Introduction
Hydrogen sulfide (H2S), a well-known
toxic gas, is regarded as the third endogenous gaseous signaling
molecule (1,2). Cystathioine γ-lyase (CSE), one of
the three enzymes in the transsulfuration pathway, is responsible
for the production of endogenous H2S using L-cysteine or
L-homocysteine as a substrate (2–5).
The generation of CSE mRNA occurs along with the generation of
H2S in the rat aorta (6,7)
that is provided with exogenous L-cysteine. Generally,
cystathionine β-synthase (CBS) is considered to play a crucial role
in the development and maintenance of the central nervous system,
and radial glia/astrocyte dysfunction may be involved in the
complex neuropathological features (8). Moreover, 3-mercaptopyruvate
sulfurtransferase (3MST), another H2S-producing enzyme,
is localized to neurons in the brain and to the vascular
endothelium (9).
Previous studies have shown that the
H2S/CSE signaling pathway is involved in the
inflammation induced by endotoxins, such as lipopolysaccharides
(LPS). H2S may represent a novel endogenous mechanism of
cytoprotection in the inflamed joint, suggesting a potential
opportunity for therapeutic intervention (10). The inhibitory effects of LPS on
endothelium-dependent relaxation resulting in pulmonary
hypertension may also be mediated by H2S (11). NaHS (an H2S donor) has
been shown to dose-dependently inhibit LPS-induced chemokine
receptor CX3CR1 expression in macrophages (12). Inhaling H2S has also
been shown to prevent inflammation and improve survival after LPS
challenge by altering sulfide metabolism in mice (13). The decrease in pulmonary
surfactant (PS) levels is the most important physiopathological
process of acute lung injury (ALI) induced by LPS. Exogenously
applied H2S can attenuate the process of ALI, possibly
since H2S can adjust the composition and secretion of PS
(14). The downregulation in
CSE/H2S expression is involved in the pathogenesis of
LPS-induced ALI, and LPS can stimulate CSE expression and
H2S production (15,16).
Nuclear factor (NF)-κB is a heterodimer involving a
variety of signaling pathways (17). The transcription factor, NF-κB,
regulates inflammatory responses by inducing the expression of a
variety of genes (18).
H2S can inhibit NF-κB activation in LPS-stimulated
macrophages (19). In a previous
study, S-propargyl-cysteine (SPRC), a novel
H2S-modulated agent, exerted beneficial effects by
inhibiting IκB-α degradation and the phosphorylation of
transcription factors associated with nuclear factor-κB p65
activation induced by LPS (20).
The NF-κB pathway can be rapidly activated by a large spectrum of
chemically diverse agents and stress conditions, including
bacterial LPS, microbial and viral pathogens, cytokines and growth
factors (18). SPRC has been
shown to exert anti-inflammatory effects in LPS-stimulated H9c2
cells partly through the CSE/H2S pathway by impairing
IκB-α/NF-κB signaling (21).
However, the transcriptional regulation of the CSE
gene in mammalian cells treated with LPS remains largely unknown.
Based on the fact that the H2S/CSE signaling pathway is
involved in the inflammation induced by endotoxins, such as LPS, in
the present study, we investigated the effects of the binding site
of the transcription factor, NF-κB, on the transcriptional
regulation of the CSE gene in mammalian cells treated with LPS.
Materials and methods
Construction of luciferase reporter under
the control of the mouse CSE promoter
A 1.5-kb DNA fragment upstream of the transcription
start site (−1453 to +25) of the mouse CSE gene was amplified by
PCR using pGL4.12-KM1716 [the construct with the mouse CSE gene
promoter previously constructed in our laboratory (22)] as the template (forward primer,
5′-CGGGG TACCGTGGAAGGGACATTCCTGTGAATAG-3′ and reverse primer,
5′-CCGCTCGAGAGGAGTGCGAGGTGTTGCTTT GGCT-3′). The thermal cycling
conditions were as follows: initial denaturation at 94°C for 3 min,
followed by 30 cycles of 95°C for 30 sec, 60°C for 45 sec, 72°C for
2 min and a final elongation step at 72°C for 8 min. The PCR
product was digested with the restriction enzymes, KpnI and
XhoI (both from Takara Biotechnology, Co., Ltd., Dalian,
China), and cloned into the promoterless pGL4.12 vector (Promega,
Madison, WI, USA) that contains a Firefly luciferase gene driven by
the inserted promoter. The resultant construct was designated as
pGL4.12-KM1478. The inserted DNA fragment was confirmed by DNA
sequencing (Biosune Biotechnology Co., Ltd., Shanghai, China). The
construction of the reporter with the mutant CSE promoter was the
same as above, the only difference being that an alternative
forward primer (5′-CGGGG TACCGTGGAAATCTCATTCCTGTGATAG-3′) was used
during PCR amplification.
Cell culture and treatments and
luciferase assay
For transfection, HEK-293 and COS-7 cells were grown
to 70–80% confluence Thereafter, 5 μg pGL4.12-KM1478 or 5 μg
pGL4.12-KM1478m together with the pRL-CMV control vector (0.032 μg
for the HEK-293 cells, 0.0032 μg for the COS-7 cells) were
transfected into the cells using Xfect™ transfection reagent
(Clontech Laboratories, Inc., Palo Alto, CA, USA) according to the
manufacturer’s instructions. Twelve hours later, the transfected
cells were trypsinized and seeded into 35-mm dishes. Following
treatment with LPS for 6 h, both Firefly lucierase and
Renilla luciferase activities were assayed using the
Dual-Luciferase® reporter assay (Promega) according to
the manufacturer’s instructions with the SpectraMax®
microplate reader (Molecular Devices Corp., Sunnyvale, CA, USA),
according to the manufacturer’s instructions. The Firefly
luciferase activity was normalized against the Renilla
luciferase activity.
Isolation of RNA and quantitative
reverse-transcription PCR (RT-qPCR)
Total RNA was isolated using TransZol Up reagent
(TransGen Biotech Co., Ltd., Beijing, China), according to the
manufacturer’s instructions, after the treated cells were rinsed
twice with 1× dPBS buffer (Thermo Scientific, Rockford, IL, USA).
The cells were directly lysed in a culture dish by the addition of
1 ml of TransZol Up to a 3.5-cm dish, and passing the cell lysate
several times through a pipette. The homogenized samples were then
incubated for 5 min at 15–30°C to permit the complete dissociation
of the nucleoprotein complexes. In total, 0.2 ml of chloroform per
1 ml of TransZol were added. The tubes were vigorously shaken by
hand for 15 sec and were then incubated at room temperature for 3
min. The samples were cetrifuged at 10,000 rpm for 15 min at 4°C.
Following centrifugation, the mixture was separated into a lower
red, phenol-chloroform phase, an interphase, and a colorless upper
aqueous phase. RNA remains exclusively in the aqueous phase. The
aqueous phase was transferred to a fresh tube. The RNA from the
aqueous phase was precipitated by mixing with isopropyl alcohol. A
total of 0.5 ml of isopropyl alcohol per 1 ml of TransZol Up was
used for the initial homogenization. The samples were incubated at
room temperature for 10 min and centrifuged at 10,000 rpm for 10
min at 4°C. The RNA precipitate, often invisible before
centrifugation, forms a gel-like pellet on the side and bottom of
the tube. The supernatant was carefully removed. The RNA pellet was
washed once with 75% ethanol, adding at least 1 ml of 75% ethanol
per 1 ml of TransZol Up used for the initial homogenization. The
sample was mixed by vortexing and centrifugation at no more than
7,000 rpm for 5 min at 4°C. At the end of the procedure, the RNA
pellet was air-dried for 5–10 min. The RNA was dissolved in
RNase-free water by passing the solution a few times through a
pipette tip, incubating for 10 min at 55–60°C and cooling to room
temperature. The RNA samples were quantified by absorbance at 260
nm (Biophoto, Eppendorf, Germany) and the integrity, purity and
amount of RNA were verified by the visualization of rRNA following
agarose gel electrophoresis. The fluorescence intensity was
determined for the RNA concentrations using a Qubit™ RNA assay kit
(Invitrogen Life Technologies, Shanghai, China) according to the
manufacturer’s instructions. First-strand cDNA was synthesized by
incubation at 42°C for 30 min using an anchored
oligo(dT)18 primer. The total volume of the the reaction
mixture was 20 μl containing 2 μg of RNA, 1 μl of anchored
oligo(dT)18, 10 μl of 2× TS Reaction mix and
TransScript™ RT/RI Enzyme mix as well as RNase-free water (TransGen
Biotech Co., Ltd.). The reaction was terminated by incubation at
85°C for 5 min and the reaction mixture was stored at −20°C and
used for RT-qPCR.
RT-qPCR was performed in a final volume of 25 μl
containing 11 μl cDNA diluted with double-distilled H2O
(1:20), 0.5 μl of 0.2 μM each primer, 0.5 μl Passive Reference Dye
II and 12.5 μl of 2× TransStart™ Green qPCR SuperMix (TransGen
Biotech Co., Ltd.). All reactions were run in triplicate on an
Agilent Mx3000P QPCR System (Agilent Technologies, Santa Clara, CA,
USA) and MxPro QPCR software (Agilent Technologies) using a
SYBR-Green fluorescence quantification system with the following
conditions: an initial denaturation step at 95°C for 10 min, and 45
cycles of 30 sec at 95°C, 30 sec at 60°C and 10 sec at 72°C. The
primer pair Q CSE Forward Primer/Q CSE Reverse Primer (Table I) was designed to determine the
relative expression of CSE. The primers specific to our PCR
template were created by Blast biological software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
The fluorescence was measured at the end of the extension step at
72°C. Controls for genomic DNA and primer contamination were
routinely performed with non-RT or no template PCR reactions,
respectively. Dissociation curves were performed for each set of
oligonucleotides to test primer specificity and to confirm the
presence of a unique PCR product. To estimate PCR efficiencies,
standard curves were performed based on 5 serial dilutions of a
cDNA stock. PCR efficiencies of all primer sets were between 95 and
100%. After verifying that the β-actin gene (ACTB) and CSE
mRNA primers had similar amplification efficiencies, the
comparative Ct method 2−ΔΔCt was used to for performing
relative quantification analysis of the mRNA levels, as previously
described (23). The relative
amount of each mRNA was normalized to the housekeeping gene,
ACTB. Each sample was run and analyzed in triplicate. The
average of the relative amount of each mRNA in the control group is
defined as 1.0. The quantity of the transcripts was estimated from
a standard line derived from 20-fold serial dilutions of cDNA
pooled from J774.1A or RAW264.7 cells treated with LPS.
 | Table IPrimers used for RT-qPCR. |
Table I
Primers used for RT-qPCR.
| Gene | GenBank accesion
no. | Forward/reverse
primer | Exon | Amplicon size |
|---|
| CSE | NM_145953.2 |
5′-TGGGCTGCCCTCTCATCCACA-3′ | 2 | 112 bp |
| |
5′-TCCTCCCAAGCTCTCGGCCA-3′ | 2 | |
| ACTB | NM_007393.3 |
5′-GCGAGCACAGAGCCTCGCCTTT-3′ | 2 | 128 bp |
| |
5′-CCTTGCACATGCCGGAGCCGT-3′ | 2 | |
Western blot analysis
For total protein extraction, 1×106 cells
were incubated in 120 μl of RIPA lysis buffer (mild) (Biomiga,
Inc., San Diego, CA, USA), supplemented with 1 mM PMSF proteinase,
0.25 U/μl Benzonase, inhibitor cocktail (Sigma-Aldrich, St. Louis,
MO, USA). The cells were incubated on ice for 30 min, and the
lysate was cleared by centrifugation at 12,000 × g at 4°C for 15
min. Following centrifugation, glycerol, 2-mercaptoethanol, 10%
sodium dodecylsulfate (SDS), 1 M Tris-HCl (pH 6.7) and bromophenol
blue were added to the supernatant, and then denatured at 95–100°C
for 10 min. The proteins were separated by electrophoresis on a 10%
[for the detection of cystathionine-γ-lyase (CSE), and α-tubulin
(B-7)(TUBA)] SDS-polyacrylamide gel (Sigma-Aldrich) and transferred
onto an polyvinylidene difluoride (PVDF) membrane (0.45 μm,
Immobilon-P; Millipore, Billerica, MA, USA). The membrane was
blocked with a blocking solution containing 5% skim milk, 137 mM
NaCl, 0.1% Tween-20 and 20 mM Tris-HCl (pH 7.6). After a wash, the
membrane was incubated overnight at 4°C with anti-CSE (30.7; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA) monoclonal
antibodies (1:1,000 dilutions) or anti-α-tubulin (B-7; Santa Cruz
Biotechnology, Inc.) monoclonal antibodies (1:15,000 dilutions).
After another wash, the membrane was incubated with a horseradish
peroxidase (HRP)-conjugated goat anti-mouse antibody (1:10,000)
(Santa Cruz Biotechnology, Inc.), for the detection of CSE and
TUBA. Positive bands for CSE or TUBA were identified around 43–47
or 52–58 kDa, respectively, by SuperSignal West Pico
Chemiluminescent Substrate (Thermo Scientific). The resulting films
(Kodak Co., Ltd., Xiamen, China) were scanned and quantified using
densitmetric analytical software (the Bio-Rad Quantity One
software; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All data are expressed as the means ± SEM of at
least 4 experiments. Statistical significance was assessed by
either one-way or two-way ANOVA for repeated measures followed by
Turkey’s test. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Effects of LPS on CSE gene expression at
the mRNA level
To determine the effects of LPS on the transcription
of the CSE gene, we examined the mRNA expression level of CSE in
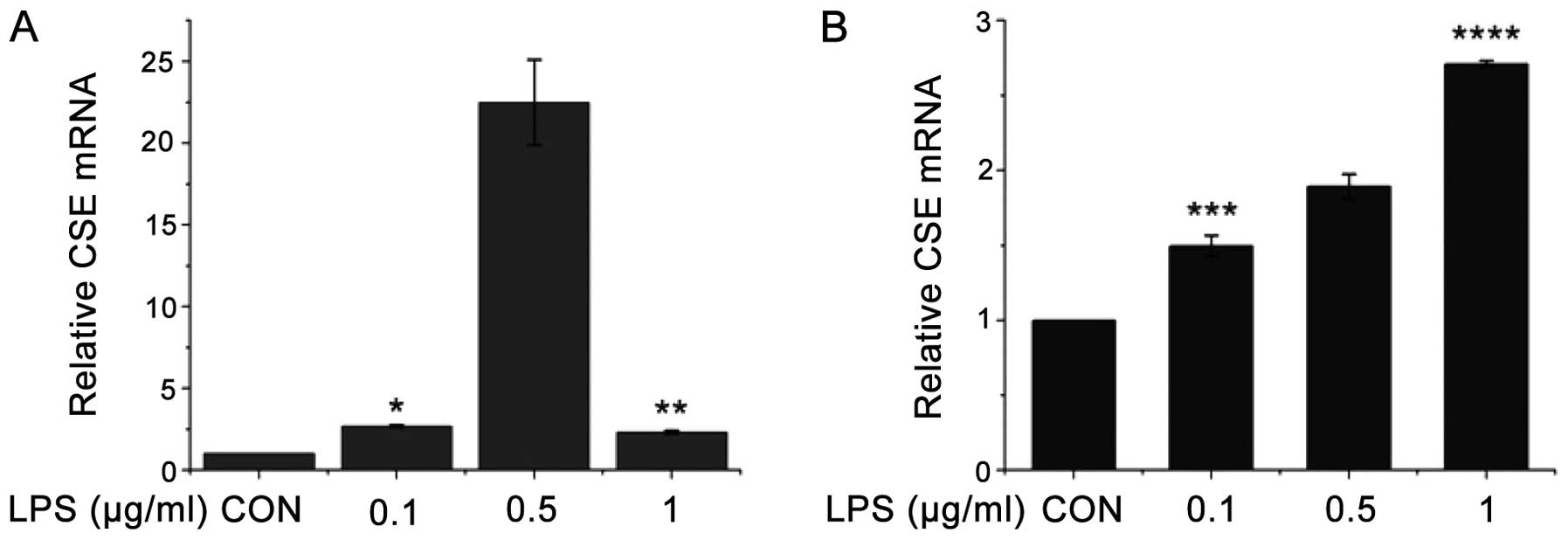
the J774.1A and RAW264.7 cells. As shown in Fig. 1A, following treatment with LPS
(0.1, 0.5 and 1 μg/ml) for 6 h, the CSE mRNA levels in the J774.1A
cells increased, at the 0.5 μg/ml dose in particular. However, high
concentrations of LPS (1.0 μg/ml) had less significant effects,
possibly caused by the toxicity of LPS to the cells. As shown in
Fig. 1B, following treatment with
LPS (0.1, 0.5 and 1 μg/ml) for 6 h, the CSE mRNA levels in the
RAW264.7 cells gradually increased in a dose-dependent manner; this
suggests that the RAW264.7 cells are possibly more tolerant to the
toxicity induced by LPS. As LPS has both immune-stimulating effects
and a toxic activity, it may exert varying effects on different
cell types.
Effects of LPS on CSE gene expression at
the protein level
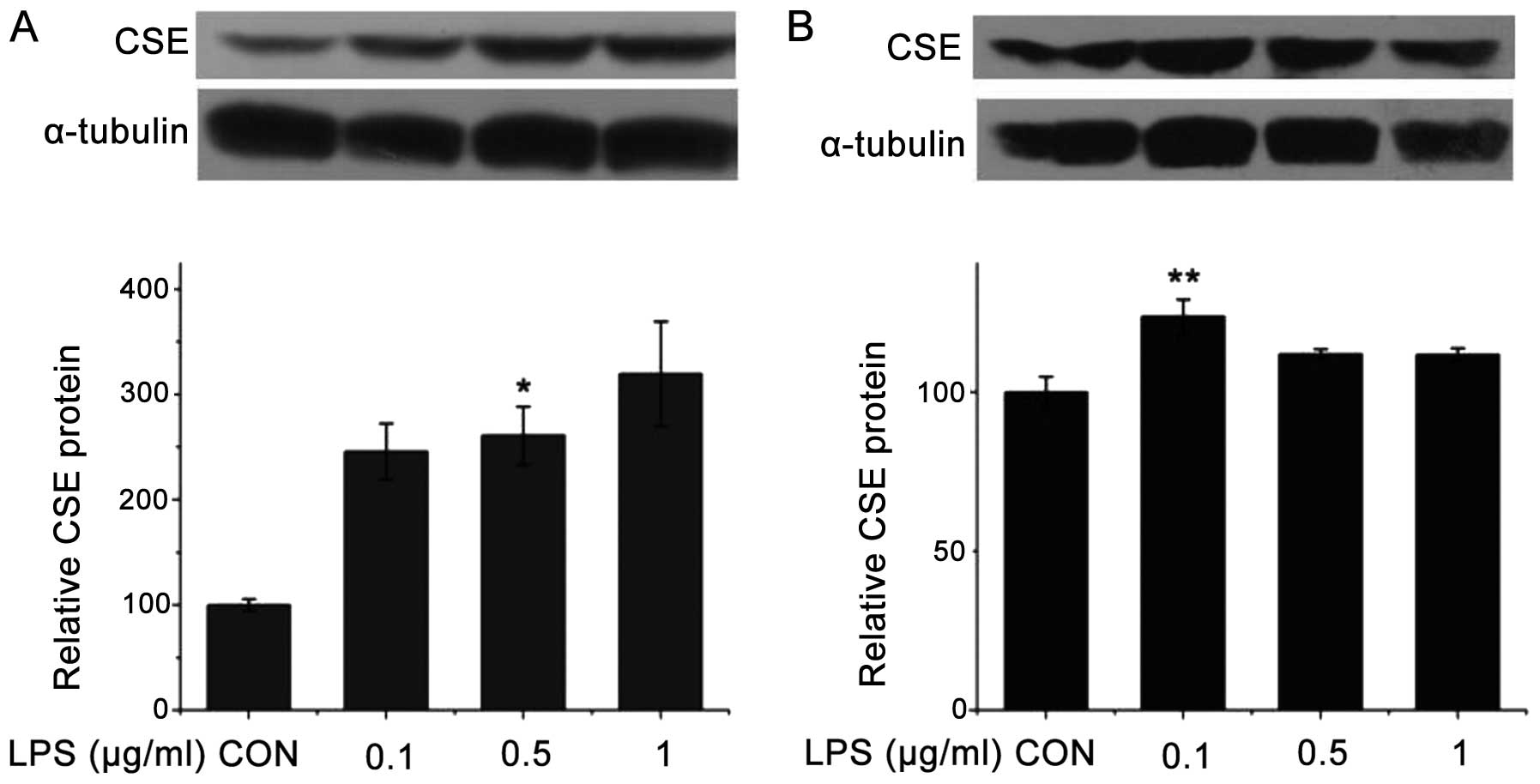
We examined the effects of LPS on the protein
expression of CSE in the J774.1A and RAW264.7 cells. As shown in
Fig. 2A, following treatment with
LPS (0.1, 0.5 and 1 μg/ml) for 6 h, the CSE protein levels in the
J774.1A cells increased, particularly at the dose of 1 μg/ml. As
shown in Fig. 2B, following
treatment with LPS (0.1, 0.5 and 1 μg/ml) for 6 h, the CSE protein
levels in the RAW264.7 cells also increased, although not as
significantly as the CSE protein levels in the J774.1A cells.
Effects of LPS on the wild-type or mutant
promoter activity of the CSE gene
We also determined the effects of LPS on the
transactivation of the promoter activity of the CSE gene using the
transient transfection experiment. As mentioned above, LPS
increased the expression of CSE in 2 lymphocyte cell lines.
Bioinformatics analysis using the search algorithm TFSEARCH:
Searching Transcription Factor Binding Sites (version 1.3;
http://mbs.cbrc.jp/research/db/TFSEARCH.html) as
previously described (24)
revealed the potential NF-κB binding site on the promoter of the
mouse CSE gene with the DNA sequences of 5′-GGGACATTCC-3′. To
determine whether NF-κB is involved in regulating the expression of
the CSE gene, we constructed a reporter assay in which the reporter
luciferase expression was either controlled by the wild-type (with
NF-κB binding site) or the mutant (without NF-κB site) control
region of the CSE gene (Fig.
3).
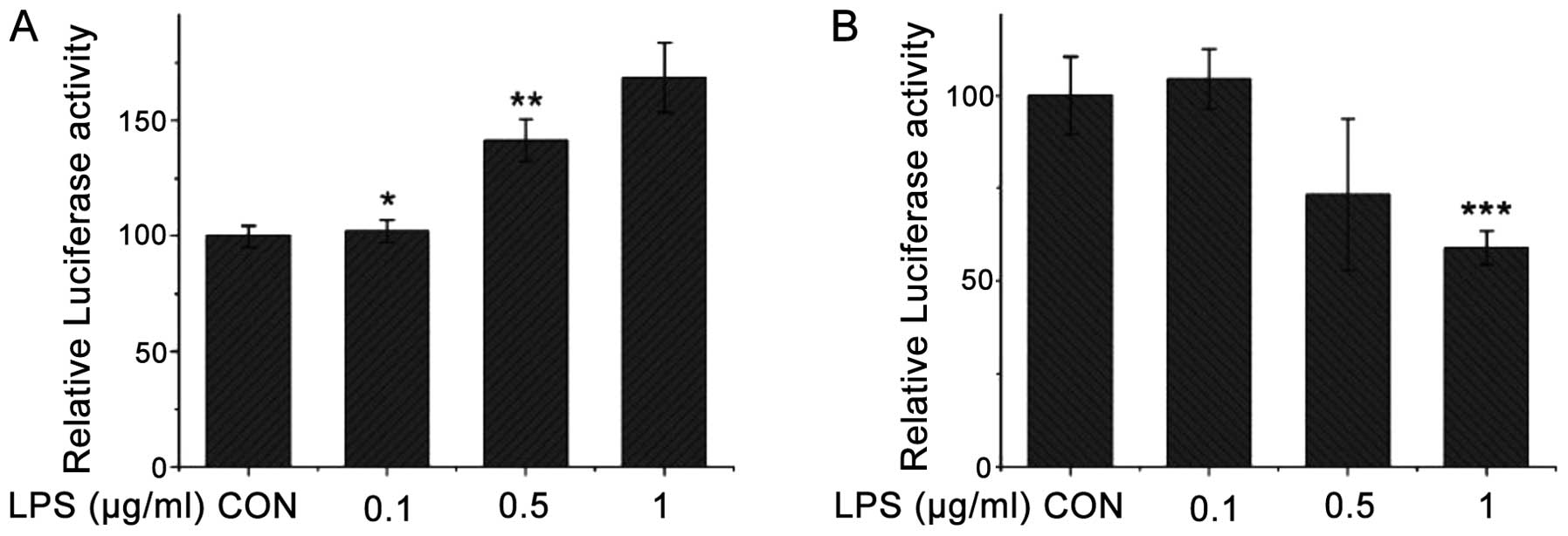
Following treatment with LPS (0.1, 0.5 and 1 μg/ml)
for 6 h, the wild-type promoter activity of the CSE gene in the
transfected COS-7 cells increased with the increment in the LPS
concentration (Fig. 4A). On the
contrary, the mutant promoter activity of the CSE gene gradually
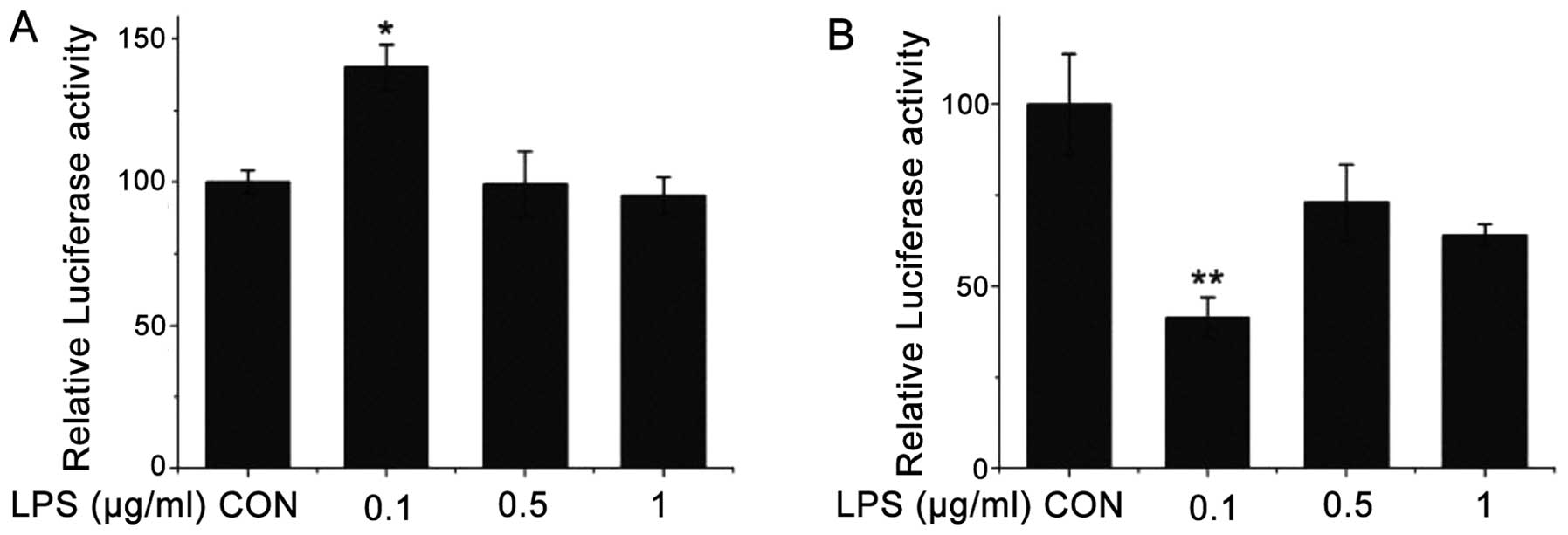
decreased with the increment in the LPS concentration (Fig. 4B). Following treatment with LPS
(0.1, 0.5 and 1 μg/ml) for 6 h, the wild-type promoter activity of
the CSE gene increased in the transfected HEK-293 cells treated
with LPS at 0.1 μg/ml (Fig. 5A).
The CSE promoter activity did not increase significantly in the
cells treated with LPS at 0.5 and 1 μg/ml. However, following
treatment with LPS (0.1, 0.5 and 1 μg/mL) for 6 h, the mutant
promoter activity of the CSE gene gradually decreased with the
increment in LPS concentration (Fig.
5B).
Discussion
H2S is regarded as the third endogenous
gaseous signaling molecule (1,2).
CSE, which produces H2S from cysteine or homocysteine
(25), is mainly expressed in the
liver, kidneys, lungs, thoracic aorta, ileum, portal vein, uterus
and the brain, as well as in pancreatic islets and the placenta in
mammals (7,26–33). A number of studies have shown that
the H2S/CSE signaling pathway is involved in the
inflammation induced by endotoxins, such as LPS (10,11,19–21,34). In the present study, our results
revealed that the transcriptional and post-transcriptional
regulation of the CSE gene is mediated by the binding site of NF-κB
on the CSE promoter in mammalian cells treated with LPS.
Both COS-7 and HEK-293 cells (two mammalian kidney
cell lines) have not only been successfully used in the study of
the CSE/H2S signaling pathway (35), but also in research on high DNA
transfection efficiency in gene transfection experiment. Some
typical mammalian cell lines, such as J774.1A and RAW264.7 cells,
are frequently used in the study of the H2S/CSE
signaling pathway (12,36–39). Therefore, both the J774.1A and
RAW264.7 cells are good models for testing the post-transcriptional
regulation of the CSE gene by exogenous H2S in mammalian
cells, and the COS-7 and HEK-293 cells for testing the
transcriptional activity.
Certain studies have indicated that H2S
plays an important role in inflammation, and LPS stimulates the
expression of the CSE gene and the H2S production rate.
An increase in the plasma H2S concentration, alongside
augmented liver H2S biosynthesis from exogenous
cysteine, is also apparent in animals 4 h after an LPS injection
(15,40). As previously demonstrated, the
serine (Ser) 276 phosphorylation of p65 is increased by the
LPS-mediated protein kinase A (PKA) activation in Raw264.7 murine
macrophages (41). SPRC exerts
beneficial effects by inhibiting IκB-α degradation and the
phosphorylation of transcription factors associated with nuclear
factor κ-B p65 activation induced by LPS (20). Our results demonstrated that LPS
significantly increased the mRNA and protein expression levels of
of the CSE gene in the J774.1A and RAW264.7 cells following
treatment with LPS for 6 h. As LPS significantly affected the
post-transcriptional regulation of the CSE gene, the binding site
of the transcription factor, NF-κB, on the promoter of the CSE gene
in mammalian cells may be involved in this regulation.
After the transfected HEK-293 and COS-7 cells were
incubated with various concentrations of LPS for 6 h, the wild-type
promoter activity of the CSE gene significantly increased. However,
after the transfected HEK-293 and COS-7 cells were incubated with
various concentrations of LPS for 6 h, the mutant-type promoter
activity of the CSE gene significantly decreased. These results
indicated that the GGGACATTCC DNA sequence on the promoter of the
CSE gene was closely associated with the transcriptional regulation
of the CSE gene in mammalian cells treated with LPS.
Taken together, our results suggest that the binding
site of the transcription factor, NF-κB, on the CSE promoter is
critical for LPS-induced CSE expression in mammalian cells. The
data presnted in this study may significantly contribute to the
further understanding of the regulatory mechanisms of the CSE gene
in the inflammation process. Furthermore, these are important
findings with potential clinical significance. However, the
regulatory mechanisms involved in the effects of LPS on the
transcriptional regulation of the CSE gene in mammalian cells via
the NF-κB pathway require further investigation.
Acknowledgements
This study was supported by the National Basic
Research Program of China (973 Program, no. 2010CB912604) and the
Startup Project of Doctor Scientific Research of Hanshan Normal
University (no. QD20140102).
References
|
1
|
Wang R: The gasotransmitter role of
hydrogen sulfide. Antioxid Redox Signal. 5:493–501. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kimura H: Hydrogen sulfide: its production
and functions. Exp Physiol. 96:833–835. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kimura H: Hydrogen sulfide as a
neuromodulator. Mol Neurobiol. 26:13–19. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang R: Hydrogen sulfide: the third
gasotransmitter in biology and medicine. Antioxid Redox Signal.
12:1061–1064. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang R: Hydrogen sulfide: a new EDRF.
Kidney Int. 76:700–704. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao W, Zhang J, Lu Y and Wang R: The
vasorelaxant effect of H(2)S as a novel endogenous gaseous K(ATP)
channel opener. EMBO J. 20:6008–6016. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hosoki R, Matsuki N and Kimura H: The
possible role of hydrogen sulfide as an endogenous smooth muscle
relaxant in synergy with nitric oxide. Biochem Biophys Res Commun.
237:527–531. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Enokido Y, Suzuki E, Iwasawa K, Namekata
K, Okazawa H and Kimura H: Cystathionine beta-synthase, a key
enzyme for homocysteine metabolism, is preferentially expressed in
the radial glia/astrocyte lineage of developing mouse CNS. FASEB J.
19:1854–1856. 2005.PubMed/NCBI
|
|
9
|
Shibuya N, Tanaka M, Yoshida M, et al:
3-Mercaptopyruvate sulfurtransferase produces hydrogen sulfide and
bound sulfane sulfur in the brain. Antioxid Redox Signal.
11:703–714. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fox B, Schantz JT, Haigh R, et al:
Inducible hydrogen sulfide synthesis in chondrocytes and
mesenchymal progenitor cells: is H2S a novel
cytoprotective mediator in the inflamed joint? J Cell Mol Med.
16:896–910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang XL, Zhou XH, Wei P, Zhang XJ, Meng
XY and Xian XH: Role of endogenous hydrogen sulfide in pulmonary
hypertension induced by lipopolysaccharide. Sheng Li Xue Bao.
60:211–215. 2008.(In Chinese).
|
|
12
|
Zhang H, Guo C, Wu D, et al: Hydrogen
sulfide inhibits the development of atherosclerosis with
suppressing CX3CR1 and CX3CL1 expression. PloS One. 7:e411472012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tokuda K, Kida K, Marutani E, et al:
Inhaled hydrogen sulfide prevents endotoxin-induced systemic
inflammation and improves survival by altering sulfide metabolism
in mice. Antioxid Redox Signal. 17:11–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang P, Zhang JX, Gong JP, Li LF, Jin PL
and Ding CM: Effects of hydrogen sulfide on pulmonary surfactant in
rats with acute lung injury induced by lipopolysccharide. Zhongguo
Ying Yong Sheng Li Xue Za Zhi. 27:485–489. 2011.(In Chinese).
|
|
15
|
Zhu XY, Liu SJ, Liu YJ, Wang S and Ni X:
Glucocorticoids suppress cystathionine gamma-lyase expression and
H2S production in lipopolysaccharide-treated
macrophages. Cell Mol Life Sci. 67:1119–1132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou XH, Huang XL, Wei P, Tian FJ and Ling
YL: Role of hydrogen sulfide/cystathionine-gamma-lyase system in
acute lung injury induced by lipopolysaccharide in rats. Zhongguo
Wei Zhong Bing Ji Jiu Yi Xue. 21:199–202. 2009.(In Chinese).
|
|
17
|
Pahl HL: Activators and target genes of
Rel/NF-kappaB transcription factors. Oncogene. 18:6853–6866. 1999.
View Article : Google Scholar
|
|
18
|
Baeuerle PA and Baltimore D: NF-kappa B:
ten years after. Cell. 87:13–20. 1996.PubMed/NCBI
|
|
19
|
Oh GS, Pae HO, Lee BS, et al: Hydrogen
sulfide inhibits nitric oxide production and nuclear factor-kappaB
via heme oxygenase-1 expression in RAW264.7 macrophages stimulated
with lipopolysaccharide. Free Radic Biol Med. 41:106–119. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gong QH, Wang Q, Pan LL, Liu XH, Xin H and
Zhu YZ: S-propargyl-cysteine, a novel hydrogen sulfide-modulated
agent, attenuates lipopolysaccharide-induced spatial learning and
memory impairment: involvement of TNF signaling and NF-kappaB
pathway in rats. Brain Behav Immun. 25:110–119. 2011. View Article : Google Scholar
|
|
21
|
Pan LL, Liu XH, Gong QH and Zhu YZ:
S-Propargyl-cysteine (SPRC) attenuated lipopolysaccharide-induced
inflammatory response in H9c2 cells involved in a hydrogen
sulfide-dependent mechanism. Amino Acids. 41:205–215. 2011.
View Article : Google Scholar
|
|
22
|
Wang M, Guo Z and Wang S: Cystathionine
gamma-lyase expression is regulated by exogenous hydrogen peroxide
in the mammalian cells. Gene Expr. 15:235–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Heinemeyer T, Wingender E, Reuter I, et
al: Databases on transcriptional regulation: TRANSFAC, TRRD and
COMPEL. Nucleic Acids Res. 26:362–367. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abe K and Kimura H: The possible role of
hydrogen sulfide as an endogenous neuromodulator. J Neurosci.
16:1066–1071. 1996.PubMed/NCBI
|
|
26
|
Diwakar L and Ravindranath V: Inhibition
of cystathionine-gamma-lyase leads to loss of glutathione and
aggravation of mitochondrial dysfunction mediated by excitatory
amino acid in the CNS. Neurochem Int. 50:418–426. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaneko Y, Kimura Y, Kimura H and Niki I:
L-cysteine inhibits insulin release from the pancreatic beta-cell:
possible involvement of metabolic production of hydrogen sulfide, a
novel gasotransmitter. Diabetes. 55:1391–1397. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patel P, Vatish M, Heptinstall J, Wang R
and Carson RJ: The endogenous production of hydrogen sulphide in
intrauterine tissues. Reprod Biol Endocrinol. 7:102009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vitvitsky V, Thomas M, Ghorpade A,
Gendelman HE and Banerjee R: A functional transsulfuration pathway
in the brain links to glutathione homeostasis. J Biol Chem.
281:35785–35793. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chunyu Z, Junbao D, Dingfang B, Hui Y,
Xiuying T and Chaoshu T: The regulatory effect of hydrogen sulfide
on hypoxic pulmonary hypertension in rats. Biochem Biophys Res
Commun. 302:810–816. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bhatia M, Wong FL, Fu D, Lau HY, Moochhala
SM and Moore PK: Role of hydrogen sulfide in acute pancreatitis and
associated lung injury. FASEB J. 19:623–625. 2005.PubMed/NCBI
|
|
32
|
Wagner F, Scheuerle A, Weber S, et al:
Cardiopulmonary, histologic, and inflammatory effects of
intravenous Na2S after blunt chest trauma-induced lung contusion in
mice. J Trauma. 71:1659–1667. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Madden JA, Ahlf SB, Dantuma MW, Olson KR
and Roerig DL: Precursors and inhibitors of hydrogen sulfide
synthesis affect acute hypoxic pulmonary vasoconstriction in the
intact lung. J Applied Physiol. 112:411–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang J, Xie Y, Xu Y and Shao C:
Suppression of endogenous hydrogen sulfide contributes to the
radiation-induced bystander effects on hypoxic HepG2 cells. Radiat
Res. 178:395–402. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ishii I, Akahoshi N, Yu XN, et al: Murine
cystathionine gamma-lyase: complete cDNA and genomic sequences,
promoter activity, tissue distribution and developmental
expression. Biochem J. 381:113–123. 2004. View Article : Google Scholar
|
|
36
|
Cao Q, Mak KM and Lieber CS: Cytochrome
P4502E1 primes macrophages to increase TNF-alpha production in
response to lipopolysaccharide. Am J Physiol Gastrointest Liver
Physiol. 289:G95–107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oh JH, Lee TJ, Park JW and Kwon TK:
Withaferin A inhibits iNOS expression and nitric oxide production
by Akt inactivation and down-regulating LPS-induced activity of
NF-kappaB in RAW 264.7 cells. Eur J Pharmacol. 599:11–17. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moon EY and Park H: B cell activating
factor (BAFF) gene promoter activity depends upon co-activator,
p300. Immunobiology. 212:637–645. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Prakash H, Luth A, Grinkina N, et al:
Sphingosine kinase-1 (SphK-1) regulates Mycobacterium smegmatis
infection in macrophages. PloS One. 5:e106572010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li L, Whiteman M and Moore PK:
Dexamethasone inhibits lipopolysaccharide-induced hydrogen sulphide
biosynthesis in intact cells and in an animal model of endotoxic
shock. J Cell Mol Med. 13:2684–2692. 2009. View Article : Google Scholar
|
|
41
|
Moon EY, Lee JH, Lee JW, Song JH and Pyo
S: ROS/Epac1-mediated Rap1/NF-kappaB activation is required for the
expression of BAFF in Raw264.7 murine macrophages. Cell Signal.
23:1479–1488. 2011. View Article : Google Scholar : PubMed/NCBI
|