Introduction
In the past two decades, epigenetic mechanisms have
been recognized as key factors in the development of complex human
disorders, including cancer, and neurodegenerative, neurological
and autoimmune diseases (1).
Changes in chromatin conformation constitute the basis of
epigenetic effects since they can alter gene expression without
depending on DNA sequence changes. Chromatin epigenetic mechanisms
include DNA methylation and post-translational covalent
modifications of histones. These modifications comprise heritable
alterations that regulate essential biological processes, such as
gene and microRNA expression, cellular differentiation,
X-chromosome inactivation and genomic imprinting. DNA methylation
is the most widely studied and best characterized epigenetic
modification of the human genome (2). This epigenetic modification is
catalyzed by DNA methyltransferases (DNMTs), which add a methyl
group to the carbon 5 of cytosines that are followed by a guanine.
Thus, DNA methylation occurs almost exclusively in the context of a
5′-CpG-3′ dinucleotide, particularly in dense CpG regions termed
CpG islands. Promoter-specific CpG island hypermethylation is known
to be associated with gene silencing due to their transcription
repression effect (3).
Recent evidence indicates that endometriosis, an
enigmatic disease in which endometrial-like tissue is detected
outside the uterus, is also an epigenetic disease (4–6).
This hypothesis was based on the abnormal DNA methylation patterns
observed in the promoter regions of specific genes (7–12)
and the higher expression levels of DNMTs in endometriotic lesions
in comparison with normal endometrium (13). The first evidence of increased
levels of DNA methylation of a specific gene came from a pioneer
study by Martini et al (7), which found aberrant DNA methylation
in the tumor-suppressor genes, cyclin-dependent kinase inhibitor 2A
(TP16) and mutL homolog 1, colon cancer, nonpolyposis type 2
(hMLH1) in one and four samples of endometriosis,
respectively. Subsequently, by using a candidate gene approach,
other studies have described aberrant methylation patterns in the
promoter region of genes encoding the transcription factor homeobox
10A (HOXA10) (8),
progesterone receptor isoform B (PGR) (9), estrogen receptor 2, ERβ
(ESR2) (10), nuclear
receptor subfamily 5, group A, member 1 (NR5A1, primarily
named SF-1 or steroidogenic factor-1) (11) and cyclooxygenase-2 (COX-2)
(12) in endometriotic lesions
derived from different anatomical sites. Furthermore, endometriotic
cells treated in vitro with the demethylating agent
5-aza-2′deoxycytidine showed an increase in the expression of
aromatase mRNA, suggesting that DNA methylation of the
CYP19A1 aromatase gene also occurs in stromal cells from
endometriotic lesions (14).
Endometriosis is believed to be an
estrogen-dependent disease characterized by the loss of
progesterone-protective signaling in endometriotic cells. Xue et
al (10) demonstrated that
this effect could be related to hypomethylation of the ESR2
gene in stromal endometriotic cells, which could lead to an
increase in the expression of estrogen receptor β (ERβ). These
authors also demonstrated the presence of DNA methylation in a
specific region of ESR2 that confers promoter activity and
co-localizes with a CpG island: in vitro methylation was
correlated with inactivation of this promoter. Subsequently, it was
proposed that the increased expression of ERβ, due to the
hypomethylation of its coding gene, could inhibit the expression of
the PGR gene (15). In
addition, hypermethylation of the PGR gene, particularly at
promoter B, was also observed in the epithelial component of
endometriotic lesions (9), with
consequent decrease in the expression of its transcript.
Overall, steroid hormone receptor genes show a
complex pattern of regulation involving multiple promoters and
alternative mRNA isoforms. The ESR1 gene has two proximal
promoters (transcripts A and B) located within ~2 kb of the
translation start site and an upstream promoter C (16,17). Similarly, the PGR gene has
two alternative transcripts regulated by two promoter-specific
regions for the PGRA and PGRB isoforms, which are
associated with well-characterized CpG islands (18). The ESR2 gene also presents
alternatively spliced transcript variants and distinct promoters
regulated by DNA methylation (19), although not associated with the
presence of classic CpG islands.
Potentially, aberrant DNA methylation can disrupt
the effects of hormone steroids due to changes in the expression
levels of its receptors and may influence the origin and
progression of endometriosis. Thus, the present study was conducted
to investigate the methylation patterns of the alternative promoter
regions of the ESR1, ESR2 and PGR genes in intestinal
deep endometriosis, which accounts for 8–12% of all endometriosis.
This condition is characterized by multifocal infiltrating and
aggressive lesions associated with pelvic pain and infertility
(20).
Materials and methods
Biological samples
Fresh endometriotic tissues were collected from 44
premenopausal patients (mean age, 35.1±6.8 years) who underwent
laparoscopy for diagnosis and surgical resection (nodule resection
or segmental resection of the rectum). Laparoscopy was indicated
after clinical evaluation and imaging methods (transvaginal
ultrasound with bowel preparation). Matched eutopic endometrial
tissue samples were collected by curettage simultaneously from 7 of
these patients. No patient had clinical diagnosis of immunological
diseases or cancer. The classification system followed the American
Society for Reproductive Medicine’s recommendations for staging
(I–IV) (Revised American Fertility Society classification of
endometriosis - ASRM, 1997) (21). After histopathological
confirmation of the diagnosis, lesions were morphologically
classified as well-differentiated glandular pattern, stromal
pattern, glandular pattern of mixed differentiation and
undifferentiated glandular pattern according to previously
described criteria (22). In
addition, the phase of the menstrual cycle was also estimated by
histological evaluation. The clinical and histopathological data
are provided in Table I. Approval
for the study was obtained from the Institutional Review Board of
the Hospital das Clínicas and the Faculdade de Medicina da
Universidade de São Paulo (CAPPesp - USP). All samples were
collected after each patient provided informed consent.
 | Table IClinical and histopathological data
of the patients with intestinal deep endometriosis. |
Table I
Clinical and histopathological data
of the patients with intestinal deep endometriosis.
| No. of cases | Percentage (%) |
|---|
| Clinical
features/symptoms |
| Infertility | 24/44 | 54 |
| Dyspareunia | 29/44 | 66 |
| Dysmenorrhea | 39/44 | 89 |
| ASMR stagea (n=44) |
| I | 7 | 16 |
| II | 6 | 14 |
| III | 3 | 7 |
| IV | 28 | 63 |
| Histological
classificationb (n=36) |
|
Well-differentiated glandular
pattern | 1 | 3 |
| Glandular of mixed
differentiation and stromal pattern | 26 | 72 |
| Undifferentiated
glandular and stromal pattern | 9 | 25 |
| Phase of the
menstrual cycle (n=39) |
| Proliferative | 34 | 87 |
| Secretory | 2 | 5 |
| Menstrual | 3 | 8 |
All tissue samples were kept on dry ice immediately
after surgical resection and stored at -80°C. Subsequently, the
frozen tissue samples were manually macrodissected using
custom-built needles (23).
DNA extraction and methylation
analysis
Genomic DNA was obtained by standard sodium dodecyl
sulfate/proteinase K digestion, followed by phenol/chloroform
extraction and ethanol precipitation. DNA conversion by sodium
bisulfite was performed using an established protocol (24). Methylation-specific polymerase
chain reaction (MS-PCR) was used to evaluated the DNA methylation
patterns of the alternative promoter regions A and B of the
PGR and ESR1 genes as well as a promoter region of
the ESR2 gene. Methylated and unmethylated sequences of each
gene were detected using specific oligonucleotides as previously
described by Sasaki et al (25). The reactions were performed in a
total volume of 25 μl containing 200 μM of each dNTP, 15 mM of
Tris-HCl pH 8.0, 50 mM of KCl and 1 unit of AmpliTaq Gold DNA
polymerase (Applied Biosystems, Foster City, CA, USA).
Amplification and additional reaction conditions are listed in
Table II. The amplified products
were visualized after electrophoresis on a 6% polyacrylamide gel,
followed by silver nitrate staining. Water blanks were included in
each assay.
 | Table IIPrimer sequences, reaction and
amplification conditions used in the MS-PCR analysis. |
Table II
Primer sequences, reaction and
amplification conditions used in the MS-PCR analysis.
| Primer | Oligonucleotide
sequence | Reaction
conditions | Amplification
conditions | Amplicon (bp) |
|---|
| PGRA-UF |
5′-ATGGGTTATTTTTTTTTTG-3′ | (MgCl2)
2.5 mM | 1 cycle: 95°C - 10
min; 35 cycles: | 99 |
| PGRA-UR |
5′-TAAAATATACACCCTCCACA-3′ | (Primers) 0.08
μM | 95°C - 1 min, 51°C
- 1 min, | |
| PGRA-MF |
5′-ACGGGTTATTTTTTTTTCG-3′ | | 72°C - 1 min; 1
cycle: 72°C - 1 min | |
| PGRA-MR |
5′-TAAAATATACGCCCTCCACG-3′ | | | |
| PGRB-UF |
5′-TGATTGTTGTTTGTAGTATG-3′ | (MgCl2)
4.0 mM | 1 cycle: 95°C - 10
min; 35 cycles: | 200 |
| PGRB-UR |
5′-CAACAATTTAATAACACACA-3′ | (Primers) 0.24
μM | 95°C - 1 min, 50°C
- 1 min, | |
| PGRB-MF |
5′-TGATTGTCGTTCGTAGTACG-3′ | | 72°C - 1 min; 1
cycle: 72°C - 1 min | |
| PGRB-UR |
5′-CGACAATTTAATAACACGCG-3′ | | | |
| ESR1A-UF |
5′-GGATATGGTTTGTATTTTGTTTGT-3′ | (MgCl2)
2.5 mM | 1 cycle: 95°C - 10
min; | 123 |
| ESR1A-UR |
5′-ACAAACAATTCAAAAACTCCAACT-3′ | (Primers) 0.16
μM | 35 cycles: 95°C - 1
min, | |
| ESR1A-MF |
5′-GATACGGTTTGTATTTTGTTCGC-3′ | | 54°C (U)/50°C (M) -
1 min, | 121 |
| ESR1A-MR |
5′-CGAACGATTCAAAAACTCCAACT-3′ | | 72°C - 1 min; 1
cycle: 72°C - 1 min | |
| ESR1B-UF |
5′-TTTATTGTTATTTATTTAGT-3′ | (MgCl2)
4.0 mM | 1 cycle: 95°C - 10
min; | 180 |
| ESR1B-UR |
5′-AAAAATATACTCACATATACA-3′ | (Primers) 0.16
μM | 35 cycles: 95°C - 1
min, | |
| ESR1B-MF |
5′-TTTATTGTTATTTATTTAGC-3′ | | 47°C (U)/49°C (M) -
1 min, | |
| ESR1B-MR |
5′-AAAAATATACTCGCATATACG-3′ | | 72°C - 1 min; 1
cycle: 72°C - 1 min | |
| ESR2-UF |
5′-TTTGGAAGGTGGGTTTGGTT-3′ | (MgCl2)
3.0 mM | 1 cycle: 95°C - 10
min; | 109 |
| ESR2-UR |
5′-CACATACAAATATAATAACTAACA-3′ | (Primers) 0.16
μM | 35 cycles: 95°C - 1
min, | |
| ESR2-MF |
5′-TTTGGAAGGTGGGTTTGGTC-3′ | | 54°C - 1 min, 72°C
- 1 min; | |
| ESR2-MR |
5′-CGCATACAAATATAATAACTAACG-3′ | | 1 cycle: 72°C - 1
min | |
For the standardization of the DNA methylation
analysis, MS-PCR assays were initially performed in three breast
cancer cell lines showing a known hormone receptor expression
profile (26). The cell lines
selected were MDA-MB-231, classified as negative for ER and PgR
protein expression, and T47D and MCF-7 cells which are positive for
both receptors. Furthermore, gene-specific methylation data for
these cell lines, available in the Cancer Methylome System
(27), were obtained in
silico and used as reference in the optimization step of the
MS-PCR assays in the present study.
Immunohistochemical analysis
ERα, ERβ and PgR protein levels were analyzed in
formalin-fixed and paraffin-embedded tissues from 7 selected
endometriotic tissue samples (cases 2, 3, 28, 35, 36, 40 and 42).
Sections were freshly cut (3 μm) and mounted on slides with
organosilane (3-aminopropyl triethoxy-silane) (Sigma-Aldrich Co.,
St. Louis, MO, USA). Slides were deparaffinized in xylene,
gradually rehydrated through a series of alcohol rinses, and washed
in phosphate-buffered saline. Intrinsic peroxidase activity was
blocked with hydrogen peroxidase, and the sections were incubated
with the primary antibodies: RTU-ER-6F11 (Novocastra, Newcastle,
UK) (dilution 1:50), monoclonal anti-human estrogen receptor β1
(Dako Cytomation, Glostrup, Denmark) (dilution 1:60), and
monoclonal anti-human progesterone receptor 1A6 (Dako, Carpinteria,
CA, USA) (dilution 1:50). After incubation for 1 h, the sections
were washed in phosphate-buffered saline, incubated for 30 min with
the secondary biotinylated antibody, and for an additional 30 min
with the streptavidin peroxidase complex (LSAB; Dako, Carpinteria,
CA, USA). Color development was obtained with 3,3′-diaminobenzidine
and counterstaining with hematoxylin and eosin (H&E). Positive
and negative controls for each marker were routinely performed
during experiments. In areas of well-preserved tissue, the staining
intensity of endometriotic lesions and the percentage of cells
showing antibody reactivity were scored as +1 (<25%), +2
(>25% and <50%), +3 (>50% and <75%), and +4
(>75%).
Statistical analysis
Descriptive mean and percentage statistics were used
to summarize patient data and the gene hypermethylation status.
Associations between DNA methylation and clinical and histological
parameters were evaluated using Fisher’s exact test with a 5% level
of significance.
Results
The set of biological samples investigated in the
present study was comprised of 44 cases of intestinal deep
endometriosis. In this group of women, the most frequent symptoms
were moderate to severe dysmenorrhea (n=39, 89%), dyspareunia
(n=29, 66%) and infertility (n=24, 54%). Moreover, the majority of
the endometriotic specimens were classified as advanced stages III
and IV (n=31, 70%). The histological classification was able to be
assessed in 36 cases indicating a predominance of glandular and
stromal pattern (n=26, 72%) and an undifferentiated glandular
pattern in 9 cases (25%). In addition, the menstrual cycle stage
was determined as proliferative in almost all of the tissue samples
evaluated for this parameter (34/39 cases, 87%).
As expected, the presence of methylation as
determined by the MS-PCR assays was inversely correlated with the
expression at the protein level in the three breast cell lines
investigated. Methylated alleles for PGRA, PGRB, ESR1A and
ESR1B were detected in the MDA-MB-231 cell line (ER- and
PgR-negative) and only unmethylated alleles were found in the T47D
breast cancer cells (ER- and PgR-positive). Similarly, in the MCF-7
cells (ER- and PgR-positive) only unmethylated alleles were
detected in the ESR1A, PGRA and PGRB promoter
regions, while both methylated and unmethylated alleles were found
in the ESR1B promoter. These methylation pattens were also
confirmed in the Cancer Methylome System, which present the
methylation profile generated by immunoprecipitation using the
methyl-CpG binding domain of the MBD2 protein followed by
high-throughput sequencing (27)
(available at http://cbbiweb.uthscsa.edu/KMethylomes/) (data not
shown).
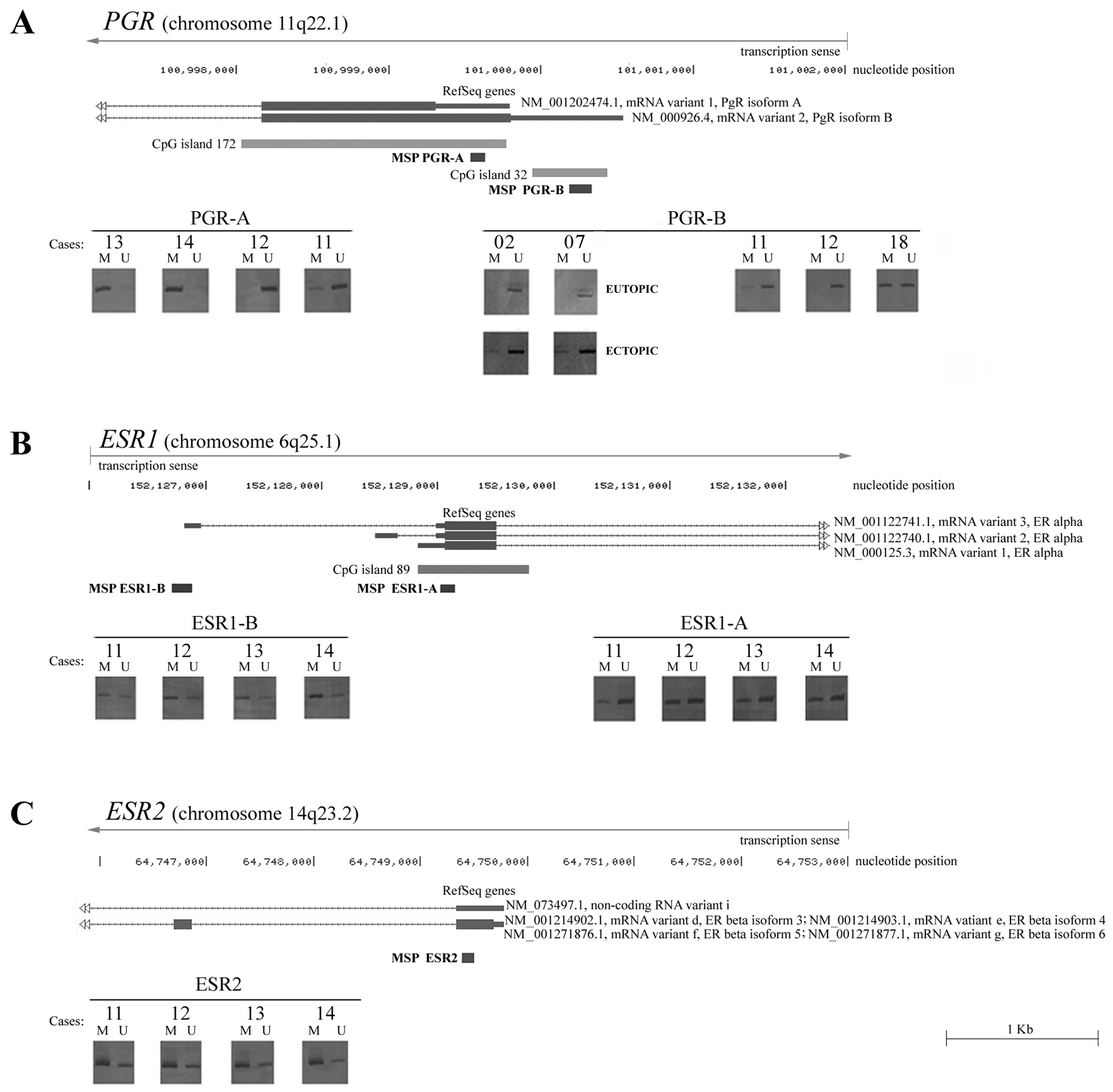
Subsequently, the PGRB and ESR2
promoter methylation patterns were obtained in the 44 cases of
infiltrating intestinal deep endometriosis. Unmethylated and
methylated alleles were observed for the ESR2 gene in all
samples analyzed. PGRB promoter methylation was observed in
39% (17/44 cases) of the endometriotic lesions. In 7 of the 44
samples, it was also possible to determine the methylation pattern
in matched endometrium obtained from the same patient. No
difference was observed in the ESR2 methylation pattern in
these cases; however, only unmethylated alleles were observed in
the PGRB promoter region. Thus, direct comparison of the
PGRB promoter region methylation between matched endometrium
and endometriotic samples revealed a differential pattern in 4
pairs since hypermethylated alleles were specifically detected in
the endometriotic tissue (Fig.
1).
In an attempt to remove the adjacent intestinal
tissue in the fragments containing the endometriotic infiltrating
lesions, macrodissection of the frozen tissue specimens was
performed, which resulted in a limited quantity of DNA for the
multiple MS-PCR tests. Thus, the MS-PCR analysis of the ESR1A,
ESR1B and PGRA promoters was possible in a subset of
only 37 out of 44 endometriotic lesions. While methylated and
unmethylated alleles were simultaneously detected for both
ESR1 promoters in all of these samples, PGRA
methylated alleles were only detected in 7 lesions (19%) (Fig. 1).
Differences in the DNA methylation frequencies in
endometriotic lesions were observed for the two promoter regions of
the PGR gene: 14 samples were methylated at the PGRB
promoter, 7 were methylated at the PGRA promoter, and 3 were
methylated at both promoters (cases 11, 13 and 14). However, no
statistically significant differences were observed between the
presence/absence of DNA methylation of the A and B
promoter regions of the PGR gene and the stage of the
lesions, histological classification, or menstrual cycle phase
(Table III).
 | Table IIIClinical and histopathological
features and DNA methylation of the A and B promoter regions of the
PGR gene. |
Table III
Clinical and histopathological
features and DNA methylation of the A and B promoter regions of the
PGR gene.
| PGRA | PGRB |
|---|
|
|
|
|---|
| Parameter | M | U | P-value | M | U | P-value |
|---|
| ASMR stage
(PGRA, n=37 and PGRB, n=44) |
| I + II | 1 | 6 | 0.64 | 6 | 7 | 0.52 |
| III + IV | 9 | 21 | 11 | 20 | | |
| Histological
classification (PGRA, n=29 and PGRB, n=36) |
| Differentiated
glandular pattern and stromal pattern | 4 | 16 | 1.0 | 9 | 18 | 1.0 |
| Undifferentiated
glandular and stromal pattern | 1 | 8 | 3 | 6 | | |
| Phase of the
menstrual cycle (PGRA, n=29 and PGRB, n=36) |
| Proliferative | 6 | 22 | 1.0 | 12 | 0 | 0.54 |
| Secretory | 0 | 1 | 22 | 2 | | |
The immunostaining of the ERα, ERβ and PgR proteins
in the endometriotic lesions exhibited a heterogeneous pattern with
the presence of both positive and negative epithelial and stromal
cells (Fig. 2). Of note, some
endometriotic samples showed positive staining for PgR only in the
stromal component, while the epithelial component were negative
(Fig. 1, case 3 and Fig. 2H). In these cases, both methylated
and unmethylated alleles were identified, probably due to the
epigenetic silencing mediated by DNA methylation of the PGR
gene in the epithelial fraction of the endometriotic lesions.
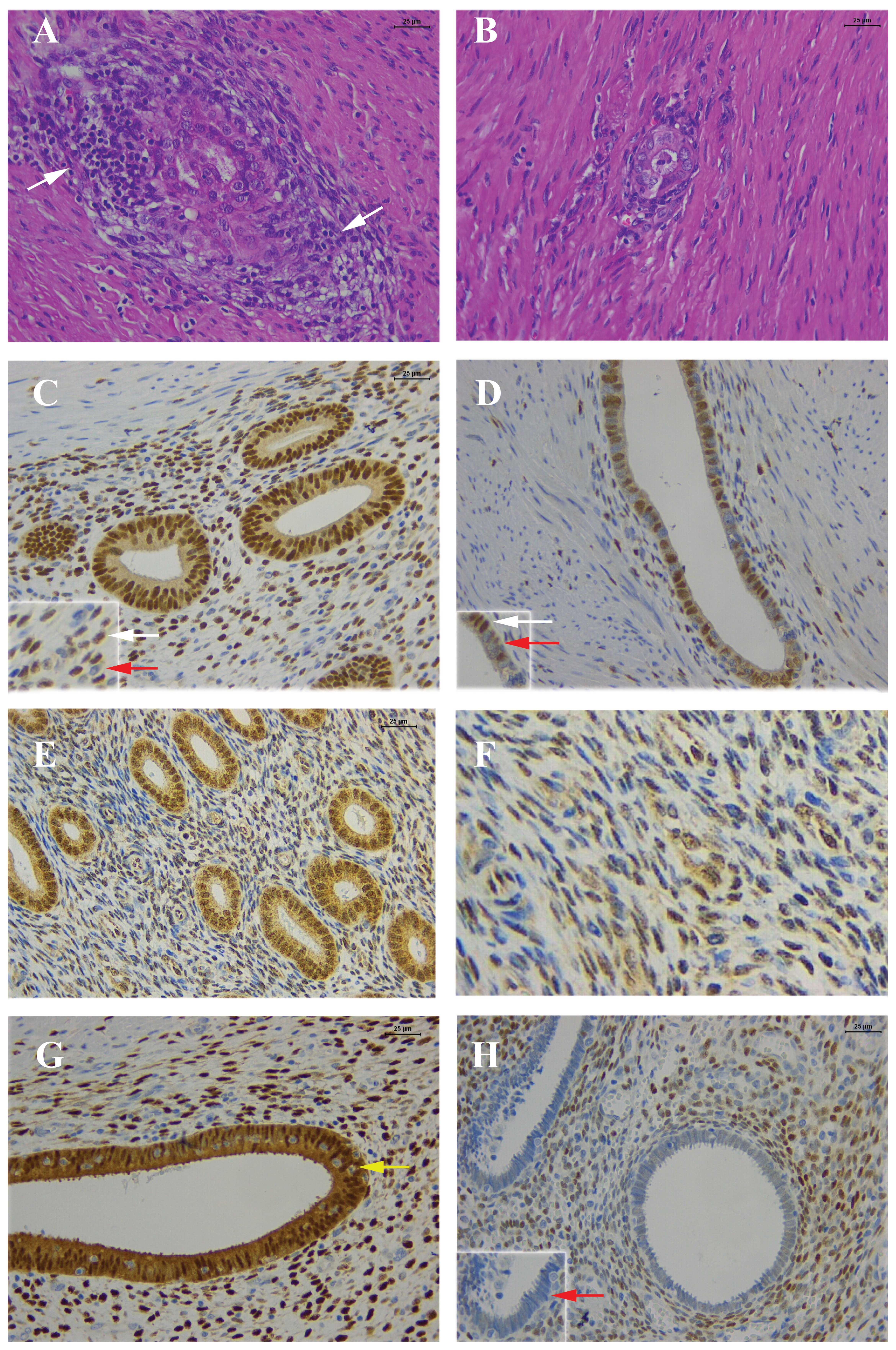 | Figure 2H&E and immunostaining of ERα,
ERβ and PgR. (A) Note the moderate inflammatory infiltrate (white
arrows) in the stroma of the endometriosis; H&E. (B)
Endometrial gland with scattered stroma and few inflammatory cells;
H&E. (C) Estrogen receptor α (ERα) in the epithelial glandular
cells (all cells are positive) and some positive stromal cells.
Inset: detail of positive (white arrow) stromal cells and negative
(red arrow) stromal cells for ERα (epithelium, score +4; stroma,
score +3). (D) ERα. Endometrial gland of an endometriotic
intestinal focus, with positive and negative nuclear ERα, and few
positive cells in the stroma. Inset: detail of positive (white
arrow) stromal cells and negative (red arrow) epithelial cells for
ERα (epithelium, score +4; stroma, score +3). (E) Estrogen receptor
β (ERβ). Positive nuclear staining in all epithelial cells and in
most of the stromal cells (epithelium and stroma, score +4); (F)
ERβ. Higher magnification of the stromal component of the
endometriotic focus, with positive and negative cells (epithelium
and stroma, score +4). (G) Progesterone receptor (PgR). Note that
all nuclei of the epithelial cells are positive for PgR and some of
the stromal cells. Inside the gland there are a number of
inflammatory cells, negative for PgR (yellow arrow) (epithelium and
stroma, score +4). (H) PgR for case 03. All epithelial cells from
the endometriotic focus are negative, and the majority of the
stromal cells are positive (epithelium, negative; stroma: 3+).
Inset: details of the negative (red arrow) epithelial cells for
PgR. Scale bar, 25 μm. |
Discussion
The major finding of our study was that promoter
region B of the PGR gene differed in regards to the
methylation status between eutopic endometrium and deep
endometriosis compromising the rectum: methylated alleles were
specifically detected in the endometriotic lesions, while
endometrium samples showed only unmethylated alleles. These data
suggest that this epigenetic alteration may be a possible biomarker
for endometriosis. Using the same MS-PCR primer set, Wu et
al (9) reported the presence
of partial methylation in this promoter in the epithelial component
of peritoneal and ovarian endometriotic implants, but not in the
promoter region that controls the expression of the A isoform. In
addition, the authors reported low levels of PGRB expression
in endometriotic epithelial cells. In the present study, we
demonstrated that both A and B promoter regions of the PGR
genes were methylated in a subset of infiltrating intestinal
endometriosis.
It has been proposed that the PGRB isoform is
crucial for mediating the inhibitory effects of progesterone on
cell growth and invasion, while PGRA has a suppressor
function (28,29). Thus, the results of the present
study indicates the importance of DNA methylation alterations of
the PGR gene in a subset of infiltrative deep endometriosis,
which is characterized by multifocal lesion sites, increased
aggressiveness and severe clinical symptoms as compared to pelvic
endometriosis, which was the type commonly investigated in previous
epigenetic studies (7,9–11,14).
Collectively, these data support the observation
that endometriotic lesions exhibit a trend of partial methylation
of the PGR gene, which does not completely inactivate gene
expression in the entire lesion focus, yet to some extent, is
responsible for a decrease in the expression of the PgR receptor
restricted to various areas or specific cells. These findings were
validated in the present study by the heterogeneous pattern
observed upon PgR immunohistochemistry (Fig. 2H), which showed positive and
negative staining for the PgR receptor coexisting in the same
endometriotic focus. This observation may explain the lower
expression level of progesterone receptors in endometriosis and may
be associated with the fact that approximately 9% of women with
endometriosis are not responsive to treatment with progestin
(5).
At present, endometriotic lesions are described as
estrogen-dependent endometrium-like tissue consisting of glands and
stroma. This abnormal tissue exhibits a unique expression profile
of steroid hormone receptors as compared to matched eutopic
endometrium or endometrium from women without endometriosis.
Previous studies have detected lower levels of ERα and higher
levels of ERβ in endometriosis (30,31). ERα appears to be the primary
mediator of estradiol-induced progesterone action in this tissue
(32), and progesterone exerts
its functions in the endometrium by binding to the nuclear
receptors PgRA and PgRB.
Epigenetic mechanisms modulate the dynamic
regulation of the estrogen receptor genes and their functions
(32). Promoter-specific DNA
methylation may be causally related to the differential expression
of the ERα and ERβ receptors in endometriosis. These receptors have
differential affinity for ligands, are differentially expressed in
a tissue-specific manner and may function antagonistically
(33). Both ER isoforms exist as
several splice variants. The alternative transcripts are
ubiquitous, although their biological significance remains poorly
understood (34). Furthermore,
alternative promoter usage leads to pre-mRNAs with a variable
length non-coding 5′ untranslated region (35–37).
In breast cancer cell lines, ESR1 expression
was previously found to be suppressed by promoter DNA methylation
(38,39) and histone hypoacetylation
(40) and was derepressed by DNA
methyltransferase (5-aza-2′deoxycytidine) and histone deacetylase
(trichostatin A) inhibitors (41,42). Promoter region hypermethylation
has also been shown to be inversely associated with ESR2
expression levels (43). In line
with this, the methylation pattern of the ESR1 gene was
investigated in endometriosis in the present study and methylated
alleles at both promoters A (ESR1A) and B (ESR1B)
were found. The consequences of DNA methylation in gene expression
are best understood at CpG island-containing promoters, however,
the two promoter regions investigated in the present study,
ESR1B and ESR2, are CpG island-free (Fig. 1). However, this also represents a
promising gene region for further DNA methylation studies in
endometriosis, since this region has been confirmed to be
differentially methylated in studies investigating other diseases,
such as prostate or breast cancer (25,44), suggesting that the regulation of
this gene by DNA methylation overlaps other complex molecular
mechanisms as recently described (45,46).
To the best of our knowledge, only one study has
evaluated the methylation status of the ESR2 gene in 8
ovarian endometriomas by bisulfite-modified DNA sequencing
(10). The authors described high
levels of ESR2 transcripts associated with hypomethylation of the
respective promoter region in comparison with endometrium. In
contrast, in the present study, MS-PCR analysis targeting a
different region of an alternative promoter of ESR2
(Fig. 1) in matched samples of
the endometria and intestinal deep endometriosis showed a more
complex pattern, with methylated and unmethylated alleles in both
tissues. Notwithstanding, the region investigated in this study
represents the beginning of numerous new transcripts identified as
expressed at premenopausal endometrium or endometriosis (GenBank,
available at http://www.ncbi.nlm.nih.gov/nuccore, accession numbers
AF074598 and AF074599, respectively). In this context, it is
tempting to speculate whether the ESR2 gene generates
multiple transcripts by triggering transcription initiation at
alternative sites that could be inactivated by tissue-specific
methylation. Therefore, interpretation of differential DNA
methylation patterns has proven difficult, in part because the
functional consequences depend on the genomic region involved, the
specific CpG dinucleotides, and inter-tissue and intra-tissue
heterogeneity. In line with this, the immunostaining performed in
this study was able to indicate the heterogeneity inherent to
endometriotic lesions, due to the different staining pattern
between epithelial and stromal cells, or even within the same
cellular component (stromal or epithelial). Furthermore, this
heterogeneity was found to be increased due to the additional
presence of other types of infiltrative cells, such as inflammatory
cells (Fig. 2A and B).
From a translational research perspective,
gene-specific methylation patterns may be useful biomarkers for
detection since alterations in DNA methylation potentially provide
a positive signal for endometriotic cell detection. MS-PCR is an
extremely sensitive PCR-based method, and it has been used to
detect hypermethylated genes in samples of limited quantity derived
from patients with various types of malignancies, such as those
obtained from fine needle aspiration, biopsies and corporal fluids
(47). In this context,
epigenetic studies may lead to the identification of the target
genes of aberrant DNA methylation in endometriosis, thus
representing a promising strategy for the monitoring of the onset
and progression of the disease. In the present study, we showed
that DNA methylation in the promoter of the PGRB gene is a
potential biomarker for endometriotic lesions as compared to
endometrium from the same patient. Moreover, aberrant PGRB
methylation was able to be identified in the entire lesion focus,
without the necessity of isolation of the glandular and stromal
components. Thus, this epigenetic change has the potential to be
considered as a useful biomarker for the pathogenesis of a subset
of endometriosis consisting of intestinal deep endometriosis.
In summary, these data indicate that abnormal
methylation patterns occur in deep endometriosis compromising the
rectum. Further studies are clearly necessary to elucidate the
possible relationship between DNA hypermethylation of the A and B
promoter regions of the PGR gene and progesterone
resistance. In addition, future epigenetic studies involving the
evaluation of steroid receptor methylation in endometriosis should
consider the histological pattern and each cellular component
separately (stromal or epithelial) in order to verify the effect of
DNA methylation on the expression of each alternative steroid
receptor transcript in this complex disorder.
Acknowledgements
The authors thank FAPESP and CAPES for their
financial support. We also thank Ana Carolina Machado Poppe for
technical support and Annacarolina F.L. da Silva and Alexandre
Fabro for assistance with the histopathological analysis and tissue
macrodissection. This study was supported by Fundação de Amparo à
Pesquisa do Estado de São Paulo (FAPESP, grants 2008/53716-5 and
2008/52270-3) and Coordenação de Aperfeiçoamento de Pessoal de
Nível Superior (CAPES).
Abbreviations:
|
ASRM
|
American Society for Reproductive
Medicine
|
|
COX-2
|
cyclooxygenase-2
|
|
CYP19A1
|
cytochrome P450, family 19, subfamily
A, polypeptide 1
|
|
DNMTs
|
DNA methyltransferases
|
|
ESR1 or ERα
|
estrogen receptor 1 or α
|
|
ESR2 or ERβ
|
estrogen receptor 2 or β
|
|
hMLH1
|
mutL homolog 1, colon cancer,
nonpolyposis type 2
|
|
H&E
|
hematoxylin and eosin
|
|
HOXA10
|
transcription factor homeobox 10A
|
|
MS-PCR
|
methylation-specific polymerase chain
reaction
|
|
NR5A1 or SF-1
|
nuclear receptor subfamily 5 or
steroidogenic factor-1
|
|
PGR or PgR
|
progesterone receptor
|
|
TP16
|
cyclin-dependent kinase inhibitor
2A
|
References
|
1
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ballestar E: An introduction to
epigenetics. Adv Exp Med Biol. 711:1–11. 2011. View Article : Google Scholar
|
|
3
|
Illingworth RS and Bird AP: CpG islands -
‘a rough guide’. FEBS Lett. 583:1713–1720. 2009.
|
|
4
|
Guo SW: Epigenetics of endometriosis. Mol
Hum Reprod. 15:587–607. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bulun SE: Endometriosis. N Engl J Med.
360:268–279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nasu K, Kawano Y, Tsukamoto Y, et al:
Aberrant DNA methylation status of endometriosis: epigenetics as
the pathogenesis, biomarker and therapeutic target. J Obstet
Gynaecol Res. 37:683–695. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Martini M, Ciccarone M, Garganese G, et
al: Possible involvement of hMLH1, p16(INK4a) and PTEN in the
malignant transformation of endometriosis. Int J Cancer.
102:398–406. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu Y, Halverson G, Basir Z, Strawn E, Yan
P and Guo SW: Aberrant methylation at HOXA10 may be responsible for
its aberrant expression in the endometrium of patients with
endometriosis. Am J Obstet Gynecol. 193:371–380. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu Y, Strawn E, Basir Z, Halverson G and
Guo SW: Promoter hypermethylation of progesterone receptor isoform
B (PR-B) in endometriosis. Epigenetics. 1:106–111. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue Q, Lin Z, Cheng YH, et al: Promoter
methylation regulates estrogen receptor 2 in human endometrium and
endometriosis. Biol Reprod. 77:681–687. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xue Q, Lin Z, Yin P, et al:
Transcriptional activation of steroidogenic factor-1 by
hypomethylation of the 5′ CpG island in endometriosis. J Clin
Endocrinol Metab. 92:3261–3267. 2007.PubMed/NCBI
|
|
12
|
Wang D, Chen Q, Zhang C, Ren F and Li T:
DNA hypomethylation of the COX-2 gene promoter is associated with
up-regulation of its mRNA expression in eutopic endometrium of
endometriosis. Eur J Med Res. 17:122012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Y, Strawn E, Basir Z, Halverson G and
Guo SW: Aberrant expression of deoxyribonucleic acid
methyltransferases DNMT1, DNMT3A, and DNMT3B in women with
endometriosis. Fertil Steril. 87:24–32. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Izawa M, Taniguchi F, Uegaki T, Takai E,
Iwabe T, Terakawa N and Harada T: Demethylation of a nonpromoter
cytosine-phosphate-guanine island in the aromatase gene may cause
the aberrant up-regulation in endometriotic tissues. Fertil Steril.
95:33–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bulun SE, Cheng YH, Pavone ME, et al:
Estrogen receptor-beta, estrogen receptor-alpha, and progesterone
resistance in endometriosis. Semin Reprod Med. 28:36–43. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grandien K: Determination of transcription
start sites in the human estrogen receptor gene and identification
of a novel, tissue-specific, estrogen receptor-mRNA isoform. Mol
Cell Endocrinol. 116:207–212. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grandien K, Berkenstam A and Gustafsson
JA: The estrogen receptor gene: promoter organization and
expression. Int J Biochem Cell Biol. 29:1343–1369. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wen DX, Xu YF, Mais DE, Goldman ME and
McDonnell DP: The A and B isoforms of the human progesterone
receptor operate through distinct signaling pathways within target
cells. Mol Cell Biol. 14:8356–8364. 1994.PubMed/NCBI
|
|
19
|
Hirata S, Shoda T, Kato J and Hoshi K: The
multiple untranslated first exons system of the human estrogen
receptor beta (ER beta) gene. J Steroid Biochem Mol Biol. 78:33–40.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wills HJ, Reid GD, Cooper MJ and Morgan M:
Fertility and pain outcomes following laparoscopic segmental bowel
resection for colorectal endometriosis: a review. Aust NZ J Obstet
Gynaecol. 48:292–295. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
No authors listed. Revised American
Society for Reproductive Medicine classification of endometriosis.
Fertil Steril. 67:817–821. 1997. View Article : Google Scholar
|
|
22
|
Abrao MS, Neme RM, Carvalho FM, Aldrighi
JM and Pinotti JA: Histological classification of endometriosis as
a predictor of response to treatment. Int J Gynaecol Obstet.
82:31–40. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pires AR, da Matta Andreiulo F and de
Souza SR: TMA for all: a new method for the construction of tissue
microarrays without recipient paraffin block using custom-built
needles. Diagn Pathol. 1:142006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Negraes PD, Favaro FP, Camargo JL,
Oliveira ML, Goldberg J, Rainho CA and Salvadori DM: DNA
methylation patterns in bladder cancer and washing cell sediments:
a perspective for tumor recurrence detection. BMC Cancer.
8:2382008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sasaki M, Tanaka Y, Perinchery G, Dharia
A, Kotcherguina I, Fujimoto S and Dahiya R: Methylation and
inactivation of estrogen, progesterone, and androgen receptors in
prostate cancer. J Natl Cancer Inst. 94:384–390. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Neve RM, Chin K, Fridlyand J, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu F, Doderer MS, Huang YW, et al: CMS: a
web-based system for visualization and analysis of genome-wide
methylation data of human cancers. PLoS One. 8:e609802013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dai D, Wolf DM, Litman ES, White MJ and
Leslie KK: Progesterone inhibits human endometrial cancer cell
growth and invasiveness: down-regulation of cellular adhesion
molecules through progesterone B receptors. Cancer Res. 62:881–886.
2002.
|
|
29
|
Wu Y, Shi X and Guo SW: The knockdown of
progesterone receptor isoform B (PR-B) promotes proliferation in
immortalized endometrial stromal cells. Fertil Steril.
90:1320–1323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brandenberger AW, Lebovic DI, Tee MK, Ryan
IP, Tseng JF, Jaffe RB and Taylor RN: Oestrogen receptor (ER)-alpha
and ER-beta isoforms in normal endometrial and
endometriosis-derived stromal cells. Mol Hum Reprod. 5:651–655.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fujimoto J, Hirose R, Sakaguchi H and
Tamaya T: Expression of oestrogen receptor-alpha and -beta in
ovarian endometriomata. Mol Hum Reprod. 8:742–747. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leader JE, Wang C, Popov VM, Fu M and
Pestell RG: Epigenetics and the estrogen receptor. Ann NY Acad Sci.
1089:73–87. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ström A, Hartman J, Foster JS, Kietz S,
Wimalasena J and Gustafsson JA: Estrogen receptor beta inhibits
17beta-estradiol-stimulated proliferation of the breast cancer cell
line T47D. Proc Natl Acad Sci USA. 101:1566–1571. 2004.PubMed/NCBI
|
|
34
|
Taylor SE, Martin-Hirsch PL and Martin FL:
Oestrogen receptor splice variants in the pathogenesis of disease.
Cancer Lett. 288:133–148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Flouriot G, Griffin C, Kenealy M,
Sonntag-Buck V and Gannon F: Differentially expressed messenger RNA
isoforms of the human estrogen receptor-alpha gene are generated by
alternative splicing and promoter usage. Mol Endocrinol.
12:1939–1954. 1998.
|
|
36
|
Lu B, Leygue E, Dotzlaw H, Murphy LJ,
Murphy LC and Watson PH: Estrogen receptor-beta mRNA variants in
human and murine tissues. Mol Cell Endocrinol. 138:199–203. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu B, Dotzlaw H, Leygue E, Murphy LJ,
Watson PH and Murphy LC: Estrogen receptor-alpha mRNA variants in
murine and human tissues. Mol Cell Endocrinol. 158:153–161. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ottaviano YL, Issa JP, Parl FF, Smith HS,
Baylin SB and Davidson NE: Methylation of the estrogen receptor
gene CpG island marks loss of estrogen receptor expression in human
breast cancer cells. Cancer Res. 54:2552–2555. 1994.PubMed/NCBI
|
|
39
|
Lapidus RG, Nass SJ and Davidson NE: The
loss of estrogen and progesterone receptor gene expression in human
breast cancer. J Mammary Gland Biol Neoplasia. 3:85–94. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sharma D, Saxena NK, Davidson NE and
Vertino PM: Restoration of tamoxifen sensitivity in estrogen
receptor-negative breast cancer cells: tamoxifen-bound reactivated
ER recruits distinctive corepressor complexes. Cancer Res.
66:6370–6378. 2006. View Article : Google Scholar
|
|
41
|
Macaluso M, Montanari M, Noto PB, Gregorio
V, Bronner C and Giordano A: Epigenetic modulation of estrogen
receptor-alpha by pRb family proteins: a novel mechanism in breast
cancer. Cancer Res. 67:7731–7737. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sharma D, Blum J, Yang X, Beaulieu N,
Macleod AR and Davidson NE: Release of methyl CpG binding proteins
and histone deacetylase 1 from the estrogen receptor alpha (ER)
promoter upon reactivation in ER-negative human breast cancer
cells. Mol Endocrinol. 19:1740–1751. 2005. View Article : Google Scholar
|
|
43
|
Rody A, Holtrich U, Solbach C, et al:
Methylation of estrogen receptor beta promoter correlates with loss
of ER-beta expression in mammary carcinoma and is an early
indication marker in premalignant lesions. Endocr Relat Cancer.
12:903–916. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao L, Yu Z, Li Y, et al: Clinical
implications of ERβ methylation on sporadic breast cancers in
Chinese women. Med Oncol. 29:1569–1575. 2012.
|
|
45
|
Han H, Cortez CC, Yang X, Nichols PW,
Jones PA and Liang G: DNA methylation directly silences genes with
non-CpG island promoters and establishes a nucleosome occupied
promoter. Hum Mol Genet. 20:4299–4310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zelenko Z, Aghajanova L, Irwin JC and
Giudice LC: Nuclear receptor, coregulator signaling, and chromatin
remodeling pathways suggest involvement of the epigenome in the
steroid hormone response of endometrium and abnormalities in
endometriosis. Reprod Sci. 19:152–162. 2012. View Article : Google Scholar
|
|
47
|
Laird PW: The power and the promise of DNA
methylation markers. Nat Rev Cancer. 3:253–266. 2003. View Article : Google Scholar : PubMed/NCBI
|