Introduction
From the treatment of diabetes to experimental
therapy in critically ill patients, insulin is a widely used drug
affecting a number of metabolic pathways in human tissue (1–5).
In the present study, we characterized the effects of insulin on
pulmonary surfactant-associated genes and proteins (SFTPs). The
expression levels of 5 genes (SFTPA1, SFTPA2, SFTPB, SFTPC and
SFTPD) and their 4 protein products (SFTPA, SFTPB, SFTPC and SFTPD)
were examined in 2 cell lines, A549 and H441. Subsequently, we
determined the feasibility of performing these measurements in
patients with lung adenocarcinoma.
The A549 and H441 cell lines are commonly used as
models of lung surfactant-producing cells. A549 is a human lung
adenocarcinoma cell line which was established in 1972. A549 cells
have been used as a model of type II pneumocytes, demonstrating
typical morphology and ultra-structural features, such as lamellar
bodies that produce lung surfactant. A549 cells are also a
pulmonary epithelial cell model for drug metabolism (6). The phospholipids produced, which are
major components of surfactant, are similar to those generated by
type II pneumocytes (7). A549
cells typically grow in confluent monolayers.
H441 is a human lung adenocarcinoma epithelial cell
line used as a model of Clara cells. Clara cells are similar to
type II pneumocytes but are located in the bronchi, where they also
produce lung surfactant (8). H441
cells signal and respond to mitogenic growth factors in a manner
similar to type II pneumocytes (9). These cell lines have previously been
used in research studies to assess the link between insulin and
SFTPs (10,11).
Lung surfactant is a mixture of phospholipids and
proteins that reduces surface tension. Without lung surfactant, the
alveoli would collapse due to rapid changes in air pressure. Lung
surfactant also controls innate immunity. Surfactant contains 4
proteins that are crucial for its function. SFTPA (SFTPA1 and
SFTPA2) and SFTPD are large, hydrophilic proteins that mediate host
immunity and are both members of the collectin family (12). The SFTPA monomers vary between 26
and 35 kDa in size, and the SFTPD monomer is 43 kDa in size
(13). SFTPA and SFTPD multimers
bind to bacterial and fungal cells to suppress growth, stimulate
cell lysis and promote phagocytosis (14–16). Both proteins also bind to IgE and
lower allergic hypersensitivity (17). SFTPA and SFTPD are also involved
in the removal of apoptotic cells (18), and both proteins inhibit the
proliferation of T cells (19,20). SFTPB and SFTPC are small,
hydrophobic proteins. SFTPB is a 17-kDa homodimer, with a
biophysical function. SFTPB facilitates surfactant adsorption on
the surface of the alveoli, and surfactant film would not be
functional without SFTPB (21–23). In infants, the lack of SFTPB is
associated with SFTPC deficiency and leads to fatal respiratory
failure (24). SFTPC is the
smallest (4 kDa) and most abundant (65%) surfactant-associated
protein. SFTPC is an integral membrane protein with recent
evolutionary derivation. SFTPC has been identified only in the lung
tissue of mammals (22,25). The proper formation of mature
SFTPC from its pro-protein depends on the presence of functional
SFTPB (26,27). SFTPC binds bacterial
lipopolysaccharide to suppress inflammation (13). SFTPC (and SFTPA) also stimulates
cells to reabsorb surfactant phospholipids.
The mechanisms underlying the regulation of SFTPs
remain elusive; however, regulation at the transcriptional level
has been demonstrated. Thyroid transcription factor 1
(TTF-1/Nk×2.1) increases the transcription of SFTPs through
interactions with their promoters (28). TTF-1 and hepatocyte nuclear factor
(HNF)-3β have been implicated in the induction of SFTP expression
(29,30). Furthermore, glucocorticoid and
thyroid hormones have been shown to promote surfactant synthesis
and protein production through increased cAMP levels (28,31,32). Nitric oxide can inhibit the
transcription of SFTPB (33).
Little is known about the regulation of SFTP translation in
vivo. Glucocorticoids can affect the stability of SFTP mRNA
(28), and SFTPA translation can
be affected through specific miRNAs (34).
In this study, we attempted to characterize SFTPs in
the A549 and H441 cell lines and lung tissue. We examined the
combined effect of corticosteroids (dexamethasone was added to
stimulate gene expression) and insulin treatment on SFTP
expression. Furthermore, we demonstrate the effects of insulin on
SFTPs in patients with lung adenocarcinoma undergoing lung
resection. The sufficient production of lung surfactant is
important in these patients since it directly affects breathing,
which is seriously impaired following lung resection. The results
obtained in this study contribute to a better understanding of
surfactant biology and reveal the limitations of the use of the
A549 and H441 cell lines, as it is crucial to know the constraints
before selecting a proper in vitro model.
Materials and methods
Cell lines
The A549 and H441 cells were obtained from the
American Type Culture Collection (ATCC; Rockville, MD, USA). The
cells were cultivated in medium (A549, DMEM; H441, RPMI-1640)
enriched with 10% fetal calf serum (FCS), 50 U/ml penicillin G, and
50 μg/ml streptomycin sulfate at 37°C in a humidified
atmosphere with 5% CO2. The cells were grown to 85%
(A549) or 70% (H441) confluence, washed once with 1X PBS, and
placed in serum-free medium for 24 h prior to the experiment.
Insulin (Humulin; Eli Lilly, Prague, Czech Republic) diluted in
water was added to the cultures.
To determine the optimal dosage of insulin, we
considered published experiments (10,11,35) and performed optimization
experiments, examining serial dilutions of insulin from 2,500 to
0.25 ng/ml. Only the 2 highest concentrations of insulin exhibited
a reproducible impact on SFTP transcription. Thus, we used an
insulin concentration of 2,500 ng/ml in the medium in the
subsequent experiments.
We also optimized the duration of the insulin
treatment (1, 3, 6, 9, 12, 24 and 48 h), using 2,500 ng/ml insulin
in the medium. The results following exposure to insulin for
periods shorter than 24 h varied greatly, and we did not consider
these results as reliable. Thus, we used a 48-h exposure in the
subsequent experiments, as the effect of insulin was generally
stronger after 48 h than after 24 h.
Surprisingly, the mRNA levels of some SFTPs were
generally low in the cultured cells. Therefore, we added
dexamethasone (corticosteroid) to stimulate mRNA expression.
Corticosteroids are typically used to stimulate fetal lung
maturation in cases of premature labor. Based on previous studies,
we used the recommended concentration of 10−7 M
dexamethasone (Sigma-Aldrich, St. Louis, MO, USA) in the media
(36,37). Dexamethasone was diluted in
ethanol.
Lung tissue samples
Samples were obtained from 10 patients undergoing
lung resection. Written informed consent for the scientific use of
biological material was obtained from all patients in accordance
with the requirements of the institutional ethics committee.
Patient cohorts were divided into 2 groups with and without insulin
administration. Insulin was administered to maintain glycemia
levels between 6.0–8.0 mmol/l during surgery. The glycemia level
was assessed every hour. The infusion of 10% glucose was
administered at approximately 50 ml/h, and the glucose uptake was
maintained at 1 mg/kg/min. Surgical resection was performed under
general anesthesia, with additional epidural anesthesia or
paravertebral block. The details and insulin dosage for each
patient are presented in Table I.
After removal, the lung tissue samples were immediately stored in
RNAlater (Ambion, Austin, TX, USA) according to the manufacturer’s
instructions until total RNA or proteins were isolated. The samples
were homogenized using a MagNA Lyser Instrument (Roche Diagnostics,
Mannheim, Germany).
 | Table I.Details of patients with
adenocarcinoma. |
Table I.
Details of patients with
adenocarcinoma.
| Patients | Gender | Age ± (mean
SD) | Insulin dose (IU,
mean ± SD) |
|---|
| Untreated | M (n=2), F
(n=3) | 61.6±6.6 | 0 |
| Treated | M (n=2), F
(n=3) | 67.0±6.4 | 22.0±6.6 |
RNA isolation
The cells were washed twice with ice-cold 1X PBS,
harvested, and subsequently total RNA was isolated using an RNeasy
kit (Qiagen, Hilden, Germany) according to the standard protocol.
The RNA concentration was determined spectrophotometrically
(NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE, USA).
Reverse transcription
cDNA was synthesized from 2 μg of RNA using a
Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA,
USA) and oligo(dT) primers (Sigma-Aldrich), according to the
manufacturer’s instructions. The RNasin RNase Inhibitor (Promega,
Mannheim, Germany) was added.
qPCR
The TaqMan® Gene Expression Master Mix
was used together with TaqMan® Gene Expression Assays:
SFTPA1, Hs01921510_s1; SFTPA2, Hs00359837_m1; SFTPB, Hs00167036_m1;
SFTPC, Hs00161628_m1; SFTPD, Hs00358340_m1 (Applied Biosystems,
Foster City, USA). In each reaction, 100 ng of cDNA was used, and a
human GAPDH endogenous control (Applied Biosystems) was added.
GAPDH was selected as a reference gene, as GAPDH has shown a
stringent association with lung cells and demonstrates the least
variation out of the 7 common reference genes (38). The PCR reaction was performed in
96-well plates (25 μl per well) on a 7500 Real-Time PCR
System (Applied Biosystems). The following temperature settings
were used: step 1 at 50°C for 2 min, step 2 at 95°C for 10 min,
step 3 at 95°C for 15 sec, and step 4 at 60°C for 1 min. Steps 3
and 4 were repeated 40 times. The expression data obtained from the
cell line samples were calculated using the ΔΔCt method and
expressed as relative units. The data obtained from the lung tissue
samples were calculated using the ΔCt method.
Protein isolation
The cells were washed twice with ice-cold 1X PBS,
harvested, lysed in 250 μl RIPA lysis buffer [150 mM sodium
chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM
Tris (pH 8.0), Complete Protease Inhibitor Cocktail Tablets (Roche
Diagnostics), PhosSTOP Phosphatase Inhibitor Cocktail Tablets
(Roche Diagnostics)], and sonicated. The protein concentration was
determined using the BCA Protein assay (Pierce Biotechnology,
Rockford, IL, USA). The samples were subsequently diluted with RIPA
buffer to a final concentration of 0.95 μg/μl and
mixed with 2X Laemmli buffer [0.125 M Tris (pH 6.8), 20% glycerol,
4% SDS, 5% β-mercaptoethanol and 0.005% bromophenol blue]. The
mixture was boiled at 100°C for 5 min and cooled before loading
onto the electrophoresis gel.
SDS-PAGE and western blot analysis
Each cell line sample contained 24 μg of
total protein and each lung tissue sample contained 4 μg of
total protein, which was loaded into a 12% Precise Protein Gel for
SDS-PAGE (Pierce Biotechnology). The Amersham Full-Range Molecular
Weight Marker RPN800E (GE Healthcare, Little Chalfont, UK) was
used. SDS-PAGE was performed according to the manufacturer’s
instructions using the Mini-PROTEAN 3 system (Bio-Rad Laboratories,
Hercules, CA, USA). The proteins were electrophoresed in
Tris-HEPES-SDS buffer at 80 V for 70 min. The primary antibodies
were diluted in 5% BSA (PAN-Biotech, Aidenbach, Germany) and
applied overnight at 4°C. The secondary antibodies were diluted in
1X TBS buffer containing 0.05% Tween-20 (Sigma-Aldrich, St. Louis,
MO, USA) and 5% non-fat milk. The blots were developed using an
Immun-Star WesternC kit (Bio-Rad Laboratories) according to the
manufacturer’s instructions. The images were captured using a G:Box
Bio-Imaging System (Syngene, Frederick, MD, USA).
Antibodies
The following primary antibodies were used: mouse
monoclonal [HYB 238-04] antibody to surfactant protein A (AB51891;
Abcam, Cambridge, UK; dilution 1:200); rabbit anti-human
prosurfactant protein B polyclonal antibody (AB3430; Millipore,
Temecula, CA, USA; dilution 1:2,000); rabbit anti-prosurfactant
protein C polyclonal antibody (AB3786; Millipore; 1:3,000); mouse
monoclonal [12G5] to surfactant protein D (AB17781; Abcam; dilution
1:5,000); and GAPDH (14C10) rabbit monoclonal antibody (#2118; Cell
Signaling Technology, Danvers, MA, USA; dilution 1:1,000). The
following secondary antibodies were used: rabbit IgG secondary
antibody, H&L (AB6721; Abcam, dilution 1:5,000); and anti-mouse
IgG (whole molecule) peroxidase conjugate (A9044; Sigma-Aldrich;
dilution 1:5,000).
Statistical analysis
The cells were cultured in biological triplicates
and each sample was loaded into 3 wells of qPCR plates. The results
were transformed using a common logarithm, and based on the results
of the f-test, two-sample Student’s t-tests with equal or unequal
variance were applied. The P-value was set to 0.05. The analysis
compared samples cultured with insulin or dexamethasone to the
control samples. The samples cultured with both insulin and
dexamethasone were compared to the samples cultured with
dexamethasone only. The patients were divided into 2 groups of 5
patients, and the results were analyzed in a similar manner.
Results
The regulation of surfactant-associated protein
expression was investigated using bronchiolar-epithelial cells
(H441) and adenocarcinoma-derived alveolar epithelial cells (A549).
The cells were exposed to insulin (2,500 ng/ml) and dexamethasone
(10−7 M), and the expression of SFTPs was determined
using qPCR. The proof of principle was tested on a small group
(5+5) of lung adenocarcinoma patients. Insulin was administered to
these patients to maintain normoglycemia during surgery. Lung
samples were obtained from the healthy tissue surrounding the
tumor.
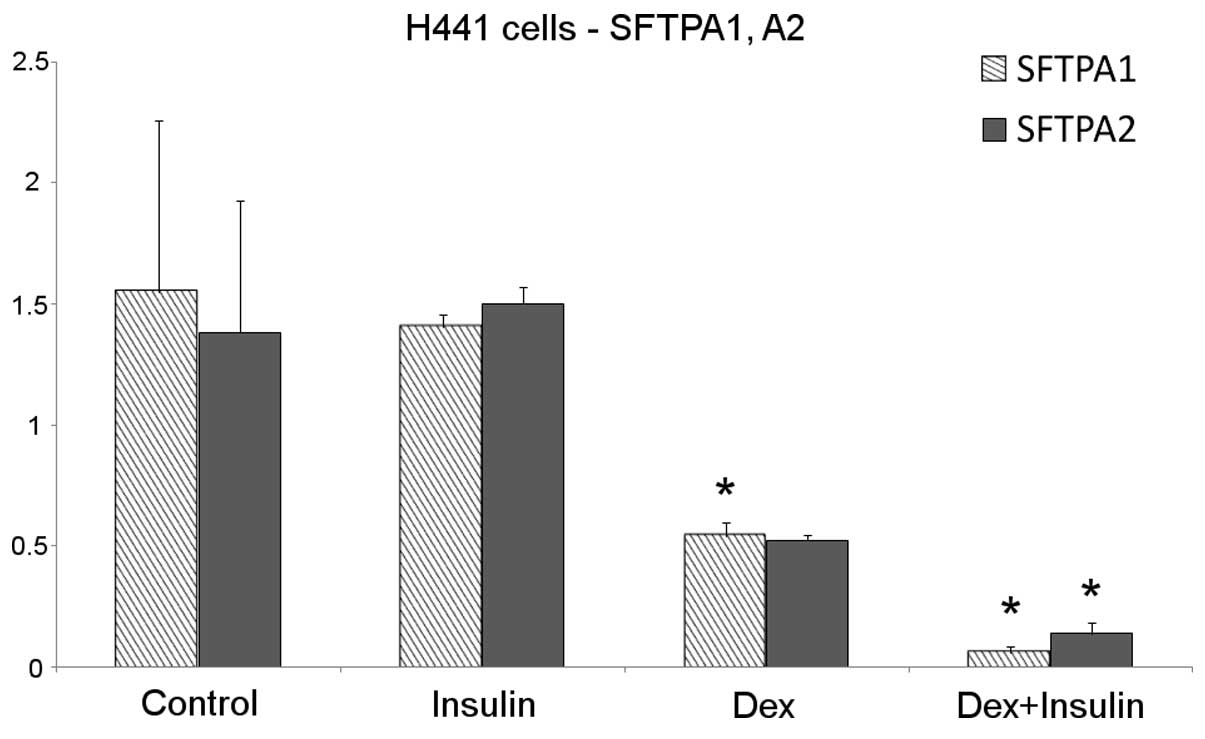
SFTPA1 and SFTPA2
No changes in the transcription of the SFTPA1 and
SFTPA2 genes were observed in the H441 cells following insulin
treatment for 48 h. Dexamethasone treatment decreased SFTPA1 and
SFTPA2 mRNA production by approximately 50%. Surprisingly, the
cells that received the combined dexamethasone and insulin
treatment showed an enhanced downregulation of SFTPA1 and SFTPA2
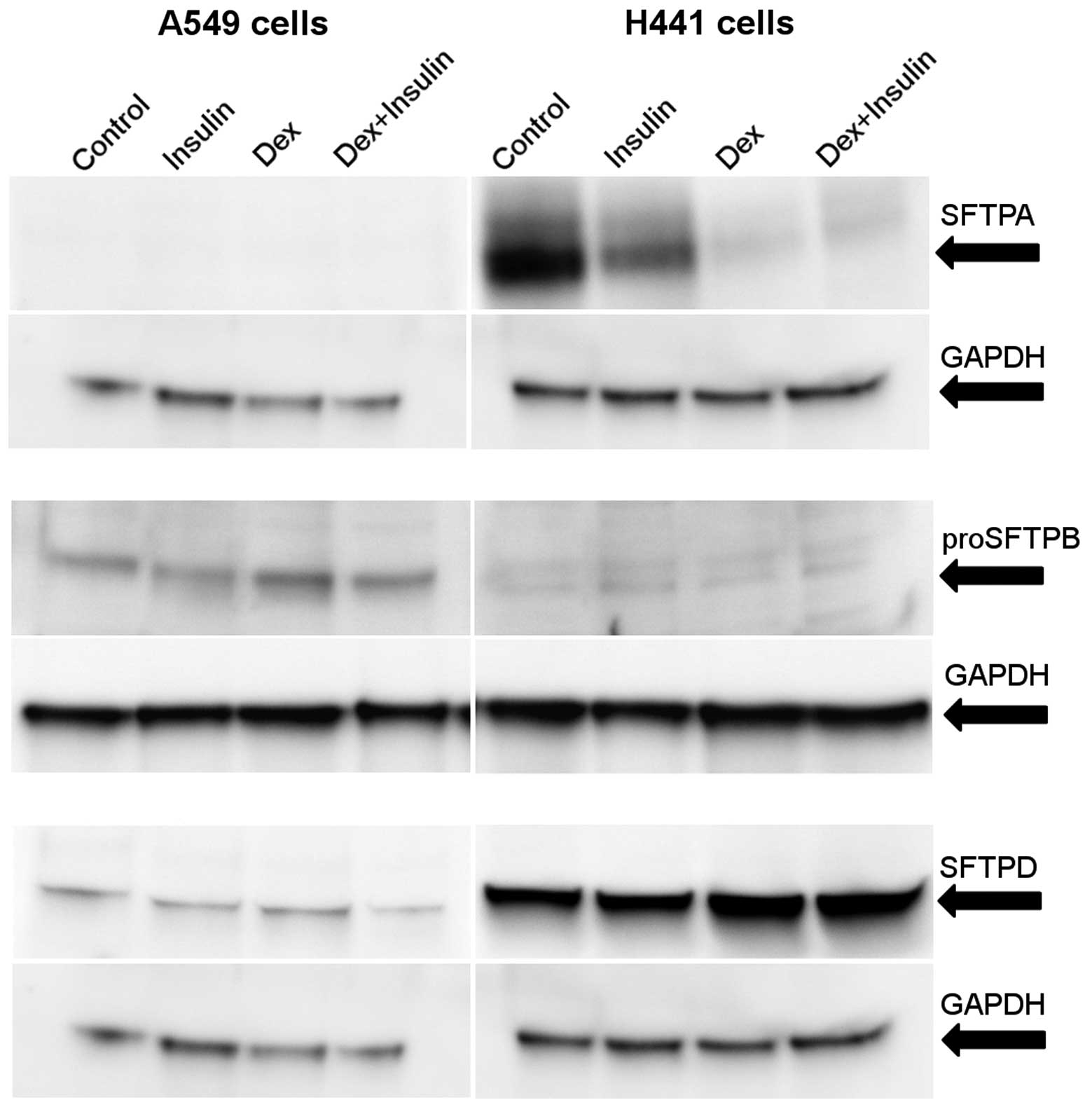
compared with the untreated cells (Fig. 1). The SFTPA protein levels were
reduced following treatment with dexamethasone or a combination of
dexamethasone and insulin. Unlike the mRNA levels, insulin alone
also caused a partial reduction in the SFTPA protein level
(Fig. 2).
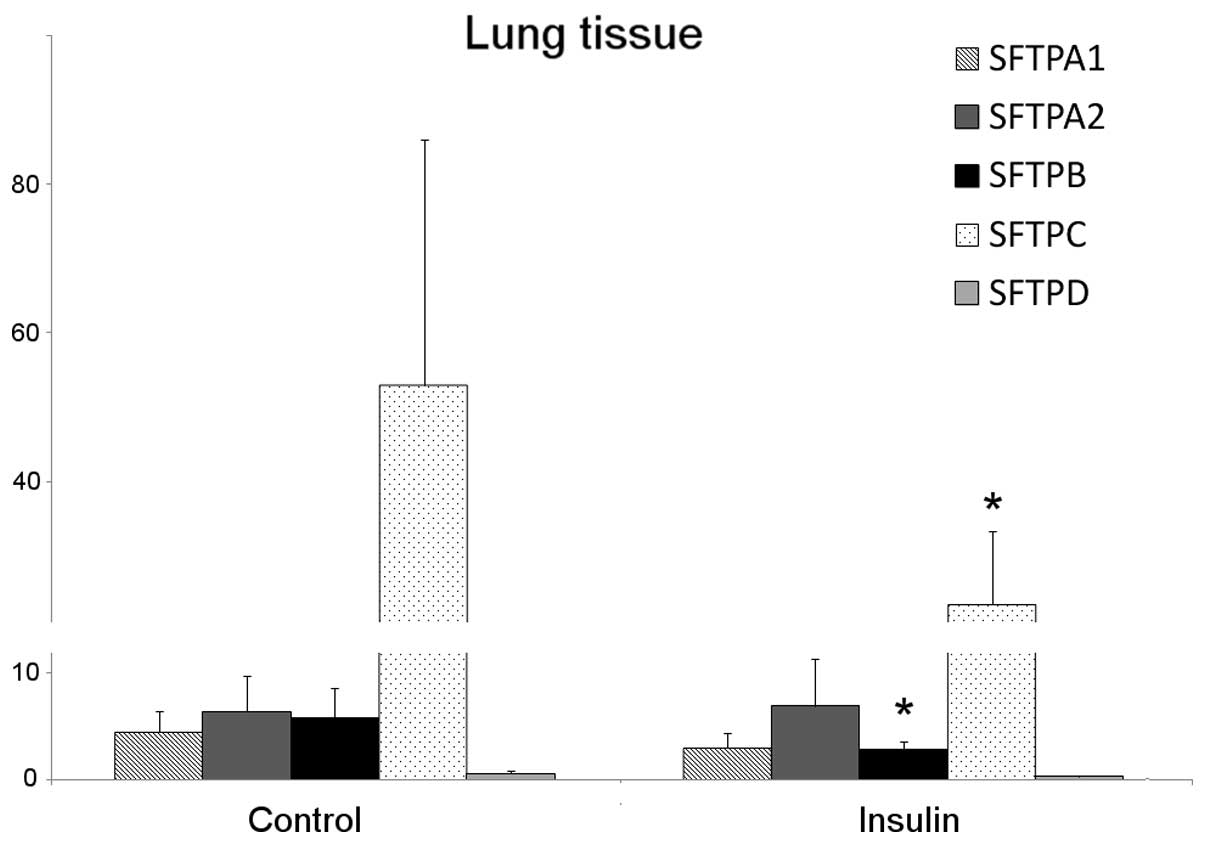
The mRNAs encoding SFTPA1 and SFTPA2 or the SFTPA
protein were not detected in the A549 cell line. In the lungs, the
SFTPA1 mRNA level was reduced by 30% following treatment with
insulin; however, this change was not statistically significant. No
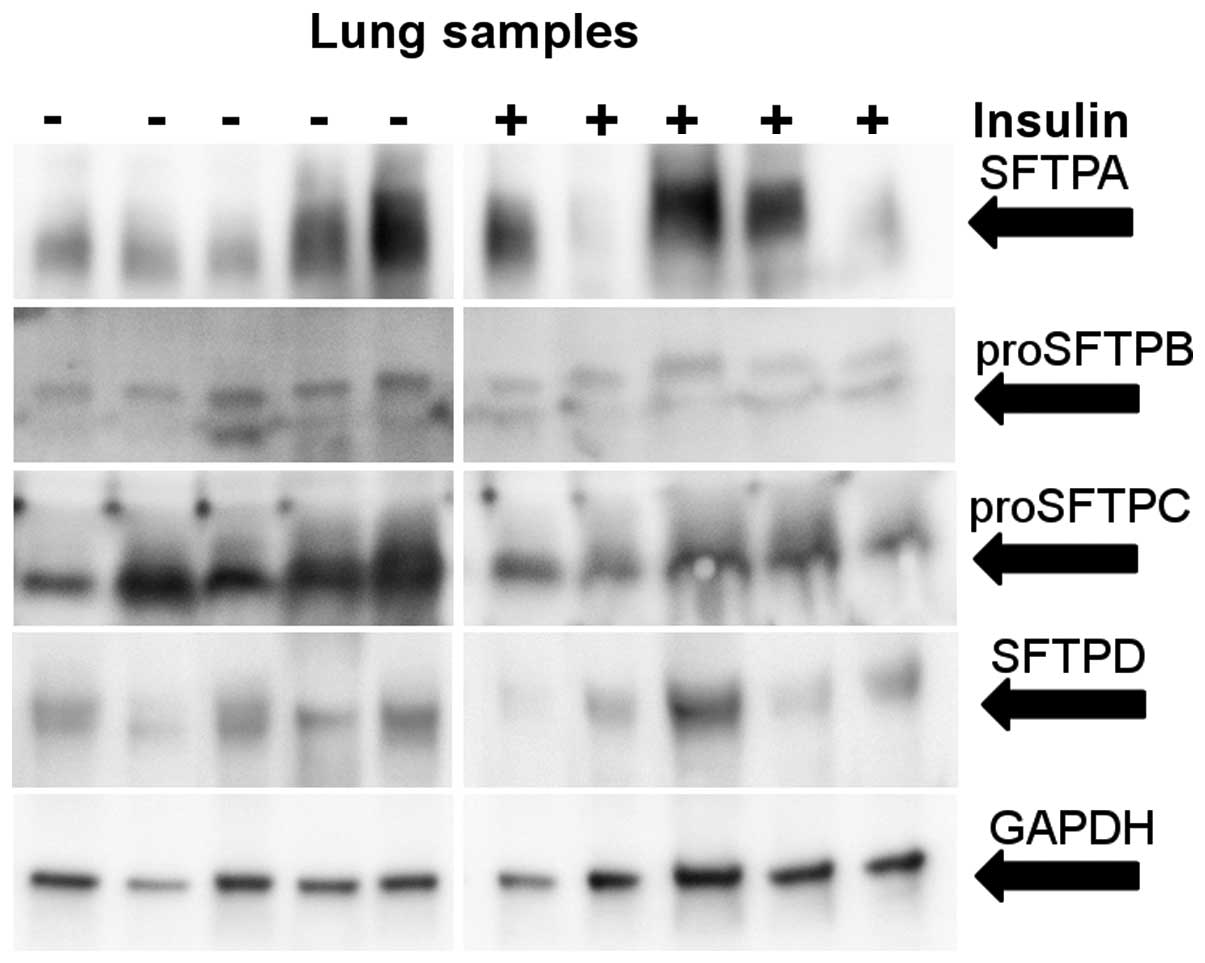
change was observed in the SFTPA2 mRNA levels (Fig. 4). The levels of total SFTPA
protein varied among the patients; however, no change was observed
in response to insulin treatment (Fig. 5).
SFTPB
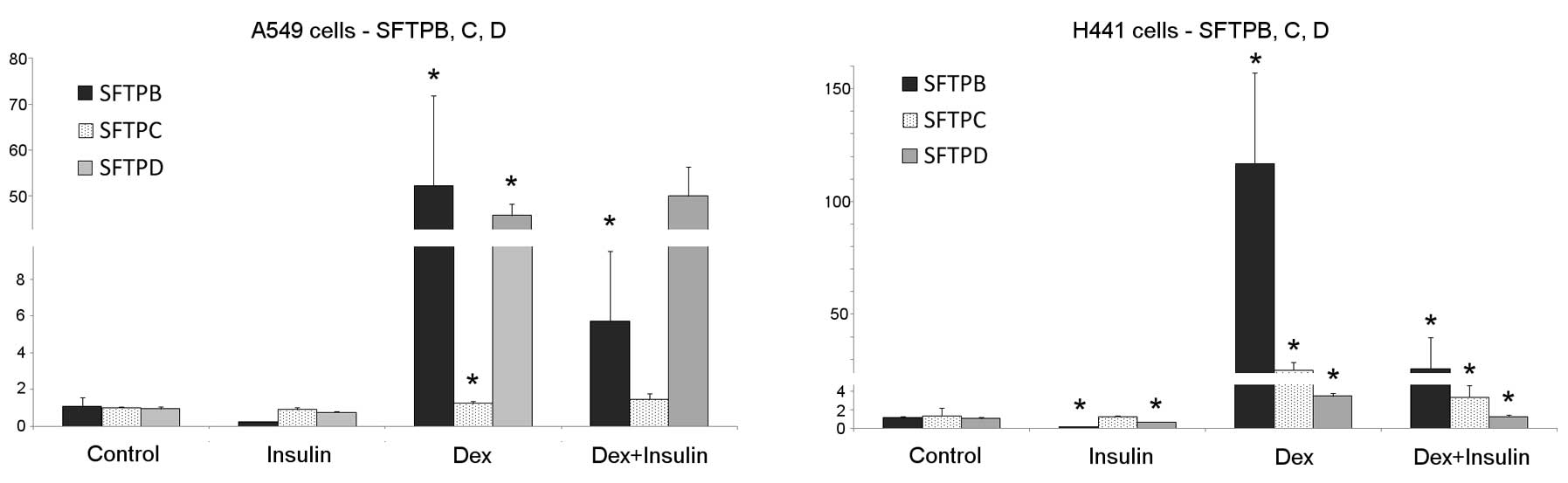
Significant changes in SFTPB expression were
detected in the H441 and A549 cell lines following treatment with
insulin or dexamethasone. Treatment with dexamethasone alone
increased the level of SFTPB mRNA in both cell lines. The cells
treated with a combination of dexamethasone and insulin exhibited a
strong inhibition of SFTPB mRNA transcription compared with cells
treated with dexamethasone alone. The rapid reduction in the SFTPB
mRNA level was observed in the H441 cells treated with insulin
alone. Untreated or insulin-treated A549 cells produced only trace
amounts of SFTPB mRNA (Fig.
3).
We then focused on the detection of the pro-form of
SFTPB, as mature SFTPB is a small molecule (8 kDa) and we were not
able to detect it in the cell lines used in the current study. The
observed levels of proSFTPB protein were low or barely detectable
in the H441 cells (Fig. 2),
although the level of SFTPB mRNA was 100,000-fold higher in the
H441 cells compared to the A549 cells (data not shown). However, in
the A549 cells, an increase in the proSFTPB protein level was
observed following treatment with dexamethasone. The combined
treatment with dexamethasone and insulin reduced the proSFTPB
protein level. In the patients, statistically significant lower
levels of SFTB mRNA (50%) and protein (partially) were observed in
the insulin-treated patients compared with the control group
(Figs. 4 and 5).
SFTPC
A significant upregulation in the SFTPC mRNA level
following treatment with dexamethasone was observed in the H441
cells. In these cells, a significant reduction in the
dexamethasone-induced expression of SFTPC was observed following 48
h of insulin co-treatment.
The A549 cells did not respond to insulin treatment,
and a minimum response to the dexamethasone treatment was observed
(Fig. 3). As both cell lines
produced only a trace amount of SFTPC mRNA, we were unable to
detect any SFTPC or proSFTPC protein expression. A statistically
significant reduction (45%) in SFTC mRNA expression in the
insulin-treated patients was observed (Fig. 4), and the level of proSFTPC
protein was also reduced in this group (Fig. 5).
SFTPD
Both H441 and A549 cells produced SFTPD, which was
further stimulated following treatment with dexamethasone.
Treatment with insulin inhibited the normal or
dexamethasone-induced transcription of SFTPD mRNA in the H441
cells, but not in the A549 cells (Fig. 3). In the A549 cells, the combined
treatment of dexamethasone and insulin reduced the SFTPD protein
levels; however, in the H441 cells, dexamethasone treatment, with
or without insulin, elevated SFTPD protein levels (Fig. 2).
These results provide evidence for the differential
regulation of mature protein through dexamethasone and insulin. In
the clinical samples, a 50% reduction in SFTPD mRNA levels
following treatment with insulin was observed; however, this result
was not statistically significant (Fig. 4). No difference in the SFTPD
protein expression was observed (Fig.
5). These results are summarized in Table II.
 | Table II.Regulation of mRNA/protein levels of
SFTPs in A549 and H441 cells following treatment with insulin,
dexamethasone or a combination of both for 48 h. |
Table II.
Regulation of mRNA/protein levels of
SFTPs in A549 and H441 cells following treatment with insulin,
dexamethasone or a combination of both for 48 h.
| Cell line | Treatment | SFTPA1a | SFTPA2a | SFTPBb | SFTPC | SFTPD |
|---|
| A549 | Insulin | -/- | -/- | ↔/↔ | ↔/- | ↔/↔ |
| Dex | -/- | -/- | ↑↑↑/↑ | ↔/- | ↑↑↑/↔ |
| Dex + insulin | -/- | -/- | ↑/↔ | ↔/↑ | ↑↑↑/↓ |
| H441 | Insulin | ↔/↓ | ↔/↓ | ↓/↔ | ↔/- | ↓/↔ |
| Dex | ↓/↓↓ | (↓)/↓↓ | ↑↑↑/↔ | ↑↑/- | ↑/↑ |
| Dex + insulin | ↓↓/↓↓ | ↓↓/↓↓ | ↑/↔ | ↑/- | ↔/↑ |
Differences in mRNA levels
Surprisingly, significant differences in the mRNA
levels were observed between the lung tissue samples and the cell
lines using the ΔΔCt method (GAPDH was used as a reference gene in
both cell lines and lung tissue). The expression of SFTPA1 and A2
in the control samples was 100- and 200-fold lower, respectively in
the H441 cell line. No SFTPA mRNA expression was detected in the
A549 cell line. The SFTPB mRNA level in the H441 cells was
approximately 2,000-fold lower than that in the lung tissue, and
approximately 10,000,000-fold lower in the A549 cells. SFTPC mRNA
was the most abundant mRNA in the lung tissue, but in both cell
lines only trace amounts were detected, with approximately
10,000,000-fold lower concentrations compared to the lung tissue.
The SFTPD mRNA concentration was 10,000-fold lower in the cell
lines compared to the lung tissue.
Discussion
The aim of this study was to examine the effects of
insulin on the transcription and translation of SFTPs in lung
surfactant-producing cells. Two cell lines, A549 and H441, are
commonly used as models of surfactant-producing cells.
Surprisingly, the 2 cell lines used in the current study differed
in several biological aspects, particularly as regards the
production of SFTPs. Moreover, there was a significant difference
in SFTP mRNA levels in these cell lines compared to those in the
lung tissue samples, making it difficult to generalize the complex
role of insulin and dexamethasone in primary surfactant-producing
lung cells.
Specifically, expression analysis revealed the
significantly reduced production of SFTPA mRNA in the H441 cells
and no production of SFTPA mRNA in the A549 cells. In addition, the
SFTPB mRNA level was low in the H441 cells, and only trace amounts
were detected in the A549 cells. The levels of SFTPC and SFTPD mRNA
were extremely low in both cell lines. The concentration of SFTPC
mRNA in the H441 cells was barely detectable, and it was originally
considered that the H441 cells do not synthesize SFTPC mRNA
(10). Accordingly, the levels of
SFTPC or proSFTPC proteins were below detection limits in the cell
lines used in the present study.
To compensate for the low quantity of SFTP mRNA and
proteins in these cell lines, dexamethasone was successfully
applied to stimulate the transcription of the SFTPB, SFTPC and
SFTPD genes. The effects of insulin treatment were accurately
measured following treatment with dexamethasone; however, the the
relatively large differences in the mRNA levels between the cell
lines and lung tissue were maintained.
Our results (Table
II) demonstrated that insulin reduced the mRNA transcription of
the SFTPB, SFTPC and SFTPD genes in the H441 cells, but did not
alter the transcription of SFTPA1 and SFTPA2, unless used in
combination with dexamethasone. Surprisingly, the combination of
insulin and dexamethasone significantly reduced the transcription
of SFTPA mRNA compared with the effects of dexamethasone alone. In
the A549 cells, only SFTPB transcription was down-regulated
following treatment with insulin. Insulin did not affect SFTPC or
SFTPD mRNA levels.
Although the SFTPA mRNA level in the H441 cells was
not affected by insulin alone, the level of SFTPA protein was
reduced in response to insulin, but not to the extent of the
reduction observed with dexamethasone treatment alone. The
treatment with dexamethasone alone, or in combination with insulin,
induced only trace amounts of the SFTPA protein in the H441 cells.
As expected, based on the absence of SFTPA1 or A2 mRNA, no SFTPA
protein was detected in the A549 cells. Our results suggested that
the stability of the proSFTPB protein was impaired in the H441
cells. However, the detection of mature SFTPB protein using western
blot analysis was not successful, as the mature SFTPB protein had a
low molecular weight. In the A549 cells, insulin counteracted the
increase in proSFTPB protein levels induced by dexamethasone. This
result was consistent with the qPCR results. Therefore, we suggest
that the increase in proSFTPB protein levels is a direct
consequence of lower SFTPB mRNA production.
Although SFTPD protein was detected in both cell
lines, this protein was more abundant in the H441 cells. This
difference was further increased following treatment with
dexamethasone. Insulin treatment did not counteract the effects of
dexamethasone on the expression of SFTPD protein. We suggest that
the stability of the SFTPD protein is greater in the H441 cells
since the SFTPD protein level is significantly higher in the H441
cells than in the A549 cells, despite the fact that both cell lines
contain the same amount of SFTPD mRNA.
SFTPB protein levels remained unresponsive to the
increased mRNA levels induced by dexamethasone in the A549 cells.
This result may reflect a reduction in the stability of this
protein in A549 cells. The combination of dexamethasone and insulin
treatment exerted inhibitory effects on the expression of SFTPB
protein in the A549 cells and SFTPA protein in the H441 cells.
In contrast to the findings of previous studies
concerning the effects of insulin in human fetal lung explants
maintained in vitro (39)
and H441 cells (10), we did not
observe any inhibitory effects of insulin on the SFTPA1 and SFTPA2
mRNA levels after 48 h of insulin treatment in the H441 cells.
However, we also observed a reduction in SFTPB mRNA levels in the
H441 cells, consistent with the findings of the aforementioned
studies. We observed a reduction in SFTPC protein levels in the
H441 cells following treatment with dexamethasone and insulin. This
result was not observed in the A549 cells. In human fetal lung
explants, no significant effect on SFTPC mRNA levels was observed
(39).
In this study, we demonstrate that dexamethasone
differentially upregulates the transcription of SFTPB, SFTPC and
SFTPD in the H441 and A549 cell lines, suggesting that this process
involves cell context-dependent transcriptional machinery. The
transcription of SFTPA1 and SFTPA2 mRNA was enhanced following
treatment with dexamethasone in the H441 cells. In non-human
primate fetal lung cells, dexamethasone treatment has also been
shown to reduce the levels of SFTPA mRNA (authors did not
distinguish the 2 isoforms), although the sensitivity to
dexamethasone treatment was reduced as the tissue matured (40). In the present study, the H441
cells showed typical characteristics of immature lung tissue.
According to Kumar and Snyder (35), treatment with dexamethasone
reduced the SFTPA1 mRNA level by 60%, and treatment with insulin
reduced the SFTPA1 mRNA level by 40% in the H441 cells. Moreover,
dexamethasone and insulin co-treatment did not affect the SFTPA2
mRNA levels. In this study, we confirmed that dexamethasone reduces
SFTPA1 transcription, and we observed an equivalent reduction
(approximately 50%) in SFTPA2 transcription. Moreover, in the study
by Kumar and Snyder, using human fetal lung explants, treatment
with dexamethasone or insulin reduced the SFTPA1 and the SFTPA2
mRNA level to the same degree (35). In the present study, we did not
detect any significant effects of insulin treatment on SFTPA1 and
SFTPA2 mRNA levels, unless insulin was combined with
dexamethasone.
The stimulatory effect of dexamethasone on the SFTPB
and SFTPC transcription has been previously established (41). This effect may reflect the
increased transcription and SFTPB mRNA stabilization, in addition
to the increased transcription and disputable effect on SFTPC mRNA
stabilization (42–46). Previous studies have shown that
SFTPD transcription is enhanced by treatment with dexamethasone,
although there are no data concerning the effect of dexamethasone
on the stability of SFTPD mRNA (47). In our study, surprisingly, the
A549 cells responded to dexamethasone treatment and displayed an
increase in the production of SFTPC mRNA. The H441 cells exhibited
a slight increase in the production of SFTPD mRNA.
Two factors challenge the suitability of the A549
and H441 cell lines as models for lung surfactant-producing cells,
type II pneumocytes and Clara cells: low levels of SFTP mRNA in the
A549 and H441 cells compared with the lung tissue and the
differences between the 2 cell lines, which were expected to
exhibit similar SFTP production. We suggest that the use of A549
and H441 cells is limited, and no solid conclusions concerning
SFTPs should be based on experiments performed only on these cell
lines.
Therefore, our study demonstrates the possibility of
measuring the effects of insulin on SFTP expression in patients
with lung adenocarcinoma. However, insulin is only administered to
maintain normoglycemia, which is disrupted through stress as a
result of surgery. Therefore, it is only possible to examine the
effects of insulin in a range of hours and not days. In this study,
we focused on insulin- and dexamethasone-induced changes in the
expression of SFTPs. We demonstrate that insulin reduces the
transcription of SFTPB, SFTPC and SFTPD in H441 cells. In the A549
cells, only SFTPB transcription was downregulated following
treatment with insulin. The expression of SFTPA1 and SFTPA2 was
only detected in the H441 cells, although insulin did not affect
their mRNA levels. Insulin significantly reduced the protein levels
of SFTPA in the H441 cells. The inhibitory effect of insulin on
proSFTPB protein levels was observed in the A549 cells.
Dexamethasone upregulated the transcription of SFTPB, SFTPC and
SFTPD genes in the A549 and H441 cells. Dexamethasone downregulated
the transcription of the SFTPA1 and SFTPA2 genes in the H441 cells.
However, the differences between the A549 and H441 cells and the
generally low levels of SFTP mRNA compared with the lung tissue,
challenge the suitability of these cell lines as models of
surfactant-producing cells. We recommend studying the effects of
insulin on SFTPs in patients, demonstrating the feasibility of
these experiments using tissues samples from patients with lung
adenocarcinoma during lung resection. We suggest that the use of
these samples may generate reliable results when the use of cell
line models is disputable.
Acknowledgements
This study was supported through
funding from the Czech Science Foundation (GACR P302/12/G157) and
the Ministry of Education, Youth and Sports of the Czech Republic
(MSM 0021622430).
References
|
1.
|
Bland D, Fankhanel Y, Langford E, et al:
Intensive versus modified conventional control of blood glucose
level in medical intensive care patients: a pilot study. Am J Crit
Care. 14:370–376. 2005.PubMed/NCBI
|
|
2.
|
Finfer S, Chittock D, Su S, et al:
Intensive versus conventional glucose control in critically ill
patients. N Engl J Med. 360:1283–1297. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Van den Berghe G, Wouters P, Weekers F, et
al: Intensive insulin therapy in the critically ill patients. N
Engl J Med. 345:1359–1367. 2001.
|
|
4.
|
Van den Berghe G: How does blood glucose
control with insulin save lives in intensive care? J Clin Invest.
114:1187–1195. 2004.PubMed/NCBI
|
|
5.
|
Van den Berghe G, Schetz M, Vlasselaers D,
et al: Clinical review: Intensive insulin therapy in critically ill
patients: NICE-SUGAR or Leuven blood glucose target? J Clin
Endocrinol Metab. 94:3163–3170. 2009.PubMed/NCBI
|
|
6.
|
Foster K, Oster C, Mayer M, Avery M and
Audus K: Characterization of the A549 cell line as a type II
pulmonary epithelial cell model for drug metabolism. Exp Cell Res.
243:359–366. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Nardone LL and Andrews SB: Cell line A549
as a model of the type II pneumocyte. Phospholipid biosynthesis
from native and organometallic precursors. Biochim Biophys Acta.
573:276–295. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Ladenburger A, Seehase M, Kramer BW, et
al: Glucocorticoids potentiate IL-6-induced SP-B expression in H441
cells by enhancing the JAK-STAT signaling pathway. Am J Physiol
Lung Cell Mol Physiol. 299:L578–L584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Chess PR, Ryan RM and Finkelstein JN: H441
pulmonary epithelial cell mitogenic effects and signaling pathways
in response to HGF and TGF-alpha. Exp Lung Res. 24:27–39. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Miakotina OL, Dekowski SA and Snyder JM:
Insulin inhibits surfactant protein A and B gene expression in the
H441 cell line. Biochim Biophys Acta. 1442:60–70. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Miakotina OL, Goss KL and Snyder JM:
Insulin utilizes the PI 3-kinase pathway to inhibit SP-A gene
expression in lung epithelial cells. Respir Res. 3:272002.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Holmskov U, Malhotra R, Sim R and
Jensenius J: Collectins: collagenous C-type lectins of the innate
immune defense system. Immunol Today. 15:67–74. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Chaby R, Garcia-Verdugo I, Espinassous Q
and Augusto LA: Interactions between LPS and lung surfactant
proteins. J Endotoxin Res. 11:181–185. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
McCormack F, Gibbons R, Ward S, Kuzmenko
A, Wu H and Deepe GJ: Macrophage-independent fungicidal action of
the pulmonary collectins. J Biol Chem. 278:36250–36256. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Wu H, Kuzmenko A, Wan S, et al: Surfactant
proteins A and D inhibit the growth of Gram-negative bacteria by
increasing membrane permeability. J Clin Invest. 111:1589–1602.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16.
|
van Iwaarden F, Welmers B, Verhoef J,
Haagsman H and van Golde L: Pulmonary surfactant protein A enhances
the host-defense mechanism of rat alveolar macrophages. Am J Respir
Cell Mol Biol. 2:91–98. 1990.PubMed/NCBI
|
|
17.
|
Deb R, Shakib F, Reid K and Clark H: Major
house dust mite allergens Dermatophagoides pteronyssinus 1
and Dermatophagoides farinae 1 degrade and inactivate lung
surfactant proteins A and D. J Biol Chem. 282:36808–36819.
2007.PubMed/NCBI
|
|
18.
|
Vandivier RW, Ogden CA, Fadok VA, et al:
Role of surfactant proteins A, D, and C1q in the clearance of
apoptotic cells in vivo and in vitro: calreticulin and CD91 as a
common collectin receptor complex. J Immunol. 169:3978–3986. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Borron P, Veldhuizen RA, Lewis JF, et al:
Surfactant associated protein-A inhibits human lymphocyte
proliferation and IL-2 production. Am J Respir Cell Mol Biol.
15:115–121. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Borron P, McCormack FX, Elhalwagi BM, et
al: Surfactant protein A inhibits T cell proliferation via its
collagen-like tail and a 210-kDa receptor. Am J Physiol.
275:L679–L686. 1998.PubMed/NCBI
|
|
21.
|
Nogee L: Genetic mechanisms of surfactant
deficiency. Biol Neonate. 85:314–318. 2004. View Article : Google Scholar
|
|
22.
|
Serrano AG and Pérez-Gil J: Protein-lipid
interactions and surface activity in the pulmonary surfactant
system. Chem Phys Lipids. 141:105–118. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Cruz A, Worthman LA, Serrano AG, Casals C,
Keough KM and Pérez-Gil J: Microstructure and dynamic surface
properties of surfactant protein
SP-B/dipalmitoylphosphatidylcholine interfacial films spread from
lipid-protein bilayers. Eur Biophys J. 29:204–213. 2000. View Article : Google Scholar
|
|
24.
|
Beers M, Hamvas A, Moxley M, et al:
Pulmonary surfactant metabolism in infants lacking surfactant
protein B. Am J Respir Cell Mol Biol. 22:380–391. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Besnard V, Wert SE, Kaestner KH and
Whitsett JA: Stage-specific regulation of respiratory epithelial
cell differentiation by Foxa1. Am J Physiol Lung Cell Mol Physiol.
289:L750–L759. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Weaver TE and Conkright JJ: Function of
surfactant proteins B and C. Annu Rev Physiol. 63:555–578. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Clark JC, Wert SE, Bachurski CJ, et al:
Targeted disruption of the surfactant protein B gene disrupts
surfactant homeostasis, causing respiratory failure in newborn
mice. Proc Natl Acad Sci USA. 92:7794–7798. 1995. View Article : Google Scholar
|
|
28.
|
Boggaram V: Regulation of lung surfactant
protein gene expression. Front Biosci. 8:d751–d764. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Zhou L, Lim L, Costa RH and Whitsett JA:
Thyroid transcription factor-1, hepatocyte nuclear factor-3beta,
surfactant protein B, C, and Clara cell secretory protein in
developing mouse lung. J Histochem Cytochem. 44:1183–1193. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Stahlman MT, Gray ME and Whitsett JA:
Temporal-spatial distribution of hepatocyte nuclear factor-3beta in
developing human lung and other foregut derivatives. J Histochem
Cytochem. 46:955–962. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Mendelson CR: Endocrinology of the Lung:
Development and Surfactant Synthesis. Humana Press; Totowa, New
Jersey: pp. 2000
|
|
32.
|
Mendelson CR and Boggaram V: Regulation of
pulmonary surfactant protein synthesis in fetal lung: a major role
of glucocorticoids and cyclic AMP. Trends Endocrinol Metab.
1:20–25. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Boggaram V, Chandru H, Gottipati KR,
Thakur V, Das A and Berhane K: Transcriptional regulation of SP-B
gene expression by nitric oxide in H441 lung epithelial cells. Am J
Physiol Lung Cell Mol Physiol. 299:L252–L262. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Silveyra P and Floros J: Genetic
complexity of the human surfactant-associated proteins SP-A1 and
SP-A2. Gene. 2012.
|
|
35.
|
Kumar A and Snyder J: Differential
regulation of SP-A1 and SP-A2 genes by cAMP, glucocorticoids, and
insulin. Am J Physiol. 274:L177–L185. 1998.PubMed/NCBI
|
|
36.
|
Mouhieddine-Gueddiche OB, Pinteur C,
Chailley-Heu B, Barlier-Mur AM, Clement A and Bourbon JR:
Dexamethasone potentiates keratinocyte growth factor-stimulated
SP-A and SP-B gene expression in alveolar epithelial cells. Pediatr
Res. 53:231–239. 2003. View Article : Google Scholar
|
|
37.
|
Salinas D, Sparkman L, Berhane K and
Boggaram V: Nitric oxide inhibits surfactant protein B gene
expression in lung epithelial cells. Am J Physiol Lung Cell Mol
Physiol. 285:L1153–L1165. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Liu D, Chen S and Liu H: Choice of
endogenous control for gene expression in nonsmall cell lung
cancer. Eur Respir J. 26:1002–1008. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Dekowski S and Snyder J: Insulin
regulation of messenger ribonucleic acid for the
surfactant-associated proteins in human fetal lung in vitro.
Endocrinology. 131:669–676. 1992.PubMed/NCBI
|
|
40.
|
Seidner S, Smith M and Mendelson C:
Developmental and hormonal regulation of SP-A gene expression in
baboon fetal lung. Am J Physiol. 271:L609–L616. 1996.PubMed/NCBI
|
|
41.
|
Liley H, White R, Warr R, Benson B,
Hawgood S and Ballard P: Regulation of messenger RNAs for the
hydrophobic surfactant proteins in human lung. J Clin Invest.
83:1191–1197. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Boggaram V and Margana R: Developmental
and hormonal regulation of surfactant protein C (SP-C) gene
expression in fetal lung. Role of transcription and mRNA stability.
J Biol Chem. 269:27767–27772. 1994.PubMed/NCBI
|
|
43.
|
Margana R and Boggaram V: Transcription
and mRNA stability regulate developmental and hormonal expression
of rabbit surfactant protein B gene. Am J Physiol. 268:L481–L490.
1995.PubMed/NCBI
|
|
44.
|
Huang H, Bi W, Jenkins G and Alcorn J:
Glucocorticoid regulation of human pulmonary surfactant protein-B
mRNA stability involves the 3’-untranslated region. Am J Respir
Cell Mol Biol. 38:473–482. 2008.PubMed/NCBI
|
|
45.
|
Planer B, Ning Y, Kumar S and Ballard P:
Transcriptional regulation of surfactant proteins SP-A and SP-B by
phorbol ester. Biochim Biophys Acta. 1353:171–179. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Venkatesh V, Iannuzzi D, Ertsey R and
Ballard P: Differential glucocorticoid regulation of the pulmonary
hydrophobic surfactant proteins SP-B and SP-C. Am J Respir Cell Mol
Biol. 8:222–228. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Mariencheck W and Crouch E: Modulation of
surfactant protein D expression by glucocorticoids in fetal rat
lung. Am J Respir Cell Mol Biol. 10:419–429. 1994. View Article : Google Scholar : PubMed/NCBI
|