Introduction
Idiopathic pulmonary fibrosis (IPF) is a common,
chronic, progressive and usually lethal fibrotic lung disease with
poor prognosis (1). This disease
is characterized by focal areas of alveolar epithelial cell injury
and the excessive proliferation of mesenchymal cells in the
interstitium, which results in the excessive deposition of
extracellular matrix (ECM) and distorted architecture leading to
impaired gas exchange (2,3). Although many pathobiological
concepts are emerging, including the role of aging and cellular
senescence, oxidative stress, endoplasmic reticulum stress,
cellular plasticity, microRNA (miRNA) and mechanotransduction, the
molecular mechanisms behind IPF are not yet completely understood
(4).
The Encyclopedia of DNA Elements (ENCODE) project,
which aimed to comprehensively characterize the human genome, has
shown that >90% of the genome has been transcribed; however,
only 1–2% of that is composed of genes (5). The majority of these transcripts are
not translated into proteins and are, therefore, termed non-coding
RNAs (ncRNAs).
Long non-coding RNAs (lncRNAs), a type of ncRNA,
vary in size from 200 bp to >100 kb, and are transcribed by RNA
polymerase II (6). They play an
important role in imprinting (7),
enhancing various biological functions (8), X chromosome inactivation (9), chromatin structure (10) and genomic rearrangement during the
generation of antibody diversity (11). Thus, lncRNAs are critical for
normal development and, in many cases, are deregulated in diseases,
such as cancer (12).
Thus far, studies on lncRNAs in IPF are limited.
Studies on differentially expressed lncRNAs are necessary for the
classification of genes regulated by lncRNAs in IPF. Among the
lncRNA profiling technologies, lncRNA microarrays offer a new tool
for understanding the biological role of lncRNAs. This study aimed
to construct lncRNA expression profiles in bleomycin-induced lung
fibrosis and normal lung tissue, in order to identify the
differentially expressed lncRNAs and to discover new molecular
targets for the therapy of IPF.
Materials and methods
Animals
Sprague Dawley (SD) rats (8 to 12 weeks old) were
provided by the Yantai Green Leaf Experimental Animal Center,
Yantai, China. A total of 20 SD rats were randomly divided into 2
groups (n=10 in each group): the normal control and lung fibrosis
model group. All animal experiments were carried out in accordance
with the principles of the National Institutes of Health Guide for
the Care and Use of Laboratory Animals.
Bleomycin administration
Rats in the model group were administered 5 mg/kg
bleomycin (Daiichi Pure Chemicals Co., Ltd., Tokyo, Japan)
dissolved in sterile phosphate-buffered saline (PBS) via a single
intratracheal instillation under anesthesia. The normal control
rats were administered an equal volume of saline. Lung tissues were
harvested on the 28th day following treatment with bleomycin.
Hydroxyproline assay
The total collagen content in the lungs was measured
using a colorimetric assay. The lung specimens were washed with
normal saline and hydrolysed with 6 ml/l hydrochloric acid at 100ºC
for 5 h. The hydrolysates were then diluted with distilled water
after neutralizing with sodium hydroxide. The hydroxyproline level
in the hydrolysates was then assessed colorimetrically at 560 nm
with p-dimethylaminobenzaldehyde, and the results were
calculated as mg hydroxyproline/g.
Masson's trichrome
Masson's trichrome method was used to show collagen
deposition. After fixing with 4% paraformaldehyde overnight,
dehydration in 70% ethanol and clearing in xylene, the lung tissues
were embedded in paraffin wax. Sections (4 μm) were prepared and
stained with Masson's trichrome. Nine random areas were examined at
a magnification of ×400. The severity of fibrosis was blindly
determined by a semiquantitative assay.
Observation of cell morphology under a
transmission electron microscope (TEM)
Lung tissues were fixed in fresh 3% glutaraldehyde
for at least 4 h at 4ºC, post-fixed in 1% osmium tetroxide for 1.5
h, dehydrated in a gradient series of ethanol, infiltrated with
Epon 812, embedded and cultured at 37, 45 and 60ºC for 24 h.
Ultrathin sections were ultracut using an ultracut E ultramicrotome
and stained with uranyl acetate and lead citrate prior to
observation under a JEM-1400 TEM (Jeol Ltd., Tokyo, Japan).
RNA isolation and quality assessment
Total RNA from lung tissues was isolated using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and the RNeasy mini
kit (Qiagen, Hilden, Germany) according to the manufacturer's
instructions. RNA quantity and quality were measured using the
NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.,
Wilmington, DE, USA) and RNA integrity was assessed by standard
denaturing agarose gel electrophoresis.
lncRNA microarray
ArrayStar, Inc. (Rockville, MD, USA) rat lncRNA
microarray was used and run by the service provider. The rat lncRNA
array was designed for profiling the lncRNAs and protein-coding
genes. Approximately 9,000 rat lncRNA candidates were identified
from the most authoritative databases, such as NCBI RefSeq, UCSC
all_mrna records and orthologs of mouse lncRNAs. Highly similar
sequences and ncRNAs shorter than 200 bp were excluded. Probes for
housekeeping genes and negative probes were printed multiple times
to ensure hybridization quality. ArrayStar designed the lncRNA
ChIP, taking into account not only the transcript already in the
database, but also with the aim of predicting new transcripts.
‘Potential lncRNA’ is a probe designed to help researchers discover
new lncRNAs and explore their functions.
RNA labeling and array hybridization
Sample labeling and array hybridization were
performed according to the Agilent One-Color Microarray-Based Gene
Expression Analysis protocol (Agilent Technologies, Inc., Santa
Clara, CA, USA). Briefly, 1 μg of total RNA from each sample was
linearly amplified and labeled with Cy3-dCTP. The labeled cRNAs
were purified using the RNeasy mini kit (Qiagen). The concentration
and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were
measured using the NanoDrop ND-1000 spectrophotometer. A total of 1
μg of each labeled cRNA was fragmented by the addition of 11 μl 10X
blocking agent and 2.2 μl of 25X fragmentation buffer, then the
mixture was heated at 60ºC for 30 min; finally, 55 μl 2X GE
hybridization buffer were added to dilute the labeled cRNA.
Hybridization solution (100 μl) was dispensed into the gasket slide
and assembled to the lncRNA expression microarray slide. The slides
were incubated for 17 h at 65ºC in an Agilent hybridization oven.
The hybridized arrays were washed, fixed and scanned using the
Agilent DNA Microarray Scanner.
Microarray data analysis
Agilent Feature Extraction software (version
10.7.3.1) was used to analyze the acquired array images. Quantile
normalization and subsequent data processing were performed using
the GeneSpring GX v11.5.1 software package (Agilent Technologies,
Inc.). Following quantile normalization of the raw data, lncRNAs
and mRNAs with at least 2 out of 2 present or marginal flags were
selected for further data analysis. Differentially expressed
lncRNAs and mRNAs were identified through fold change filtering.
Hierarchical clustering was performed using the Agilent GeneSpring
GX software (version 11.5.1). Gene Ontology (GO) analysis and
pathway analysis were performed using the standard enrichment
computation method.
In situ hybridization
The fixed lung tissues were dehydrated in ethanol,
cleared in xylene, transferred to paraffin and sectioned at 5 μm.
Paraffin sections were treated with Triton X-100 to enhance probe
penetration following conventional dewaxing with water. The slides
were washed in PBS and fixed again in 4% paraformaldehyde. After
washing in PBS and pre-hybridization for 4 h at 40ºC, the slides
were hybridized with digoxin-labeled RNA oligonucleotide probes at
40ºC overnight. The following day, the lung tissue sections were
washed with various concentrations of saline sodium citrate (SSC)
at 50ºC. After the addition of blocking solution made with sheep
serum for 1 h at 37ºC, the slides were incubated with
anti-digoxigenin-AP antibody (Roche Diagnostics GmbH, Mannheim,
Germany) overnight at 4ºC. Finally, the slides were stained with
NBT/BCIP solution (Roche Diagnostics GmbH) avoiding light after
being washed with Tris-Nacl buffer. The RNA oligonucleotide probe
of AJ005396 was as follows:
AATGTCCTTTGGAGGAAGGAGATATGAATTTTATCAATAAATCAAGTCTTGTCTA CCTGG. The
RNA oligonucleotide probe of S69206 was as follows:
TGCACGAGTCAGAGTCTCCAAGCTAGAGAA CTCTT TTGATATCCCTTGGGATCAACAAG.
Statistical analysis
All data are expressed as the means ± SD.
Statistical analysis was performed using SPSS11.0 software by
one-way ANOVA. A P-value <0.05 was considered to indicate a
statistically significant difference.
Results
Detection of bleomycin-induced lung
fibrosis
As the hydroxyproline content can indirectly reflect
collagen content, we determined the levels of hydroxyproline to
evaluate the degree of fibrosis. The hydroxyproline content in the
lungs was significantly increased in the model group compared with
the normal group (Fig. 1H). We
also used Masson's trichrome staining and TEM to observe
histopathological changes and collagen deposition following
bleomycin infusion. The results revealed a clear pulmonary
structure with an integral air-blood barrier and lower collagen
levels in the normal group (Fig. 1A,
C and E). By contrast, the structure of the lung tissue was
disordered, the air-blood barrier was severely damaged, the
pulmonary interalveolar septum was thickened and a significant
number of collagen fibers were deposited in the model group
(Fig. 1B, D, F and G). These
results indicated that we successfully established a model of
bleomycin-induced lung fibrosis.
RNA quality control (QC)
The integrity of the RNA was assessed by
electrophoresis on a denaturing agarose gel. The RNA run on the
denaturing gel had sharp 28S and 18S rRNA bands. The 28S rRNA band
was approximately twice as intense as the 18S rRNA band. This 2:1
intensity ratio indicated that the RNA was intact. A NanoDrop
ND-1000 spectrophotometer was then used to accurately measure the
RNA concentrations (OD260), protein contamination (ratio
of OD260/OD280) and organic compound
contamination (ratio of OD260/OD230). The OD
A260/A280 ratio was close to 2.0. The OD
A260/A230 ratio was calculated as
>1.8.
Differentially expressed lncRNAs and
mRNAs
To examine the potential biological functions of
lncRNAs in lung fibrosis, we determined the lncRNA and mRNA
expression profiles in bleomycin-induced lung fibrosis through
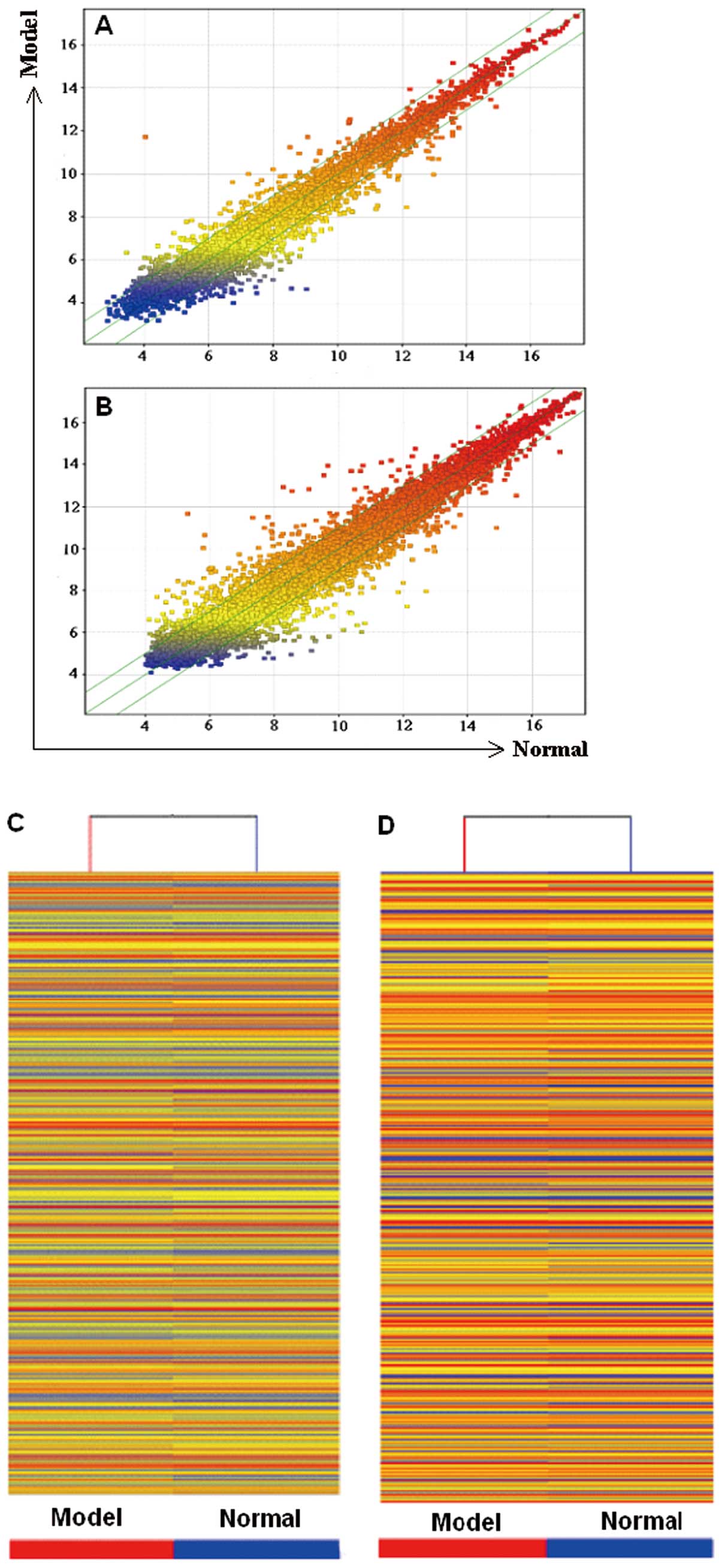
microarray analysis (Fig. 2). Up
to 568 lncRNAs were differentially expressed in the
bleomycin-treated lung samples compared with the normal control
group, among which 210 were upregulated, whereas 358 were
downregulated. A total of 945 mRNAs were differentially expressed.
Among these mRNAs, 415 were upregulated, whereas 530 were
downregulated. The fold change threshold was ≥2.0. The list only
shows the partial results for the differentially expressed lncRNAs
(Table I) and mRNAs (Table II) in the model vs. the normal
control groups.
 | Table IScreening of differentially expressed
lncRNAs. |
Table I
Screening of differentially expressed
lncRNAs.
| A, Upregulated
lncRNAs |
|---|
|
|---|
| SeqID | Fold change | Log fold
change | Absolute fold
change | Regulation | Model (raw) | Normal (raw) | Model
(normalized) | Normal
(normalized) |
|---|
| S69206 | 210.7381700 | 7.719308 | 210.73817 | Up | 3141.4421 | 18.151241 | 11.734264 | 4.0149565 |
| MRAK053938 | 7.0106416 | 2.8095465 | 7.0106416 | Up | 946.3922 | 172.075 | 10.021786 | 7.2122393 |
| AJ005396 | 6.756214 | 2.756215 | 6.756214 | Up | 2243.4438 | 409.89282 | 11.255636 | 8.499421 |
| MRAK143591 | 5.094425 | 2.3489194 | 5.094425 | Up | 132.47684 | 33.959503 | 7.2617292 | 4.91281 |
| BC091243 | 4.1729445 | 2.0610657 | 4.1729445 | Up | 4966.646 | 1405.8405 | 12.391487 | 10.330421 |
| MRAK081523 | 4.068016 | 2.0243254 | 4.068016 | Up | 349.46616 | 114.143684 | 8.62955 | 6.6052246 |
| MRAK158582 | 3.3001428 | 1.7225285 | 3.3001428 | Up | 52.645744 | 20.9094 | 5.929387 | 4.2068586 |
| AY007370 | 3.1992314 | 1.6777253 | 3.1992314 | Up | 159.31357 | 66.472755 | 7.5263243 | 5.848599 |
| XR_008064 | 3.097738 | 1.6312151 | 3.097738 | Up | 56.149826 | 23.58112 | 6.0165567 | 4.3853416 |
| MRuc007mlw | 2.9334714 | 1.552609 | 2.9334714 | Up | 84.11148 | 37.512997 | 6.623669 | 5.07106 |
| M77361 | 2.8096433 | 1.490387 | 2.8096433 | Up | 43.492096 | 20.006016 | 5.639081 | 4.148694 |
| MRAK089766 | 2.0438807 | 1.031311 | 2.0438807 | Up | 24.99775 | 15.457541 | 4.81422 | 3.782909 |
| MRAK029230 | 2.6328747 | 1.3966389 | 2.6328747 | Up | 60.88167 | 30.110182 | 6.138603 | 4.7419643 |
| MRAK045258 | 2.0882866 | 1.0623198 | 2.0882866 | Up | 32.666325 | 19.7309 | 5.194111 | 4.131791 |
| XR_007265 | 3.2810228 | 1.7141457 | 3.2810228 | Up | 14708.692 | 5233.981 | 13.952381 | 12.238235 |
| uc.161+ | 2.5087821 | 1.3269873 | 2.5087821 | Up | 116.44161 | 61.64021 | 7.0759335 | 5.748946 |
| BC166600 | 2.0669036 | 1.047471 | 2.0669036 | Up | 47.62331 | 29.747671 | 5.7733784 | 4.7259073 |
| AM293774 | 2.5931861 | 1.3747258 | 2.5931861 | Up | 299.42767 | 151.79784 | 8.409706 | 7.0349803 |
| Z93366 | 3.409905 | 1.7697315 | 3.409905 | Up | 163.78604 | 63.930973 | 7.566777 | 5.7970457 |
| XR_007062 | 2.5269985 | 1.3374248 | 2.5269985 | Up | 100.54225 | 53.123276 | 6.8767934 | 5.5393686 |
|
| B, Downregulated
lncRNAs |
|
| SeqID | Fold change | Log fold
change | Absolute fold
change | Regulation | Model (raw) | Normal (raw) | Model
(normalized) | Normal
(normalized) |
|
| XR_005532 | −20.369864 | −4.3483644 | 20.369864 | Down | 22.40888 | 577.1471 | 4.6590877 | 9.007452 |
| AF159100 | −12.764141 | −3.6740246 | 12.764141 | Down | 311.6397 | 4852.019 | 8.463682 | 12.137707 |
| MRAK018459 | −6.187315 | −2.6293135 | 6.187315 | Down | 62.63327 | 504.11053 | 6.1816454 | 8.810959 |
| MRAK153573 | −5.0353293 | −2.332086 | 5.0353293 | Down | 58.95229 | 389.28036 | 6.090506 | 8.422592 |
| MRAK040987 | −4.682474 | −2.227271 | 4.682474 | Down | 68.1992 | 422.10565 | 6.3133345 | 8.540606 |
| MRAK033233 | −4.4625397 | −2.157865 | 4.4625397 | Down | 45.224556 | 266.30106 | 5.6959915 | 7.8538566 |
| MRuc008euj | −4.306261 | −2.1064358 | 4.306261 | Down | 48.580467 | 275.86755 | 5.8036256 | 7.9100614 |
| BC088752 | −3.3241138 | −1.7329698 | 3.3241138 | Down | 44.798225 | 197.15247 | 5.683831 | 7.416801 |
| AF352172 | −3.1898289 | −1.6734791 | 3.1898289 | Down | 30.87954 | 128.38947 | 5.107747 | 6.781226 |
| MRAK036098 | −3.0710256 | −1.6187205 | 3.0710256 | Down | 23.849337 | 96.428856 | 4.751513 | 6.3702335 |
| AB072248 | −2.826814 | −1.499177 | 2.826814 | Down | 95.15817 | 356.84283 | 6.7976427 | 8.29682 |
| MRAK031681 | −2.5653043 | −1.3591299 | 2.5653043 | Down | 184.00778 | 610.2933 | 7.723872 | 9.083002 |
| XR_007761 | −2.4044764 | −1.2657228 | 2.4044764 | Down | 84.096306 | 272.36227 | 6.623521 | 7.8892436 |
| MRAK081707 | −2.5626018 | −1.3576093 | 2.5626018 | Down | 27.748964 | 93.19641 | 4.9642982 | 6.3219075 |
| MRAK134949 | −2.3397248 | −1.2263389 | 2.3397248 | Down | 15.463284 | 46.174065 | 4.121881 | 5.34822 |
| MRAK080342 | −2.2991571 | −1.2011051 | 2.2991571 | Down | 1250.1749 | 3400.0752 | 10.417191 | 11.618296 |
| MRuc007wwx | −2.3716562 | −1.2458949 | 2.3716562 | Down | 216.22974 | 656.4996 | 7.94957 | 9.195465 |
| MRAK042040 | −2.2730455 | −1.1846266 | 2.2730455 | Down | 94.70941 | 288.2609 | 6.7916226 | 7.976249 |
| XR_007395 | −2.0279934 | −1.0200529 | 2.0279934 | Down | 632.9264 | 1555.753 | 9.455593 | 10.475646 |
| MRAK042427 | −2.4156244 | −1.2723961 | 2.4156244 | Down | 475.61267 | 1413.5208 | 9.065523 | 10.337919 |
 | Table IIScreening of differentially expressed
mRNAs. |
Table II
Screening of differentially expressed
mRNAs.
| A, Upregulated
mRNAs |
|---|
|
|---|
| SeqID | Fold change | Log fold
change | Absolute fold
change | Regulation | Model (raw) | Normal (raw) | Model
(normalized) | Normal
(normalized) |
|---|
| NM_019322 | 84.40361 | 6.399233 | 84.40361 | Up | 3035.9275 | 44.22444 | 11.6874275 | 5.2881947 |
| NM_053605 | 19.508001 | 4.285994 | 19.508001 | Up | 967.8983 | 62.61611 | 10.053534 | 5.7675395 |
| NM_031808 | 11.300015 | 3.4982529 | 11.300015 | Up | 314.0429 | 35.48023 | 8.475712 | 4.977459 |
| NM_001024763 | 8.801827 | 3.137803 | 8.801827 | Up | 281.56186 | 40.555855 | 8.315399 | 5.177596 |
| NM_001113781 | 8.117821 | 3.0210924 | 8.117821 | Up | 481.5752 | 76.96868 | 9.081482 | 6.0603895 |
| NM_134329 | 7.2603674 | 2.8600426 | 7.2603674 | Up | 418.20984 | 74.544266 | 8.874704 | 6.014662 |
| NM_053750 | 6.7114744 | 2.7466297 | 6.7114744 | Up | 208.96045 | 40.024292 | 7.9063206 | 5.159691 |
| NM_001009919 | 5.0369353 | 2.3325462 | 5.0369353 | Up | 207.11671 | 54.101795 | 7.8946905 | 5.5621443 |
| NM_031073 | 4.622809 | 2.2087698 | 4.622809 | Up | 445.40768 | 126.902695 | 8.972042 | 6.7632723 |
| NM_145093 | 4.364931 | 2.125959 | 4.364931 | Up | 384.9827 | 115.6088 | 8.749628 | 6.623669 |
| NM_012845 | 4.2602963 | 2.0909538 | 4.2602963 | Up | 261.5588 | 80.66606 | 8.215511 | 6.1245575 |
| NM_001106425 | 4.04188 | 2.0150266 | 4.04188 | Up | 189.40274 | 61.544144 | 7.761023 | 5.7459965 |
|
| B, Downregulated
mRNAs |
|
| SeqID | Fold change | Log fold
change | Absolute fold
change | Regulation | Model (raw) | Normal (raw) | Model
(normalized) | Normal
(normalized) |
|
| NM_012589 | −28.815973 | −4.848797 | 28.815973 | Down | 132.6186 | 4777.8477 | 7.264641 | 12.113438 |
| NM_053624 | −11.540494 | −3.528633 | 11.540494 | Down | 28.109798 | 413.18723 | 4.983698 | 8.512331 |
| NM_001013145 | −10.523788 | −3.3955822 | 10.523788 | Down | 47.613003 | 643.6955 | 5.773218 | 9.1688 |
| NM_001024890 | −7.6730404 | −2.9397984 | 7.6730404 | Down | 43.61209 | 434.4988 | 5.6424265 | 8.582225 |
| NM_001109233 | −7.4248977 | −2.8923712 | 7.4248977 | Down | 24.390635 | 235.72002 | 4.7840214 | 7.6763926 |
| NM_022280 | −6.217645 | −2.6363683 | 6.217645 | Down | 43.506603 | 351.66534 | 5.639379 | 8.275747 |
| NM_017226 | −5.3901715 | −2.4303312 | 5.3901715 | Down | 103.21507 | 721.87415 | 6.9101744 | 9.340506 |
| NM_001108195 | −5.0052595 | −2.3234448 | 5.0052595 | Down | 44.6552 | 294.27753 | 5.6805167 | 8.003962 |
| NM_001107444 | −4.81651 | −2.2679882 | 4.81651 | Down | 57.48932 | 360.80945 | 6.049778 | 8.317766 |
| NM_133288 | −4.4379377 | −2.1498895 | 4.4379377 | Down | 40.90932 | 238.50754 | 5.5452423 | 7.695132 |
| NM_001042354 | −4.3010206 | −2.104679 | 4.3010206 | Down | 81.92266 | 465.0587 | 6.5834494 | 8.688128 |
| NM_201420 | −4.1225066 | −2.043522 | 4.1225066 | Down | 81.31964 | 443.837 | 6.572609 | 8.616131 |
Location and expression of AJ005396 and
S69206
To validate the microarray data and to explore the
location of lncRNAs, 2 of the upregulated lncRNAs (AJ005396 and
S69206) selected by the fold change and the raw intensities
(Table IA) were analyzed by ISH.
A blue-violet color indicated a positive reaction. The results
revealed that the alveoli have clear hollow cavities, and that the
alveolar membranes were thinner in the normal group compared to the
model group. AJ005396 and S69206 were observed in the cytoplasm of
the interstitial lung cells. Their expression was significantly
increased in the model group compared with the normal control group
which was consistent with our microarray data (Fig. 3).
Pathway analysis
Pathway analysis was carried out based on the latest
Kyoto Encyclopedia of Genes and Genomes (KEGG) database (13). This analysis was used to determine
the biological pathways associated with the most differentially
expressed mRNAs in lung fibrosis. Up to 22 downregulated and 16
upregulated pathways were identified associated with
cytokine-cytokine receptor interaction, chemokine signaling
pathway, cell adhesion molecules, Jak/STAT signaling pathway, cell
cycle, complement and coagulation cascades, the peroxisome
proliferator-activated receptor (PPAR) signaling pathway, as well
as others. The predominant pathways are summarized in Fig. 4.
GO analysis
GO analysis is a functional analysis that associates
differentially expressed mRNAs. The GO categories were derived from
the Gene Ontology website (www.geneontology.org) and comprised of 3 structured
networks: biological processes, cellular components and molecular
function. According to the GO annotation tool, the differentially
expressed genes were principally enriched for GO terms related to
immune response, cell differentiation, tyrosine phosphorylation of
STAT3 protein linked with biological processes associated with
extracellular structure organization, ECM, cytoskeleton, fibrillar
collagen involved in cellular components, as well as chemokine
activity, chemokine receptor binding, immunoglobulin binding and
insulin-like growth factor (IGF) binding in molecular functions.
The chart shows the top 10 counts of the significant enrichment
terms with the most number of differentially expressed genes
(Fig. 5).
Discussion
lncRNAs participate in a wide range of biological
processes. Almost every step in the life cycle of genes from
transcription to mRNA splicing, RNA decay and translation, is
influenced by lncRNAs (14–16). Previous studies have demonstrated
that the abnormal expression of lncRNAs contributes to numerous
diseases (17–21). However, the profile and the
biological function of lncRNAs in lung fibrosis remain unknown. In
the present study, we provide new information as to the expression
of lncRNAs using a rat model of bleomycin-induced lung fibrosis. To
explore the role of lncRNAs in lung fibrosis, a microarray analysis
was used to verify the lncRNA expression profiles. Furthermore, we
selected 2 lncRNAs, AJ005396 and S69206, to validate the microarray
data and explore their location for further research.
lncRNAs can be roughly classified according to the
positional relationship of the protein-coding genes in the
chromosome. In a recent study, Sui et al divided lncRNAs
into 4 groups by analyzing complex transcriptional loci that
include lncRNAs and their associated protein-coding genes, namely,
cis-antisense lncRNAs, intronic lncRNAs, promoter-associated
lncRNAs, and bidirectional lncRNAs (22). According to our microarray
results, lncRNAs can also be partially subjected to a similar
classification system: MRAK089766, MRAK029230 and MRAK081707, among
others, belong to the sense_exon_overlap (the exon of the lncRNA
overlaps with a coding transcript exon on the same genomic strand);
MRAK045258, MRAK134949 and MRAK080342, among others, represent the
sense_intron_overlap (the lncRNA overlaps with the intron of a
coding transcript on the same genomic strand); XR_007265,
MRuc007wwx and MRAK042040 are categorized under the
antisense_exon_overlap (the lncRNA is transcribed from the
antisense strand and overlaps with a coding transcript); uc.161+
and XR_007395 comprise the antisense_intron_overlap (the lncRNA is
transcribed from the antisense strand without sharing overlapping
exons); BC166600 and MRAK042427 are bidirectional (the lncRNA is
oriented head to head to a coding transcript within 1,000 bp); and
AM293774, Z93366 and XR_007062, among others, are intergenic (there
are no coding transcripts within 30 kb of the lncRNA) (Table I).
Although research on lncRNAs has gradually
increased, only a few lncRNAs have official names and clear
functions. In our microarray results, a vast majority of the
differentially expressed lncRNAs in bleomycin-induced lung fibrosis
have official names and clear functions, apart from H19, RNA
component of mitochondrial RNA processing endoribonuclease (RMRP),
and telomerase RNA component (TERC). A previous study demonstrated
that H19 regulates biological processes on at least 3 separate
levels of the highest significance: in imprinting, as an lncRNA
transcript, and as the miR-675 host (23). Another study showed that it can
affect the expression of IGF (24). In the present study, H19 was
upregulated, whereas IGF binding protein 11 was downregulated. GO
analysis showed that some differentially expressed genes were
involved in IGF binding. However, whether H19 has a negative
regulatory effect on IGF and the regulatory mechanisms involved in
IPF require further verification.
RMRP in rats measures 257 nucleotides in length.
Maida et al discovered that human telomerase reverse
transcriptase (TERT) and RMRP form a distinct ribonucleoprotein
complex that has RNA-dependent RNA polymerase activity and produces
double-stranded RNAs that can be processed into small interfering
RNA in a Dicer (also known as DICER1)-dependent manner (25). TERT mutations have been reported
in aplastic anemia and IPF (26).
Moreover, the catalytic TERT and TERC are the minimal components of
active telomerase (27). TERC not
only serves as a template for telomeric DNA synthesis but also
plays an important role in catalysis, accumulation, localization
and holoenzyme assembly (28–30). Some mutations in TERC can result
in a significant change in enzyme activity in vivo and in
vitro (30). Recently, Calado
et al (31) reported that
human telomere disease includes pulmonary fibrosis due to the
disruption of the CCAAT box of the TERC promoter, as described by
Aalbers et al (32).
Accordingly, it can be hypothesized that RMRP and TERC are both
crucial to the development process of lung fibrosis.
Two more differentially expressed lncRNAs were
detected by in situ hybridization for paraffin-embedded lung
tissue: AJ005396 and S69206. To date, limited studies have applied
in situ hybridization to measure lncRNA levels in
formalin-fixed paraffin-embedded tissue samples. By querying the
NCBI database, we found that AJ005396 is an mRNA for collagen α 1
type XI (col11a1) and that S69206 is protease mRNA. In addition,
the UCSC database revealed that the AJ005396 and S69206 sequences
overlapped with col11a1 and rat mast cell protease 1 precursor
(RMCP-1), respectively. In the present study, AJ005396 and S69206
were upregulated, but there was no signficant change in col11a1 and
RMCP-1 between the differently expressed mRNAs. Although the
database displayed that they contained a potential CDS region, it
only predicted protein sequences that must be confirmed with
further experiments. Thus, ArrayStar regards them as potential
lncRNAs. Moreover, previous studies have shown that parts of
lncRNAs overlapped with protein-coding gene sequences, such as MVIH
[an lncRNA located within the intron of the ribosomal protein S24
(RPS24) gene] (33) and RERT (an
lncRNA whose sequence overlaps with Ras-related GTP-binding protein
4b and EGLN2) (34).
Subsequently, they proved using the Open Reading Frame (ORF) Finder
and codon substitution frequency analysis that the overexpressed
lncRNA, MVIH, in hepatocellular carcinoma (HCC) is a non-coding RNA
transcribed independently of the RPS24 gene and that no correlation
existed between the transcriptional levels of RPS24 and MVIH
(33). Another study showed that
the overexpression of the RERT lncRNA upregulated EGLN2 (34). Dharap et al found 62
stroke-responsive lncRNAs showing 90% sequence homology with exons
of protein-coding genes in their study on lncRNA expression
profiles in focal ischemia (35).
Similarly, Ziats et al reported that most differentially
expressed lncRNAs in autism spectrum disorders (ASD) were from
intergenic regions (~60%), from antisense to protein-coding loci
(~15%), or within introns of protein-coding genes (~10%), with the
others representing overlapping transcripts from exons or introns
in both sense and antisense directions (36). Another complicating factor is the
presence of bifunctional RNAs, which are transcripts that function
as non-coding transcripts under certain conditions and are
translated into functional proteins in other situations (37,38). In view of this, in a subsequent
study, we hope to confirm whether AJ005396 and S69206 are lncRNAs
and transcribed independently of their overlapping protein-coding
RNAs and whether an interaction between their expressions
exists.
Furthermore, our array contained probes for known
protein-coding transcripts and carried out pathway analysis as well
as GO analysis based on differentially expressed mRNAs. Before
this, numerous studies of the mRNA or miRNA transcriptome in
pulmonary fibrosis have been performed. Research has shown that
genes related to the immune system, structural constituents of the
cytoskeleton, cellular adhesion, metabolism of the ECM, chemokines
and tissue remodeling are overexpressed in fibrotic lesions
(39,40), which is consistent with our
results. Another study focusing on the KEGG pathway to identify
fibrosis-associated genes targeted by miRNAs in the late phase
after bleomycin infusion, discovered that they mainly involved
cytokine-cytokine receptor interaction, focal adhesion and the
Jak/STAT signaling pathway (41).
Consistent with this article, cytokine-cytokine receptor
interaction exhibited a similarly significant change in our pathway
analysis. In addition, the cell cycle, the renin-angiotensin
system, the PPAR signaling pathway, the chemokine signaling
pathway, cell adhesion molecules, the Jak/STAT signaling pathway
and other signaling pathways were involved in the bleomycin-induced
lung fibrosis. Recent studies have indicated that PPAR-γ ligands
inhibit TGFβ signaling by affecting two pro-survival pathways that
culminate in myofibroblast differentiation. Further investigation
of PPAR-γ ligands and small electrophilic molecules may lead to a
new generation of anti-fibrotic therapeutics (42,43).
In conclusion, the current study used microarray
analysis to systematically and comprehensively examine global
lncRNA expression in a rat model of bleomycin-induced lung fibrosis
and in a normal control group. lncRNA target prediction and further
functional characterization may help to elucidate their specific
roles in lung injury and fibrosis. The related experiments are
currently in progress in our laboratory. Further studies on lncRNA
should expand our understanding of the genomic regulatory networks
in lung fibrosis and may provide new potential therapeutic targets
in lung fibrosis.
Acknowledgements
This study was supported by the ‘Taishan scholar’
position and the National Natural Science Foundation of China (no.
81273957), the Important Project of Science and Technology of
Shandong Province (nos. 2010GWZ20254 and 2011GHY11501), the Natural
Science Foundation of Shandong Province (nos. ZR2009EM006 and
ZR2012HQ042) and the Project of Science and Technology of the
Education Department of Shandong Province (no. J11FL87).
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
IPF
|
idiopathic pulmonary fibrosis
|
|
ECM
|
extracellular matrix
|
|
TEM
|
transmission electron microscope
|
|
IGF
|
insulin-like growth factor
|
|
RMRP
|
RNA component of mitochondrial RNA
processing endoribonuclease
|
|
TERC
|
telomerase RNA component
|
|
TERT
|
telomerase reverse transcriptase
|
|
RMCP-1
|
rat mast cell protease 1 precursor
|
References
|
1
|
American Thoracic Society. Idiopathic
pulmonary fibrosis: diagnosis and treatment. International
consensus statement American Thoracic Society (ATS), and the
European Respiratory Society (ERS). Am J Respir Crit Care Med.
161:646–664. 2000. View Article : Google Scholar
|
|
2
|
Noble PW and Homer RJ: Back to the future:
historical perspective on the pathogenesis of idiopathic pulmonary
fibrosis. Am J Respir Cell Mol Biol. 33:113–120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Noble PW and Homer RJ: Idiopathic
pulmonary fibrosis: new insights into pathogenesis. Clin Chest Med.
25:749–758. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ding Q, Luckhardt T, Hecker L, et al: New
insights into the pathogenesis and treatment of idiopathic
pulmonary fibrosis. Drugs. 71:981–1001. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
ENCODE Project Consortium. Birney E,
Stamatoyannopoulos JA, Dutta A, et al: Identification and analysis
of functional elements in 1% of the human genome by the ENCODE
pilot project. Nature. 447:799–816. 2007.
|
|
6
|
Guttman M, Amit I, Garber M, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs inmammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagano T and Fraser P: Emerging
similarities in epigenetic gene silencing by long noncoding RNAs.
Mamm Genome. 20:557–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim A, Zhao H, Ifrim I and Dean A:
Beta-globin intergenic transcription and histone acetylation
dependent on an enhancer. Mol Cell Biol. 27:2980–2986. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee JT: Lessons from X-chromosome
inactivation: long ncRNA as guides and tethers to the epigenome.
Genes Dev. 23:1831–1842. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rinn JL, Kertesz M, Wang JK, et al:
Functional demarcation of active and silent chromatin domains in
human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Krangel MS: T cell development: better
living through chromatin. Nat Immunol. 8:687–694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Perez DS, Hoage TR, Pritchett JR, et al:
Long, abundantly expressed non-coding transcripts are altered in
cancer. Hum Mol Genet. 17:642–655. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar
|
|
15
|
Mattick JS: The genetic signatures of
noncoding RNAs. PLoS Genet. 5:e10004592009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar
|
|
17
|
Hindorff LA, Sethupathy P, Junkins HA,
Ramos EM, Mehta JP, Collins FS and Manolio TA: Potential etiologic
and functional implications of genome-wide association loci for
human diseases and traits. Proc Natl Acad Sci USA. 106:9362–9367.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocelluar carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qureshi IA, Mattick JS and Mehler MF: Long
non-coding RNAs in nervous system function and disease. Brain Res.
1338:20–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tufarelli C, Stanley JA, Garrick D, Sharpe
JA, Ayyub H, Wood WG and Higgs DR: Transcription of antisense RNA
leading to gene silencing and methylation as a novel cause of human
genetic disease. Nat Genet. 34:157–165. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sui W, Yan Q, Li H, Liu J, Chen J, Li L
and Dai Y: Genome-wide analysis of long noncoding RNA expression in
peripheral blood mononuclear cells of uremia patients. J Nephrol.
Oct 24–2012.(Epub ahead of print).
|
|
23
|
Steck E, Boeuf S, Gabler J, Werth N,
Schnatzer P, Diederichs S and Richter W: Regulation of H19 and its
encoded microRNA-675 in osteoarthritis and under anabolic and
catabolic in vitro conditions. J Mol Med (Berl). 90:1185–1195.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tran VG, Court F, Duputié A, Antoine E, et
al: H19 antisense RNA can up-regulate Igf2 transcription by
activation of a novel promoter in mouse myoblasts. PLoS One.
7:e379232012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maida Y, Yasukawa M, Furuuchi M, et al: An
RNA-dependent RNA polymerase formed by TERT and the RMRP RNA.
Nature. 461:230–235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Calado RT and Young NS: Telomere
maintenance and human bone marrow failure. Blood. 111:4446–4455.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weinrich SL, Pruzan R, Ma L, et al:
Reconstitution of human telomerase with the template RNA component
hTR and the catalytic protein subunit hTRT. Nat Genet. 17:498–502.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Q, Kim NK and Feigon J: Architecture
of human telomerase RNA. Proc Natl Acad Sci USA. 108:20325–20332.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cong Y and Shay JW: Actions of human
telomerase beyond telomeres. Cell Res. 18:725–732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zvereva MI, Shcherbakova DM and Dontsova
OA: Telomerase: structure, functions, and activity regulation.
Biochemistry (Mosc). 75:1563–1583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Calado RT and Young NS: Telomere diseases.
N Engl J Med. 361:2353–2365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aalbers AM, Kajigaya S, van den
Heuvel-Eibrink MM, van der Velden VH, Calado RT and Young NS: Human
telomere disease due to disruption of the CCAAT box of the TERC
promoter. Blood. 119:3060–3063. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuan SX, Yang F, Yang Y, et al: Long
noncoding RNA associated with microvascular invasion in
hepatocellular carcinoma promotes angiogenesis and serves as a
predictor for hepatocellular carcinoma patients' poor
recurrence-free survival after hepatectomy. Hepatology.
56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu Z, Gao X, He Y, et al: An
insertion/deletion polymorphism within RERT-lncRNA modulates
hepatocellular carcinoma risk. Cancer Res. 72:6163–6172. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dharap A, Nakka VP and Vemuganti R: Effect
of focal ischemia on long noncoding RNAs. Stroke. 43:2800–2802.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ziats MN and Rennert OM: Aberrant
epxpression of long noncoding RNAs in autistic brain. J Mol
Neurosci. 49:589–593. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dinger ME, Pang KC, Mercer TR and Mattick
JS: Differentiating protein-coding and noncoding RNA: challenges
and ambiguities. PLoS Comput Biol. 4:e10001762008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ulveling D, Francastel C and Hube F: When
one is better than two: RNA with dual functions. Biochimie.
93:633–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hanaoka M, Ito M, Droma Y, Ushiki A,
Kitaguchi Y, Yasuo M and Kubo K: Comparison of gene expression
profiling between lung fibrotic and emphysematous tissues sampled
from patients with combined pulmonary fibrosis and emphysema.
Fibrogenesis Tissue Repair. 5:172012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Konishi K, Gibson KF, Lindell KO, et al:
Gene expression profiles of acute exacerbations of idiopathic
pulmonary fibrosis. Am J Respir Crit Care Med. 180:167–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie T, Liang J, Guo R, Liu N, Noble PW and
Jiang D: Comprehensive microRNA analysis in bleomycin-induced
pulmonary fibrosis identifies multiple sites of molecular
regulation. Physiol Genomics. 43:479–487. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kulkarni AA, Thatcher TH, Olsen KC,
Maggirwar SB, Phipps RP and Sime PJ: PPAR-γ ligands repress
TGFβ-induced myofibroblast differentiation by targeting the
PI3K/Akt pathway: implications for therapy of fibrosis. PLoS One.
6:e159092011.
|
|
43
|
Ferguson HE, Kulkarni A, Lehmann GM, et
al: Electrophilic peroxisome proliferator-activated receptor-γ
ligands have potent antifibrotic effects in human lung fibroblasts.
Am J Respir Cell Mol Biol. 41:722–730. 2009.
|