Introduction
Patients are becoming more educated regarding their
health care and studies have shown that both diet and ethnicity
with genetic variants influence cardiovascular disease (CVD)
development (1-4).
Since CVD is considered the leading cause of mortality worldwide,
preventing, reducing and lessening the development of the disease
is critical for improving mortality and morbidity rates (5). Lifestyle and daily diet vary
considerably between countries and continents, and this has been
shown to affect the development of CVD (6-11).
Patients who are aware of this wish to know what effect they will
have from a certain treatment, medicine or lifestyle change. From a
medical professional point of view, it is crucial to have
information on whether a recommended treatment or lifestyle change
will be effective for an individual patient. Variations in the
effects of drugs between patients have been observed and even more
will be discovered in the future (12,13).
From a health-care planning perspective, finances can be
distributed more accurately there is available knowledge on whether
a patient will experience the desired effect of a drug considered
for prescription. Personalized medicine is currently being more
commonly requested by patients, medical professionals and
politicians.
Garlic (Allium sativum L.) has played a
paramount role in traditional medicine, dating back to the ancient
Egyptians. Garlic extracts have been shown to increase nitric oxide
synthase activity, a critical part of vascular function and
angiogenesis (14). Aged garlic
extract (AGE), a garlic supplement, contains a number of active
ingredients, one of these being S-allylcysteine (SAC) (15). SAC has antioxidant properties, it
prevents lipid and protein oxidation, and is the most extensively
studied component of AGE (16-18).
Ahmadi et al (19)
demonstrated that AGE was associated with a reduction in the levels
of an inflammatory biomarker, a lack of progression of coronary
artery calcification (CAC) and with increased vascular function. In
their studies, Budoff et al (20-22)
also demonstrated an association of AGE with a favorable
improvement in oxidative biomarkers, vascular function and the
reduced progression of atherosclerosis measured by CAC progression.
Steiner et al (23)
revealed that AGE exerted a beneficial effect on lipid profiles and
blood pressure. The authors have previously demonstrated an
association between AGE and improved microcirculation, a decrease
in blood pressure and a positive effect on inflammatory biomarkers
(24-26).
A method for predicting the outcome of various drugs
and medical treatments is known as data mining. It is performed by
analyzing large blocks of information and trying to find patterns,
similarities or trends. Translating a large set of data to a trend
or a new-found pattern can make the data easier to interpret and
lead to new and useful information. The method is based on
combining visualization of diagrams and different algorithms to
finding patterns or trends. There are two starting positions,
either driven by subject-specific information and hypotheses using
data mining to confirm the theory or using data mining to generate
data-driven hypotheses. These two approaches should be combined to
avoid flawed conclusions. Probably the most well-known approach is
the cross-industry standard process for data mining (CRISP-DM)
(27-29).
Using this method and approach, the authors previously developed an
algorithm for predicting which patient will have a more favorable
effect on CAC scores and blood pressure following the intake of AGE
capsules (25). The AGE algorithm
was developed using a European cohort. The aim of the present study
was to validate the method of constructing an algorithm for
prediction of which patient will have a more favorable effect of
AGE on CAC and systolic blood pressure (SBP) using data accumulated
in the USA.
Materials and methods
Study outcome
The primary objective was to validate a method for
developing a predictive model with the aim of identifying patients
who would have a more favorable effect of AGE on CAC and SBP after
12 months of AGE intake.
Study population
The data of the patients in the present study were
collected from five previously published studies. Of these studies,
one study was completed in Sweden on a European cohort (25) and four studies were completed in
the USA (20,30-32).
All patients in these studies had an increased risk of developing
CVD. The Swedish cohort was used solely for the initial model
development in the previous study by the authors (25) and was not included in the model
training, testing, or analysis in the present validation study. The
present study exclusively applied the developed method to an
independent US patient cohort to assess external model performance
(n=78). In the studies examined, all patients were sent for a
cardiac computed tomography (CT) for the definition of the CAC
score, blood pressure was measured, and blood samples were
collected and analyzed using standard techniques in the beginning
of the study, at 0 months and after 12 months. All patients were
administered AGE (Kyolic; Wakunaga of America Co. Ltd.) daily for
the duration of 12 months. Upon receiving the data from the studies
in the USA, the inclusion and exclusion criteria from the Swedish
study were applied; hence, patients with a CAC score >1,000, CAC
score <1 and a body mass index (BMI) >40 in the USA cohort
were excluded. Further details and baseline values of the cohorts
are presented in Table I. More
detailed information about the study populations is available in
the previously published studies (20,25,30-32).
 | Table IBaseline data of all the patients
included in the study. |
Table I
Baseline data of all the patients
included in the study.
| | Sweden (n=46) | USA (n=78) | |
|---|
| Parameter | Mean | SD | Mean | SD | P-value |
|---|
| Age (years) | 64.4 | 6.4 | 59.9 | 8.7 | <0.05 |
| BMI | 27.6 | 3.7 | 27.9 | 4.0 | 0.653 |
| CAC | 207.3 | 237.7 | 194.7 | 213.8 | 0.761 |
| CRP
(mg/l)a | 2.2 | 3.6 | 1.0 | 1.4 | <0.05 |
| Triglycerides
(mmol/l) | 1.4 | 0.7 | 1.3 | 0.7 | 0.758 |
| Cholesterol
(mmol/l) | 5.2 | 1.3 | 4.6 | 1.2 | <0.05 |
| HDL (mmol/l) | 1.6 | 0.5 | 1.3 | 0.4 | <0.05 |
| LDL (mmol/l) | 3.4 | 1.1 | 2.7 | 1.0 | <0.05 |
| Il-6
(ng/l)a | 4.5 | 3.7 | 2.0 | 1.6 | <0.05 |
| Homocysteine
(µmol/l)a | 13.4 | 3.8 | 10.7 | 2.4 | <0.05 |
| DBP | 88 | 9 | 82 | 10 | <0.05 |
| SBP | 148 | 19 | 135 | 15 | <0.05 |
Development of the AGE algorithm
The AGE algorithm developed in the Swedish study was
created using CRISP-DM (25,28).
The target variables, i.e., the progression of CAC and SBP, were
created using the measured progression of CAC and SBP following 12
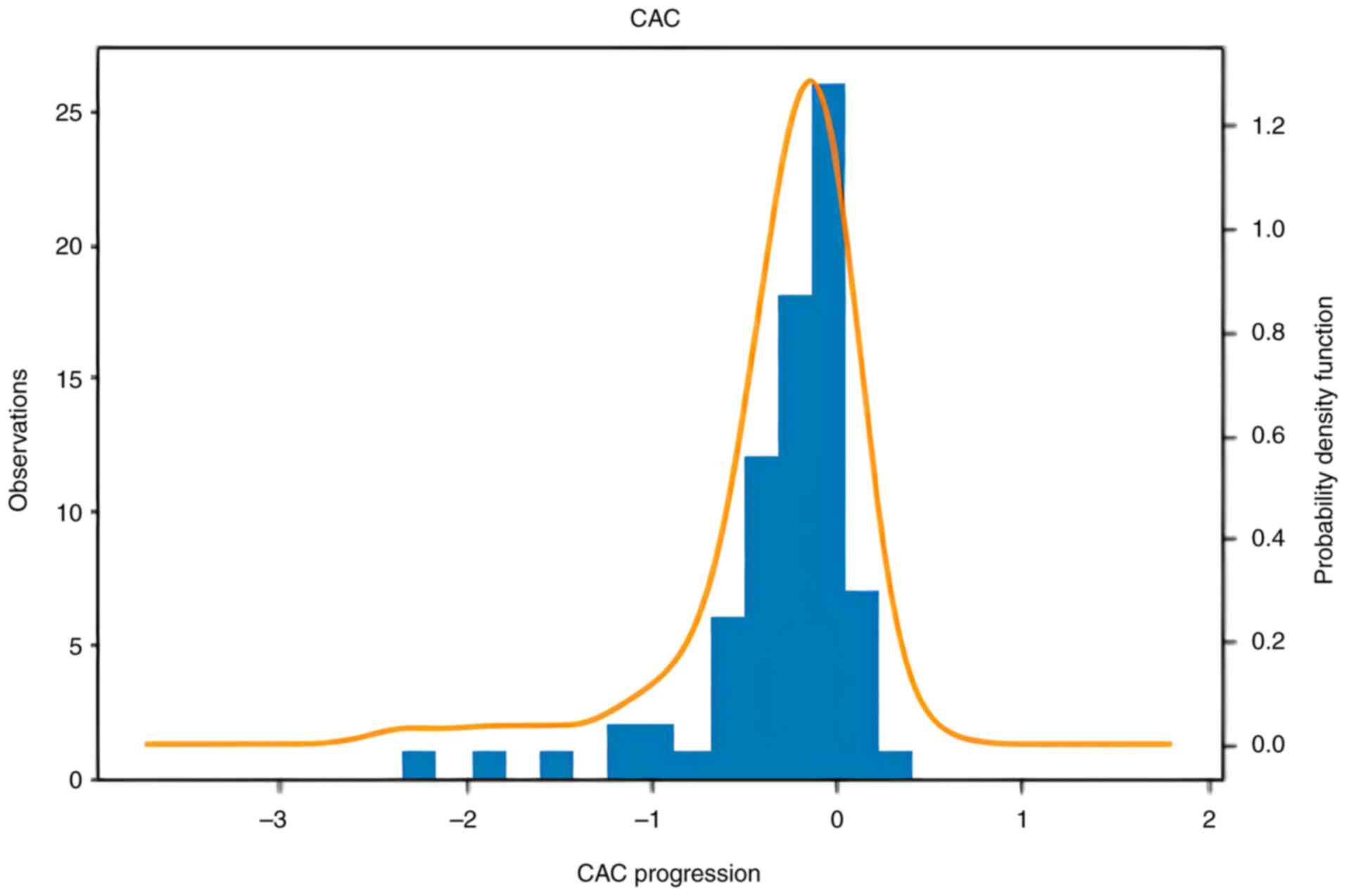
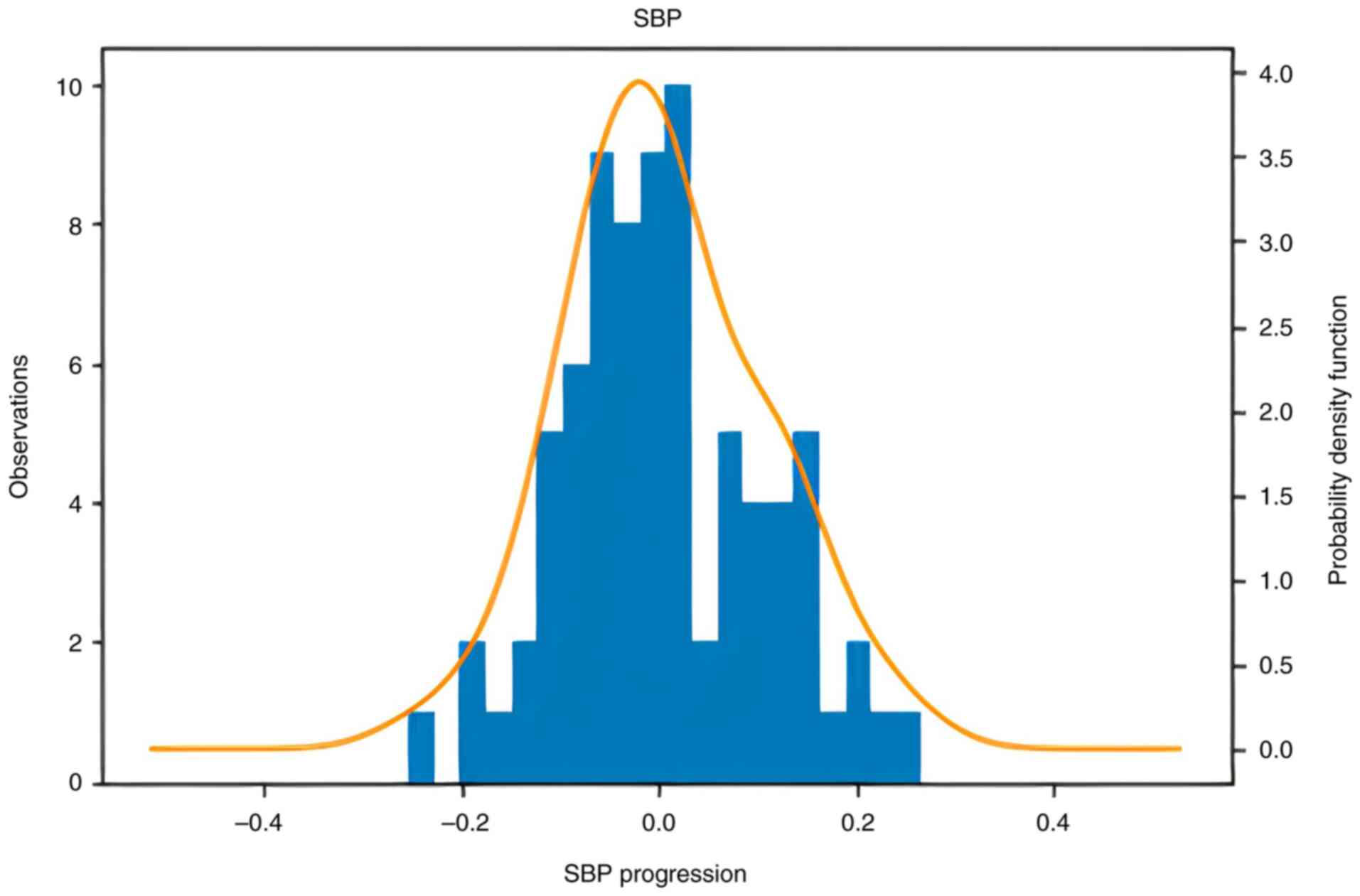
months of treatment with AGE. Information of the distribution of
the progression of CAC and SBP is presented in Figs. 1 and 2. The formula used for measuring the
progression for each target variable was the following:
where ‘B’ represents the baseline measure at the
start of the study and ‘F’ represents the follow-up measure after
12 months of AGE supplementation. To validate the developed method
of engineering an algorithm, new data were acquired from the USA.
The patients in the USA cohort were divided into two groups by the
median, depending on the amount of progression of the target
variables. Patients with a CAC progression larger than the median
were assigned the value 0 and patients with a CAC progression below
the median were assigned the value 1. Patients with a decrease in
SBP larger than the median were assigned the value 1 and the
remainder were assigned the value 0. As a result, 50% of the
patients who benefited most from AGE supplement treatment were
assigned the value 1 and the remainder were assigned the value
0.
Preprocessing and predictor
variables
The following parameters functioned as predictor
variables in the modeling phase: Sex, age, BMI, CAC, cholesterol,
SBP, diastolic blood pressure (DBP), high-density lipoprotein
(HDL), low-density lipoprotein (LDL), homocysteine, interleukin-6
(IL-6), triglycerides and C-reactive protein (CRP). The categorical
variable ‘sex’ was encoded, converting each sex category to 1=male
and 0=female. All parameters were measured at baseline, i.e., at 0
months. In the USA cohort there are missing values of CRP (n=38),
Il-6 (n=41) and homocysteine (n=19). These were completed with the
mean value of the remainder of the cohort for each of the
variables. In the machine-learning algorithm, variables function as
features and were measured ones, i.e. blood samples, clinical
measurements, such as blood pressure, as well as generated ones
from the previously mentioned studies (20,25,30-32).
This was achieved by the addition of polynomial features of the
second-degree and interaction features to capture both non-linear
effects and interactions. Interactions are variables that are
multiplied with other variables and second-degree polynomials are
squared variables. At the end of the preprocessing step the new set
of predictor variables, a total of 105, including the added
features, were standardized. This feature scaling was performed for
the modeling phase in order for the regularization, loss of
function, in the logistic regression to function correctly.
Modeling: Feature selection and
parameter tuning
Following the preprocessing step, the initial
predictor model set consisted of an algorithm with blanks for the
variables. In order to both optimize for the model accuracy and to
reduce overfitting, a feature selection step was performed. The
selected feature selection function was recursive feature
elimination (RFE). This searches for a subset of features by
starting with all features in the training set and removing
features until the desired number remains. RFE functions as a
‘wrapper’ by fitting the given machine-learning algorithm used in
the core of the model, in this case, logistic regression, ranking
features by importance, discarding the least important features and
re-fitting the model. The process is repeated until a specified
number of features remains. Following the feature selection step, a
grid search is performed to optimize for the tuning of the
developed model by searching over a set of hyperparameter settings.
The results from the feature selection and grid search step are
presented in Table II for CAC and
in Table III for SBP.
 | Table IIIncluded features of the CAC
model. |
Table II
Included features of the CAC
model.
| Coefficients | Predictor variables
after RFE for CAC |
|---|
| 3.08 | LDL |
| 1.69 | Ses x
cholesterol |
| -2.05 | Sex x SBP |
| -1.17 | Age x TG |
| -0.95 | BMI x LDL |
| -1.31 | CAC2 |
| 1.73 | CAC x
homocysteine |
| -3.83 | Cholesterol x
LDL |
| 2.75 | Cholesterol x
TG |
| 1.93 | IL-6 x SBP |
| -2.09 | IL-6 x TG |
| 0.12 | LDL2 |
 | Table IIIIncluded features SBP model. |
Table III
Included features SBP model.
| Coefficients | Predictor variables
from RFE for SBP |
|---|
| -1.98 | Sex x DBP |
| 2.08 | Sex x HDL |
| -1.69 | Age x IL-6 |
| 1.55 | Age x CRP |
| 1.41 | CAC x IL-6 |
| -1.77 | CAC x CRP |
| 1.29 | DBP2 |
| -3.30 | HDL x SBP |
| 6.93 | HDL x CRP |
| 1.44 | Homocysteine x
IL-6 |
| -2.41 | LDL x CRP |
| -10.25 | CRP2 |
Validation of the prediction
model
Leave-one-out cross validation (LOOCV), was selected
as the validation approach. It provides an estimate as to how the
algorithm will perform on unseen data. LOOCV is performed by
training and testing the algorithm on the N-1 patient, N
representing the total number of patients in the cohort, and a
prediction is made for the excluded patient using the parameters of
that patient. This process is repeated until all observations have
been tested (a visualization of the method is presented in Table IV). When the prediction model was
tested on all 78 patients, a mean key performance indicator score
from all tests was obtained.
 | Table IVValidation. |
Table IV
Validation.
| | Patient 1 | Patient 2 | Patient 3 | … | Patient 77 | Patient 78 |
|---|
| Model 1 | Train | Train | Train | … | Train | Test |
| Model 2 | Train | Train | Train | … | Test | Train |
| Model 3 | Train | Train | Train | … | Train | Train |
| … | … | … | … | … | … | … |
| Model 77 | Train | Test | Train | … | Train | Train |
| Model 78 | Test | Train | Train | … | Train | Train |
Statistical analyses
The models were created in Python 3.9.0 (Van Rossum,
G. & Drake, F.L., 2009. Python 3 Reference Manual, Scotts
Valley, CA: CreateSpace) utilizing the following listed libraries:
scikit-learn (version 0.23.2) (https://pypi.org/project/scikit-learn/0.23.2/), Pandas
(1.1.3) (https://pypi.org/project/pandas/1.1.3/) and NumPy
(1.19.2) (https://pypi.org/project/numpy/1.19.2/). All
continuous data are presented as the mean ± SD. The Student's
t-test was used to assess differences between groups. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
In a previously published article by the authors, we
developed the AGE algorithm on 46 patients in the Swedish cohort.
To validate the method of developing the AGE algorithm in the
present study, data from four studies were retrieved, performed in
the USA, termed the USA cohort. The USA cohort consisted of a total
of 78 patients who received AGE who were included in the present
study. Following the aforementioned developmental steps of the
prediction model, the model was validated using LOOCV. For each of
the 78 subjects in the dataset, the authors computed the estimated
probability of the i:th subject being in the 50th percentile with
the optimal progression following 12 months of AGE supplement
treatment. i:th represents the subject who is being tested on among
the N observations. The following mean modeling key performance
indicators were provided by the LOOCV: CAC: Accuracy, 65%;
precision, 64%; recall, 69%; and SBP: Accuracy, 65%; precision,
66%; recall, 64% (Table V). In
total, 12 features were selected to be in the model for sufficient
accuracy, but to reduce overfitting. For CAC, the 12 features were
the following: LDL, sex x cholesterol, sex x SBP, age x
triglycerides (TG), BMI x LDL, CAC2, CAC x homocysteine,
cholesterol x LDL, cholesterol x TG, Il-6 x SBP, IL-6 x SBP, IL-6 x
TG and LDL2. An overview and each feature of the associated
coefficient are provided in Table
II. For SBP the 12 features were the following: Sex x DBP, sex
x HDL, age x IL-6, age x CRP, CAC x IL-6, CAC x CRP, DBP2, HDL x
SBP, HDL x CRP, homocysteine x IL-6, LDL x CRP and CRP2. An
overview and each feature of the associated coefficient are
provided in Table III.
 | Table VPerformance metrics. |
Table V
Performance metrics.
| Algorithm | Accuracy (%) | Precision (%) | Recall (%) |
|---|
| CAC algorithm | 65 | 64 | 69 |
| SBP algorithm | 65 | 66 | 64 |
Discussion
The terms ‘personalized medicine’, ‘precision
medicine’ and ‘stratified medicine’ are all used to describe the
same concept. The majority of articles, reports and researchers use
these terms interchangeably. The concept is to customize and tailor
medical treatments, drugs and recommendations to each individual
patient depending on the predicted response or risk of disease.
Personalized medicine often uses diagnostic testing with molecular
or cellular analysis to allocate patients to groups. In one aspect
of personalized medicine, pharmacogenomics or pharmacogenetics, the
genetic information of an individual is used to tailor medical
treatments. Although this may appear as a very modern approach,
medical professionals have been making decisions such as these for
decades. It is well established that a number of diseases have a
large hereditary component and patients are questioned about this
for the purpose of determining whether they have a smaller or
larger risk of developing a certain disease or condition. The
medical professional then reaches a decision based on this
information, prescribing the recommended medicine as
‘one-size-fits-all’, meaning that the majority of patients are then
prescribed the same drug and dose. The decision behind this is that
the majority of studies are based on broad population averages.
However, not many patients fit into this description and numerous
drugs will not function in the same manner for everyone. The
majority of drugs are also prescribed along the basis of ‘trial and
error’. Treatment begins with the most common and popular drug and
then in the case that the patient develops side-effects or does not
respond in as expected, the drug is changed to the second drug of
choice on the list for that condition. This often results in
frustrated patients and tired doctors. The notion behind
personalized medicine is to avoid this waste of time and money by
selecting the right medicine or recommendation to start with.
The present study used this notion to create an
algorithm that could be applied to patients who wish to know
whether AGE will have a positive effect on their health. As
described previously, patients with diverse lifestyles, certain
genes and diets respond differently to drugs and dietary
supplements (1-4,6-11).
The authors have previously studied patients who are at an
intermediate-to-high risk of developing cardiovascular events and
created an algorithm, the AGE algorithm, to predict the effects of
AGE on CAC progression and blood pressure of an individual
(25). The algorithm is based on
the CAC score, blood lipids and blood pressure and could predict,
with 80% precision, which patient will have a significantly reduced
CAC progression after 12 months of AGE supplement (25). The same type of algorithm, the one
developed in the present study, could predict with a 64% precision
score, which patients will have a significantly reduced CAC
progression after 12 months of AGE supplementation. The algorithm
could predict with a 66% precision score which patients will have a
significant reduction in blood pressure after 12 months of AGE
supplementation. Unlike existing cardiovascular risk calculators,
such as the Framingham Risk Score (https://www.mdcalc.com/calc/38/framingham-risk-score-hard-coronary-heart-disease)
or the ASCVD Risk Estimator (https://www.mdcalc.com/calc/3398/ascvd-atherosclerotic-cardiovascular-disease-2013-risk-calculator-aha-acc),
which predict the likelihood of future cardiovascular events based
on broad population data, the model used herein is specifically
designed to predict individual patient response to a defined
intervention, AGE supplementation. While previous studies have
demonstrated average cohort-level effects of AGE, to the best of
our knowledge, no previous study to date has developed or validated
a personalized prediction tool estimating individual treatment
responses (33,34). By applying data science
methodologies incorporating interaction terms and non-linear
relationships, this model represents a novel, practical step toward
personalized, supplement-based cardiovascular prevention. The
algorithm demonstrated a precision of 64% for CAC and 66% for SBP,
which is notable given the modest sample size. To the best of our
knowledge, this is the first externally validated prediction tool
tailored to identify which patients are most likely to benefit from
AGE, providing a novel contribution to precision prevention
strategies in cardiovascular care. It is clear there is a potential
in predicting which patients would benefit the most from AGE
supplement treatment using a limited sample (25). Therefore, further validation of the
method of developing the algorithm on another cohort is required to
broaden the concept of using it in day-to-day medical consultations
with patients interested in dietary supplements.
When developing the model, a combination of
variables and engineered features was made available for the model
to use. A balance between model complexity and performance was
sought to maintain the number of features to a minimum, to avoid
overfitting and increase model stability. Initially, a model with
10% of the available features was tested, i.e., 10 out of 105
features in total. The model was performing well; however, a model
with an additional two features was tested, which was found to
outperform the model with 10 features. As a result, a model with 12
features was selected as the final model. Logistic regression was
selected for its interpretability and suitability for binary
outcomes in clinical applications. While other machine learning
methods could be used, logistic regression provided a practical
balance of transparency and predictive performance, particularly in
a limited sample size.
Overall, the models generalized well on the test
data. When selecting the validation method, 10-fold cross
validation was explored; however, it was deemed to be too sensitive
to the random sampling effect when creating the folds. It is
important to realize that in a 10-fold cross validation, 10% of the
sample is used as test, while only having 78 patients, the test
fold consists of only 8 patients. With a limited sample size, the
probability of a test set being skewed is high. LOOCV is a reliable
method that produces an unbiased estimate of the performance of a
model, and it is recommended that it is used in situations where
data are limited. Although LOOCV is computationally expensive, the
limited number of observations render the computational cost
negligible. Therefore, with limited data, the random sampling
effects using 10-fold cross validation, limiting bias and enabling
testing on unseen data renders it the optimal validation method to
use.
As aforementioned, the prediction model performs
better on unseen data, i.e., the more data it is used on.
Generally, the effect from the amount of study data available can
be broken down into two parts. First, the negative random effect on
the partitioning of data into training sets and test sets
diminishes the more data is available, i.e., the representativity
of both the training and test set increases. Second, more data
allow for the addition of more categorical predictor features to
the modeling phase. Increased clinical information, such as
lifestyle, certain genes and dietary intake in categorical form
would increase the performance of the model. This is due to the
fact that a model is built on a sample with a higher variance,
i.e., a larger sample, which results in a model with an intrinsic
ability to generalize more effectively on unseen data. Overall, the
present study indicates the feasibility of developing predictive
models classifying patients into who would have the best results
from the AGE supplement or not. This predictive modeling framework
could also be applied to other areas of biomedical research. Recent
advances in AI-driven treatment prediction, organ-on-a-chip
platforms for drug screening, engineered cardiac organoids for
cardiovascular disease modeling and artificial vascular graft
technologies illustrate the expanding opportunities for integrating
predictive models into precision medicine and translational
cardiovascular research (35-38).
It is important to note that baseline differences
between the Swedish and USA cohorts, particularly in age, LDL and
SBP, could influence model performance across populations. Although
the model incorporated interaction terms and non-linear features to
account for these variations, further validation in larger, more
diverse cohorts will be necessary to confirm broader applicability.
The observed decline in model performance when applied to the USA
cohort is likely explained by differences in baseline variable
distributions, particularly in age, LDL, SBP and CAC scores, which
can alter predictor-outcome relationships. Additionally, as feature
selection was based on the Swedish cohort, the relative importance
of certain variables and interactions may have differed in the USA
population, contributing to reduced accuracy and precision. These
findings highlight the importance of accounting for
population-specific variability in model transportability and
support the need for broader validation in diverse clinical
populations.
The requirements of gathering data to assess who
would benefit the most from AGE supplement treatment are not high:
ordinary blood tests and a CT scan can be utilized to gather the
necessary variables. However, further studies, tests and modeling
are necessary before a model can be used to assess which patient
will have a favorable effect of AGE supplement treatment or
not.
The present study had certain limitations which
should be mentioned. The data in the present study were acquired
from four studies performed previously. Given the variation in
collected variables and the limited number of patients, these could
potentially affect the scores. Furthermore, the relatively small
sample size, with 46 patients in the initial model and 78 in the
validation cohort, may limit the power of the model and increase
the risk of overfitting despite the use of recursive feature
elimination and leave-one-out cross-validation. Thus, larger,
prospective studies are warranted to further validate and improve
the predictive accuracy and generalizability of the model. A larger
study cohort would probably reduce the random effect of
within-group variation. CAC progression was defined as the absolute
change in CAC. A potential site variation in regard to different
brands of the CT scan itself and the analyzing software might also
inflict some variations. The interpreting radiologist differed,
which could be observed as a variance of weakness, but also a
strength. The serum level of SAC was not measured to ensure patient
compliance in the studies. All patients were advised not to change
their lifestyle and diet apart from taking the AGE capsules;
however, studies have indicated that patients enrolled in studies
are more likely to adopt lifestyle changes (39,40).
The lifestyle changes were not assessed formally to evaluate the
effect on the endpoint. A randomized trial of diet and lifestyle
would be needed to better assess these potential interventions.
In conclusion, the present study demonstrates that
it is possible and realistic to develop predictive models,
classifying patients into who would have the optimal results from
AGE supplement treatment or not. The algorithm was able to predict
with 64% precision which patient would have a significant reduction
of CAC progression using the AGE supplement. With the same type of
model, we could also predict with 66% precision which patient would
have a significant blood pressure-lowering effect using the AGE
supplement.
Acknowledgements
The authors would like to thank the statistical
engineers, Mr. Andreas Timglas (Securitas Security Services, New
Jersey, USA) and Mr. Roy Ollila (Fanwl Consulting, Bjärred, Sweden)
for assisting with the development of the prediction model.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MW, MJB, EB and SL conceptualized the study and were
involved in the study methodology. MW and SL were involved in data
validation. MW, MM and SL were involved in the formal analysis. AK,
MJB and MW were involved in the investigative aspects of the study.
SL provided monetary and material resources. AK, MJB and MW were
involved in data curation. MW, EB, MM and SL were involved in the
writing and preparation of the original draft of the manuscript. EB
and SL were involved in writing, review and editing the manuscript.
MW and EB were involved in preparation of figures. SL supervised
the study and was involved in project administration. All authors
have read and agreed to the published version of the manuscript. SL
and MW confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was based on aggregated data from
previously published studies (20,25,30-32).
The previously published studies have all been approved by ethical
boards, and further details are described in the individual
published studies (20,25,30-32).
No individual level data were accessed at any step in the analysis,
and no indirect identification of the study subjects was possible;
additional ethical vetting was not applicable for the present
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brown AW, Kaiser KA, Keitt A, Fontaine K,
Gibson M, Gower BA, Shikany JM, Vorland CJ, Beitz DC, Bier DM, et
al: Science dialogue mapping of knowledge and knowledge gaps
related to the effects of dairy intake on human cardiovascular
health and disease. Crit Rev Food Sci Nutr. 61:179–195.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Astrup A: Yogurt and dairy product
consumption to prevent cardiometabolic diseases: Epidemiologic and
experimental studies. Am J Clin Nutr. 99 (5 Suppl):1235S–1242S.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Diaz-Lopez A, Bullo M, Martinez-Gonzalez
MA, Corella D, Estruch R, Fito M, Gómez-Gracia E, Fiol M, García de
la Corte FJ, Ros E, et al: Dairy product consumption and risk of
type 2 diabetes in an elderly Spanish Mediterranean population at
high cardiovascular risk. Eur J Nutr. 55:349–360. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Leigh JA, Alvarez M and Rodriguez CJ:
Ethnic minorities and coronary heart disease: An update and future
directions. Curr Atheroscler Rep. 18(9)2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
World Health Organization (WHO):
Cardiovascular diseases (CVDs). WHO, Geneva, 2021. https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds).
Accessed July 18, 2021.
|
|
6
|
Prentice RL, Aragaki AK, Van Horn L,
Thomson CA, Beresford SA, Robinson J, Snetselaar L, Anderson GL,
Manson JE, Allison MA, et al: Low-fat dietary pattern and
cardiovascular disease: Results from the Women's Health Initiative
randomized controlled trial. Am J Clin Nutr. 106:35–43.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rees K, Hartley L, Flowers N, Clarke A,
Hooper L, Thorogood M and Stranges S: ‘Mediterranean’ dietary
pattern for the primary prevention of cardiovascular disease.
Cochrane Database Syst Rev CD009825, 2013.
|
|
8
|
Denke MA: Diet and lifestyle modification
and its relationship to atherosclerosis. Med Clin North Am.
78:197–223. 1994.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ruan Y, Huang Y, Zhang Q, Qin S, Du X and
Sun Y: Association between dietary patterns and hypertension among
Han and multi-ethnic population in southwest China. BMC Public
Health. 18(1106)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang CJ, Shen YX and Liu Y: Empirically
derived dietary patterns and hypertension likelihood: A
meta-analysis. Kidney Blood Press Res. 41:570–581. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang X, Ding N, Tucker KL, Weisskopf MG,
Sparrow D, Hu H and Park SK: A Western diet pattern is associated
with higher concentrations of blood and bone lead among middle-aged
and elderly men. J Nutr. 147:1374–1383. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dean L and Kane M: Codeine Therapy and
CYP2D6 Genotype. In: Medical Genetics Summaries. Pratt VM, Scott
SA, Pirmohamed M, et al (eds). National Center for
Biotechnology Information (US), Bethesda, MD, 2012.
|
|
13
|
Torkamani A, Pham P, Libiger O, Bansal V,
Zhang G, Scott-Van Zeeland AA, Tewhey R, Topol EJ and Schork NJ:
Clinical implications of human population differences in
genome-wide rates of functional genotypes. Front Genet.
3(211)2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Das I, Khan NS and Sooranna SR: Potent
activation of nitric oxide synthase by garlic: A basis for its
therapeutic applications. Curr Med Res Opin. 13:257–263.
1995.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zeb I, Ahmadi N, Nasir K, Kadakia J,
Larijani VN, Flores F, Li D and Budoff MJ: Aged garlic extract and
coenzyme Q10 have favorable effect on inflammatory markers and
coronary atherosclerosis progression: A randomized clinical trial.
J Cardiovasc Dis Res. 3:185–190. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ide N, Matsuura H and Itakura Y:
Scavenging effect of aged garlic extract and its constituents on
active oxygen species. Phytotherapy Res. 10:340–341. 1996.
|
|
17
|
Numagami Y and Ohnishi ST: S-allylcysteine
inhibits free radical production, lipid peroxidation and neuronal
damage in rat brain ischemia. J Nutr. 131 (3s):1100S–1105S.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Maldonado PD, Barrera D, Rivero I, Mata R,
Medina-Campos ON, Hernandez-Pando R and Pedraza-Chaverrí J:
Antioxidant S-allylcysteine prevents gentamicin-induced oxidative
stress and renal damage. Free Radic Biol Med. 35:317–324.
2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ahmadi N, Nabavi V, Hajsadeghi F, Zeb I,
Flores F, Ebrahimi R and Budoff M: Aged garlic extract with
supplement is associated with increase in brown adipose, decrease
in white adipose tissue and predict lack of progression in coronary
atherosclerosis. Int J Cardiol. 168:2310–2314. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Budoff MJ, Ahmadi N, Gul KM, Liu ST,
Flores FR, Tiano J, Miller E and Tsimikas S: Aged garlic extract
supplemented with B vitamins, folic acid and L-arginine retards the
progression of subclinical atherosclerosis: A randomized clinical
trial. Prev Med. 49:101–107. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Budoff M: Aged garlic extract retards
progression of coronary artery calcification. J Nutr. 136 (3
Suppl):741S–744S. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Budoff MJ, Takasu J, Flores FR, Niihara Y,
Lu B, Lau BH, Rosen RT and Amagase H: Inhibiting progression of
coronary calcification using Aged Garlic Extract in patients
receiving statin therapy: A preliminary study. Prev Med.
39:985–991. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Steiner M, Khan AH, Holbert D and Lin RI:
A double-blind crossover study in moderately hypercholesterolemic
men that compared the effect of aged garlic extract and placebo
administration on blood lipids. Am J Clin Nutr. 64:866–870.
1996.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wlosinska M, Nilsson AC, Hlebowicz J,
Malmsjo M, Fakhro M and Lindstedt S: Aged garlic extract preserves
cutaneous microcirculation in patients with increased risk for
cardiovascular diseases: A double-blinded placebo-controlled study.
Int Wound J. 16:1487–1493. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wlosinska M, Nilsson AC, Hlebowicz J,
Hauggaard A, Kjellin M, Fakhro M and Lindstedt S: The effect of
aged garlic extract on the atherosclerotic process-a randomized
double-blind placebo-controlled trial. BMC Complement Med Ther.
20(132)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wlosinska M, Nilsson AC, Hlebowicz J,
Fakhro M, Malmsjo M and Lindstedt S: Aged garlic extract reduces
IL-6: A double-blind placebo-controlled trial in females with a low
risk of cardiovascular disease. Evid Based Complement Alternat Med.
2021(6636875)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rivo E, de la Fuente J, Rivo A,
Garcia-Fontan E, Canizares MA and Gil P: Cross-industry standard
process for data mining is applicable to the lung cancer surgery
domain, improving decision making as well as knowledge and quality
management. Clin Transl Oncol. 14:73–79. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Harper G and Pickett SD: Methods for
mining HTS data. Drug Discov Today. 11:694–699. 2006.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cruz-Bermudez JL, Parejo C, Martinez-Ruiz
F, Sanchez-Gonzalez JC, Ramos Martin-Vegue A, Royuela A,
Rodríguez-González A, Menasalvas-Ruiz E and Provencio M: Applying
data science methods and tools to unveil healthcare use of lung
cancer patients in a teaching hospital in Spain. Clin Transl Oncol.
21:1472–1481. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Larijani VN, Ahmadi N, Zeb I, Khan F,
Flores F and Budoff M: Beneficial effects of aged garlic extract
and coenzyme Q10 on vascular elasticity and endothelial function:
The FAITH randomized clinical trial. Nutrition. 29:71–75.
2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shaikh K, Kinninger A, Cherukuri L,
Birudaraju D, Nakanishi R, Almeida S, Jayawardena E, Shekar C,
Flores F, Hamal S, et al: Aged garlic extract reduces low
attenuation plaque in coronary arteries of patients with diabetes:
A randomized, double-blind, placebo-controlled study. Exp Ther Med.
19:1457–1461. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Matsumoto S, Nakanishi R, Li D, Alani A,
Rezaeian P, Prabhu S, Abraham J, Fahmy MA, Dailing C, Flores F, et
al: Aged garlic extract reduces low attenuation plaque in coronary
arteries of patients with metabolic syndrome in a prospective
randomized double-blind study. J Nutr. 146:427S–432S.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ried K, Frank OR and Stocks NP: Aged
garlic extract reduces blood pressure in hypertensives: A
dose-response trial. Eur J Clin Nutr. 67:64–70. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ried K, Travica N and Sali A: The effect
of aged garlic extract on blood pressure and other cardiovascular
risk factors in uncontrolled hypertensives: The AGE at Heart trial.
Integr Blood Press Control. 9:9–21. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bai L, Jing Y, Reis RL, Chen X, Su J and
Liu C: Organoid research: Theory, technology, and therapeutics. OR.
1(025040007)2025.
|
|
36
|
Zhao J, Shen F, Sheng S, Wang J, Wang M,
Wang F, Jiang Y, Jing Y, Xu K and Su J: Cartilage-on-chip for
osteoarthritis drug screening. Organoid Res. 1(8461)2025.
|
|
37
|
Hu K, Li Y, Ke Z, Yang H, Lu C, Li Y, Guo
Y and Wang W: History, progress and future challenges of artificial
blood vessels: A narrative review. Biomater Transl. 3:81–98.
2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang Y, Hou Y, Hao T, Garcia-Contreras M,
Li G, Cretoiu D and Xiao J: Model construction and clinical
therapeutic potential of engineered cardiac organoids for
cardiovascular diseases. Biomater Transl. 5:337–354.
2024.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Schmidt SK, Hemmestad L, MacDonald CS,
Langberg H and Valentiner LS: Motivation and barriers to
maintaining lifestyle changes in patients with type 2 diabetes
after an intensive lifestyle intervention (The U-TURN Trial): A
longitudinal qualitative study. Int J Environ Res Public Health.
17(7454)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Jones DE, Carson KA, Bleich SN and Cooper
LA: Patient trust in physicians and adoption of lifestyle behaviors
to control high blood pressure. Patient Educ Couns. 89:57–62.
2012.PubMed/NCBI View Article : Google Scholar
|