Introduction
Breast cancer is the most common cancer in women and
one of the leading causes of morbidity in women between the ages of
40 and 44. This malignancy represents a heterogeneous group of
tumors with varied morphology, behavior and response to therapy.
Nek2 is a serine/threonine kinase of the NIMA-related kinase family
that localizes to the centrosomes (1–4),
the microtubule-organizing centers of a cell. It is involved in
cell division and mitotic regulation by centrosome splitting
(1,2,4).
When compared to normal breast tissue (NBT), breast carcinoma cells
have a much higher frequency of centrosome defects, including
amplification of centrosome number, increased volume, and
supernumerary centrioles (5,6).
According to Liu et al, the elevation of Nek2 contributes to
chromosome instability and promotes aneuploidy by disrupting the
control of the mitotic checkpoint (7). Nek2 is upregulated in a number of
human cancer cell lines, including breast, cervical and prostate
carcinomas, and its downregulation inhibits cell proliferation
(8–10). Nek2 has 3 splice variants in
humans, namely, Nek2A, Nek2B and Nek2C. Nek2A is 445 amino acids in
length (48 kDa) and is considered as a full-length protein. Nek2B
is 384 amino acids long (44 kDa) and arises through the use of an
alternative polyadenylation site within the terminal intron. These
characteristics indicate that Nek2A and Nek2B are identical up to
residue 370 but differ in their extreme C-termini. Nek2C arises
from an alternative splicing event, excising 8 amino acids
(371–378) from the C-terminal domain of the full-length protein
(11,12). Similar to many protein kinases
with roles in mitosis, Nek2C is overexpressed in breast carcinoma
in a tumor-specific manner. However, the role of this protein in
breast carcinomas has not been fully elucidated.
A comprehensive and consistent depiction of the
genetic changes that underlie breast cancer initiation, development
and progression has not yet been provided. The MCF10 model includes
a series of cell lines, such as the normal immortalized breast cell
line (MCF10A), atypical ductal hyperplasia (ADH; MCF10AT), ductal
carcinoma in situ (DCIS; MCF10DCIS.com) and invasive
carcinoma (MCF10CA1a) cell lines. These 4 cell lines resemble the
initiation, development and progression steps of breast carcinoma,
respectively. Their cytogenetic and molecular variation may help to
reveal genetic changes which are relevant to breast cancer
(13). MCF10A (a non-transformed,
near diploid, spontaneously immortalized human mammary epithelial
cell line) has been used as a normal immortalized breast epithelium
control for studies of human breast cell lines (14).
The above studies reported the function of Nek2A and
Nek2B in tumor cell lines, however, few studies have focused on the
role of Nek2C in breast cancer MCF10 cell lines. In this study, we
detected Nek2C mRNA levels not only in MCF10 cell lines (MCF10A,
MCF10AT, MCF10DCIS.com and MCF10CA1a), but also in breast tissue,
including NBT, ADH, DCIS and invasive ductal carcinoma (IDC)
tissues. This study provides information regarding Nek2C mRNA
expression in breast tissue and suggests a correlation between
Nek2C mRNA expression and the development and progression of breast
carcinoma. The results from our study provide evidence that Nek2C
is a novel potential biomarker for the diagnosis and treatment of
human breast cancer.
Materials and methods
Cell lines and cultures
The MCF10A (normal immortalized breast epithelial
cell line), MCF10AT (ADH) and MCF10CA1a (malignant breast
epithelial) cell lines were purchased from the Barbara Ann Karmanos
Cancer Institute of Wayne State University, Detroit, MI, USA. The
MCF10DCIS.com cell line (DCIS) was purchased from an Asterand
Business Development Representative. The breast cancer cell lines,
MCF10DCIS.com and MCF10CA1a, were grown in DMEM/F12 (1:1) with 5%
horse serum, 0.029 M sodium bicarbonate, 10 mM HEPES, penicillin
and streptomycin. The culture media for the MCF10A and MCF10AT
series were similar but also included insulin (10 μg/ml),
EGF (20 ng/ml), hydrocortisone (0.5 μg/ml), and cholera
toxin (100 ng/ml). All cell lines were cultured as recommended by
the suppliers, and all cell assays were conducted in
serum-containing growth medium unless otherwise specified. All cell
lines were passaged for <6 months in this study.
siRNA interference
The siRNAs targeting Nek2C, obtained from Invitrogen
(Beijing, China), are referred to as Nek2C siRNA-1, siRNA-2 and
siRNA-3 as follows: Nek2C siRNA-1 sense, UUUGUAAUUACACUAGCCAGAUCCC
and antisense, GGGAUCUGGCUAGUGUAAUUACAAA; Nek2C siRNA-2 sense,
UUAAUAUUCUAGCUAGCCCAAAGUC and antisense, GACUUUGGGCUAGCUAGAAUAUUAA;
Nek2C siRNA-3 sense, CAUUAAUGCACAUAACUCAU ACAGC and antisense,
GCUGUAUGAGUUAUGUGCAUU AAUG. Control siRNAs containing non-specific
sequences without homologs in the human genome were also provided
by Invitrogen (Cat no. 12935-300). The transfection procedure was
achieved using Lipofectamine™ RNAiMAX reagent (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
Seventy-two hours after transfection using the Lipofectamine™ PLUS
transfection reagent (Invitrogen), MCF10DCIS.com and MCF10CA1a
cells were harvested and used for further experiments.
Plasmid transfection
The Nek2C expression plasmid (pcDNA-mycNek2C) was
generated by cloning its coding region into the pcDNA3.0-myc vector
(Invitrogen, Beijing, China) and the pcDNA3.0-myc vector was used
as the control. For DNA transfections, MCF10A and MCF10AT cells
were plated at 300,000 cells/well in 6-well plates 2 days prior to
transfection and cultured to 70% confluence the following day in 5%
horse serum. The following day, cells were transfected with
pcDNA3.1-Nek2C or with the empty vector as the control according to
the manufacturer’s instructions using Lipofectamine 2000
(Invitrogen). Seventy-two hours after transfection, the cells were
harvested and used for further experiments.
Semiquantitative reverse
transcription-PCR (RT-PCR)
Total-RNA was isolated from cultured cells using TRI
reagent (Invitrogen) according to the manufacturer’s instructions.
RNA was treated with DNase I RNase-free (Roche) and purified over
an RNeasy column. The first-strand complementary DNA was
synthesized with oligo(dT) primer by using the Reverse
Transcription System (Promega Biotech, Beijing, China). PCR
amplification of Nek2C-specific fragments of 129 bp was performed
in a 20 μl reaction using TaqDNA polymerase (Invitrogen),
with 1 μl of the first-strand cDNA synthesis mixture as the
template, and the following primers: 1077F,
5′-GGAACGGAAGTTCCTGTC-3′; and 1229R, 5′-CACTTGGACTTAGATGTGA-3′. The
conditions for PCR reactions were as follows: 95°C for 5 min, 95°C
for 15 sec, 62.8°C for 15 sec, 72°C for 20 sec and 72°C for 5 min,
performed for 35 cycles. The expression of GAPDH was used as the
internal control.
Western blot analysis
Seventy-two hours after transfection with the
plasmid or siRNA, the cells were harvested and washed with ice-cold
phosphate buffer. Cells were lysed in RIPA buffer (150 mmol/l NaCl,
50 mmol/l Tris-HCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 0.5%
sodium deoxycholate and 1 mmol/l phenylmethylsulfonyl fluoride).
Protein concentrations were determined using the BCA method (Thermo
Scientific-Pierce Biotechnology, Inc.). Proteins were resolved in
sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and
transferred onto polyvinylidene fluoride membranes (Millipore,
Bedford, MA). Membranes were incubated with the following primary
antibodies overnight at 4°C: Nek2 (1:500; Sigma, St. Louis, MO),
γ-tubulin (1:1,000, Sigma), myc (1:2,000; Cell Signaling
Technology, Inc.) and β-actin (1:2,000; Santa Cruz Biotechnology,
Inc.). After incubation with secondary antibodies, immunostained
bands were detected by ECL (Thermo Scientific-Pierce Biotechnology,
Inc.) western blotting reagents. The expression of β-actin was used
as the internal control.
Cell growth analysis
Cells were cultured in 96-well plates at a density
of 1×104 cells/well. After transfection, the quantity of
viable cells was estimated by a colorimetric assay using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT).
MTT (10 μl of 5 mg/ml solution, Sigma) was added to each
well of the titration plate and incubated at 37°C for 4 h. The
cells were then treated with DMSO (40 μl/well) and incubated
at 37°C for 1 h. The absorbance of each well was determined at 570
nm. The percentage viability was defined as the relative absorbance
of the transfected vs. blank control cells. All experiments were
performed with 5-wells/experiment and repeated at least 3
times.
Assessment of percentage of apoptotic
cells
To detect apoptotic cells, the cells were stained
with DNA binding dye Hoechst 33342 (Sigma). Cells with the
indicated treatment were fixed with 4% formaldehyde in
phosphate-buffered saline (PBS) for 10 min at 4°C and then washed
with PBS. Cells were incubated for 20 min with 5 μg/ml of
Hoechst 33342 (Sigma) to stain the nuclei. After washing with PBS,
the apoptotic cells were observed under a confocal fluorescence
microscope (Leica TCS SMD FCS, Leica, Wetzlar, Germany) (x100).
Cells exhibiting condensed chromatin and fragmented nuclei (Hoechst
33342 stain, blue) were scored as apoptotic cells. For each Hoechst
experiment, at least 200 cells in 5 random scope fields were
counted for apoptotic rate.
In vitro cell invasion and migration
assay
Invasion assay with a Matrigel-coated membrane and
migration assay with a Matrigel-uncoated membrane were performed
using a 24-well chamber system (BD Biosciences, Bedford, MA),
according to the manufacturer’s instructions. The cells were
trypsinized and seeded in the upper chamber at 2.5×105
cells/well in serum-free medium. Medium supplemented with 50% FBS
was placed at the bottom of the wells. Incubation was carried out
for 24 h at 37°C in humidified air with a 5% CO2
atmosphere. The cells were allowed to migrate through a porous,
Matrigel-coated or uncoated membrane (BD Biosciences). Following
incubation, the chambers were removed, and invading cells on the
bottom side of the membrane were fixed with methanol and stained
with Giemsa. The number of invading cells or migrating cells were
determined by counting 5 high-power fields (x400) on each membrane
and calculated as the mean number of cells/field.
Immunofluorescence microscopy
Cells grown on glass coverslips were fixed with cold
methanol for 5 min. Cells were then blocked by 2% bovine serum
albumin (BSA) in PBS for 30 min. Cells were then fixed and
double-stained with anti-myc (1:2,000; Cell Signaling Technology,
Inc.) or anti-γ-tubulin (1:1,000; Sigma) or anti-centrin 2
antibodies (1:50; Santa Cruz Biotechnology, Inc.). The cells were
then incubated with Alexa Fluor 594 goat anti-mouse or Alexa Fluor
488 goat anti-rabbit (1 g/ml; Invitrogen) as the secondary
antibodies, followed by staining with 4′,6-diamidino-2-phenylindole
(DAPI; Biosource) for 5 min. The fluorescence staining intensity
was then examined by immunofluorescence microscopy.
Tissue specimens
All cases of breast surgical specimens, including 10
NBT, 10 ADH, 10 DCIS and 10 IDC, were anonymized after collection
from the Breast Pathology Department, Tianjin Tumor Hospital.
Tissues of 10 NBT cases were obtained from the quadrant far away
from the original foci in the surgical specimens of 10 cases of
IDC. The pathological diagnosis was confirmed by 2 senior
pathologists according to the 2003 WHO histological classification
of tumors of the breast. Tissue collection and analysis in this
study were approved by the Ethics Committee of the Tianjin Medical
University Cancer Institute and Hospital, Tianjin, China. Informed
consent was obtained from all patients prior to surgery and
examination of the specimens.
Statistical analysis
All in vitro experiments were repeated 3
times. Differences were statistically evaluated using the Student’s
t-test. A P-value <0.05 was considered to indicate a
statistically significant difference.
Results
Effect of Nek2C expression on breast
tumor cells
Breast cancer cells (MCF10DCIS.com and MCF10CA1a)
were transfected with Nek2C siRNA and a scrambled siRNA, as the
negative control (siControl), to study the function of Nek2C. The
transcriptional levels of Nek2C were examined by RT-PCR. The
reduced expression of Nek2C was observed in the MCF10DCIS.com
cells, which were transfected with Nek2C siRNA (siRNA-2), compared
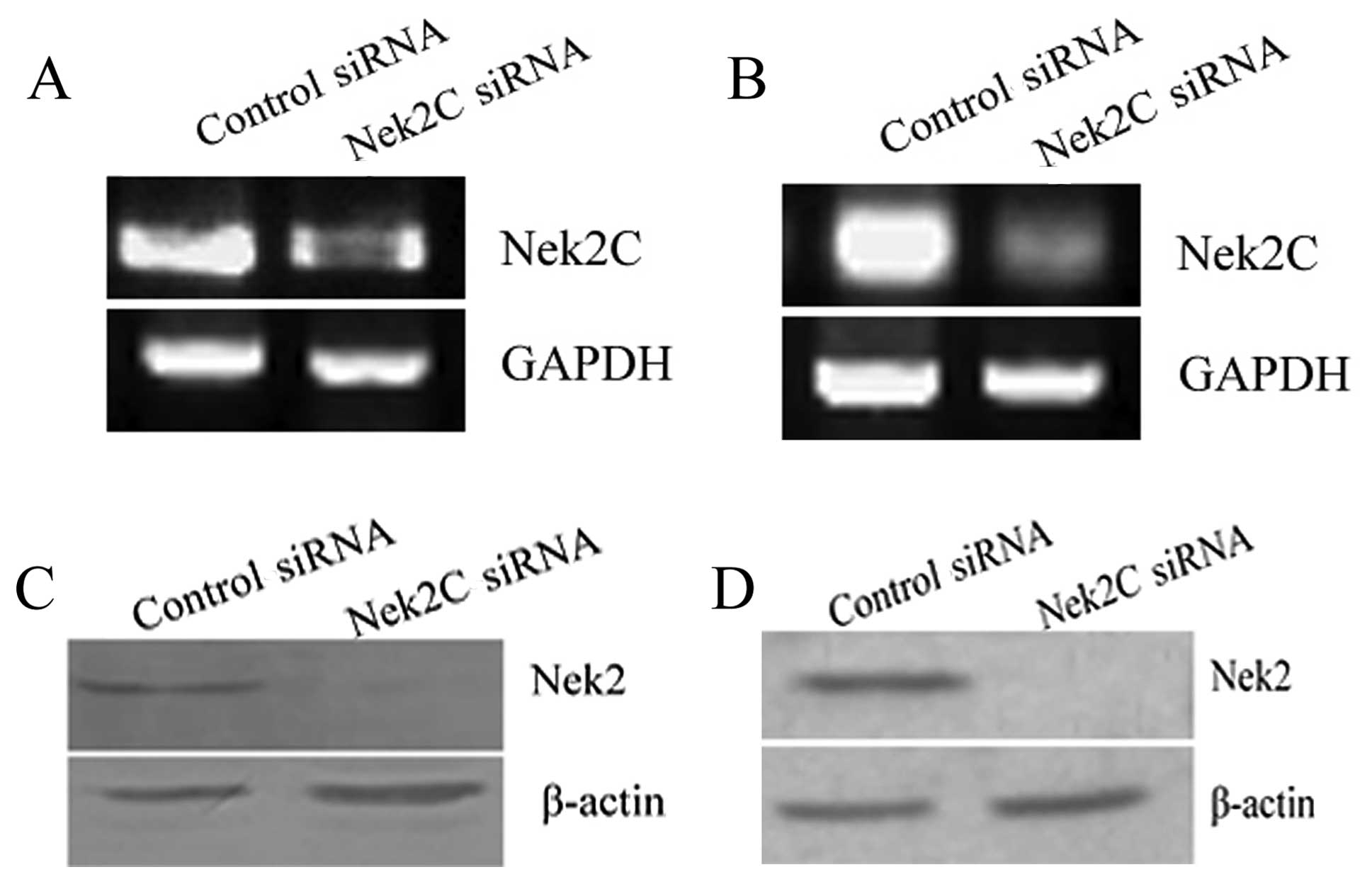
to the cells transfected with control siRNA (Fig. 1A). The reduction rates in Nek2C
signals were almost similar to those in MCF10CA1a cells (Fig. 1B). Hence, siRNA inhibits Nek2C
expression. Western blot analysis was carried out to detect the
protein expression of Nek2 in these breast carcinoma cell lines.
The protein levels of Nek2 in the breast cancer cell lines
(MCF10DCIS.com and MCF10CA1a), which were transfected with Nek2C
siRNA (siRNA-2), were reduced compared to those in the control
siRNA-transfected cells (Fig. 1C and
D).
We then examined the effect of siRNA treatment on
the growth of breast carcinoma cells to determine the effect of the
Nek2C knockdown. We found that Nek2C knockdown by RNAi remarkably
affected the viability of the MCF10DCIS. com and MCF10CA1a cell
lines. The downregulation of Nek2C using Nek2C siRNA blocked cell
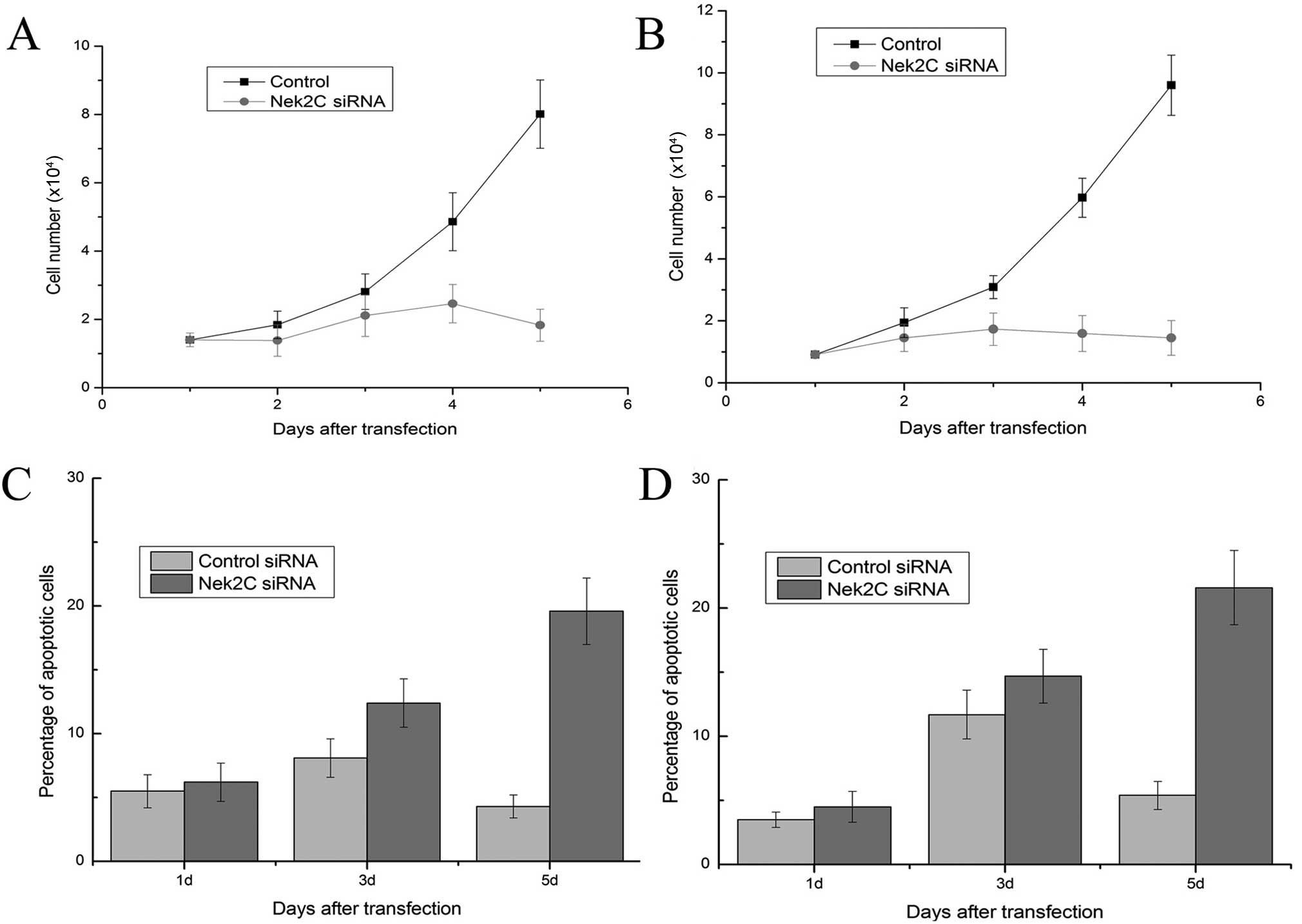
growth. Marked growth inhibition was evident at 96 h post siRNA
transfection in the MCF10DCIS.com cells (Fig. 2A). The growth of MCF10CA1a cells
was also suppressed by Nek2C siRNA treatment to levels similar to
those of MCF10DCIS.com cells (Fig.
2B). By contrast, transfection with control siRNA did not alter
cell growth significantly. Increased levels of apoptosis were
observed in MCF10DCIS.com and MCF10CA1a cell lines after 3–5 days
in the absence of Nek2C (Fig. 2C and
D). Furthermore, typical apoptotic changes, such as nuclear
fragmentation in the Nek2C siRNA-transfected cells were noted, and
the number of apoptotic cells was significantly greater compared to
the cells transfected with the control siRNA. Thus, apoptosis is
possibly the mechanism by which cell numbers are reduced.
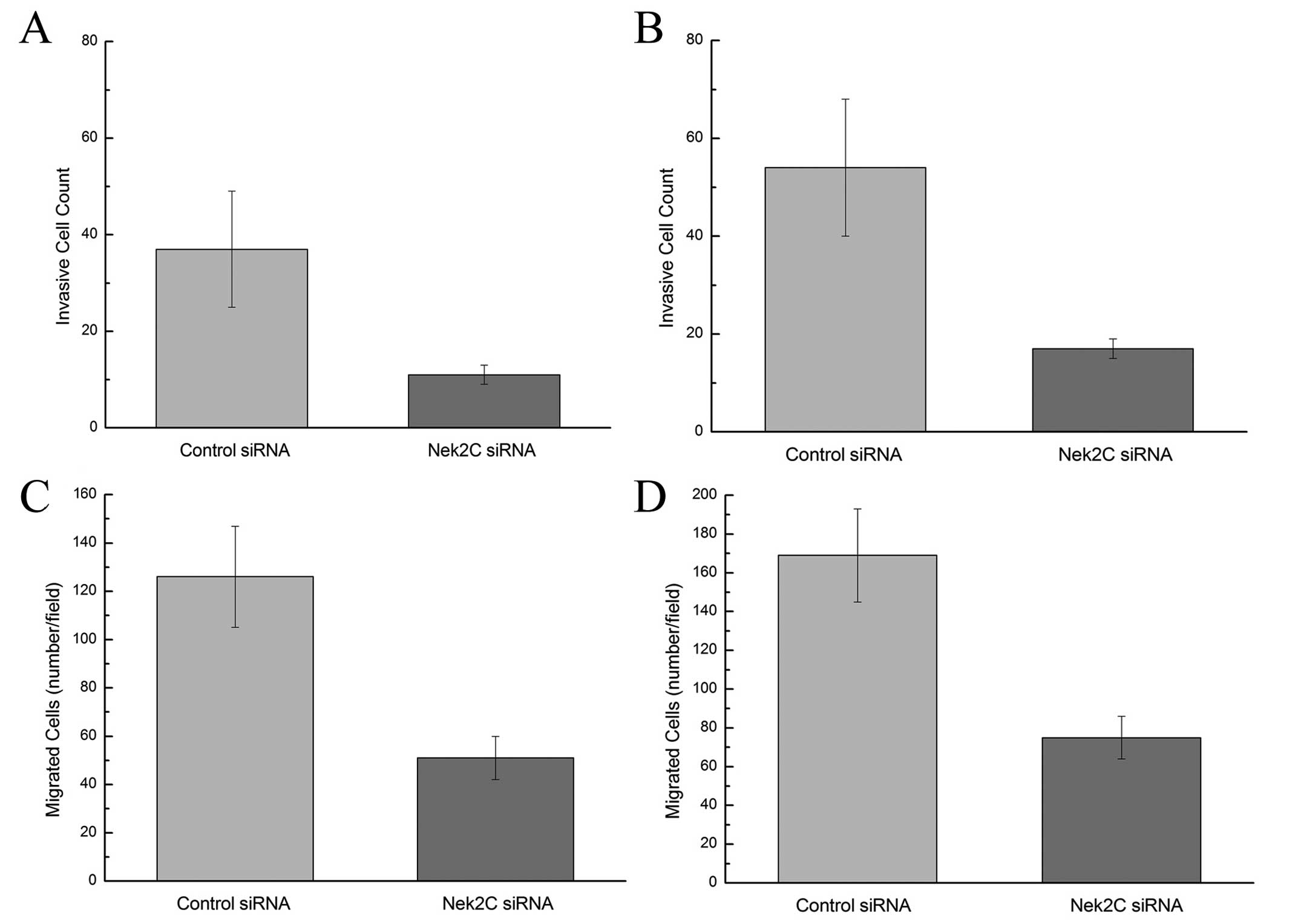
Moreover, after 24-h invasion, the invasion of
breast cancer cells was significantly attenuated in the
MCF10DCIS.com and MCF10CA1a cell lines which were transfected with
Nek2C siRNA compared to the control (Fig. 3A and B) (P<0.05). Furthermore,
the forced downregulation of Nek2C in these cell lines
significantly suppressed their migration through the Transwell
membrane (Fig. 3C and D)
(P<0.05). Collectively, these results imply a pivotal role of
Nek2C in tumor progression and aggressiveness. The knockdown of
Nek2C significantly suppresses tumor cell invasion and migration.
Thus, the reduction in Nek2C expression attenuates the progression
to an invasive tumor and impairs the growth of tumors that
ultimately form.
Role of the Nek2C plasmid in breast
cells
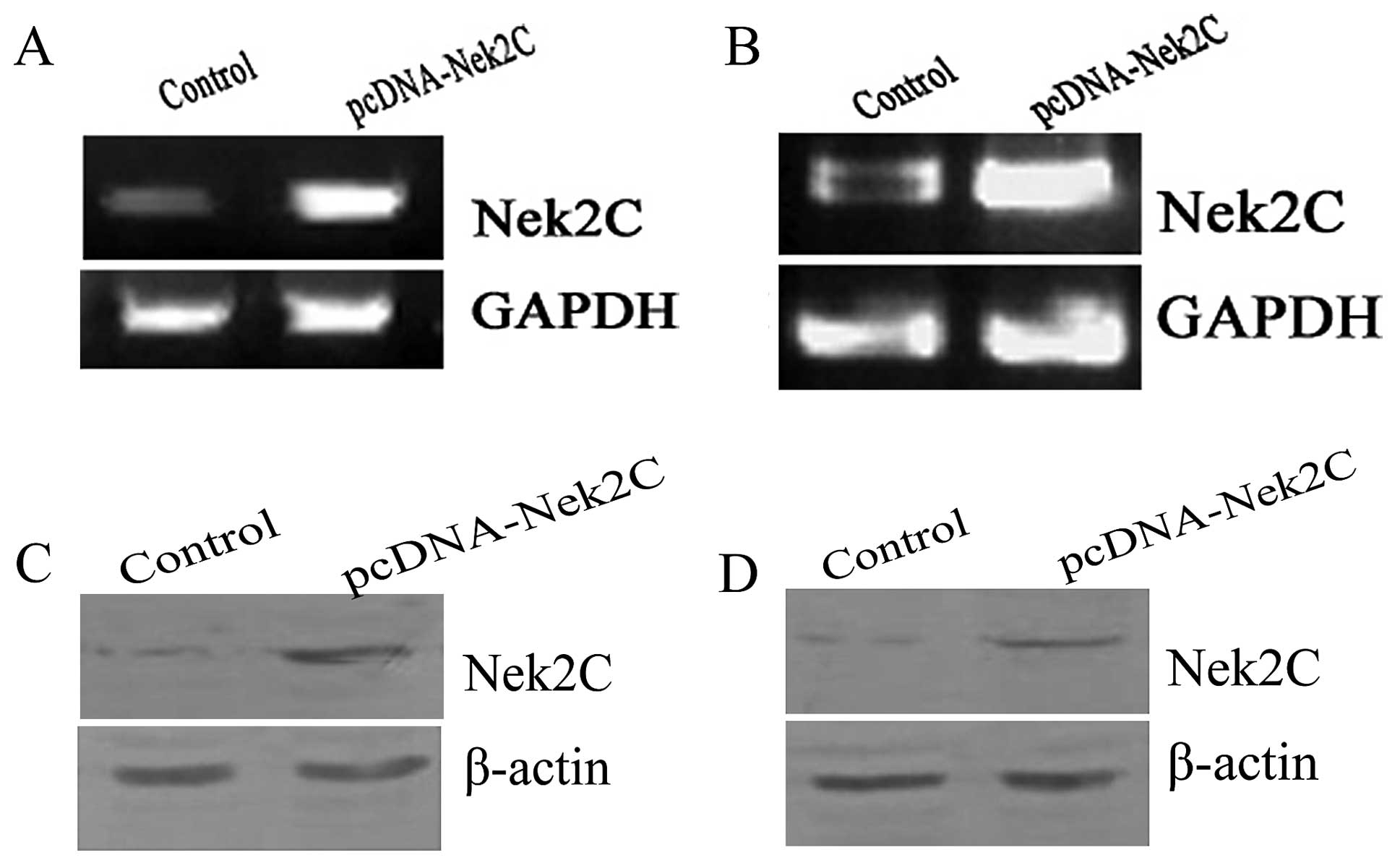
The pcDNA-mycNek2C and pcDNA3.0-myc vectors were
transfected into the breast cell lines, MCF10A (Fig. 4C) and MCF10AT (Fig. 4D). First, we examined the
expression levels of Nek2C in MCF10A and MCF10AT cells by
semiquantitative RT-PCR. The mRNA expression levels of Nek2C were
significantly upregulated in Nek2C-transfected MCF10A and MCF10AT
cells compared to those of the control cells (Fig. 5A and B). The protein expression
levels of Nek2C were then detected by western blot analysis in
these cells. Similarily, the expression levels of Nek2C proteins
were markedly higher in the Nek2C-transfected MCF10A and MCF10AT
cells than in the control cells (Fig.
5C and D). We focused on the role of Nek2C in subsequent
experiments.
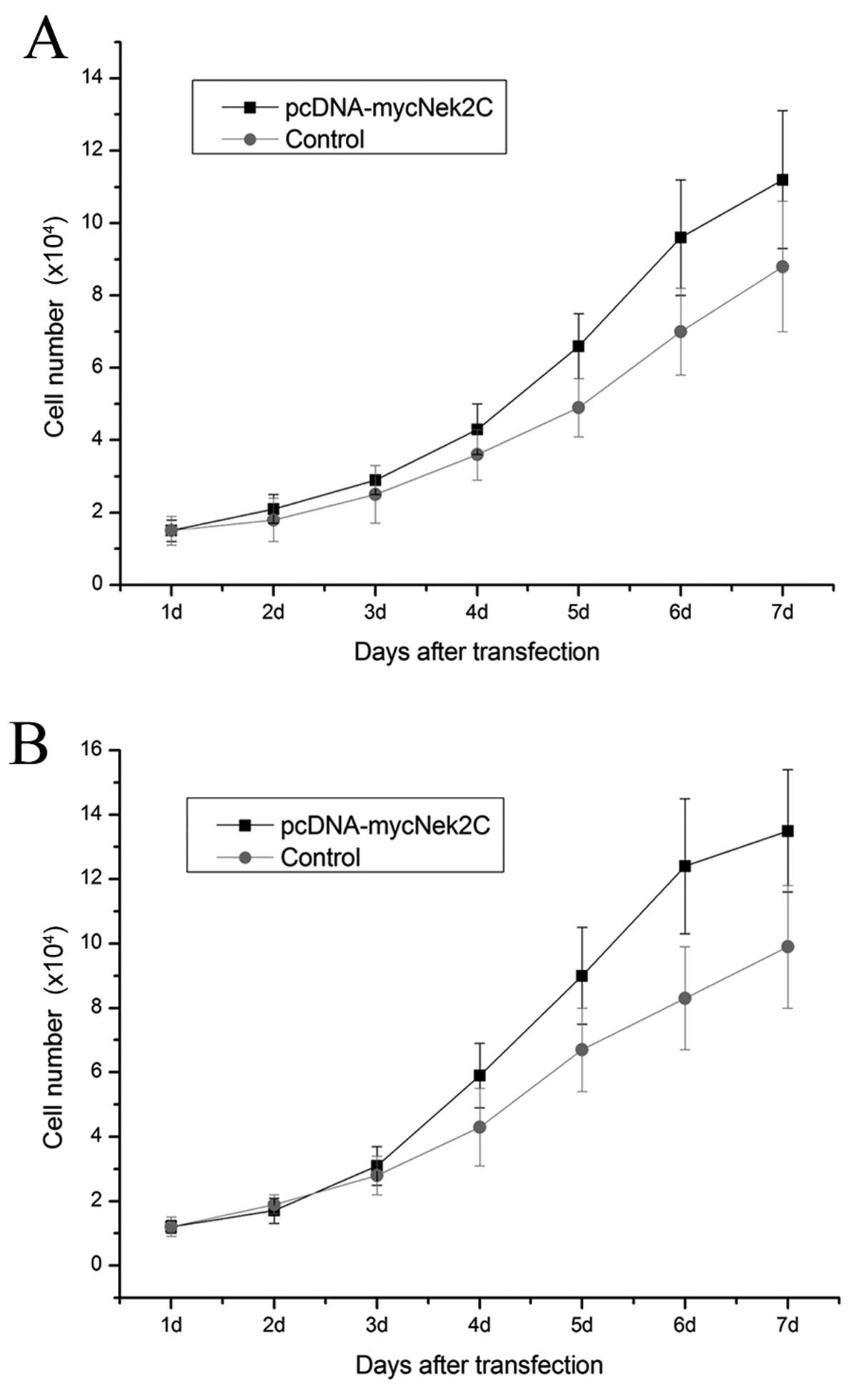
In parallel, cell viability was measured for 7 days
following transfection. pcDNA3.1-Nek2C had a detectable effect on
the viability of the breast cancer cell lines. The growth of MCF10A
cells was substantially promoted by the Nek2C plasmid transfection
compared with that of the cells transfected with the control empty
vector (Fig. 6A). The growth of
MCF10AT cells was also increased by the Nek2C plasmid to levels
similar to those of MCF10A cells (Fig. 6B). These findings suggest that
Nek2C plays an important role in the growth and survival of MCF10A
and MCF10AT cells.
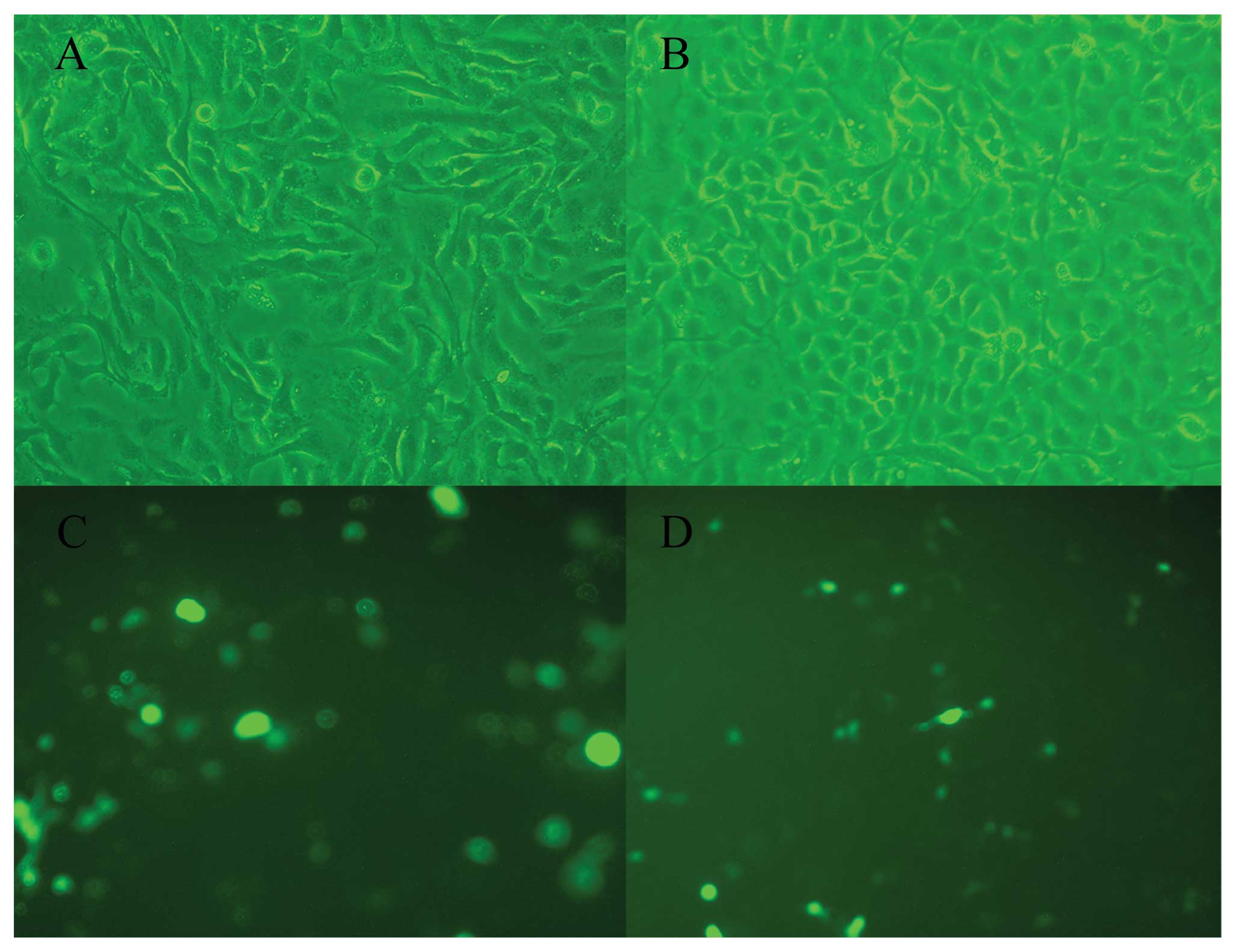
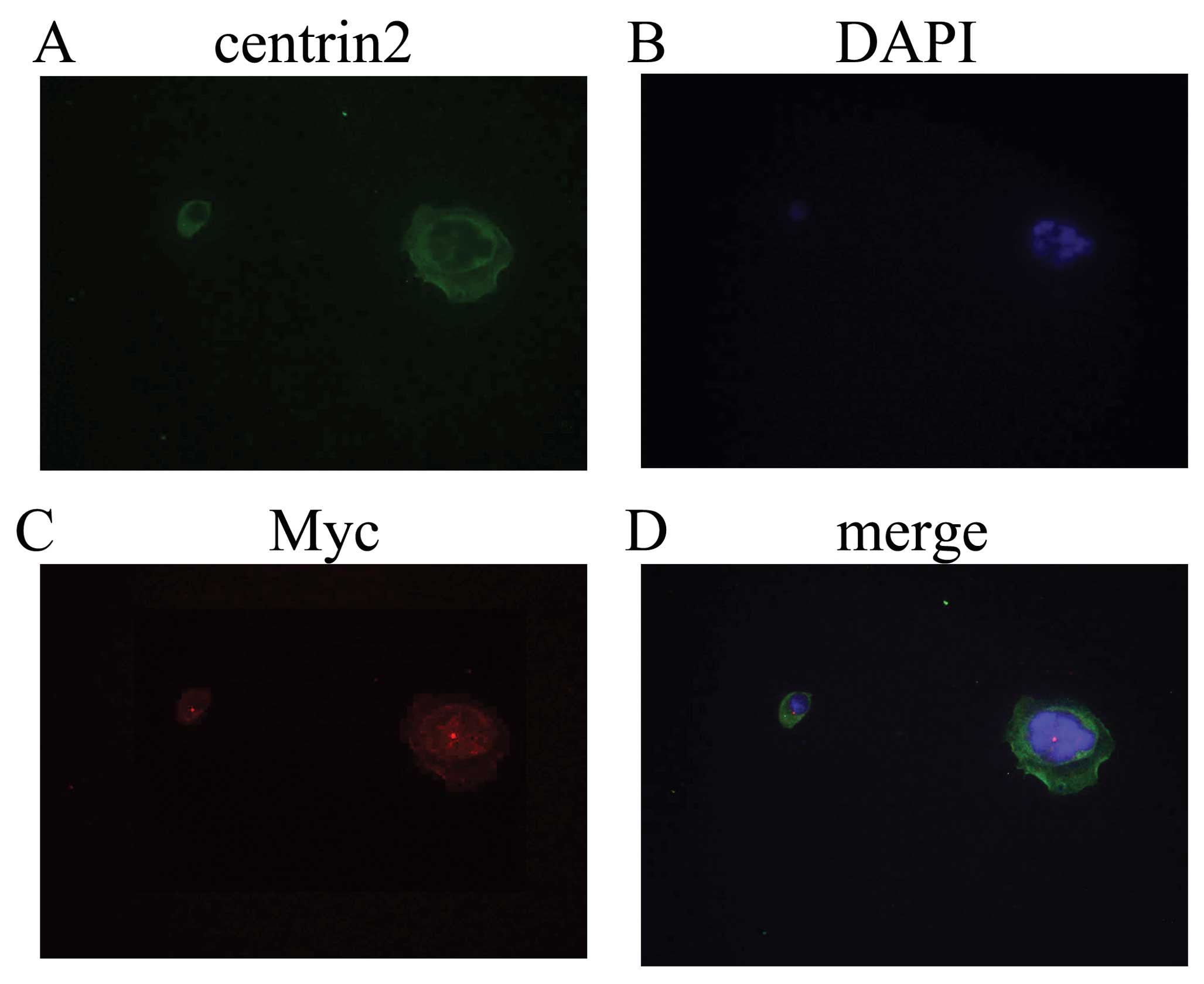
A peculiar feature of the expression of Nek2C in
human breast cells was its predominantly nuclear localization. To
further investigate the localization of Nek2C in human breast cell
lines, we used the MCF10AT cell line, which was transfected with
the Nek2C plasmid. First, western blot analysis confirmed that the
Nek2C protein was upregulated in Nek2C-transfected MCF10AT cells
with respect to the non-transformed Nek2C cells. Moreover,
subcellular localizations of the proteins were determined by
immunocytochemistry with the specific antibodies.
Immunofluorescence analysis confirmed that the Nek2C protein was
predominantly present in the nucleus in Nek2C-transfected MCF10AT
cells (Fig. 7).
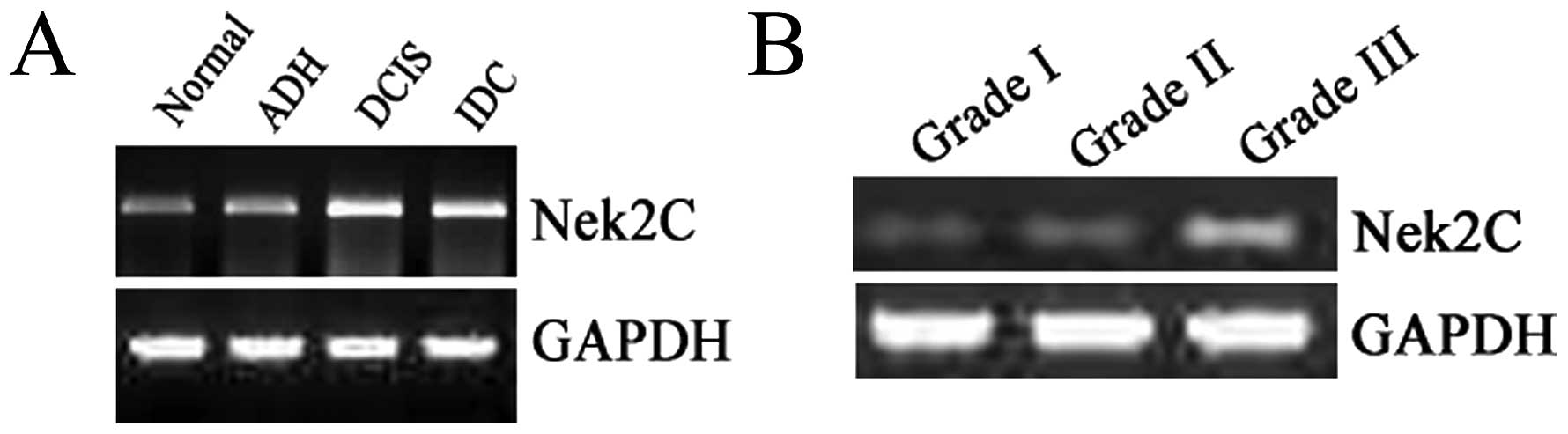
Expression of Nek2C in breast tissue
The MCF10DCIS.com and MCF10CA1a cell lines displayed
similar expression levels of Nek2C-mRNA, with Nek2C consistently
expressed at high levels. We analyzed 40 breast samples, including
10 NBT, 10 ADH, 10 DCIS and 10 IDC samples in order to determine
whether the mRNA expression levels of the Nek2C kinase were altered
in human breast tissue. Both tumor and normal tissues were examined
to compare the mRNA expression levels using RT-PCR. We found that
Nek2C-mRNA was more abundant in extracts obtained from IDC and DCIS
samples than in those obtained from NBT or samples from a patient
affected by ADH. Nine samples (90%) exhibited a significantly
elevated Nek2C mRNA expression in IDC, whereas 1 (10%) showed no
significant difference with the normal tissue. In the DCIS cases, 9
samples (90%) showed an elevated Nek2C mRNA expression levels
compared with NBT. Thus, human Nek2C is expressed at high levels in
DCIS and IDC, in contrast to its low levels in NBT (Fig. 8A). These results indicate that
Nek2C mRNA is frequently upregulated in human breast cancer. We
also used RT-PCR to evaluate the quantity of Nek2C in correlation
with tumor grade. The expression of Nek2C in high-grade (grade III)
tumor tissue (Fig. 8B) was
significantly higher than in low-grade (grade I and II) tumor
tissue. Hence, Nek2C affects tumorigenesis and progression in
breast cancer.
Discussion
Breast carcinoma is one of the leading causes of
morbidity in women. The prognosis for this disease is generally
very poor, with a high mortality rate. Nek2 in humans is expressed
as at least 3 splice variants, namely, Nek2A, Nek2B and Nek2C.
Nek2A is one of the main splice variants of Nek2, and is considered
of great importance in revealing the molecular mechanisms of
carcinogenesis (15,16). Nek2C (originally known as Nek2A-T)
was identified after yeast two-hybrid screening of a testis
library, with PP1γ1 and PP1γ2 as bait (12). Nek2C differs from Nek2A, as the
former lacks 8 amino acids (371–378) in the middle of the
non-catalytic domain. However, as our results suggest, this small
difference has significant consequences on subcellular
localization, which in turn may allow the different splice variants
to undertake distinct functions during cell division.
In breast carcinoma, Her-2/neu (17), Myc (18) and Wnt (19) are potential candidate genes for
oncogene addiction. In this study, we demonstrate that the Nek2C
pathway is necessary to maintain the tumorigenic growth of breast
carcinoma cells, suggesting Nek2C to be another candidate gene for
oncogene addiction. The effectiveness of Nek2C as a
chemotherapeutic target is worth considering. Nek2C expression is
frequently elevated in cancer cells, possibly due to gene
amplification or loss of transcriptional control. Moreover,
immunofluorescence analysis has confirmed that Nek2C is located in
the nuclear region (15). Hence,
an additional role of Nek2C in the regulation of nuclear events in
breast cancer cells is suggested. The resulting protein contains a
novel nuclear localization sequence that is absent in Nek2A and
Nek2B. Although experiments with recombinant proteins have shown
that Nek2A also partially accumulates in the nuclei, the
non-centrosomal pool of Nek2C accumulates in the nuclei more
efficiently. Given the similar size and the lack of specific
antibodies, endogenous Nek2A and Nek2C cannot be distinguished at
the protein level. The preferential uptake of Nek2C into the
nucleus is evident, whereas Nek2A is more evenly distributed
throughout the cell. This phenomenon can be explained by the loss
of the 8 amino acids from Nek2C resulting from splicing. Thus, a
specific antibody that can distinguish between Nek2A and Nek2C is
required to further investigate the localization of Nek2C.
RNA interference has emerged as a natural and highly
efficient mechanism for gene silencing (20–22). A number of previous studies have
reported on the consequences of Nek2 depletion using RNA
interference strategies (1,23–25). In this study, we treated
MCF10DCIS.com and MCF10CA1a cells with double-stranded siRNA
oligonucleotides against Nek2C to study the function of Nek2C. The
expression of Nek2C in MCF10DCIS.com and MCF10CA1a cells was
substantially suppressed by Nek2C siRNA treatment, but not by
control siRNA. We also found that the treatment of MCF10DCIS.com
and MCF10CA1a cells with Nek2C siRNA caused concomitant suppression
of cell growth, invasion and migration compared to the control
siRNA-treated cells. These findings bear some similarities to the
results of Tsunoda et al (26) and Westwood et al (27). The results showed that breast
carcinoma cells have a specific dependence on Nek2C for their
tumorigenic growth. Patients with breast cancer generally receive
chemotherapy treatment with paclitaxel, platinum-based agents, or a
combination of both. Hence, developing a novel strategy for breast
cancer treatment is necessary and urgent. To the best of our
knowledge, this study has demonstrated for the first time that
Nek2C plays a pivotal role in tumor progression, and that Nek2C
inhibition can significantly suppress tumorigenesis and progression
in breast cancer, which may imply a novel therapeutic target. Nek2C
depletion suppresses not only the anchorage-independent growth but
also the invasiveness of breast carcinoma. Although the effect of
Nek2C siRNA in breast carcinoma is quite promising, whether the
effect of siRNA is limited to breast carcinoma remains unclear.
We used MCF10A and MCF10AT breast cancer cell lines
that maintain the features of the original breast cells. Myc-tagged
Nek2C was transfected into these 2 cell lines. RT-PCR analysis
confirmed that Nek2C mRNA was upregulated in the transfected MCF10A
and MCF10AT cells compared to the control cells. The 2 types of
breast cells transfected with the Nek2C plasmid showed very similar
expression levels of Nek2C proteins, with Nek2C consistently
expressed at higher levels than in the non-transfected cells. We
examined the effect of exogenous Nek2C expression in ‘normal’
immortalized breast cells (MCF10A) and (MCF10AT) on the growth of
breast cancer cells and found that the expression of Nek2C
stimulated the growth of these cells.
We therefore analyzed Nek2C mRNA expression in
samples of human breast tissue using RT-PCR. Strikingly, Nek2C mRNA
expression was elevated significantly in most of these breast tumor
samples. Elevated Nek2C expression was clearly detected in DCIS and
IDC tissues compared with those in NBT or ADH. Moreover, analysis
of IDC tissue also revealed a significant increase in the
expression of Nek2C mRNA with tumor grade increase. Thus, Nek2C may
be a contributory factor in cancer malignant progression.
In this study, we show that Nek2C is abundantly
expressed in breast cancer cells. This protein is required for the
tumorigenic growth and survival of these cells. To the best of our
knowledge, we show for the first time that siRNA against Nek2C
substantially suppresses the growth, in vitro invasiveness
and migration of breast carcinoma cell lines. Although the precise
mechanism by which Nek2C siRNA suppresses tumor growth remains
unclear, our results strongly suggest the necessity of Nek2C in the
tumorigenesis of breast carcinoma cells. To the best of our
knowledge, this is the first report demonstrating that Nek2C plays
a critical role in carcinogenesis, tumor invasion and the
tumorigenic growth of breast carcinoma. Nek2C mRNA was
overexpressed in both breast cancer cell lines and patient tissue
samples. The contribution of this upregulation in tumor progression
should be elucidated as the elevated expression of Nek2C may drive
tumorigenesis, and a correlation exists between increased Nek2C and
poor patient outcome. Therefore, further research is required to
determine the role of Nek2C in human breast cancer.
Acknowledgements
This study was financially supported
by the National Science Foundation of China (30872519); the
Scientific and Technological Development Fund of the Tianjin
Scientific and Technological Committee (09JCYBJC10100); the Program
for Chang-jiang Scholars and Innovative Research Team in University
(IRT0743).
References
|
1.
|
L FletcherGJ CernigliaEA NiggTJ YendRJ
MuschelInhibition of centrosome separation after DNA damage: a role
for Nek2Radiat Res162128135200410.1667/RR321115387139
|
|
2.
|
AM FryThe Nek2 protein kinase: a novel
regulator of centrosome
structureOncogene2161846194200210.1038/sj.onc.120571112214248
|
|
3.
|
MJ O’ConnellMJ KrienT HunterNever say
neverThe NIMA-related protein kinases in mitotic control Trends
Cell Biol132212282003
|
|
4.
|
AM FryP MeraldiEA NiggA centrosomal
function for the human Nek2 protein kinase, a member of the NIMA
family of cell cycle regulatorsEMBO
J17470481199810.1093/emboj/17.2.4709430639
|
|
5.
|
HQ GuoM GaoJ MaT XiaoLL ZhaoY GaoQJ
PanAnalysis of the cellular centrosome in fine-needle aspirations
of the breastBreast Cancer Res9R48200710.1186/bcr175217662154
|
|
6.
|
WL LingleWH LutzJN IngleNJ MaihleJL
SalisburyCentrosome hypertrophy in human breast tumors:
Implications for genomic stability and cell polarityProc Natl Acad
Sci USA9529502955199810.1073/pnas.95.6.29509501196
|
|
7.
|
Q LiuY HirohashiX DuMI GreeneQ WangNek2
targets the mitotic checkpoint proteins Mad2 and Cdc20: a mechanism
for aneuploidy in cancerExp Mol
Pathol88225233201010.1016/j.yexmp.2009.12.00420034488
|
|
8.
|
DG HaywardAM FryNek2 kinase in chromosome
instability and cancerCancer
Lett237155166200610.1016/j.canlet.2005.06.01716084011
|
|
9.
|
DG HaywardRB ClarkeAJ FaragherMR PillaiIM
HaganAM FryThe centrosomal kinase Nek2 displays elevated levels of
protein expression in human breast cancerCancer
Res6473707376200410.1158/0008-5472.CAN-04-096015492258
|
|
10.
|
S de VosWK HofmannTM GroganGene expression
profile of serial samples of transformed B-cell lymphomasLab
Invest83271285200312594241
|
|
11.
|
K UtoN NakajoN SagataTwo structural
variants of Nek2 kinase, termed Nek2A and Nek2B, are differentially
expressed in Xenopus tissues and developmentDev
Biol208456464199910.1006/dbio.1999.923110191058
|
|
12.
|
M FardilhaW WuR SaAlternatively spliced
protein variants as potential therapeutic targets for male
infertility and contraceptionAnn NY Acad
Sci1030468478200410.1196/annals.1329.05915659832
|
|
13.
|
MJ WorshamG PalsJP SchoutenF MillerN
TiwariR van SpaendonkSR WolmanHigh-resolution mapping of molecular
events associated with immortalization, transformation, and
progression to breast cancer in the MCF10 modelBreast Cancer Res
Treat96177186200610.1007/s10549-005-9077-816319984
|
|
14.
|
A Vazquez-MartinR ColomerJ BrunetR LupuJA
MenendezOverexpression of fatty acid synthase gene activates
HER1/HER2 tyrosine kinase receptors in human breast epithelial
cellsCell
Prolif415985200810.1111/j.1365-2184.2007.00498.x18211286
|
|
15.
|
W WuJE BaxterSL WattamAlternative splicing
controls nuclear translocation of the cell cycle-regulated Nek2
kinaseJ Biol
Chem2822643126440200710.1074/jbc.M70496920017626005
|
|
16.
|
S WangW LiN LiuNek2A contributes to
tumorigenic growth and possibly functions as potential therapeutic
target for human breast cancerJ Cell
Biochem11319041914201210.1002/jcb.2405922234886
|
|
17.
|
SE MoodyD PerezTC PanThe transcriptional
repressor Snail promotes mammary tumor recurrenceCancer
Cell8197209200510.1016/j.ccr.2005.07.00916169465
|
|
18.
|
CM D’CruzEJ GuntherRB Boxerc-MYC induces
mammary tumorigenesis by means of a preferred pathway involving
spontaneous Kras2 mutationsNat Med7235239200111175856
|
|
19.
|
EJ GuntherSE MoodyGK BelkaImpact of p53
loss on reversal and recurrence of conditional Wnt-induced
tumorigenesisGenes Dev17488501200310.1101/gad.105160312600942
|
|
20.
|
TC KaragiannisA El-OstaRNA interference
and potential therapeutic applications of short interfering
RNAsCancer Gene Ther12787795200510.1038/sj.cgt.770085715891770
|
|
21.
|
PY LuF XieMC WoodleIn vivo application of
RNA interference: from functional genomics to therapeuticsAdv
Gen54117142200516096010
|
|
22.
|
M Ameyar-ZazouaV GuasconiS Ait-Si-AlisiRNA
as a route to new cancer therapiesExpert Opin Biol
Ther5221224200510.1517/14712598.5.2.22115757383
|
|
23.
|
L FletcherGJ CernigliaTJ YenRJ MuschelLive
cell imaging reveals distinct roles in cell cycle regulation for
Nek2A and Nek2BBiochem Biophys
Acta17448992200510.1016/j.bbamcr.2005.01.00715950749
|
|
24.
|
Y LouJ YaoA ZereshkiNEK2A interacts with
MAD1 and possibly functions as a novel integrator of the spindle
checkpoint signalingJ Biol
Chem2792004920057200410.1074/jbc.M31420520014978040
|
|
25.
|
J YaoC FuX DingNek2A kinase regulates the
localization of numatrin to centrosome in mitosisFEBS
Lett575112118200410.1016/j.febslet.2004.08.04715388344
|
|
26.
|
N TsunodaT KokuryoK OdaNek2 as a novel
molecular target for the treatment of breast carcinomaCancer
Sci100111116200910.1111/j.1349-7006.2008.01007.x19038001
|
|
27.
|
I WestwoodDM ChearyJE BaxterMW RichardsRL
van MontfortAM FryR BaylissInsights into the conformational
variability and regulation of human Nek2 kinaseJ Mol
Biol386476485200910.1016/j.jmb.2008.12.03319124027
|