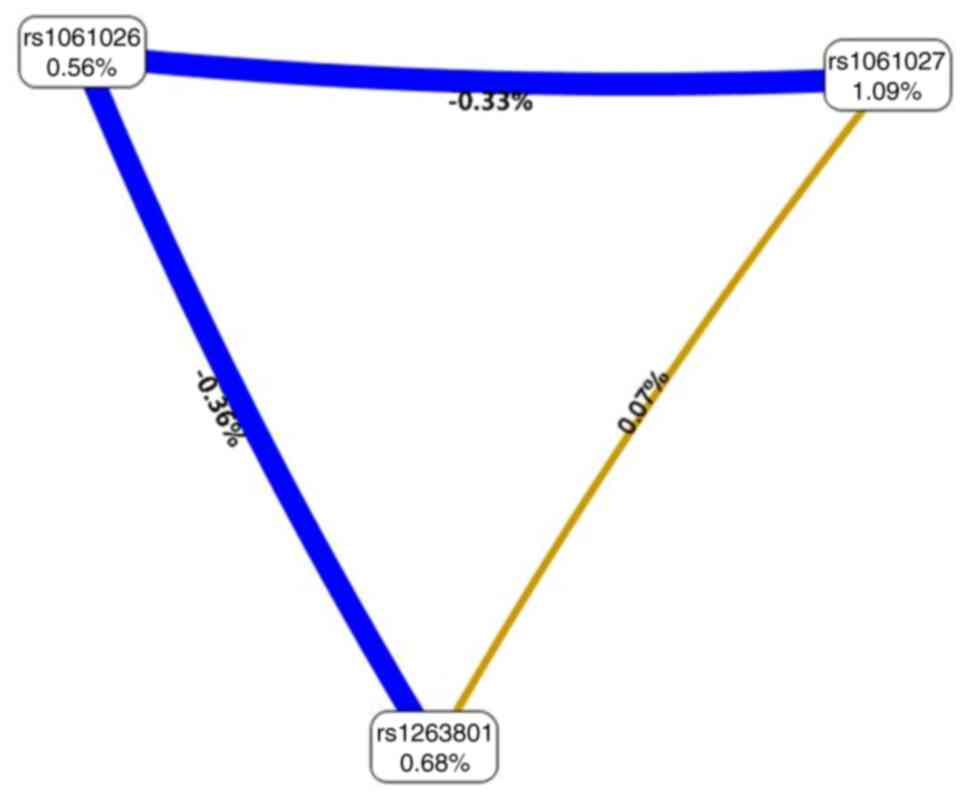
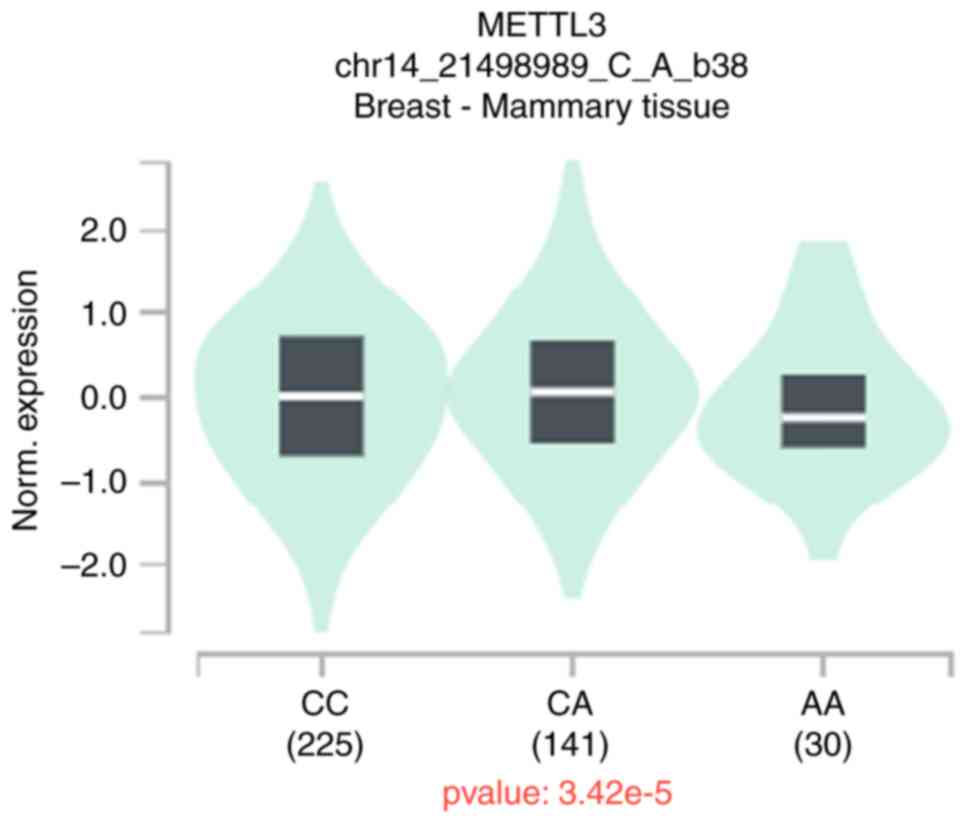
|
1
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cho KR and Shih I: Ovarian cancer. Annu
Rev Pathol. 4:287–313. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N6-methyladenosine modification in cancers:
Current status and perspectives. Cell Res. 28:507–517.
2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zheng Q, Ma C, Ullah I, Hu K, Ma RJ, Zhang
N and Sun ZG: Roles of N6-methyladenosine demethylase FTO in
malignant tumors progression. Onco Targets Ther. 14:4837–4846.
2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhuo Z, Hua R, Chen Z, Zhu J, Wang M, Yang
Z, Zhang J, Li Y, Li L, Li S, et al: WTAP gene variants confer
hepatoblastoma susceptibility: A Seven-center Case-control study.
Mol Ther Oncolytics. 18:118–125. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang X, Guan D, Wang D, Liu H, Wu Y, Gong
W, Du M, Chu H, Qian J and Zhang Z: Genetic variants in
m6A regulators are associated with gastric cancer risk.
Arch Toxicol. 95:1081–1098. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ying P, Li Y, Yang N, Wang X, Wang H, He
H, Li B, Peng X, Zou D, Zhu Y, et al: Identification of genetic
variants in m6A modification genes associated with
pancreatic cancer risk in the Chinese population. Arch Toxicol.
95:1117–1128. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lv J, Song Q, Bai K, Han J, Yu H, Li K,
Zhuang J, Yang X, Yang H and Lu Q: N6-methyladenosine-related
single-nucleotide polymorphism analyses identify oncogene RNFT2 in
bladder cancer. Cancer Cell Int. 22(301)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bian J, Zhuo Z, Zhu J, Yang Z, Jiao Z, Li
Y, Cheng J, Zhou H, Li S, Li L, et al: Association between METTL3
gene polymorphisms and neuroblastoma susceptibility: A nine-centre
case-control study. J Cell Mol Med. 24:9280–9286. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lin A, Zhou M, Hua RX, Zhang J, Zhou H, Li
S, Cheng J, Xia H, Fu W and He J: METTL3 polymorphisms and Wilms
tumor susceptibility in Chinese children: A five-center
case-control study. J Gene Med. 22(e3255)2020.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Chen H, Duan F, Wang M, Zhu J, Zhang J,
Cheng J, Li L, Li S, Li Y, Yang Z, et al: Polymorphisms in METTL3
gene and hepatoblastoma risk in Chinese children: A seven-center
case-control study. Gene. 800(145834)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13(117)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu Y, Song M, Hong Z, Chen W, Zhang Q,
Zhou J, Yang C, He Z, Yu J, Peng X, et al: The N6-methyladenosine
METTL3 regulates tumorigenesis and glycolysis by mediating m6A
methylation of the tumor suppressor LATS1 in breast cancer. J Exp
Clin Cancer Res. 42(10)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang J, Ling D, Shi L, Li H, Peng M, Wen
H, Liu T, Liang R, Lin Y, Wei L, et al: METTL3-mediated m6A
methylation regulates ovarian cancer progression by recruiting
myeloid-derived suppressor cells. Cell Biosci.
13(202)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L,
Liang X and Yang Y: METTL3 promotes the initiation and metastasis
of ovarian cancer by inhibiting CCNG2 expression via promoting the
maturation of pri-microRNA-1246. Cell Death Discov.
7(237)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L,
Liang X and Yang Y: METTL3-mediated maturation of miR-126-5p
promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR
pathway. Cancer Gene Ther. 28:335–349. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xu Z and Taylor JA: SNPinfo: Integrating
GWAS and candidate gene information into functional SNP selection
for genetic association studies. Nucleic Acids Res. 37:W600–W605.
2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kucukgoz Gulec U, Gumurdulu D, Guzel AB,
Paydas S, Seydaoglu G, Acikalin A, Khatib G, Zeren H, Vardar MA and
Altintas A: Prognostic importance of survivin, Ki-67, and
topoisomerase IIα in ovarian carcinoma. Arch Gynecol Obstet.
289:393–398. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Harlozinska A, Bar JK, Sedlaczek P and
Gerber J: Expression of p53 protein and Ki-67 reactivity in ovarian
neoplasms. Correlation with histopathology. Am J Clin Pathol.
105:334–340. 1996.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Walerych D, Napoli M, Collavin L and Del
Sal G: The rebel angel: Mutant p53 as the driving oncogene in
breast cancer. Carcinogenesis. 33:2007–2017. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Walerych D, Lisek K and Del Sal G:
Multi-omics reveals global effects of mutant p53 gain-of-function.
Cell Cycle. 15:3009–3010. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chaves-Moreira D, Morin PJ and Drapkin R:
Unraveling the mysteries of PAX8 in reproductive tract cancers.
Cancer Res. 81:806–810. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Di Palma T and Zannini M: PAX8 as a
potential target for ovarian cancer: What We Know so Far. Onco
Targets Ther. 15:1273–1280. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhou Q, Li H, Cheng Y, Ma X, Tang S and
Tang C: Pax-8: Molecular biology, pathophysiology, and potential
pathogenesis. Biofactors. 50:408–421. 2024.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kim J, Kim NY, Pyo J, Min K and Kang D:
Diagnostic roles of PAX8 immunohistochemistry in ovarian tumors.
Pathol Res Pract. 250(154822)2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang Y, Chen J, Tian J, Zhou Y and Liu Y:
Role and function of plakophilin 3 in cancer progression and skin
disease. Cancer Sci. 115:17–23. 2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Infante M, Arranz-Ledo M, Lastra E,
Olaverri A, Ferreira R, Orozco M, Hernández L, Martínez N and Durán
M: Profiling of the genetic features of patients with breast,
ovarian, colorectal and extracolonic cancers: Association to CHEK2
and PALB2 germline mutations. Clin Chim Acta.
552(117695)2024.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Nanamiya T, Takane K, Yamaguchi K, Okawara
Y, Arakawa M, Saku A, Ikenoue T, Fujiyuki T, Yoneda M, Kai C and
Furukawa Y: Expression of PVRL4, a molecular target for cancer
treatment, is transcriptionally regulated by FOS. Oncol Rep.
51(17)2024.PubMed/NCBI View Article : Google Scholar
|