.15) | 0.0198 | 0.634
(0.432-0.931) | 0.0202 | 0.629
(0.428-0.925) | 0.0185 |
|
Recessive | 203 (93.55) | 255 (94.44) | 0.6779 | 1.172
(0.553-2.4850) | 0.6782 | 1.195
(0.563-2.538) | 0.6424 |
| rs1061026 T>G
(HWE=0.31) | | | | | | | |
|
TT | 190 (84.82) | 208 (77.32) | | 1 | | 1 | |
|
TG | 31 (13.84) | 55 (20.45) | | 0.577
(0.357-0.932) | 0.0246 | 0.580
(0.359-0.937) | 0.0262 |
|
GG | 3 (1.34) | 6 (2.23) | | 0.512
(0.126-2.074) | 0.3482 | 0.502
(0.124-2.038) | 0.3352 |
|
Additive | | | 0.1085 | 0.652
(0.432-0.983) | 0.0414 | 0.653
(0.433-0.986) | 0.0426 |
|
Dominant | 34 (15.18) | 61 (22.68) | 0.0356 | 0.610
(0.384-0.970) | 0.0366 | 0.614
(0.386-0.977) | 0.0395 |
|
Recessive | 221 (98.66) | 263 (97.77) | 0.4618 | 0.595
(0.147-2.4070 | 0.4666 | 0.573
(0.141-2.323) | 0.4351 |
Stratified analysis
The rs1263801 allele variant reduced the incidence
of ovarian cancer in patients aged >51 years (adjusted OR,
0.398; 95% CI, 0.159-0.996; P=0.0489) (Table II).
 | Table IIStratification analysis of METTL3
polymorphisms with susceptibility to ovarian cancer in rs1263801
G>C. |
Table II
Stratification analysis of METTL3
polymorphisms with susceptibility to ovarian cancer in rs1263801
G>C.
| | rs1263801 G>C
(cases/controls) | |
|---|
| Variables | GG/GC | CC | Adjusted
ORa (95% CI) |
P-valuea |
|---|
| Age, years | | | | |
|
≤51 | 108 | 6 | 0.641
(0.230-1.792) | 0.3969 |
|
>51 | 118 | 7 | 0.398
(0.159-0.996) | 0.0489 |
| Metastasis | | | | |
|
Yes | 81/241 | 4/28 | 0.403
(0.136-1.188) | 0.0993 |
|
No | 128/241 | 9/28 | 0.609
(0.279-1.331) | 0.2141 |
| Clinical stage | | | | |
|
1 | 46/241 | 5/28 | 0.982
(0.358-2.693) | 0.9717 |
|
2 | 41/241 | 2/28 | 0.437
(0.100-1.915) | 0.2722 |
|
3 | 76/241 | 3/28 | 0.325
(0.096-1.102) | 0.0712 |
|
4 | 20/241 | 1/28 | 0.407
(0.052-3.160) | 0.3897 |
| No. of times
pregnant | | | | |
|
≥3 | 92/241 | 5/28 | 0.449
(0.168-1.203) | 0.1113 |
|
<3 | 134/241 | 8/28 | 0.511
(0.226-1.155) | 0.1068 |
| Menopause | | | | |
|
Post-menopause | 146/241 | 9/28 | 0.476
(0.215-1.052) | 0.0665 |
|
Pre-menopause | 74/241 | 4/28 | 0.536
(0.176-1.632) | 0.272 |
| ER | | | | |
|
Low | 27/241 | 2/28 | 0.587
(0.131-2.618) | 0.4846 |
|
High | 60/241 | 3/28 | 0.437
(0.128-1.489) | 0.1859 |
| PR | | | | |
|
Low | 26/241 | 2/28 | 0.670
(0.150-2.990) | 0.5996 |
|
High | 35/242 | 2/28 | 0.507
(0.115-2.226) | 0.3679 |
| PAX8 | | | | |
|
Low | 26/241 | 2/28 | 0.654
(0.147-2.920) | 0.5783 |
|
High | 54/241 | 1/28 | 0.155
(0.021-1.163) | 0.0698 |
| WTI | | | | |
|
Low | 31/241 | 3/28 | 0.752
(0.214-2.646) | 0.6576 |
|
High | 71/241 | 2/28 | 0.236
(0.055-1.017) | 0.0526 |
| p16 | | | | |
|
Low | 37/241 | 3/28 | 0.765
(0.218-2.686) | 0.6759 |
|
High | 61/241 | 2/28 | 0.276
(0.064-1.193) | 0.0847 |
| Ki-67 | | | | |
|
Low | 36/241 | 2/28 | 0.506
(0.115-2.229) | 0.3683 |
|
High | 69/241 | 3/28 | 0.370
(0.109-1.256) | 0.1108 |
| Wild-type p53 | | | | |
|
Positive | 51/241 | 1/28 | 0.168
(0.022-1.267) | 0.0835 |
|
Negative | 166/241 | 11/28 | 0.559
(0.270-1.155) | 0.1164 |
| Mutant p53 | | | | |
|
Positive | 98/241 | 5/28 | 0.436
(0.163-1.163) | 0.0972 |
|
Negative | 119/241 | 7/28 | 0.492
(0.208-1.161) | 0.1054 |
For the rs1061027 gene polymorphism, compared with
the CC genotype, the CA/AA genotype reduced the risk of developing
ovarian cancer in patients aged ≤51 years (adjusted OR, 0.544; 95%
CI, 0.308-0.960; P=0.0357), in those without metastases (adjusted
OR, 0.576; 95% CI, 0.359-0.924; P=0.0222), those with clinical
stage III disease (adjusted OR, 0.542; 95% CI, 0.308-0.954;
P=0.0338), those who had been pregnant three times or more
(adjusted OR, 0.525; 95% CI, 0.305-0.903 P=0.0200), those at
post-menopause (adjusted OR, 0.578; 95% CI, 0.367-0.911; P=0.0182),
those who were estrogen receptor (ER) strongly positive (adjusted
OR, 0.517; 95% CI, 0.270-0.991; P=0.0470), progesterone receptor
(PR) weakly positive (adjusted OR, 0.297; 95% CI, 0.099-0.886;
P=0.0295), those who were weakly positive for paired box 8 (PAX8)
(adjusted OR, 0.281; 95% CI, 0.094-0.836; P=0.0225), weakly
positive for WT1 (adjusted OR, 0.368l 95% CI, 0.146-0.928;
P=0.0341), strongly positive for p16 (adjusted OR, 0.498; 95% CI,
0.255-0.972; P=0.0411), weakly positive for p16 (adjusted OR,
0.284; 95% CI, 0.106-0.761; P=0.0123), weakly positive for Ki-67
(adjusted OR, 0.196; 95% CI, 0.058-0.667; P=0.0091), wild-type
p53-negative (adjusted OR, 0.583; 95% CI, 0.378-0.900; P=0.0148)
and mutant p53-positive (adjusted OR, 0.560; 95% CI, 0.333-0.939,
P=0.0280) (Table III).
 | Table IIIStratification analysis of METTL3
polymorphisms with susceptibility to ovarian cancer in rs1061027
C>A. |
Table III
Stratification analysis of METTL3
polymorphisms with susceptibility to ovarian cancer in rs1061027
C>A.
| | rs1061027 C>A
(cases/contorls) | |
|---|
| Variables | CC | CA/AA | Adjusted
ORa (95% CI) |
P-valuea |
|---|
| Age, years | | | | |
|
≤51 | 76/86 | 25/52 | 0.544
(0.308-0.960) | 0.0357 |
|
>51 | 80/81 | 36/51 | 0.715
(0.422-1.210) | 0.2114 |
| Metastasis | | | | |
|
Yes | 55/167 | 24/103 | 0.695
(0.405-1.194) | 0.1876 |
|
No | 90/167 | 32/103 | 0.576
(0.359-0.924) | 0.0222 |
| Clinical stage | | | | |
|
1 | 24/167 | 14/103 | 0.952
(0.470-1.931) | 0.8925 |
|
2 | 30/167 | 10/103 | 0.543
(0.255-1.157) | 0.1136 |
|
3 | 59/167 | 20/103 | 0.542
(0.308-0.954) | 0.0338 |
|
4 | 13/167 | 8/103 | 0.993
(0.397-2.482) | 0.9872 |
| No. of times
pregnant | | | | |
|
≥3 | 67/167 | 22/103 | 0.525
(0.305-0.903) | 0.0200 |
|
<3 | 89/167 | 39/103 | 0.709
(0.452-1.112) | 0.1343 |
| Menopause | | | | |
|
Post-menopause | 104/167 | 39/103 | 0.578
(0.367-0.911) | 0.0182 |
|
Pre-menopause | 48/167 | 20/103 | 0.676
(0.371-1.231) | 0.2002 |
| ER | | | | |
|
Low | 18/167 | 5/103 | 0.444
(0.160-1.237) | 0.1205 |
|
High | 44/167 | 14/103 | 0.517
(0.270-0.991) | 0.0470 |
| PR | | | | |
|
Low | 22/167 | 4/103 | 0.297
(0.099-0.886) | 0.0295 |
|
High | 26/167 | 10/103 | 0.627
(0.290-1.354) | 0.2345 |
| PAX8 | | | | |
|
Low | 23/167 | 4/103 | 0.281
(0.094-0.836) | 0.0225 |
|
High | 36/167 | 15/103 | 0.668
(0.348-1.281) | 0.2247 |
| WTI | | | | |
|
Low | 26/167 | 6/103 | 0.368
(0.146-0.928) | 0.0341 |
|
High | 48/167 | 18/103 | 0.598
(0.330-1.087) | 0.0915 |
| p16 | | | | |
|
Low | 28/167 | 5/103 | 0.284
(0.106-0.761) | 0.0123 |
|
High | 42/167 | 13/103 | 0.498
(0.255-0.972) | 0.0411 |
| Ki-67 | | | | |
|
Low | 25/167 | 3/103 | 0.196
(0.058-0.667) | 0.0091 |
|
High | 43/167 | 21/103 | 0.790
(0.444-1.406) | 0.4227 |
| Wild-type p53 | | | | |
|
Positive | 37/167 | 16/103 | 0.700
(0.370-1.323) | 0.2723 |
|
Negative | 114/167 | 41/103 | 0.583
(0.378-0.900) | 0.0148 |
| Mutant p53 | | | | |
|
Positive | 72/167 | 25/103 | 0.560
(0.333-0.939) | 0.0280 |
|
Negative | 79/167 | 32/103 | 0.655
(0.405-1.057) | 0.0834 |
For the rs1061026 gene polymorphism, the TG/GG
genotype reduced the risk of developing ovarian cancer compared
with the TT genotype in those with clinical staged stage I disease
(adjusted OR, 0.253; 95% CI, 0.075-0.850; P=0.0262), those at
post-menopause (adjusted OR, 0.548; 95% CI, 0.312-0.963; P=0.0366),
those who were weakly positive for Ki-67 (adjusted OR, 0.105; 95%
CI, 0.014-0.783; P=0.0279), those who were wild-type p53-negative
(adjusted OR, 0.455; 95% CI, 0.260-0.795; P=0.0057) and mutant
p53-negative (OR, 0.487; 95% CI, 0.264-0.898; P=0.0211) (Table IV).
 | Table IVStratification analysis of METTL3
polymorphisms with susceptibility to ovarian cancer in rs1061026
T>G. |
Table IV
Stratification analysis of METTL3
polymorphisms with susceptibility to ovarian cancer in rs1061026
T>G.
| | rs1061026 T>G
(cases/controls) | |
|---|
| Variables | TT | TG/GG | Adjusted
ORa (95% CI) |
P-valuea |
|---|
| Age, years | | | | |
|
≤51 | 87/104 | 15/34 | 0.527
(0.270-1.032) | 0.0617 |
|
>51 | 103/104 | 19/27 | 0.711
(0.372-1.357) | 0.3006 |
| Metastasis | | | | |
|
Yes | 72/208 | 13/61 | 0.624
(0.323-1.204) | 0.1597 |
|
No | 105/208 | 18/61 | 0.585
(0.329-1.041) | 0.0684 |
| Clinical stage | | | | |
|
1 | 39/208 | 3/61 | 0.253
(0.075-0.850) | 0.0262 |
|
2 | 34/208 | 6/61 | 0.597
(0.239-1.490) | 0.2693 |
|
3 | 66/208 | 13/61 | 0.679
(0.351-1.314) | 0.2506 |
|
4 | 14/208 | 7/61 | 1.742
(0.671-4.527) | 0.2545 |
| No. of times
pregnan | | | | |
|
≥3 | 80/208 | 14/61 | 0.602
(0.318-1.137) | 0.1178 |
|
<3 | 110/208 | 20/61 | 0.625
(0.359-1.090) | 0.0979 |
| Menopause | | | | |
|
Post-menopause | 127/208 | 20/61 | 0.548
(0.312-0.963) | 0.0366 |
|
Pre-menopause | 29/208 | 12/61 | 0.634
(0.313-1.284) | 0.2058 |
| ER | | | | |
|
Low | 24/208 | 3/61 | 0.443
(0.129-1.528) | 0.1976 |
|
High | 48/208 | 10/61 | 0.708
(0.338-1.482) | 0.3592 |
| PR | | | | |
|
Low | 19/208 | 7/61 | 1.252
(0.502-3.119) | 0.6296 |
|
High | 31/208 | 5/61 | 0.541
(0.201-1.454) | 0.2233 |
| PAX8 | | | | |
|
Low | 33/208 | 5/61 | 0.743
(0.271-2.040) | 0.5646 |
|
High | 44/208 | 10/61 | 0.783
(0.372-1.650) | 0.5204 |
| WTI | | | | |
|
Low | 28/208 | 6/61 | 0.764
(0.301-1.939) | 0.5707 |
|
High | 57/208 | 11/61 | 0.667
(0.329-1.353) | 0.2621 |
| p16 | | | | |
|
Low | 29/208 | 5/61 | 0.610
(0.226-1.650) | 0.3307 |
|
High | 49/208 | 10/61 | 0.702
(0.335-1.469_ | 0.3473 |
| Ki-67 | | | | |
|
Low | 32/208 | 1/61 | 0.105
(0.014-0.783) | 0.0279 |
|
High | 54/208 | 12/61 | 0.765
(0.284-1.522) | 0.4447 |
| Wild-type p53 | | | | |
|
Positive | 39/208 | 13/61 |
1.135(0.569-2.264) | 0.7183 |
|
Negative | 144/208 | 19/61 |
0.455(0.260-0.795) | 0.0057 |
| Mutant p53 | | | | |
|
Positive | 77/208 | 17/61 | 0.761
(0.418-1.384) | 0.3707 |
|
Negative | 106/208 | 15/61 | 0.487
(0.264-0.898) | 0.0211 |
METTL3 haplotype analysis
Polymorphisms of rs1263801, rs1061027 and rs1061026
were selected for haplotype analysis, as demonstrated in Table V; haplotype GCT was used as a
control. It was found that the risk of developing ovarian cancer
was significantly reduced in subjects with haplotype CAT (adjusted
OR, 0.638; 95% CI, 0.434-0.940; P=0.023) and haplotype CAG
(adjusted OR, 0.285; 95% CI, 0.095-0.858; P=0.026).
 | Table VAssociation between inferred
haplotypes of the METTL3 genes and the risk of developing ovarian
cancer. |
Table V
Association between inferred
haplotypes of the METTL3 genes and the risk of developing ovarian
cancer.
| Haplotypes | Cases (n=412), n
(%) | Controls (n=538), n
(%) | Crude OR (95%
CI) |
P-valuea | Adjusted OR (95%
CI) |
P-valueb |
|---|
| CT | 288 (69.90) | 352 (65.43) | 1.000 | | 1.000 | |
| CAT | 47 (11.41) | 89 (16.54) | 0.645
(0.439-0.950) | 0.026 | 0.638
(0.434-0.940) | 0.023 |
| CCG | 26 (6.31) | 43 (7.99) | 0.739
(0.443-1.232) | 0.246 | 0.748
(0.448-1.248) | 0.266 |
| CCT | 30 (7.28) | 20 (3.72) | 1.833
(1.019-3.297) | 0.043 | 1.796
(0.997-3.234) | 0.051 |
| GAT | 12 (2.91) | 10 (1.86) | 1.467
(0.625-3.444) | 0.379 | 1.454
(0.618-3.419) | 0.391 |
| CAG | 4 (0.97) | 17 (3.16) | 0.288
(0.096-0.864) | 0.026 | 0.285
(0.095-0.858) | 0.026 |
| GCG | 5 (1.21) | 5 (0.93) | 1.222
(0.350-4.263) | 0.753 | 1.141
(0.326-3.995) | 0.836 |
| GAG | 0 | 2 (0.37) | / | 0.980 | / | 0.981 |
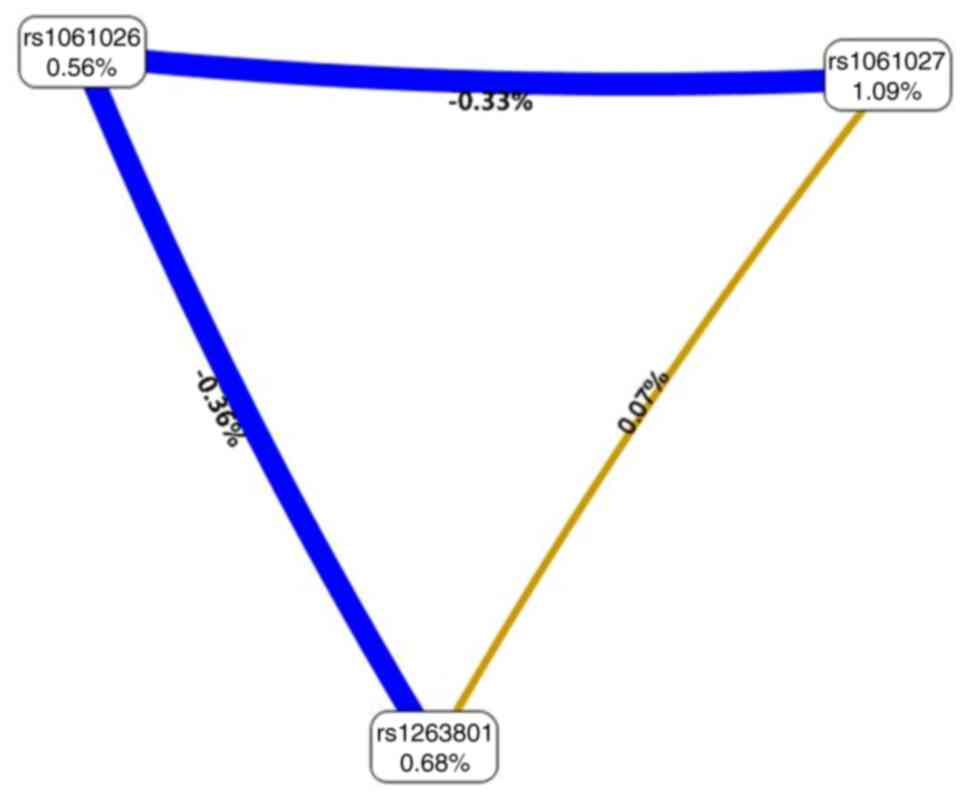
SNP-SNP interactions
The MDR analysis revealed that the CVC value of the
rs1061027 polymorphism as a single factor model in the METTL3 gene
was 10/10, with a testing accuracy of 0.5242, 95% CI, 0.4495-5.5072
and OR, 1.5734. The interaction models rs1263801 x rs1061027 and
rs1263801 x rs1061027 x rs1061026 were not statistically
significant (Table VI). The
interaction map revealed rs1061026 x rs1263801>rs1061026 x
rs1061027>rs1061027 x rs1263801 with negative entropy or
independence (0.36, 0.33 and 0.07%, respectively, indicated in blue
and yellow) (Fig. 1).
 | Table VIOptimal multifactor dimensionality
reduction interaction models. |
Table VI
Optimal multifactor dimensionality
reduction interaction models.
| Locus number | Testing
accuracy | CVC | OR | 95% CI | P-value |
|---|
| rs1061027 | 0.5242 | 10/10 | 1.5734 |
(0.4495-5.5072) | 0.4769 |
| rs1263801,
rs1061027 | 0.4947 | 7/10 | 1.1874 |
(0.3455-4.0813) | 0.785 |
| rs1263801,
rs1061027, rs1061026 | 0.5813 | 10/10 | 1.7637 |
(0.5393-5.7677) | 0.346 |
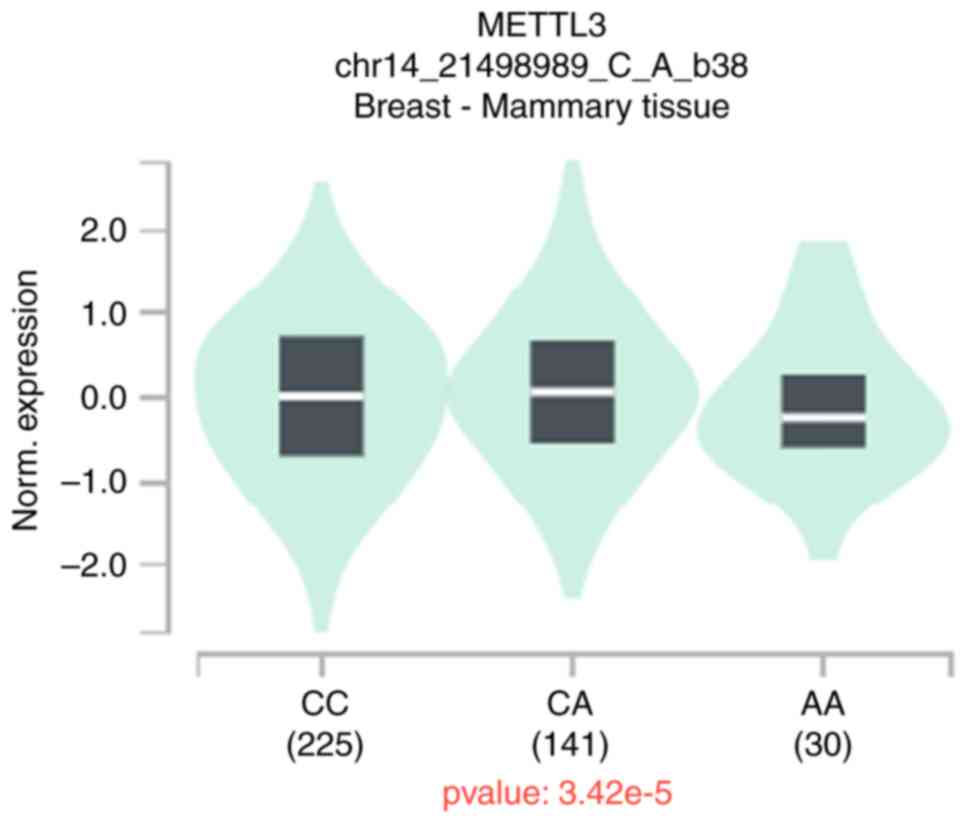
eQTL analysis
To further analyze the functional relevance of
rs1263801 G>C, rs1061027 C>A and rs1061026 T>G, eQTL
analysis was performed using data published by GTEx. The expression
of the appeal locus was not found in the patients with ovarian
cancer; however, it was found that patients with breast cancer who
carry the rs1061027 A genotype have a decreased expression of
METTL3 (Fig. 2).
Discussion
m6A has been reported to be involved in
the regulation of specific developmental processes in eukaryotes.
METTL3 with 580 amino acids is composed of a zinc finger structural
domain and a methyltransferase structural domain. When combined
with METTL14, METTL3 exerts methyltransferase activity and plays a
key role in cancer development as an oncogene or an oncogene
suppressor (13,14). It has been reported that mice
transplanted with ovarian cancer cells accompanied by
myeloid-specific METTL3 knockout exhibited increased tumor growth
(15). Moreover, it has been
reported that METTL3 targeting miR-1246 promotes the proliferation
and migration, and inhibits the apoptosis of ovarian cancer cells
(16). The silencing of METTL3
inhibits miR-126-5p to block the PI3K/Akt/mTOR pathway and inhibit
the development of ovarian cancer (17).
In the present multicenter large sample case-control
study, it was investigated whether METTL3 polymorphisms are
associated with the development of ovarian cancer. First, four SNPs
were screened, among which rs1263801 may affect the binding force
of transcription factors, rs1139130 is located at the splicing
site, and rs1061026 and rs1061027 are the binding sites of miRNAs
(12,18). The present study revealed that the
CC genotype of rs1263801 was a protective factor against ovarian
cancer and was closely related to the risk of developing ovarian
cancer in women aged >51 years. The CA genotype of rs1061027 was
also a protective factor for ovarian cancer, and the results
revealed that compared with the CC genotype, the prevalence of the
CA/AA genotype was lower in women aged <51 years, those who were
pregnant three times or more and those at post-menopause. The same
findings were found in patients with clinical stage III disease and
without metastasis. For rs1061026, it was found that the TG
genotype was associated with a reduced risk of developing ovarian
cancer. Further stratified analysis demonstrated that compared with
the TT genotype, the TG/GG genotype reduced the risk of ovarian
cancer in patients with clinical stage I disease and in
post-menopausal women.
Ki-67 suggests that proliferation is associated with
the prognosis of ovarian cancer (19). It has been reported that Ki-67 and
p53 expression are significantly elevated in ovarian cancer stages
III and IV compared with stages I and II (20). p53 mutations are the most common
mutated genes in ovarian cancer, the majority of which are missense
mutations, resulting in the loss of tumor suppressor function and
enhancing oncogenic function (21,22).
The present study demonstrated that the rs1061027 A
allele and rs1061026 G allele were protective factors against
ovarian cancer in women with a low Ki-67 expression and in
wild-type p53-negative women. However, as regards rs1061027, the
CA/AA genotype reduced the risk of ovarian cancer in mutant
p53-positive women compared with the CC genotype. As regards
rs1061026, the TG/GG genotype reduced the risk of developing
ovarian cancer in mutant p53-negative women compared with the TT
genotype.
PAX8 belongs to the paired-box gene family and plays
a role in tumor growth and participates in multiple oncogenic
pathways (23-25).
It has been reported PAX8 expression is higher in primary ovarian
cancer than in metastatic ovarian cancer; the downregulation of
PAX8 can decrease ovarian cancer cell migration and invasion,
leading to apoptosis (26). The
present study identified rs1061027A as a protective factor for
ovarian cancer in women with a low PAX8 expression.
The present study did not find an association
between rs1263801 G>C, rs1061027C>A and rs1061026 T>G with
METTL3 expression in ovarian tissues by analyzing the data released
by GTEx. Considering that ovarian and breast cancer share the same
cancer-related genes, such as BRCA1/2, p53, Ki-67, PKP3, CHEK2,
PALB2, and PVRL4 (27-29)
Furthermore, it was found that patients with breast cancer who
carry the rs1061027A genotype have a decreased expression of
METTL3; it was thus inferred that rs1061027 affected ovarian cancer
development by influencing the expression of METTL3. However,
further studies are required to confirm these finidngs.
The present study collected cases from three
hospitals; however, as a genetic polymorphism study, the sample
size remains relatively small. Ovarian cancer exhibits multiple
pathological subtypes, each with distinct mechanisms of onset and
prognosis. However, the present study did not standardize for
pathological subtypes. Despite the limitations of the present
study, it was found that METTL3 gene polymorphisms are associated
with susceptibility to ovarian cancer, and the three SNP loci of
the METTL3 gene are independent risk factors. The findings may
provide new insight for the early diagnosis of ovarian cancer.
Further studies are warranted however, to include more cases
meeting the inclusion criteria, classify pathological types and
conduct external validation.
Supplementary Material
Frequency distribution of selected
characteristics in OC cases and cancer-free controls.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National College
Students Innovation and Entrepreneurship Training Program (grant
no. CX18024) and the Science and Technology Projects in Guangzhou
(grant no. 202102010134).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
SS and DJ designed the study. FL performed the
statistical analysis. LL and HW collected the clinical samples. SS
and DJ confirm the authenticity of all the raw data. DJ edited the
manuscript. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the First Affiliated Hospital of Jinan University (KY-2022-233),
the Ethics Committee of Guangzhou Medical University Women and
Children's Medical Center (117A01), and the Ethics Committee of
Shunde Hospital, Southern Medical University (KYLS20220903) and
written informed consent was obtained by all enrolled patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cho KR and Shih I: Ovarian cancer. Annu
Rev Pathol. 4:287–313. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N6-methyladenosine modification in cancers:
Current status and perspectives. Cell Res. 28:507–517.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zheng Q, Ma C, Ullah I, Hu K, Ma RJ, Zhang
N and Sun ZG: Roles of N6-methyladenosine demethylase FTO in
malignant tumors progression. Onco Targets Ther. 14:4837–4846.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhuo Z, Hua R, Chen Z, Zhu J, Wang M, Yang
Z, Zhang J, Li Y, Li L, Li S, et al: WTAP gene variants confer
hepatoblastoma susceptibility: A Seven-center Case-control study.
Mol Ther Oncolytics. 18:118–125. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang X, Guan D, Wang D, Liu H, Wu Y, Gong
W, Du M, Chu H, Qian J and Zhang Z: Genetic variants in
m6A regulators are associated with gastric cancer risk.
Arch Toxicol. 95:1081–1098. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ying P, Li Y, Yang N, Wang X, Wang H, He
H, Li B, Peng X, Zou D, Zhu Y, et al: Identification of genetic
variants in m6A modification genes associated with
pancreatic cancer risk in the Chinese population. Arch Toxicol.
95:1117–1128. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lv J, Song Q, Bai K, Han J, Yu H, Li K,
Zhuang J, Yang X, Yang H and Lu Q: N6-methyladenosine-related
single-nucleotide polymorphism analyses identify oncogene RNFT2 in
bladder cancer. Cancer Cell Int. 22(301)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bian J, Zhuo Z, Zhu J, Yang Z, Jiao Z, Li
Y, Cheng J, Zhou H, Li S, Li L, et al: Association between METTL3
gene polymorphisms and neuroblastoma susceptibility: A nine-centre
case-control study. J Cell Mol Med. 24:9280–9286. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lin A, Zhou M, Hua RX, Zhang J, Zhou H, Li
S, Cheng J, Xia H, Fu W and He J: METTL3 polymorphisms and Wilms
tumor susceptibility in Chinese children: A five-center
case-control study. J Gene Med. 22(e3255)2020.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Chen H, Duan F, Wang M, Zhu J, Zhang J,
Cheng J, Li L, Li S, Li Y, Yang Z, et al: Polymorphisms in METTL3
gene and hepatoblastoma risk in Chinese children: A seven-center
case-control study. Gene. 800(145834)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13(117)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu Y, Song M, Hong Z, Chen W, Zhang Q,
Zhou J, Yang C, He Z, Yu J, Peng X, et al: The N6-methyladenosine
METTL3 regulates tumorigenesis and glycolysis by mediating m6A
methylation of the tumor suppressor LATS1 in breast cancer. J Exp
Clin Cancer Res. 42(10)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang J, Ling D, Shi L, Li H, Peng M, Wen
H, Liu T, Liang R, Lin Y, Wei L, et al: METTL3-mediated m6A
methylation regulates ovarian cancer progression by recruiting
myeloid-derived suppressor cells. Cell Biosci.
13(202)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L,
Liang X and Yang Y: METTL3 promotes the initiation and metastasis
of ovarian cancer by inhibiting CCNG2 expression via promoting the
maturation of pri-microRNA-1246. Cell Death Discov.
7(237)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L,
Liang X and Yang Y: METTL3-mediated maturation of miR-126-5p
promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR
pathway. Cancer Gene Ther. 28:335–349. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xu Z and Taylor JA: SNPinfo: Integrating
GWAS and candidate gene information into functional SNP selection
for genetic association studies. Nucleic Acids Res. 37:W600–W605.
2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kucukgoz Gulec U, Gumurdulu D, Guzel AB,
Paydas S, Seydaoglu G, Acikalin A, Khatib G, Zeren H, Vardar MA and
Altintas A: Prognostic importance of survivin, Ki-67, and
topoisomerase IIα in ovarian carcinoma. Arch Gynecol Obstet.
289:393–398. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Harlozinska A, Bar JK, Sedlaczek P and
Gerber J: Expression of p53 protein and Ki-67 reactivity in ovarian
neoplasms. Correlation with histopathology. Am J Clin Pathol.
105:334–340. 1996.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Walerych D, Napoli M, Collavin L and Del
Sal G: The rebel angel: Mutant p53 as the driving oncogene in
breast cancer. Carcinogenesis. 33:2007–2017. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Walerych D, Lisek K and Del Sal G:
Multi-omics reveals global effects of mutant p53 gain-of-function.
Cell Cycle. 15:3009–3010. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chaves-Moreira D, Morin PJ and Drapkin R:
Unraveling the mysteries of PAX8 in reproductive tract cancers.
Cancer Res. 81:806–810. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Di Palma T and Zannini M: PAX8 as a
potential target for ovarian cancer: What We Know so Far. Onco
Targets Ther. 15:1273–1280. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhou Q, Li H, Cheng Y, Ma X, Tang S and
Tang C: Pax-8: Molecular biology, pathophysiology, and potential
pathogenesis. Biofactors. 50:408–421. 2024.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kim J, Kim NY, Pyo J, Min K and Kang D:
Diagnostic roles of PAX8 immunohistochemistry in ovarian tumors.
Pathol Res Pract. 250(154822)2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang Y, Chen J, Tian J, Zhou Y and Liu Y:
Role and function of plakophilin 3 in cancer progression and skin
disease. Cancer Sci. 115:17–23. 2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Infante M, Arranz-Ledo M, Lastra E,
Olaverri A, Ferreira R, Orozco M, Hernández L, Martínez N and Durán
M: Profiling of the genetic features of patients with breast,
ovarian, colorectal and extracolonic cancers: Association to CHEK2
and PALB2 germline mutations. Clin Chim Acta.
552(117695)2024.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Nanamiya T, Takane K, Yamaguchi K, Okawara
Y, Arakawa M, Saku A, Ikenoue T, Fujiyuki T, Yoneda M, Kai C and
Furukawa Y: Expression of PVRL4, a molecular target for cancer
treatment, is transcriptionally regulated by FOS. Oncol Rep.
51(17)2024.PubMed/NCBI View Article : Google Scholar
|