Introduction
Heart failure caused by cardiac cell death is a
growing health issue worldwide. Despite various treatment options,
the rising prevalence of this disease renders it one of the leading
causes of morbidity and mortality. Stem cell transplantation, a
promising treatment for heart failure, has been successfully
applied on experimental animals to enhance myocardial regeneration
and improve cardiac function (1).
Over the years, a number of experimental trails have
focused on mesenchymal stem cells (MSCs) as they are easily
available and have the capacity to differentiate into
cardiomyocyte-like cells (2–4).
Studies have indicated that the regulation of histone acetylation
and deacetylation plays an important role in the process of MSC
transformation into cardiomyocyte-like cells (5–7).
Histone acetylation or deacetylation enhances the expression of
cardiac-specific genes (8,9).
The exact mechanisms responsible for histone
acetylation and deacetylation during the differentiation of MSCs
into cardiomyocyte-like cells has not been determined yet. A number
of studies have reported that the histone acetylases (HATs) and
deacetylases (HADCs) lack the cardiac-specific gene binding sites
(10,11). Therefore, HATs and HADCs cannot
specifically bind to cardiac-specific genes and regulate their
expression directly. However, Islet-1, a subtype of the
LIM-homeodomain transcription factor (LIM-HD) subfamily, contains a
DNA binding site and two LIM domains, and is able to bind with
GCN5, which is a member of the HAT family involved in
cardiomyocyte, hepatocyte, as well as bone and neuron
differentiation (12–14). Several studies have demonstrated
that Islet-1 is crucial to cardiac development and cardiomyocyte
differentiation (15–16). Thus, we hypothesized that Islet-1
is a key factor during the process of the specific differentiation
of MSCs into cardiomyocyte-like cells and that it exerts its
effects possibly through modifications in histone acetylation and
deacetylation. In this study, in order to verify our hypothesis we
used C3H10T1/2 cells overexpressing Islet-1 to determine cell
differentiation and the expression of heart development-related
genes in these cells. Our data indicate that Islet-1 enhances
cardiac cell differentiation and increases the expression of heart
development-related genes, possibly through the regulation of
histone acetylation.
Materials and methods
Construction of lentiviral vectors
carrying the Islet-1 gene (Lenti-Islet-1)
Mouse Islet-1 cDNA (NM_021459) was obtained through
the reverse transcription polymerase chain reaction (RT-PCR)
(RT-PCR system; Bio-Rad, Hercules, CA, USA) of total RNA isolated
from 13.5-day-old mouse hearts using the RNA extraction kit (RP120;
BioTeke, Beijing, China), SYBR Premix Ex Taq II and PrimeScript RT
reagent kit (Takara, Dalian, China). The Islet-1 gene primer
(5′-cggatcctacagatatggg agacatgggcgatc-3′ and
5′-cgtcgactcctcatgccctcaataggactgg-3′), was designed by Primer
Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA) and
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). RT-PCR
was performed as follows: 95ºC (10 min), 50ºC (30 sec) and 72ºC (1
min), 35 cycles.
The Islet-1 gene and pWPI-green fluorescent protein
(GFP) plasmid (Invitrogen, Grand Island, NY, USA) were double
digested with restriction endonuclease (Takara). The product was
purified, recombined directly and transformed into E.
coli-competent cells (DH5α; Invitrogen). The positive clone
with the Islet-1 gene was connected to the right vector and
identified by PCR. The pWPI-GFP-Islet-1 plasmid was sequenced and
compared with the mouse Islet-1 gene.
The packaging and production of the lentiviral
vector were carried out as previously described (17). Briefly, the lentiviral shuttle
plasmid and auxiliary packaging plasmid were constructed. The 293T
cells [Institute of Biochemistry and Cell Biology (SIBS) of the
Chinese Academy of Sciences (CAS, Shanghai, China)] were
co-transfected with the lentivirus shuttle plasmid and auxiliary
packaging plasmid. The culture medium was replaced by Dulbecco’s
modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham,
MA, USA) 8 h following transfection. Lentiviral vectors were
collected from the supernatant 48 h after the medium exchange.
Cell culture and lentiviral vector
transfection
The C3H10T1/2 cells (University of Chicago Molecular
Oncology Laboratory, Chicago, IL, USA) were grown in DMEM
supplemented with 10% fetal bovine serum (FBS; HyClone,
Philadelphia, PA, USA).
Lentiviral vectors [vectors with pWPI-GFP-Islet-1
plasmid (Lenti-Islet-1) or vectors with pWPI-GFP plasmid alone
(Lenti-N), respectively, MOI=20] and polybrene were added to the
C3H10T1/2 cells dissociated with trypsinase (Invitrogen), while the
density of the cells was 60%, with a concentration of 8 mg/l. The
culture medium was replaced by DMEM with 10% FBS after 12 h of
incubation at 37ºC, 5% CO2. Fluorescence microscopy
(BX51; Olympus, Tokyo, Japan) was used to observe GFP expression
after 3 days. Flow cytometry (FCM) (BD Canto II Flow Cytometer; BD
Biosciences, San Jose, CA, USA) was used to detect the transfection
efficiency.
RNA isolation, RT-PCR and qPCR
RNA from the untreated C3H10T1/2 cells, the
C3H10T1/2 cells transfected with Lenti-N and the C3H10T1/2 cells
transfected with Lenti-Islet-1 was isolated using the RNA
extraction kit (RP120; BioTeke). The concentration of RNA was
determined using a NanoDrop 1000 spectrophotometer (NanoDrop 1000;
NanoDrop Products, Wilmington, DE, USA). cDNA was generated using
the PrimeScript RT reagent kit (Takara) for RT-PCR (Invitrogen).
RT-PCR was performed at 30ºC (10 min), 42ºC (30 min), 99ºC (5 min)
and 5ºC (5 min). cDNA was analyzed by qPCR (CFX 96 Real-Time
System; Bio-Rad, Hercules, CA, USA) using SYBR-Green RealMasterMix
(Tiangen Biotech, Co., Inc., Beijing, China). The primer sequences,
product size and annealing temperatures are presented in Table I.
 | Table IPrimer sequences, product size and
annealing temperatures used in qPCR. |
Table I
Primer sequences, product size and
annealing temperatures used in qPCR.
| Gene | Primer sequence | Product size
(bp) | Annealing temperature
(ºC) |
|---|
| Islet-1 |
5′-acaccttgggcggacctgctatg-3′
5′-tgaaaccacactcggatgactctg-3′ | 123 | 60 |
| Gata4 |
5′-ctgtggcctctaccacaagatgaa-3′
5′-gtctggcagttggcacagga-3′ | 180 | 60 |
| Nkx2.5 |
5′-acttgtctcctcggtgcttctg-3′
5′-cacagggttagggtgggactatg-3′ | 173 | 63.7 |
| Mef2c |
5′-gcgaaagttcggattgatgaaga-3′
5′-gtggatgtcagtgctggcgta-3′ | 133 | 60 |
| AFP |
5′-ggaagatggtgagcattg-3′
5′-tgttggaatacgaagagttg-3′ | 168 | 60 |
| BGP |
5′-aacgcatctacggtatca-3′
5′-gctgtgacatccatacttg-3′ | 134 | 63 |
| Nestin |
5′-atgagcagatgacagtga-3′
5′-tccagtgattctatgttctct-3′ | 181 | 60 |
| β-actin |
5′-ggagattactgccctggctccta-3′
5′-gactcatcgtactcctgcttgctg-3′ | 50 | 60 |
qPCR was performed, including an initial
denaturation at 98ºC (5 min), followed by denaturation at 94ºC (10
sec) and annealing at the annealing temperature indicated in
Table I (10 sec). This was
finally followed by a renaturation at 72ºC (15 sec) and then the
process was repeated for 40 cycles.
Western blot analysis and
immunofluorescence
Proteins (20 μg) were loaded onto 8%
SDS-polyacrylamide gels for electrophoresis and then the proteins
were then transferred onto nitrocellulose membranes. The
transferred nitrocellulose membranes were blocked with 5% dried
skim milk for 1 h at room temperature, then incubated with primary
antibodies at 4ºC for 12 h, then incubated with the secondary
antibodies in blocking buffer for 1 h. The bands were revealed by
enhanced chemiluminescence reagents (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) for 1 min and analyzed using Image-Pro Plus
5.1 software (Media Cybernetics, Inc., Rockville, MD, USA). The
antibodies used were as follows: anti-Islet-1 (sc-23590; Santa Cruz
Biotechnology, Inc.), mouse monoclonal to cardiac troponin T (cTnT;
ab33589), albumin (ALB), bone-specific alkaline phosphatase (BALP),
glial fibrillary acidic protein (GFAP) and rabbit polyclonal to
histone H3-ChIP Grade (ab1791) (all from Abcam, Cambridge, UK).
Cells (4.4×105/well) were plated in
6-well plates on 1×1 cm2 glass coverslips. They were
later fixed in 4ºC acetone in 15 min. Following 3 washes in PBS,
the cells on glass coverslips were blocked with goat serum (1:20),
washed again, then primary Islet-1 monoclonal antibody (1:100;
Santa Cruz Biotechnology, Inc.) was added for overnight incubation
at 4ºC. The cells were then washed with PBS and secondary
antibodies (1:150; CoWin Bioscience, Beijing, China) conjugated
with TRITC were added followed by incubation for 1 h at 37ºC. DAPI
was then added for 3 min. After the final wash, images were
acdquired under a fluorescence microscope (BX51; Olympus).
Epigallocatechin gallate (EGCG)
treatment
EGCG (120 μmol/l) (Sigma-Aldrich, St. Louis, USA)
was added to the C3H10T1/2 cells transfacted with Lenti-Islet-1 2
weeks following transfection. Total RNA from these cells was
isolated 3, 6 and 12 h following treatment with EGCG. RT-PCR and
qPCR were performed to detect GATA binding protein 4 (Gata4), NK2
homeobox 5 (Nkx2.5) and myocyte enhancer factor 2C (Mef2c)
expression.
Chromatin immunoprecipitation
(ChIP)-qPCR
Chromatin samples were prepared from the cells in
the C3H10 group (untransfected cells), the negative control group
(cells transfected with Lenti-N) and the experimental group (cells
transfected with Lenti-Islet-1). ChIP assay was performed as
previously described in the study by Zsindely et al
(18). Briefly, chromatin samples
were cross-linked with 1% formaldehyde then fragmented by
sonication (Ultrasonic Disruptor UD-201; CS Bio Co., Menlo Park,
CA, USA). Agarose gel electrophoresis was carried out to verify the
length of the DNA fragments. Immunoprecipitation was performed
using rabbit polyclonal to histone H3-ChIP Grade antibody (ab1791;
Abcam). The chromatin-antibody complexes were then washed, reverse
cross-linked and purified. The ChIP process was performed using the
Chromatin Immunoprecipitation kit (Millipore, Billerica, MA, USA).
The amount of extracted DNA was determined by qPCR. ChIP-qPCR
primers were designed using Primer Premier 5.0 software and
synthesized by Shanghai DNA Biotechnologies Co., Ltd. The primer
sequences, product size and annealing temperatures of the ChIP-qPCR
reaction are presented in Table
II.
 | Table IIPrimer sequences, product size and
annealing temperatures used in ChIP-qPCR. |
Table II
Primer sequences, product size and
annealing temperatures used in ChIP-qPCR.
| Gene | Primer sequences | Product size
(bp) | Annealing
temperature (ºC) |
|---|
| Gata4 |
5′-cactgacgccgactccaaactaa-3′
5′-cgactggggtccaatcaaaag-3′ | 140 | 60 |
| Nkx2.5 |
5′-cttctggctttcaatccatcctca-3′
5′-cgggcagttctgcgtcaccta-3′ | 289 | 60 |
| Mef2c |
5′-cacgcatctcaccgcttgacg-3′
5′-caccagtgcctttctgcttctcc-3′ | 217 | 68 |
Statistical analysis
All the data are expressed as the means ± standard
error of the mean (SEM) and were analyzed with repeated measures
ANOVA (TCDD data). A test for linear trend and Dunnett’s test
(comparison of all treated groups with controls) were used as
post-tests in ANOVA. SPSS 17.0 software (SPSS Inc., Armonk, NY,
USA) was used for statistical analyses. A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
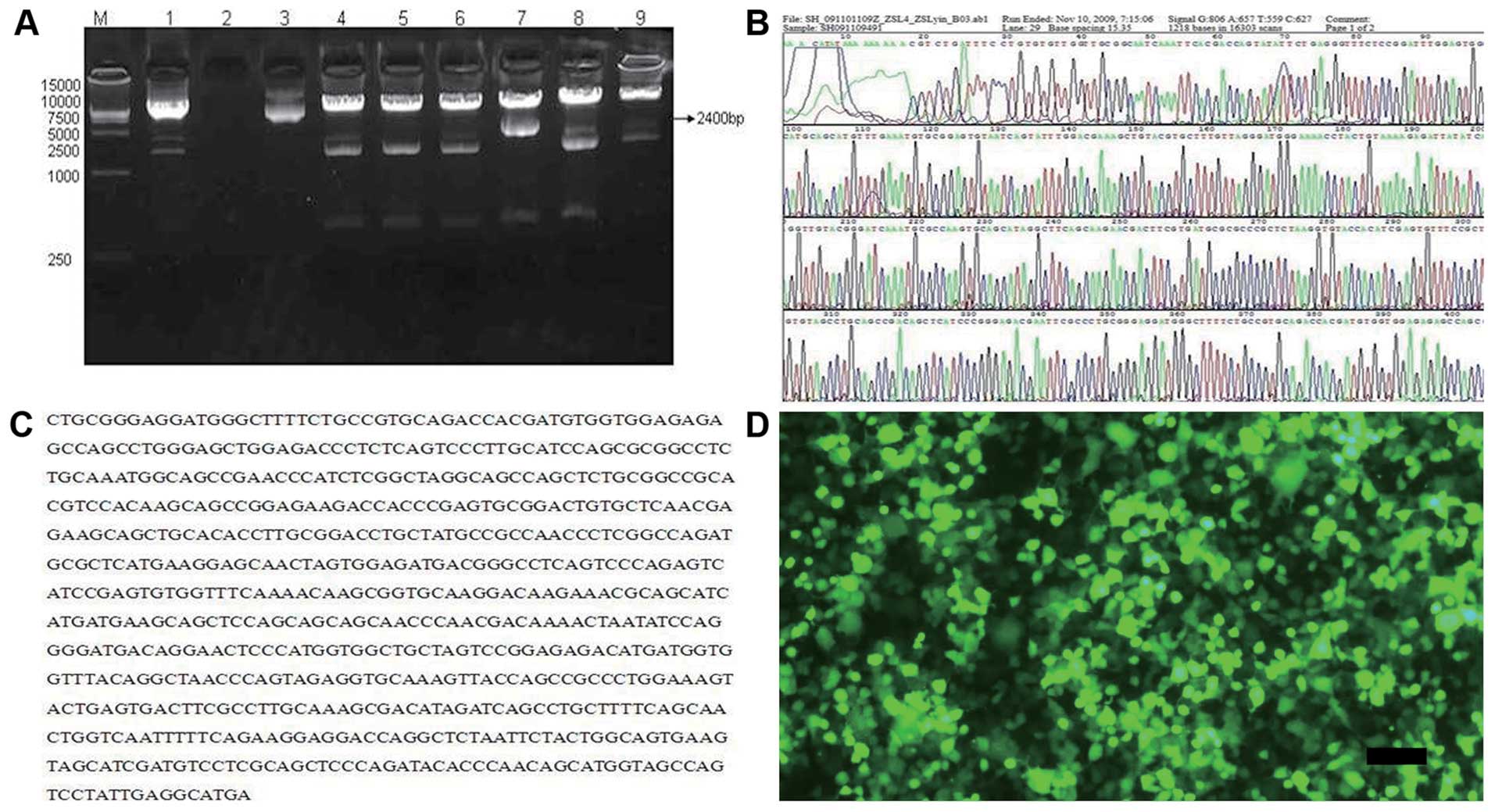
Construction of lentiviral vectors
Following double digestion with the pWPI vector, the
negative fragment was in the vicinity of 1,400 bp and the positive
fragment was 2,400 bp. Fragment 7 (2,400 bp) was the positive
clone, identified by PCR (Fig.
1A). Sequencing analysis displayed the positive clone insertion
into the pWPI vector (Fig. 1B and
C). On the 4th day after Lenti-Islet-1 transfection, GFP
expression could be detected in the 293T cells (Fig. 1D).
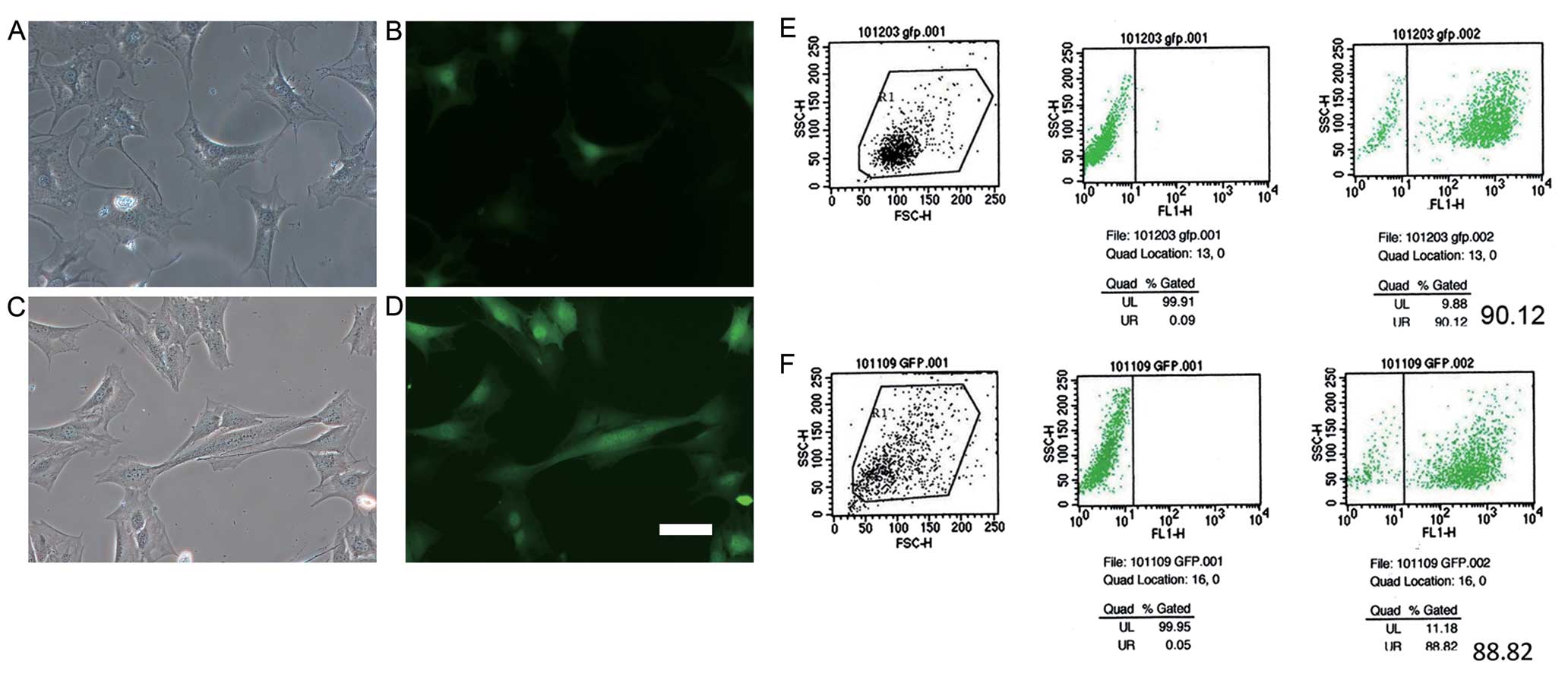
Transfection efficiency and Islet-1
expression
Since the vectors carried the pWPI-GFP plasmid, GFP
could be observed under a fluorescence microscope in both the
Lenti-Islet-1- and Lenti-N-transfected cells. Our data indicated
that GFP could be observed in the C3H10T1/2 cells transfected with
Lenti-N (Fig. 2A and B) and
Lenti-Islet-1 (Fig. 2C and D) 3
days after transfection. The transfection efficiencies of the
C3H10T1/2 cells transfected with Lenti-N and Lenti-Islet-1 were
determined by FCM. The transfection efficiency of the C3H10T1/2
cells transfected with Lenti-N was 90.12% (Fig. 2E) and that of the C3H10T1/2 cells
transfected with Lenti-Islet-1 was 88.82% (Fig. 2F).
The results from PCR revealed that the expression of
the Islet-1 gene in the Lenti-Islet-1-transfected cells was higher
than that in the untransfected cells (C3H10T1/2 cells) and those
transfected with Lenti-N (P<0.05) (Fig. 3A). Islet-1 protein was expressed
in the cytoplasm. The fluorescence intensity in the C3H10T1/2 cells
transfected with Lenti-Islet-1 was higher than that in the
untransfected cells (C3H10T1/2 cells) and the C3H10T1/2 cells
transfected with Lenti-N (Fig.
3B). The expression of Islet-1 protein in the
Lenti-Islet-1-transfected cells increased progressively with the
highest expression observed at 3 and 4 weeks after transfection
(Fig. 3C). Western blot analysis
revealed that the expression of Islet-1 protein in the
Lenti-Islet-1-transfected cells was higher than that in the
untransfected C3H10T1/2 cells and those transfected with Lenti-N
(Fig. 3D).
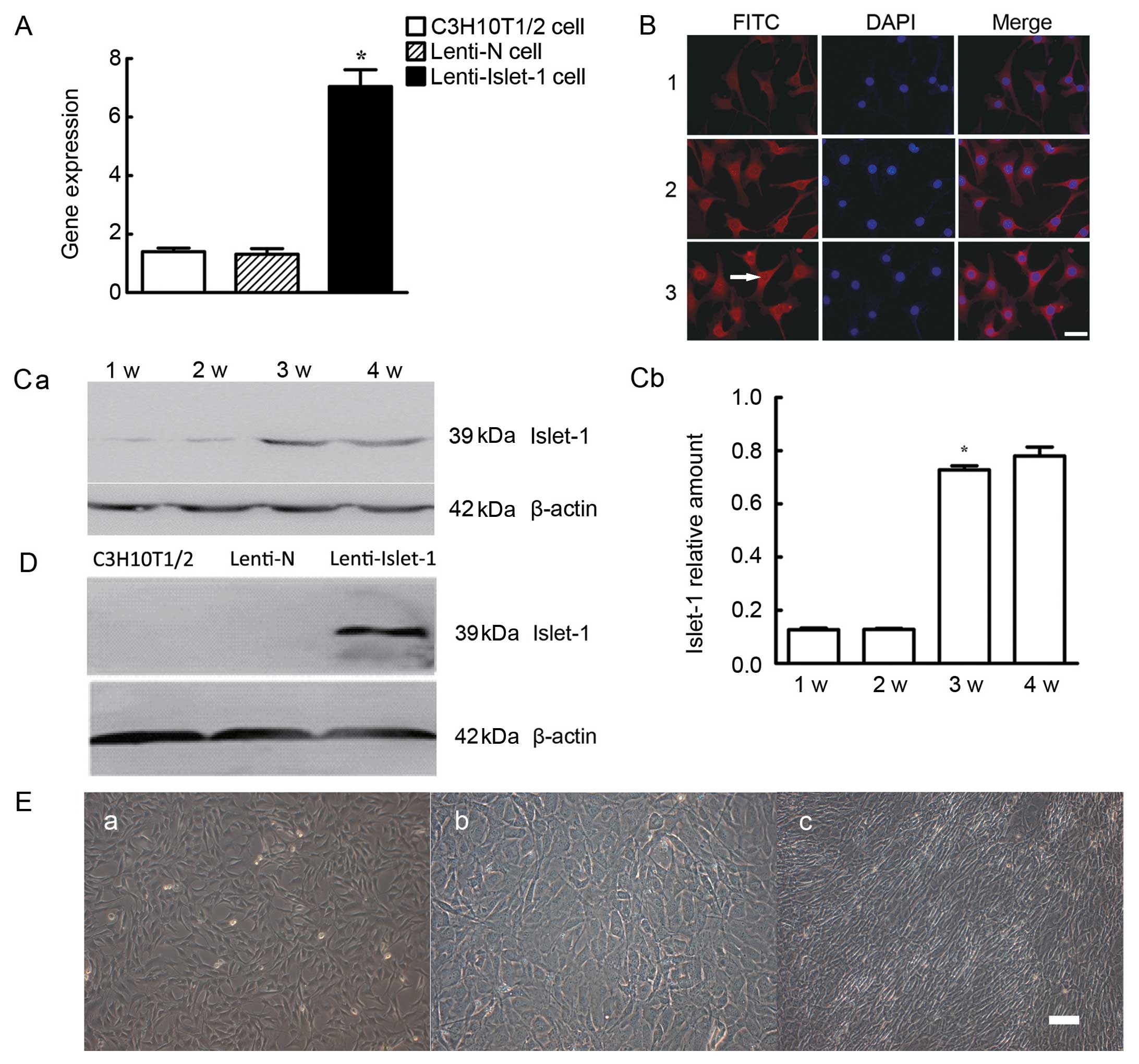 | Figure 3(A) Islet-1 gene expression in the
C3H10T1/2 cells transfected with Lenti-Islet-1 was higher
(*P<0.05) than that in the other C3H10T1/2 cells. (B)
The expression of Islet-1 was detected by immunofluorescence in
each group: Panel 1, untransfected C3H10T1/2 cells; panel 2,
C3H10T1/2 cells transfected with Lenti-N; panel 3, C3H10T1/2 cells
transfected with Lenti-Islet-1. FITC staining of Islet-1 protein
showing its location. DAPI staining of cell nucleus. Merge image
was obtained by overlapping FITC and DAPI images (magnification,
×20). Scale bar, 20 μm. (C-a) The expression of Islet-1 at
different time points in the C3H10T1/2 cells transfected with
Lenti-Islet-1: 1w, 1st week; 2w, 2nd week; 3w, 3rd week; 4w, 4th
week. (C-b) Islet-1 protein expression in the C3H10T1/2 cells
transfected with Lenti-Islet-1 was higher during the 3rd week than
in the first 2 weeks (*P<0.05). (D) Islet-1 protein
expression in the C3H10T1/2 cells transfected with Lenti-Islet-1
was higher than that in the untransfected C3H10T1/2 cells and the
C3H10T1/2 cells transfected with Lenti-N. (E) Morphological changes
of C3H10T1/2 cells transfected with lentiviral vectors with
pWPI-GFP plasmid or lentiviral vectors with pWPI-GFP-Islet-1
plasmid observed under a microscope. No difference was observed
between the untransfected C3H10T1/2 cells (magnification, ×10)
(E-a) and the C3H10T1/2 cells transfected with Lenti-N
(magnification, ×10) (E-b), whereas the C3H10T1/2 cells transfected
with Lenti-Islet-1 (×10) (E-c) turned into fibroblast-like cells
and were arranged toward the same direction. Scale bar, 100 μm. |
Islet-1 promotes the specific
differentiation of C3H10T1/2 cells into cardiomyocyte-like
cells
Under microscopic observation, no difference in cell
morphology was observed between the C3H10T1/2 cells not transfected
with lentivirus (Fig. 3E-a) and
those transfected with Lenti-N (Fig.
3E-b). However, the C3H10T1/2 cells transfected with
Lenti-Islet-1 (Fig. 3E-c) turned
into fibroblast-like cells and were arranged in the same
direction.
Cardiac-specific genes, such as Gata4, Nkx2.5 and
Mef2c, were detected by qPCR following transfection with the
lentiviral vectors. The expression of these genes in the C3H10T1/2
cells transfected with Lenti-Islet-1 was higher during the 2nd week
following transfection than the 1st week and 3rd week (Fig. 4A). The peak expression of the
cardiac-specific genes in the C3H10T1/2 cells transfected with
Lenti-Islet-1 was markedly higher than that in the untransfected
C3H10T1/2 cells and those transfected with Lenti-N 2 weeks
following transfection (Fig. 4B).
The expression of cTnT increased from the 3rd week following
transfection and not during the first 2 weeks (Fig. 4C). The expression of cTnT was
higher in the C3H10T1/2 cells transfected with Lenti-Islet-1 than
in the other 2 groups at 3 weeks following transfection (Fig. 4D). cTnT in the C3H10T1/2 cells
transfected with Lenti-Islet-1 was located in the cell nucleus and
cytoplasm (Fig. 4E).
 | Figure 4(A) The expression of Gata4, Nkx2.5
and Mef2c gene at different time points in C3H10T1/2 cells
transfected with Lenti-Islet-1. 2d, 2nd day; 1w, 1st week; 2w, 2nd
week; 3w, 3rd week. The peak expression of cardiac-specific genes
in the C3H10T1/2 cells transfected with Lenti-Islet-1 was
significantly higher than that in the untransfected C3H10T1/2 cells
and the C3H10T1/2 cells transfected with Lenti-N at 2 weeks after
transfection (*P<0.05). (B) The expression of
cardiac-specific genes in the C3H10T1/2 cells transfected with
Lenti-Islet-1 was higher in the 2nd week than the other time points
(*P<0.05). (C) The protein expression of cTnT at
different time points in the C3H10T1/2 cells transfected with
Lenti-Islet-1. 1w, 1st week; 2w, 2nd week; 3w, 3rd week; 4w, 4th
week. cTnT expression increased from the 3rd week following
transfection (*P<0.05). (D) cTnT expression in the
untransfected C3H10T1/2 cells, the C3H10T1/2 cells transfected with
Lenti-N and the C3H10T1/2 cells transfected with Lenti-Islet-1 at 3
weeks after transfection. cTnT expression was higher in the
C3H10T1/2 cells transfected with Lenti-Islet-1 than the other 2
groups of cells. (E) cTnT expression in the C3H10T1/2 cells
transfected with Lenti-Islet-1 was located in the cell nucleus and
cytoplasm (magnification, ×20). Scale bar, 20 μm. (F) Expression of
hepatocyte-, bone- and neuronal-specific markers. Albumin (ALB),
bone-specific alkaline phosphatase (BALP) and glial fibrillary
acidic protein (GFAP) expression was detected by western blot
analysis in the untransfected C3H10T1/2 cells, the C3H10T1/2 cells
transfected with Lenti-N and the C3H10T1/2 cells transfected with
Lenti-Islet-1 and the positive control group. There was no ALB,
BALP and GFAP expression observed in the untransfected C3H10T1/2
cells, the C3H10T1/2 cells transfected with Lenti-N and the
C3H10T1/2 cells transfected with Lenti-Islet-1. (G) AFP, bone Gla
protein (BGP) and nestin expression detected by qPCR. The
expression of BGP, AFP and nestin was did not differ significantly
between the 3 groups. |
The hepatocyte-specific markers, α-fetoprotein (AFP)
and ALB; the bone-specific markers, bone Gla protein (BGP) and
BALP; the neuronal specific markers, nestin and GFAP, were detected
by western blot analysis or qPCR. There was no ALB, BALP or GFAP
expression observed in the C3H10T1/2 cells, the C3H10T1/2 cells
transfected with Lenti-N and the C3H10T1/2 cells transfected with
Lenti-Islet-1 (Fig. 4F). The
expression of BGP, AFP and nestin did not differ between the 3
groups of cells (Fig. 4G).
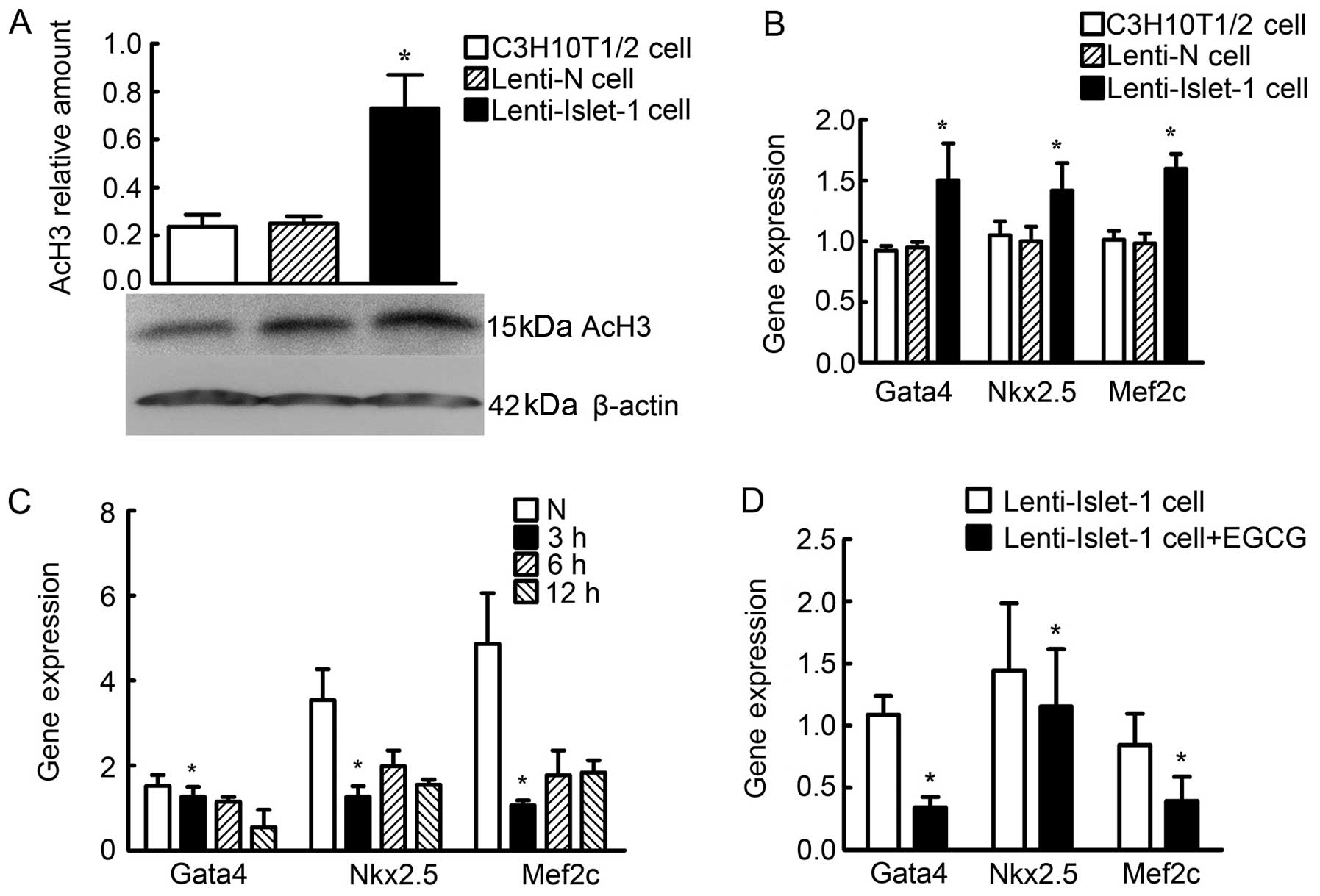
Changes in histone acetylation status
induced by Islet-1 overexpression
Acetylated histone H3 (AcH3) was detected by western
blot analysis in the untransfected C3H10T1/2 cells (controls), the
C3H10T1/2 cells transfected with Lenti-N and the C3H10T1/2 cells
transfected with Lenti-Islet-1. The relative amount of AcH3 in the
C3H10T1/2 cells transfected with Lenti-Islet-1 was higher than that
of the other cells (Fig. 5A).
ChIP and qPCR were performed to determine the acetylation levels of
histone H3 at the cardiac-specific genes, Gata4, Nkx2.5 and Mef2c,
at their peak expression times (2 weeks after transfection). The
expression of Gata4, Nkx2.5 and Mef2c combined with AcH3 in the
C3H10T1/2 cells transfected with Lenti-Islet-1 was higher than that
in the untransfected C3H10T1/2 cells and in the C3H10T1/2 cells
transfected with Lenti-N (Fig.
5B). Gata4, Nkx2.5 and Mef2c expression was found to be reduced
3 h following treatment with 120 μmol/l EGCG (Fig. 5C). Gata4, Nkx2.5 and Mef2c
expression in the C3H10T1/2 cells transfected with Lenti-Islet-1
and treated with EGCG was lower compared with the cells not treated
with EGCG (Fig. 5D).
Discussion
Stem cells have multiple differentiation potencies
and immunological features that render them promising candidates in
cell transplantation as a therapeutic option. Stem cells can
specifically differentiate into various cell types under different
treatment conditions. For example, MSCs treated with 5-azacytidine
can differentiate into cardiomyocyte-like cells (19). Endothelial progenitor cells (EPCs)
induced by vascular endothelial growth factor A (VEGF-A) can
differentiate into vascular endothelial cells (20). Stem cell-specific differentiation
is a complex process, and the mechanisms involved are still
unknown.
Histone acetylation and deacetylation are critical
to the modification of chromatin structure associated with the
regulation of gene expression (21). Acetylation and deacetylation is
involved in various developmental processes, including heart
development. A number of studies have revealed that histone
acetylation and deacetylation play an important role in the
differentiation of stem cells (22). The expression of cardiac-related
genes, such as Gata4, Nkx2.5, Mef2cj and cTnT is reduced following
interference of GCN5, a key histone acetyltransferase (23). Previous studies have demonstrated
that the expression of cardiac-related genes (both at the
transcriptional and translational level) is increased in C3H10T1/2
cells following treatment with trichostatin A [TSA, a histone
deacetylase (HADC) inhibitor] (24,25).
However, neither GCN5 nor other HATs/HADCs posses
DNA binding sites. The regulatory mechanism through histone
acetylation or deacetylation seems not to be specific. GCN5
generally binds to the specific target DNA sequence requiring other
factors that have DNA-banding domains (26).
To reveal the factors that are associated with HATs
and HADCs, studies have screened and analyzed the GCN5 protein
complexes in the process when MSCs specifically differentiate into
cardiomyocyte-like cells. LIM-HD, fibroblast growth factor-14
(FGF-14), leucine zipper protein 1 (LUZP1), cyclin-L1, NF-κB
inhibitor α and Kruppel-like factor 10 have been suggested as the
co-factors of GCN5, indicating that GCN5 is involved in
cardiomyocyte, hepatocyte, bone and neuronal differentiation
(27,28).
Islet-1, which contains one DNA binding site and two
LIM domains is a subtype of the LIM-HD subfamily, which is critical
for heart development. Islet-1 is located in the second heart field
and the outflow tract in the embryonic heart. Various types of
congenital heart disease can be caused by Islet-1 insufficiency
(14,15,29). Islet-1 has also been recognized as
a marker of cardiovascular progenitors (16,30). Islet-1-positive progenitors can
differentiate into diverse cardiovascular cell lineages.
In the present study, Islet-1 expression vectors
were generated and then successfully transfected into C3H10T1/2
cells. Our data indicate that Islet-1 is a key factor in
acetylation during the process of heart development and the
MSC-specific differentiation into cardiomyocyte-like cells. Islet-1
specifically promotes the differentiation of C3H10T1/2 cells into
cardiomyocyte-like cells through histone acetylation. C3H10T1/2
cells can be specifically induced to differentiate into
cardiomyocyte-like cells by Islet-1 overexpression. However, the
expression of hepatocyte-, bone- and neuronal-specific markers is
not affected by Islet-1. At least one of the mechanisms responsible
for the Islet-1-induced differentiation of C3H10T1/2 into
cardiomyocyte-like cells is the regulation of histone acetylation.
Islet-1, an important co-factor in histone acetylation, may promote
the cardiac-specific differentiation of stem cells. The results
obtained from the present study provide useful information
regarding the clinical application of stem cells in cell
transplantation therapy and may offer unique opportunities for the
prevention and treatment of heart diseases caused by cardiac cell
death.
Acknowledgements
This study was supported by the National Science
Foundation of China (grant no. 30973219).
References
|
1
|
Christoforou N and Gearhart JD: Stem cells
and their potential in cell-based cardiac therapies. Prog
Cardiovasc Dis. 49:396–413. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bianco P, Robey PG and Simmons PJ:
Mesenchymal stem cells: revisiting history, concepts, and assays.
Cell Stem Cell. 2:313–319. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pittenger MF, Mackay AM, Beck SC, et al:
Multilineage potential of adult human mesenchymal stem cells.
Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qin JJ, Xian SX, Huang XW and Sun JH:
Differentiation of mesenchymal stem cells into cardiomyocyte-like
cells in vitro: Drug, microenvironment and method. Zhongguo Zuzhi
Gongcheng Yanjiu Yu Linchuang Kangfu. 15:139–142. 2011.(In
Chinese).
|
|
5
|
Mafi1 P, Hindocha S, Mafi R, Griffin M and
Khan WS: Adult mesenchymal stem cells and cell surface
characterization - a systematic review of the literature. Open
Orthop J. 5:253–260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Psaltis PJ, Zannettino AC, Worthley SG and
Gronthos S: Concise review: mesenchymal stromal cells: potential
for cardiovascular repair. Stem Cells. 26:2201–2210. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Verdone L, Caserta M and Di Mauro E: Role
of histone acetylation in the control of gene expression.
Biochemistry and cell biology. Biochem Cell Biol. 83:344–353. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gupta MP, Samant SA, Smith SH and Shroff
SG: HDAC4 and PCAF bind to cardiac sarcomeres and play a role in
regulating myofilament contractile activity. J Biol Chem.
283:10135–10146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang G, Tian J, Feng C, Zhao LL, Liu Z and
Zhu J: Trichostatin a promotes cardiomyocyte differentiation of rat
mesenchymal stem cells after 5-azacytidine induction or during
coculture with neonatal cardiomyocytes via a mechanism independent
of histone deacetylase inhibition. Cell Transplant. 21:985–996.
2012. View Article : Google Scholar
|
|
10
|
Shahbazian MD and Grunstein M: Functions
of site-specific histone acetylation and deacetylation. Annu Rev
Biochem. 76:75–100. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Zang C, Cui K, et al: Genome-wide
mapping of HATs and HDACs reveals distinct functions in active and
inactive genes. Cell. 138:1019–1031. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brade T, Gessert S, Kühl M and Pandur P:
The amphibian second heart field: Xenopus islet-1 is required for
cardiovascular development. Dev Biol. 311:297–310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bu L, Jiang X, Martin-Puig S, et al: Human
ISL1 heart progenitors generate diverse multipotent cardiovascular
cell lineages. Nature. 460:113–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Cai CL, Lin L, et al: Isl1Cre
reveals a common Bmp pathway in heart and limb development.
Development. 133:1575–1585. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Laugwitz KL, Moretti A, Caron L, Nakano A
and Chien KR: Islet1 cardiovascular progenitors: a single source
for heart lineages? Development. 135:193–205. 2008.PubMed/NCBI
|
|
16
|
Nakano A, Nakano H and Chien KR:
Multipotent islet-1 cardiovascular progenitors in development and
disease. Cold Spring Harb Symp Quant Biol. 73:297–306. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Toscano MG, Frecha C, Ortega C, Santamaria
M, Martin F and Molina IJ: Efficient lentiviral transduction of
Herpesvirus saimiri immortalized T cells as a model for gene
therapy in primary immunodeficiencies. Gene Ther. 11:956–961.
2004.
|
|
18
|
Zsindely N, Pankotai T, Ujfaludi Z, et al:
The loss of histone H3 lysine 9 acetylation due to dSAGA-specific
dAda2b mutation influences the expression of only a small subset of
genes. Nucleic Acid Res. 37:6665–6680. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carvalho PH, Daibert AP, Monteiro BS, et
al: Differentiation of adipose tissue-derived mesenchymal stem
cells into cardiomyocytes. Arq Bras Cardiol. 100:82–89.
2013.PubMed/NCBI
|
|
20
|
Haberzettl P, Lee J, Duggineni D, et al:
Exposure to ambient air fine particulate matter prevents
VEGF-induced mobilization of endothelial progenitor cells from the
bone marrow. Environ Health Perspect. 120:848–856. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Chu JS, Kurpinski K, et al:
Biophysical regulation of histone acetylation in mesenchymal stem
cells. Biophys J. 100:1902–1909. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Passier R, Van Laake LW and Mummery CL:
Stem-cell-based therapy and lessons from the heart. Nature.
453:322–329. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Zhu J, Tian J, Liu X and Feng C: A
role for Gcn5 in cardiomyocyte differentiation of rat mesenchymal
stem cells. Mol Cell Biochem. 345:309–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Colussi C, Berni R, Rosati J, et al: The
histone deacetylase inhibitor suberoylanilide hydroxamic acid
reduces cardiac arrhythmias in dystrophic mice. Cardiovasc Res.
87:73–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pufahl L, Katryniok C, Schnur N, et al:
Trichostatin A induces 5-lipoxygenase promoter activity and mRNA
expression via inhibition of histone deacetylase 2 and 3. J Cell
Mol Med. 16:1461–1473. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grant PA, Duggan L, Côté J, et al: Yeast
Gcn5 functions in two multisubunit complexes to acetylate
nucleosomal histones: characterization of an Ada complex and the
SAGA (Spt/Ada) complex. Genes Dev. 11:1640–1650. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chakraborty A, Paul BD and Nagaraja V:
Bacteriophage Mu C protein is a new member of unusual leucine
zipper-HTH class of proteins. Protein Eng Des Sel. 20:1–5. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kosaka N, Kodama M, Sasaki H, et al: FGF-4
regulates neural progenitor cell proliferation and neuronal
differentiation. FASEB J. 20:1484–1485. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mishra R, Vijayan K, Colletti EJ, et al:
Characterization and functionality of cardiac progenitor cells in
congenital heart patients. Circulation. 123:364–373. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moretti A, Bellin M, Jung CB, et al: Mouse
and human induced pluripotent stem cells as a source for
multipotent Isl1+cardiovascular progenitors. FASEB J.
24:700–711. 2010.PubMed/NCBI
|