Introduction
Lung cancer is one of the most common human cancers
worldwide (1). Despite continuous
improvement in treatments, lung cancer remains the main cause of
cancer-related deaths (2).
Therefore, it is essential to understand the molecular mechanisms
of this carcinogenesis, so that effective therapeutic strategies
may be developed.
Emerging evidence has shown that tumorigenesis is a
complex multi-step process associated with genetic alterations
(3). Identification of genes that
are involved in cancer development is essential for lung cancer
diagnosis and treatment.
Lung cancer metastasis-related protein 1 (LCMR1) is
a novel gene cloned from PLA-801, a poorly differentiated human
large-cell lung carcinoma cell line, using a differential display
polymerase chain reaction technique in our laboratory in 2002
(4). Information regarding this
gene has been submitted to the National Center for Biotechnology
Information (NCBI), and a Genbank accession number (AY148462) has
been assigned. LCMR1 is located on human 11q12.1 chromosome locus
and is comprised of 949 nucleotides with an open reading frame
(ORF) encoding for a peptide with 177 amino acids. In the human
genome, LCMR1 is also known as the mediator complex subunit 19
(Med19). Med19 is a component of the mediator complex, which is a
coactivator for DNA-binding factors that activates a transcription
via RNA polymerase II (5).
The distribution of LCMR1 has been demonstrated as
site-dependent (4). A high level
of LCMR1 expression is present in the heart, skeletal muscle,
kidney, liver and placental tissues. However, the expression levels
are low in the brain, colon, thymus, spleen, small intestine, lung
and peripheral blood leukocytes. To explore the relation between
LCMR1 and lung cancer, we previously examined LCMR1 expression in
84 human non-small cell lung cancer (NSCLC) tissues using
immunohistochemistry. LCMR1 was overexpressed in NSCLC and its
expression was significantly associated with the clinical stage of
the disease. These results suggest that LCMR1 plays a critical role
in the oncogenesis of lung cancer (4).
Recent studies have reported that LCMR1 is
implicated in several important cellular processes including cell
proliferation, cell cycle and oncogenesis of cancer (6–8).
However, the function of LCMR1 in the regulation of apoptosis
remains unclear. In the present study, we employed RNA interference
(RNAi) technique to knock down LCMR1 expression and we investigated
the regulatory role of LCMR1 in the generation of lung cancer cell
apoptosis. Our results demonstrated that LCMR1 participated in the
regulation of the apoptosis of lung cancer cells. This effect was
p53-dependent and was associated with Bax and Mcl-1. These findings
suggest that LCMR1 may serve as a potential molecular target for
lung cancer therapies.
Materials and methods
Cell lines and culture
The 95D cell line, subcloned from a poorly
differentiated human large-cell lung carcinoma cell line PLA-801,
was kindly provided by Dr Lezhen Chen (Department of Pathology,
Chinese PLA General Hospital, China). A549 and H1299 cell lines
were a gift from Professor ZhiHua Liu (Chinese Academy of Medical
Sciences and Peking Union Medical College, China). 95D cells were
cultured in Roswell Park Memorial institute medium RPMI-1640. A549
and H1299 cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM). Both media were supplemented with 10% fetal bovine serum,
penicillin (100 μg/ml) and streptomycin (100 μg/ml).
The cells were maintained in an incubator with a humidified
atmosphere of 5% CO2 at 37°C.
Small interfering RNA and cell
infection
The human LCMR1-specific small interfering RNA
(siRNA) sequence, 5′-GAGAGAGAGGGACAUGCUU-3′, was designed using an
online siRNA tool provided by Invitrogen Life Technologies (USA)
with the LCMR1 sequence (GenBank code: AY148462) as a reference.
The negative control (NC) sequence was 5′-CCUACGCCACCAAUUUCGU-3′.
For each cell line, there were 3 experimental groups: cells without
infection (CON group), cells infected with the negative control
siRNA (NC group) and cells infected with the LCMR1-siRNA (RNAi
group). Cells were seeded in a 6-well plates and transfected at
∼30–40% confluency using Lipofectamine 2000 (Invitrogen) according
to the manufacturer’s protocol.
Quantitative real-time PCR
Total RNA was extracted from the cells using Trizol
reagent (Invitrogen Life Technologies) and was reverse transcribed
using MMLV Reverse Transcriptase (Promega Corporation, Madison, WI,
USA) according to the manufacturer’s protocol. Quantitative
real-time PCR reactions were prepared in a volume of 20 μl
containing 0.5 μl cDNA sample, 0.5 μl 10 μM
primers, 10 μl 2X SYBR Premix Ex Taq (Takara, Osaka, Japan)
and 9 μl ddH2O. The primers used were as follows:
β-actin, 5′-CATGTACGTTGCTATCCAGGC-3′ (forward) and
5′-CTCCTTAATGTCACGCACGAT-3′ (reverse); LCMR1,
5′-AACAGAGCCGTACCCAGGAT-3′ (forward) and 5′-GGGTGGTCTGGACATTGTC-3′
(reverse); Bax, 5′-TGGAGCTGCAGAGGATGATTG-3′ (forward) and
5′-GAAGTTGCCGTCAGAAAACATG-3′ (reverse); Mcl-1,
5′-CTCATTTCTTTTGGTGCCTTT-3′ (forward) and
5′-CCAGTCCCGTTTTGTCCTTAC-3′ (reverse). Quantitative real-time
RT-PCR analysis was performed on MyiQ2 (Bio-Rad, USA). The relative
quantity of mRNA was calculated using the 2−ΔΔCT method
with the β-actin mRNA level as the reference for normalization. All
experiments were repeated at least 3 times.
Protein extraction
The total protein of the cells was extracted by
addition of a lysis buffer (50 mM Tris pH 7.6, 10 mM EDTA, 150 mM
NaCl, 0.1% NP-40, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl
fluoride, 0.7 μg/ml pepstatin A, 10 μg/ml leupeptin
and 1 μg/ml aprotinin) for 50 min at 4°C. The lysates were
then centrifuged at 12,000 rpm for 30 min at 4°C. The soluble
protein concentrations in the lysates were determined using a BCA
Protein Assay kit (Pierce Biotechnology, Inc., Rockford, IL,
USA).
Western blotting
For western blot analyses, a 25–50 μg of
total protein was separated on a 12% SDS-PAGE gel and was
transferred onto a PVDF membrane. The membrane was blocked in 5%
dried milk for 1 h at room temperature and was then incubated with
a specific primary antibody at 4°C overnight. The membrane was then
washed three times with Tris-buffered saline Tween-20 (TBST),
followed by incubation with a horseradish peroxidase-conjugated
secondary antibody at room temperature for 1 h. Following another
round of washing with TBST, the membrane was developed using
enhanced chemiluminescence (ECL) (Amersham Life Sciences, Amersham,
UK). The staining intensity of the bands was quantitated by
densitometry, using image analyzing software (Multi Gauge Ver 3.2;
Japan).
The antibodies used were as follows: the primary
antibodies for β-actin (Santa Cruz Biotechnology, Inc., USA),
LCMR1/MED19 (Abcam, UK), caspase-3 (New England Biolabs, USA), Bax
(Abcam, USA) and Mcl-1 (Santa Cruz Biotechnology, Inc.) were
employed at dilutions of 1:500, 1:1000, 1:1000, 1:500 and 1:500,
respectively. The secondary antibodies were utilized at working
concentrations of 1:5000, 1:5000 and 1:6000 for anti-goat,
anti-rabbit and anti-mouse IgG (all were from Santa Cruz
Biotechnology, Inc.), respectively.
Flow cytometric analysis
Cells were harvested 72 h after infection by
centrifugation at 1,000 rpm for 5 min and fixed in 70% cold ethanol
overnight. The next day, cells were centrifuged at 1,000 rpm for 5
min, resuspended in PBS, then filtered through a 400-mesh membrane,
stained with propidium iodide (PI) in the dark at 4°C for 30 min
and analyzed using flow cytometry. All experiments were conducted
in triplicate.
Statistical analysis
SPSS 13.0 statistical package was utilized for
statistical analysis. The data are expressed as the means ± SD.
Significance was calculated using a Student’s t-test. A P-value
<0.05 was considered to indicate a statistically significant
difference.
Results
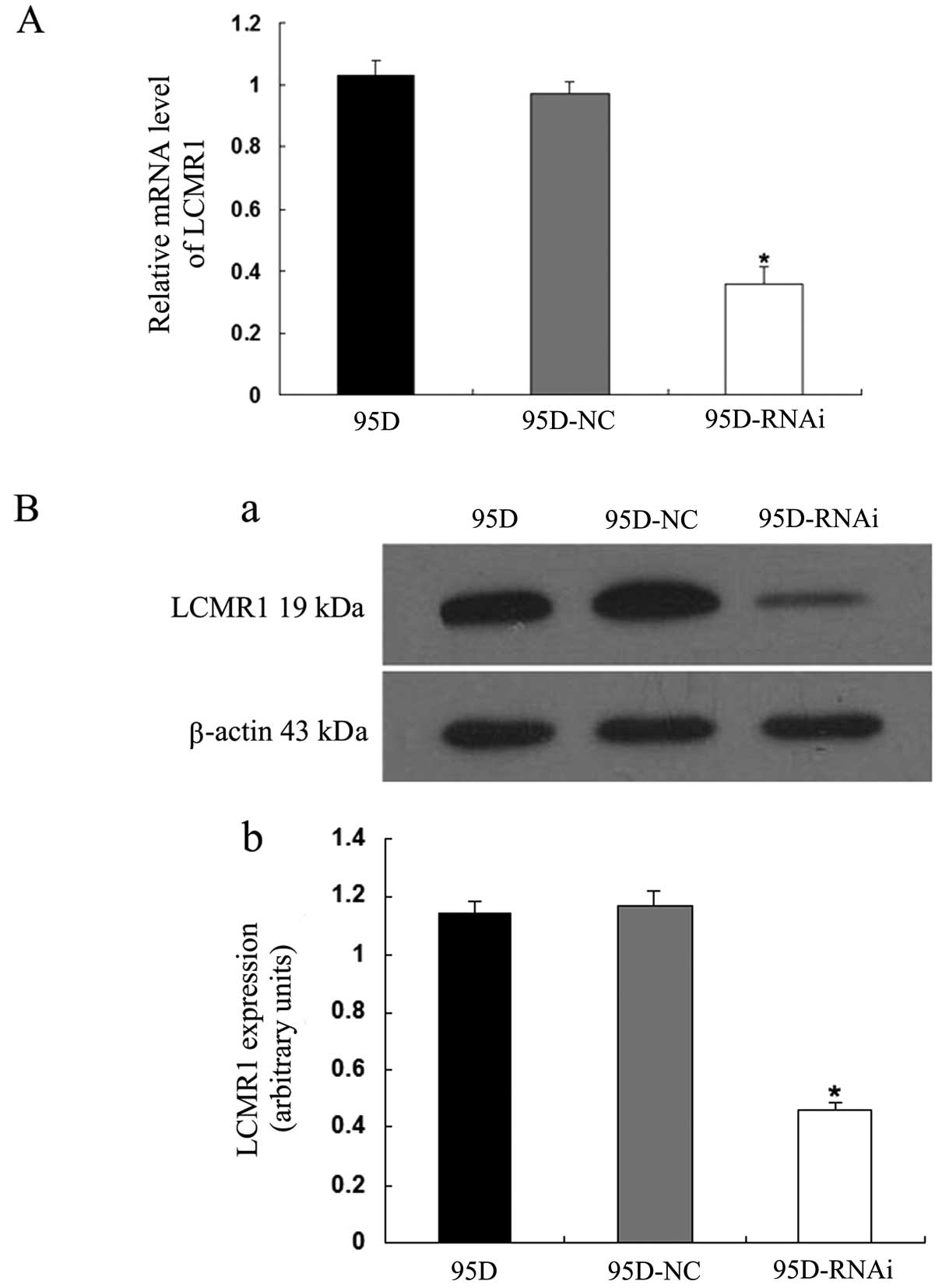
Efficacy of RNAi for LCMR1 knockdown
To determine the effectiveness of the RNAi for
silencing the LCMR1 expression in 95D cells, we examined the
changes in the expression levels of LCMR1 3 days after the
infection. As displayed in Fig.
1A, the transcriptional expression level of LCMR1, evaluated by
qRT-PCR, in the 95D-RNAi cells was significantly lower compared to
levels in the parent 95D and 95D-NC cells (P<0.05). The protein
expression level of LCMR1, assessed by western blotting, was also
decreased in the 95D-RNAi cells (P<0.05) (Fig. 1B), which is consistent with the
transcriptional change in this gene. These results indicated that
the LCMR1-siRNA used in the current study effectively reduced the
LCMR1 expression at both the mRNA and protein levels.
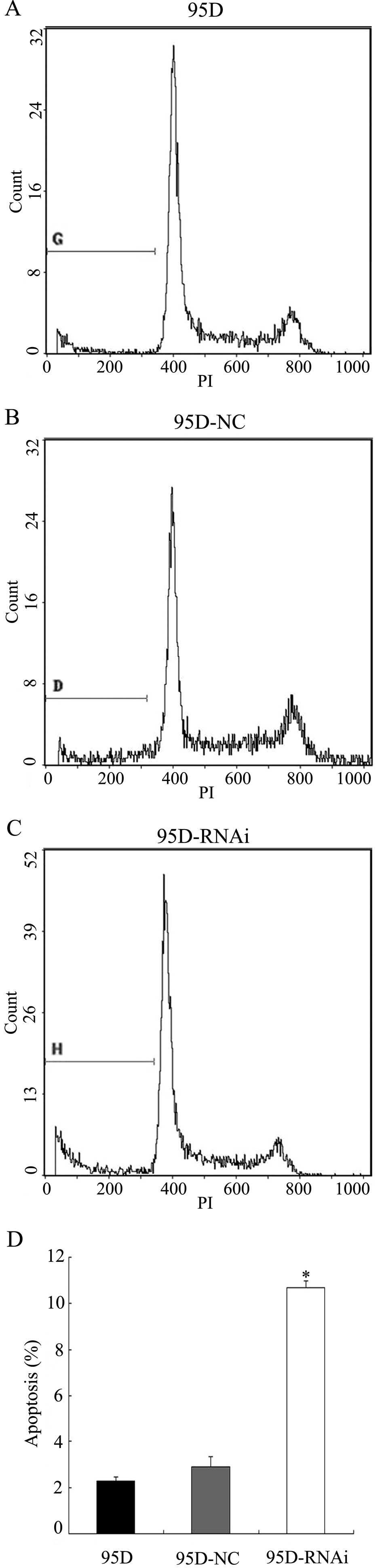
LCMR1 knockdown enhances apoptotic
activity
To determine whether LCMR1 knockdown potentiates the
apoptotic activity in 95D cells, we examined the percentages of
apoptotic cells in the 95D, 95D-NC and 95D-RNAi cell groups using
flow cytometry 72 h after the infection. As demonstrated in
Fig. 2, the percentage of
apoptotic cells in the 95D-NC cell group was similar to that in the
95D cell group (P>0.05), whereas the percentage in the 95D-RNAi
cell group was significantly higher compared to the 95D and 95D-NC
cell groups (P<0.05). These results suggest that LCMR1 knockdown
enhances apoptotic activity in 95D cells.
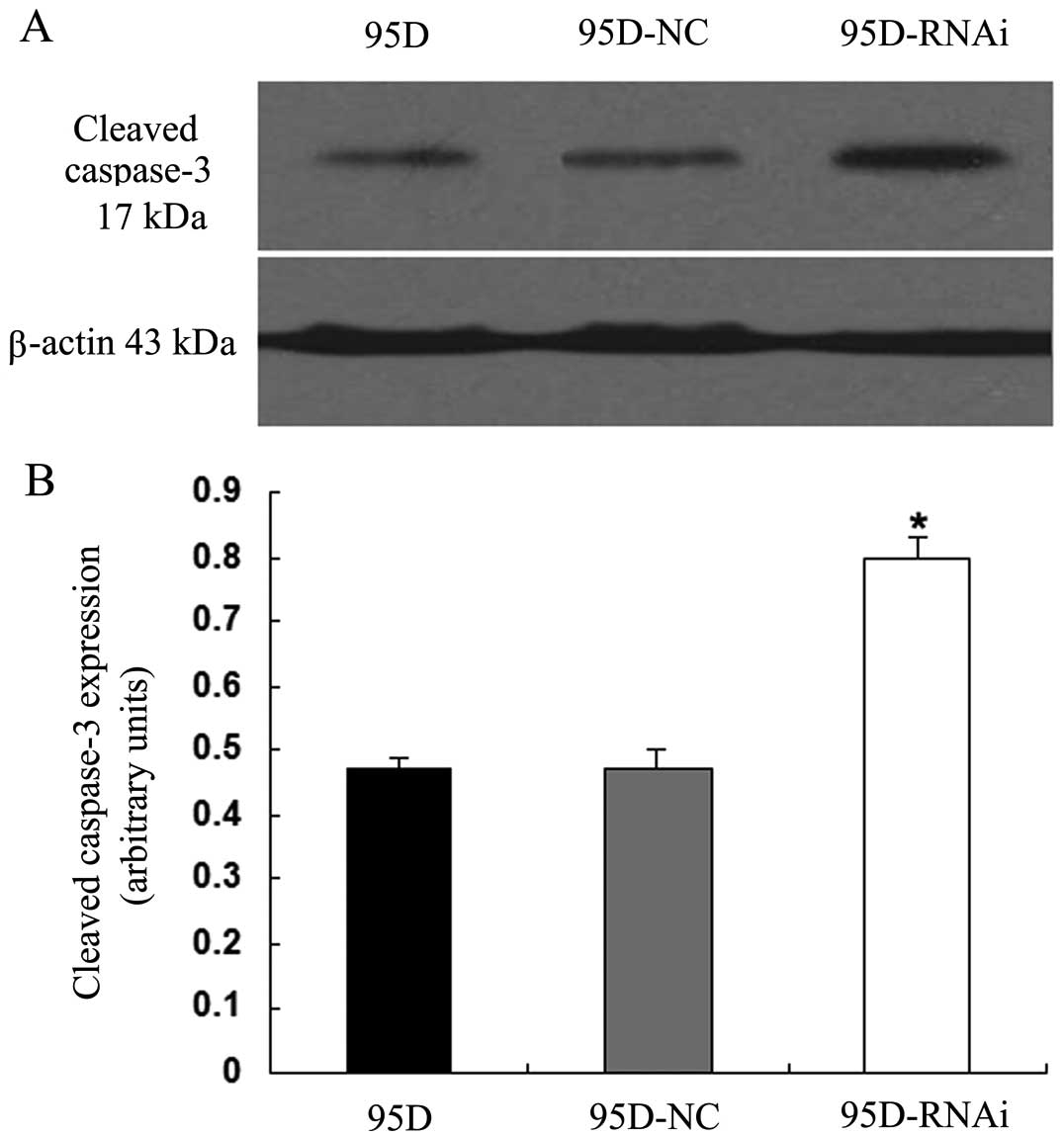
To further confirm the action of LCMR1 knockdown in
apoptosis, we compared the expression levels of cleaved caspase-3,
a biological marker of apoptosis, among the three groups using
western blot assay. The expression level of cleaved caspase-3
protein in the 95D-RNAi cells was significantly higher when
compared to expression levels in the 95D and 95D-NC cells (Fig. 3), suggesting that the suppression
of LCMR1 increases the expression of cleaved caspases-3.
Collectively, these findings suggest that the downregulation of
LCMR1 promotes apoptotic activity in lung cancer cells.
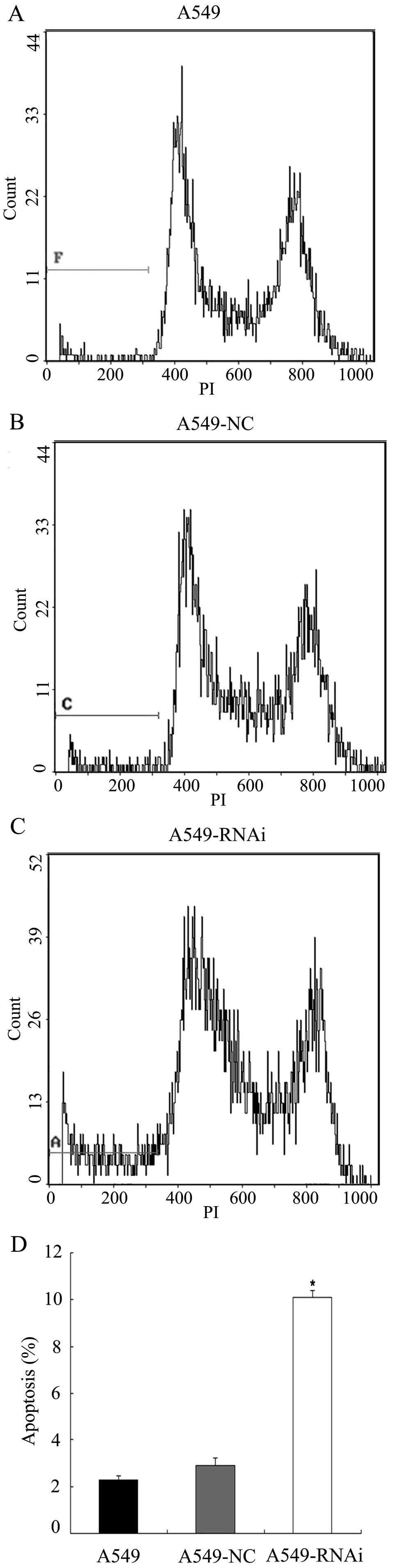
LCMR1-mediated apoptosis is associated
with the p53 pathway
Apoptosis has been ascribed as being p53-dependent
or p53-independent (9). To
determine whether p53 is involved in LCMR1-related apoptosis, we
examined the apoptotic activity in two human lung carcinoma cell
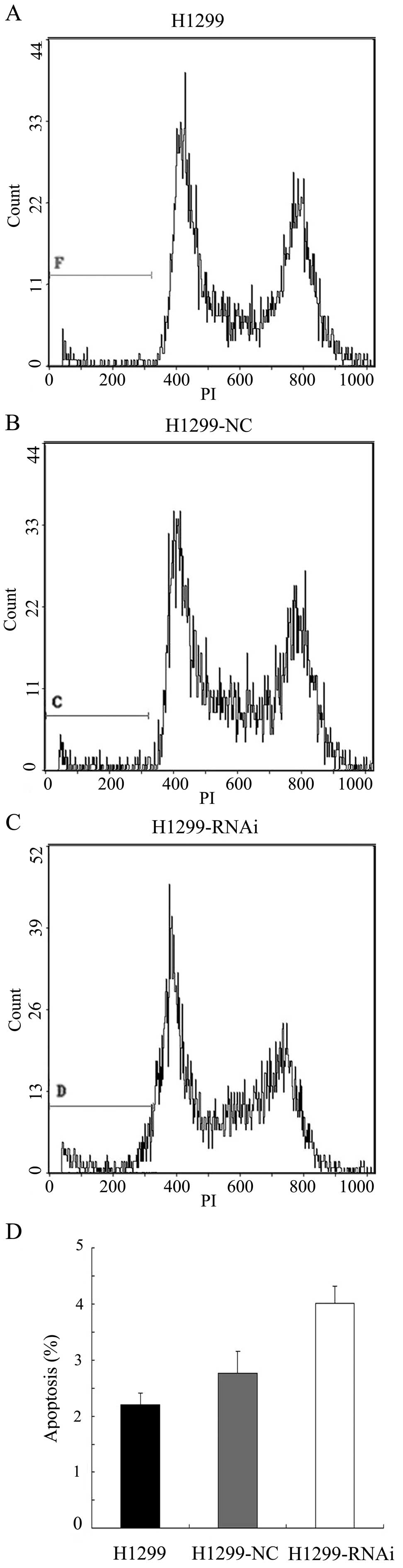
lines: A549 (p53 wild-type cancer cells) and H1299 (p53-deficient
cancer cells). A549, A549-NC, A549-RNAi, H1299, H1299-NC and
H1299-RNAi cells were subjected to flow cytometric analysis 72 h
after infection. As demonstrated in Fig. 4, the percentage of apoptotic cells
in the A549-RNAi group was significantly higher compared to the
levels in the A549 and A549-NC groups (P<0.05). By contrast, the
percentage of apoptotic cells in the H1299-RNAi group was not
significantly different from those in the H1299 or H1299-NC group
(P>0.05) (Fig. 5). These
results suggest that LCMR1-mediated apoptosis is associated with
the p53 pathway.
Bax and Mcl-1 are involved in
LCMR1-related cell apoptosis
p53 protein regulates the apoptotic pathway through
the transcriptional activation of apoptosis-related genes, such as
Bax, a pro-apoptotic gene and Mcl-1, an anti-apoptotic gene
(10). To determine whether the
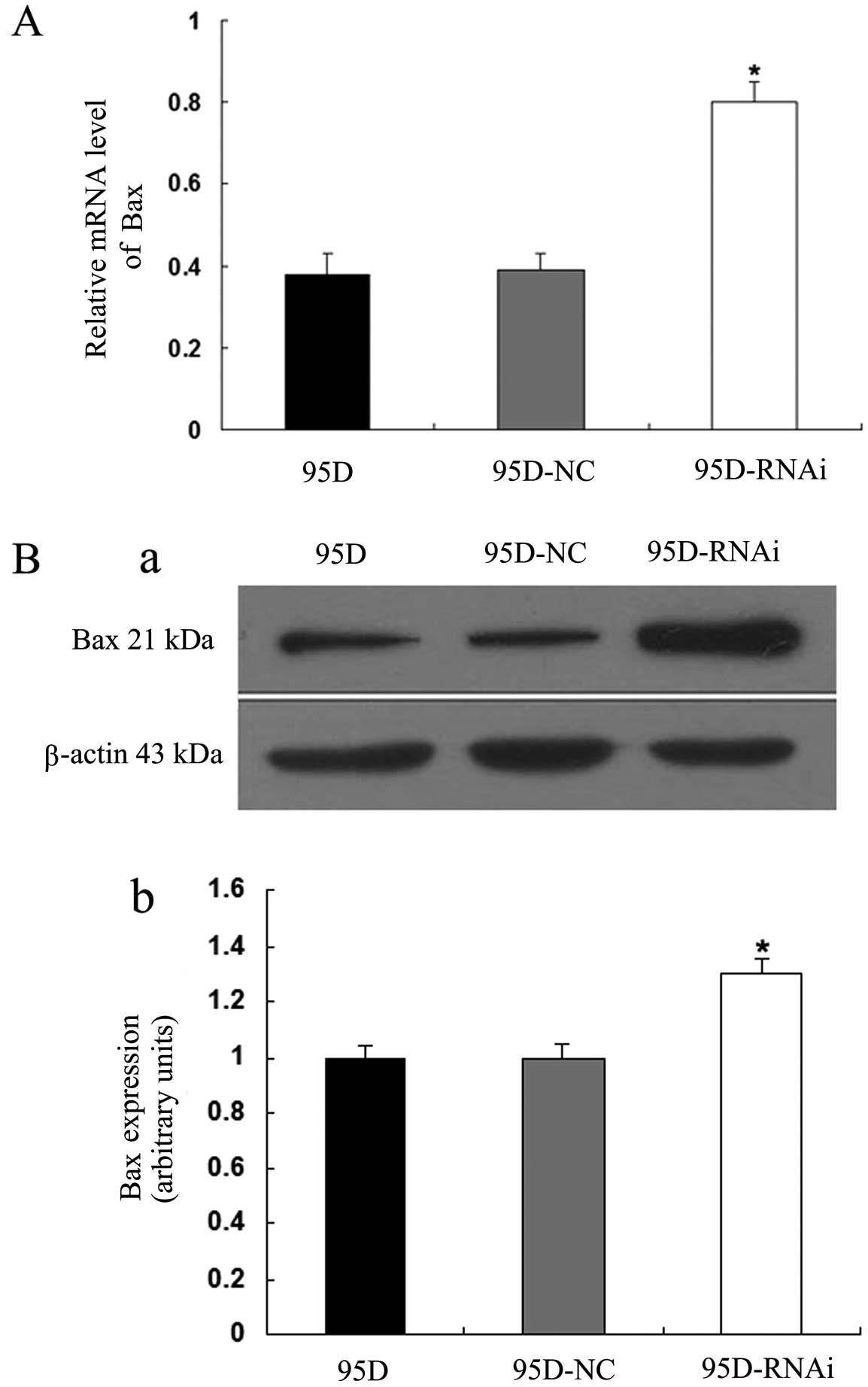
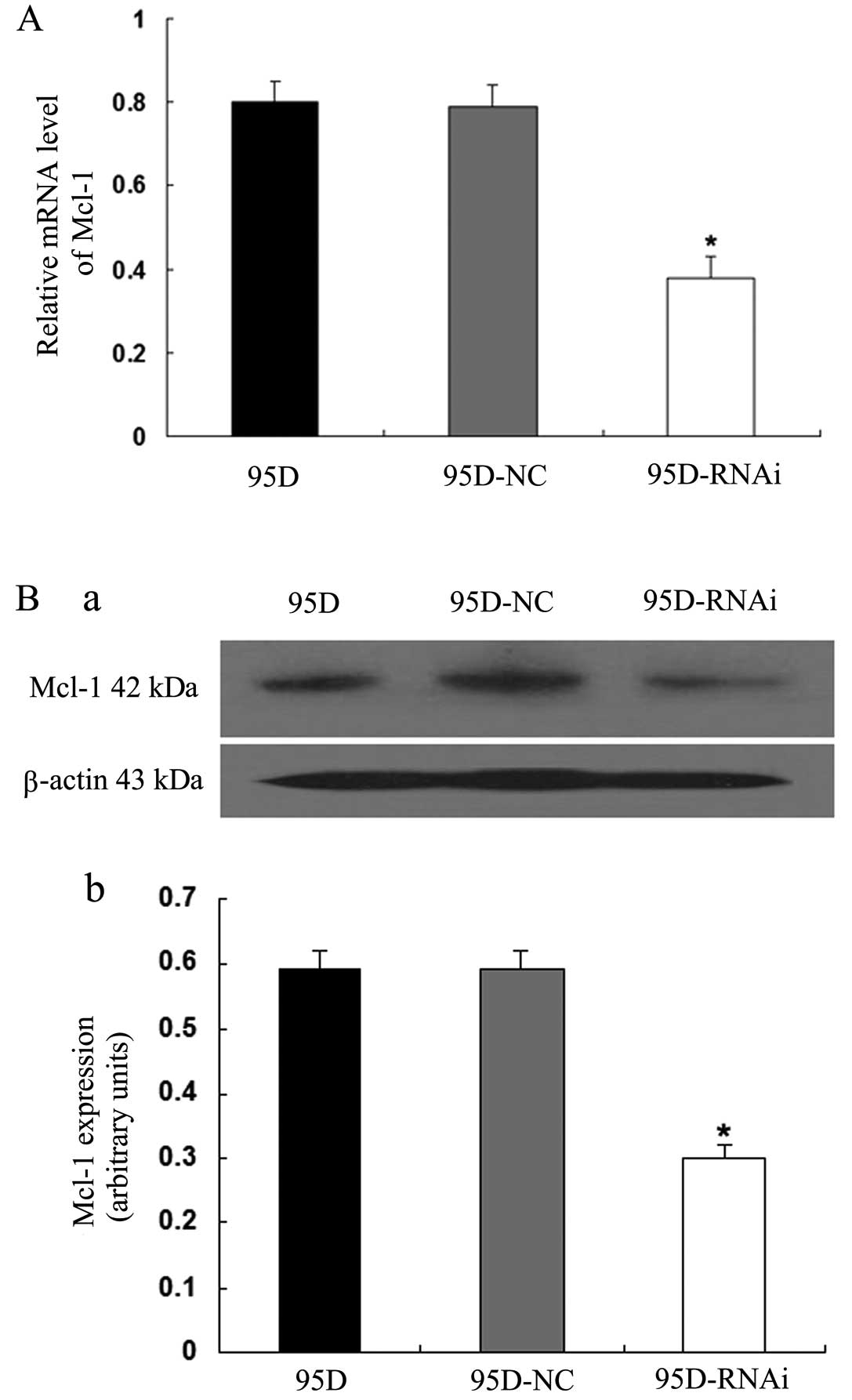
apoptosis induced by LCMR1 knockdown is related to Bax and Mcl-1,
we examined the expression levels of these two genes in the 95D,
95D-NC and 95D-RNAi cell groups using quantitative real-time RT-PCR
and western blot analyses. Bax expression was upregulated at both
the mRNA and protein levels in the 95D-RNAi cells compared to
levels in the 95D and 95D-NC cells (P<0.05) (Fig. 6). By contrast, Mcl-1 expression
was downregulated at both the mRNA and protein levels in the
95D-RNAi cells (P<0.05) (Fig.
7). These results suggest that downregulation of LCMR1
expression leads to a differential expression change in these
proand anti-apoptotic genes.
Discussion
LCMR1 and apoptosis
Cancer has a number of ‘mission critical’ events
that propel tumor cells into uncontrolled expansion and invasion
(11). The balance of
proliferation and apoptosis plays an important role in the control
of tumor growth (12). LCMR1 is a
novel lung cancer-related gene that has been implicated in cell
proliferation. However, the function of LCMR1 in the regulation of
the apoptotic process remains unclear. In this study, we employed
an RNAi approach to target LCMR1 in order to determine whether
LCMR1 participates in the regulation of lung cancer cell apoptosis.
Quantitative real-time RT-PCR and western blot analysis revealed
that LCMR1 was successfully knocked down by RNAi at both the mRNA
and protein levels. Using flow cytometry, we discovered that LCMR1
knockdown enhanced apoptotic activity in lung cancer cells. Western
blot analysis demonstrated that LCMR1 knockdown increased the
expression of cleaved caspase-3, an effector caspase activated in
approximately all apoptotic cells. These findings suggest that
LCMR1 regulates apoptotic activity in lung cancer cells, possibly
acting as an apoptosis suppressor.
p53 and LCMR1-related apoptosis
p53 is a tumor suppressor gene which mediates
apoptotic cell death in a variety of cell types (13–16). Lung cancer cell lines, A549
(p53+/+) and H1299 (p53−/−), are widely used
in the study of p53. In the current study, we applied these two
cell lines to determine the role of p53 in LCMR1-related apoptosis.
Our results demonstrated that the suppression of LCMR1 promoted
strong apoptotic activity in p53 wild-type A549 cells, but only
mild apoptosis in p53-deficient H1299 cells. These results suggest
that LCMR1-related apoptosis requires the involvement of p53.
Bax and Mcl-1 in LCMR1-related
apoptosis
The finding that LCMR1-mediated apoptosis is
associated with the p53 pathway prompted us to explore how p53
affects LCMR1-related apoptosis. p53 may exert its functions by
causing the direct transcriptional induction of specific target
genes such as the B-cell lymphoma 2 (Bcl-2) family. The Bcl-2
family members are central regulators of the intracellular
apoptotic signaling cascades (17). Bax (a pro-apoptotic gene) and its
suppressor Mcl-1 are members of the Bcl-2 family (18). p53 directly interacts with Bax to
promote Bax oligomerization (19). Oligomeric Bax induces the release
of cytochrome c from mitochondria, which in turn causes the
activation of caspases, ultimately leading to apoptosis (20). Mcl-1 blocks apoptosis by binding
and sequestering the pro-apoptotic protein Bax (21). Our results revealed that the
downregulation of LCMR1 led to an increase in Bax expression and a
decrease in Mcl-1 expression, suggesting a contribution of Bax and
Mcl-1 in LCMR1-related apoptosis. Bax and Mcl-1 have been shown to
participate in the mitochondrial signaling pathway of apoptosis
(22). The question whether LCMR1
modulates apoptosis via the same signaling apoptosis pathway
requires further research.
In conclusion, to the best of our knowledge, our
study for the first time reveals that LCMR1 suppresses apoptosis in
lung cancer cells. This effect is mediated via the p53 pathway and
is associated with apoptosis-related proteins Bax and Mcl-1. Hence,
this study indicates that LCMR1 may serve as a potential molecular
target for lung cancer therapy.
Abbreviations:
|
LCMR1
|
lung cancer metastasis-related protein
1
|
|
RNAi
|
RNA interference
|
|
NCBI
|
National Center for Biotechnology
Information
|
|
ORF
|
open reading frame
|
|
Med19
|
mediator complex subunit 19
|
|
NSCLC
|
non-small cell lung cancer
|
|
RPMI-1640
|
Roswell Park Memorial Institute medium
1640
|
|
DMEM
|
Dulbecco’s modified Eagle’s medium
|
|
siRNA
|
small interfering RNA
|
|
TBST
|
Tris-buffered saline Tween-20
|
|
PI
|
propidium iodide
|
|
Bcl-2
|
B-cell lymphoma 2
|
Acknowledgements
We thank Dr Bohua Hu for his comments
and editorial assistance. This study was supported by the National
Natural Science Foundation of China (no. 30370616).
References
|
1.
|
A JemalR SiegelE WardY HaoJ XuT MurrayMJ
ThunCancer statistics, 2008CA Cancer J
Clin587196200810.3322/CA.2007.0010
|
|
2.
|
SV SharmaDW BellJ SettlemanDA
HaberEpidermal growth factor receptor mutations in lung cancerNat
Rev Cancer7169181200710.1038/nrc208817318210
|
|
3.
|
T SantariusJ ShipleyD BrewerMR StrattonCS
CooperA census of amplified and overexpressed human cancer genesNat
Rev Cancer105964201010.1038/nrc277120029424
|
|
4.
|
L ChenZ LiangQ TianOverexpression of LCMR1
is significantly associated with clinical stage in human NSCLCJ Exp
Clin Cancer Res3018201110.1186/1756-9966-30-1821306606
|
|
5.
|
S SatoC Tomomori-SatoTJ ParmelyA set of
consensus mammalian mediator subunits identified by
multidimensional protein identification technologyMol
Cell14685691200410.1016/j.molcel.2004.05.006
|
|
6.
|
E Ji-FuJJ XingLQ HaoCG FuSuppression of
lung cancer metastasis-related protein 1 (LCMR1) inhibits the
growth of colorectal cancer cellsMol Biol
Rep3936753681201210.1007/s11033-011-1142-221732059
|
|
7.
|
M SunR JiangJD LiMED19 promotes
proliferation and tumorigenesis of lung cancerMol Cell
Biochem3552733201110.1007/s11010-011-0835-021519921
|
|
8.
|
SW ZouKX AiZG WangZ YuanJ YanQ ZhengThe
role of Med19 in the proliferation and tumorigenesis of human
hepatocellular carcinoma cellsActa Pharmacol
Sin32354360201110.1038/aps.2010.22321372827
|
|
9.
|
DA LiebermannB HoffmanRA SteinmanMolecular
controls of growth arrest and apoptosis: p53-dependent and
independent pathwaysOncogene1119921019957624128
|
|
10.
|
A BasuS HaldarThe relationship between
BcI2, Bax and p53: consequences for cell cycle progression and cell
deathMol Hum Reprod410991109199810.1093/molehr/4.12.10999872359
|
|
11.
|
GI EvanKH VousdenProliferation, cell cycle
and apoptosis in
cancerNature411342348200110.1038/3507721311357141
|
|
12.
|
J MatternM VolmImbalance of cell
proliferation and apoptosis during progression of lung
carcinomasAnticancer Res2442434246200415736479
|
|
13.
|
WM CongA BakkerPA SwalskyMultiple genetic
alterations involved in the tumorigenesis of human
cholangiocarcinoma: a molecular genetic and clinicopathological
studyJ Cancer Res Clin
Oncol127187192200110.1007/s00432000019411260864
|
|
14.
|
TM GottliebM Orenp53 and apoptosisSemin
Cancer Biol8359368199810.1006/scbi.1998.0098
|
|
15.
|
JD AmaralJM XavierCJ SteerCM RodriguesThe
role of p53 in apoptosisDiscov Med91451522010
|
|
16.
|
JS FridmanSW LoweControl of apoptosis by
p53Oncogene2290309040200310.1038/sj.onc.120711614663481
|
|
17.
|
NN DanialBCL-2 family proteins: critical
checkpoints of apoptotic cell deathClin Cancer
Res1372547263200710.1158/1078-0432.CCR-07-159818094405
|
|
18.
|
RW CraigThe bcl-2 gene familySemin Cancer
Biol6354319957548840
|
|
19.
|
JE ChipukT KuwanaL Bouchier-HayesNM
DroinDD NewmeyerM SchulerDR GreenDirect activation of Bax by p53
mediates mitochondrial membrane permeabilization and
apoptosisScience30310101014200410.1126/science.109273414963330
|
|
20.
|
B AntonssonS MontessuitS LauperR EskesJC
MartinouBax oligomerization is required for channel-forming
activity in liposomes and to trigger cytochrome c release from
mitochondriaBiochem
J345271278200010.1042/0264-6021:345027110620504
|
|
21.
|
LW ThomasC LamSW EdwardsMcl-1; the
molecular regulation of protein functionFEBS
Lett58429812989201010.1016/j.febslet.2010.05.06120540941
|
|
22.
|
S SomeyaJ XuK KondoAge-related hearing
loss in C57BL/6J mice is mediated by Bak-dependent mitochondrial
apoptosisProc Natl Acad Sci
USA1061943219437200910.1073/pnas.090878610619901338
|