Introduction
Lysyl oxidase (LOX) is a copper-dependent amine
oxidase that is responsible for the post-translational modification
of collagen and elastin by oxidizing peptidyl lysines to
α-aminoadipic Δ-semialdehydes, also known as allysines (1,2).
The peptidyl aldehydes then undergo spontaneous condensation with
unreacted ɛ-amino groups or neighboring aldehyde groups of the
lysine residues to form covalent cross-linkages found in collagen
and elastin fibers (1,2). This essential role of LOX in the
biogenesis of connective tissues qualifies the enzyme as a
potentially pivotal target for the development of therapeutic
strategies for diseases associated with extracellular matrix
remodeling.
Five proteins of the LOX family (LOX, LOXL1, LOXL2,
LOXL3 and LOXL4) have been identified in humans, each containing
the characteristic C-terminal domains of the LOX family, such as a
copper-binding domain, residues for lysyl-tyrosyl quinone (LTQ),
and a cytokine receptor-like (CRL) domain (3–7).
In addition to the crosslinking activity on collagen and elastin,
several novel functions of the LOX family have been revealed,
including tumor suppression (8,9),
tumor progression (10–12), chemotaxis (13) and the modification of histone
proteins (14–16). Thus, the presence of a LOX
gene family suggests that the differential regulation of the
LOX paralogs may lead to diverse functions, most of which
are currently assigned to LOX.
In search for more human LOX paralogs, we
identified several expressed sequence tag (EST) clones that showed
an alternative exon-intron splice pattern from LOX. These
ESTs corresponded to a novel variant, LOX transcript variant
2 (LOX-v2), which lacks exon 1 of LOX and encodes a
188 amino acid-long polypeptide of 22 kDa. In this study, we report
that LOX-v2 functions as an amine oxidase toward collagen and
elastin, but with distinct tissue specificity from LOX. An
alternative promoter element present in the exon 1 region of
LOX may be responsible for the differential transcriptional
regulation of LOX-v2.
Materials and methods
EST database search
The human LOX cDNA sequence (GenBank
accession no. NM_002317) was used to search the human EST database
using the BLASTN program (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST).
RT-PCR analysis in human tissues
Human Multiple Tissue cDNA panels (Clontech,
Mountain View, CA, USA) were PCR-amplified with primers specific to
LOX and LOX-v2 using Ex Taq polymerase (Takara Bio,
Inc., Shiga, Japan). The LOX-specific forward primer was
5′-GATACGGCACTGG CTACTTC-3′ and the LOX-v2-specific forward
primer was 5′-AGGAGTAAGGGACCTAGAGG-3′. A common reverse primer
(5′-GAATATCTTGGTCGGCTGGG-3′) was used for both LOX and
LOX-v2 specific amplicons. The LOX-specific primers
were designed to amplify the region corresponding to the nucleotide
sequence 980–1229 of LOX, while the LOX-v2-specific
primers were designed to amplify the region which corresponded to
the 136–445 bp region of the LOX-v2 nucleotide sequence. The
PCR conditions consisted of 30 cycles at 94°C for 30 sec, 58°C for
30 sec and 72°C for 30 sec with a pre- denaturation at 94°C for 4
min and a final extension at 72°C for 5 min. The amplified PCR
products were analyzed by electrophoresis on a 2% agarose gel. The
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was
used as an internal control. All RT-PCR analyses were performed in
the linear range of amplification, and quantification was performed
using a Kodak Gel Logic Imaging System (Eastman Kodak, Rochester,
NY, USA).
Expression and purification of LOX and
LOX-v2 proteins
Using the previously reported pET21a-LOX-p construct
(17) as a PCR template, the
entire coding region of LOX-v2 was PCR-amplified by
Pfu turbo polymerase (Agilent Technologies, Santa Clara, CA,
USA) according to the instructions of the manufacturer. The
sequences of the oligonucleotide primers used for the construction
of the expression plasmids were 5′-GCGGCTAGCATGTCCATGTACAACCTGAG-3′
and 5′-GCGAAGCTTATACGGTGAAATTGTGCAGC-3′. A unique restriction site,
either NheI or HindIII, was introduced in each primer
for convenient subcloning. The PCR conditions consisted of 30
cycles at 94°C for 60 sec, 56°C for 60 sec and 72°C for 60 sec with
a predenaturation at 94°C for 4 min and a final extension at 72°C
for 7 min. The sequence of the resulting expression constructs were
confirmed by DNA-sequencing analysis using a Cycle Sequencing Ready
Reaction kit (Life Technologies, Paisley, UK) according to the
manufacturer’s instructions. The E. coli strain BL21 (DE3)
(Novagen, Madison, WI, USA) was used for transformation of the LOX
or LOX-v2 expression constructs. The recombinant proteins were
expressed, purified, and refolded into an enzymatically active form
as previously reported (17).
Amine oxidase assays
The amine oxidase activity of the LOX and LOX-v2
recombinant proteins was assessed using a peroxidase-coupled
fluorometric assay with the Amplex red hydrogen peroxide assay kit
(Molecular Probes, Eugene, OR, USA) as previously described
(18). Each reaction contained 10
μg of purified LOX or LOX-v2 recombinant protein and 20 pmol of
substrate in a reaction volume of 200 μl. Parallel assays were
performed in the absence or presence of 1 mM β-aminopropionitrile
(BAPN) for 1 h at 37°C. Bovine neck ligament elastin and calfskin
type I collagen (Sigma-Aldrich, St. Louis, MO, USA) were used as
substrates for the amine oxidase assays. Fluorescence was measured
using a fluorescence spectrophotometer (Molecular Devices,
Sunnyvale, CA, USA) with excitation and emission wavelengths of 500
and 650 nm, respectively. Total amine oxidase activity was
expressed as nM of H2O2 produced per μg of
LOX or LOX-v2 recombinant protein, calculated by interpolation with
fluorescence values from an H2O2 calibration
curve.
Promoter analysis
For the construction of reporter plasmids, the
5′-flanking regions of exons 1 and 2 of LOX were separately
PCR-amplified from human genomic DNA using Ex Taq polymerase
(Takara Bio, Inc.). For LOX promoter assays, a 1,424 bp
fragment from nucleotide position +112 to −1312 was amplified. For
LOX-v2 promoter assays, a series of DNA fragments of 1,044,
792 and 169 bp, each corresponding to the 5′-flanking region of
exon 2 of LOX, was generated. The sequences of the
oligonucleotide primers used are available upon request. The
PCR-amplified genomic DNA fragments were ligated into a
promoterless luciferase reporter plasmid pGL3-basic (Promega,
Madison, WI, USA). Promoter assays were performed on HEK 293 cells
as previously described (19).
The pGL3-SV plasmid containing the strong T-antigen promoter of
SV40 was used as a positive control for the assays, while a
promoterless plasmid pGL3-Basic was used as a negative control. The
luciferase activity of each construct was expressed as a ratio to
that of the promoterless control. All transfections were performed
in quadruplicate. The results are expressed as the means ± standard
deviation (SD).
Results
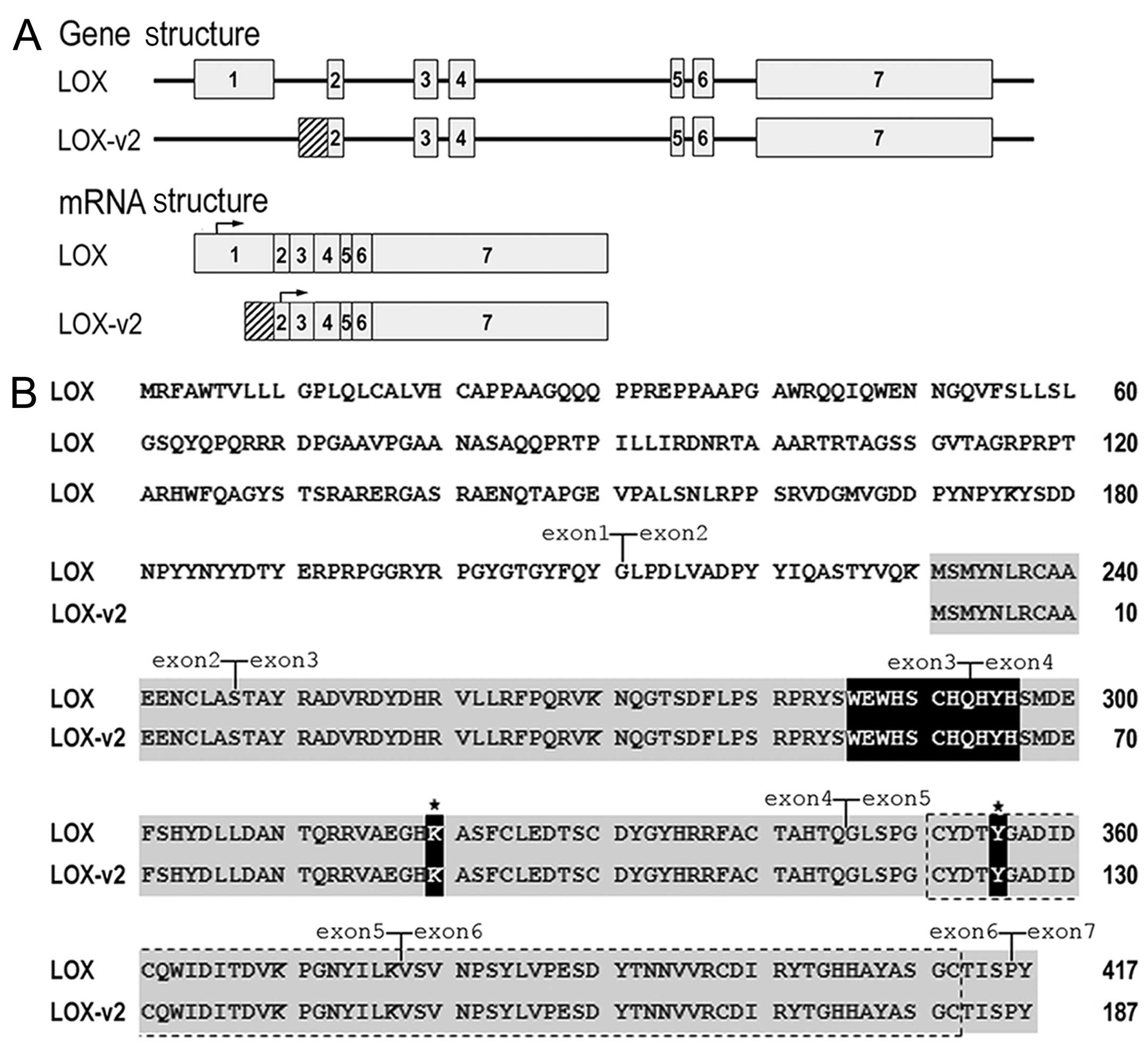
Distinct exon-intron structure of LOX-v2
from LOX
A search of the human EST databases for sequences
similar to the human LOX gene resulted in the identification
of several ESTs (AW958146, AL598579, BP205326, DA655672, DA720490
and DA883598). These ESTs showed a distinct exon-intron splice
pattern from LOX and corresponded to human LOX-v2
that was recently reported in GenBank (accession no. NM_001178102).
Exon-intron structure analysis of LOX-v2 revealed that this
novel variant is composed of 6 exons, lacking exon 1 of LOX
(Fig. 1A). At the 5′-end,
LOX-v2 mRNA contains an additional 222 bp sequence from the
5′-flanking intronic region of exon 2 of LOX (Fig. 1A).
The LOX-v2 mRNA is at least 4,393 bp in
length, encoding a 187 amino acid-long polypeptide with a
calculated molecular mass of 21.6 kDa. The 5′-UTR of the
LOX-v2 mRNA is at least 281 bp in length. The coding region
is 564 bp and the 3′-UTR is 3,548 bp with at least 3 poly(A)
signals. The methionine at codon 231 in LOX was used as the
initiation codon in the deduced amino acid sequence of LOX-v2
(Fig. 1B). The deduced LOX-v2
polypeptide contains the characteristic C-terminal domains of the
LOX family, including the copper-binding domain, the residues for
the LTQ cofactor, and the CRL domain, but does not contain the
N-terminal propeptide region that corresponds to codons
Met1 to Gly168 (Fig. 1B).
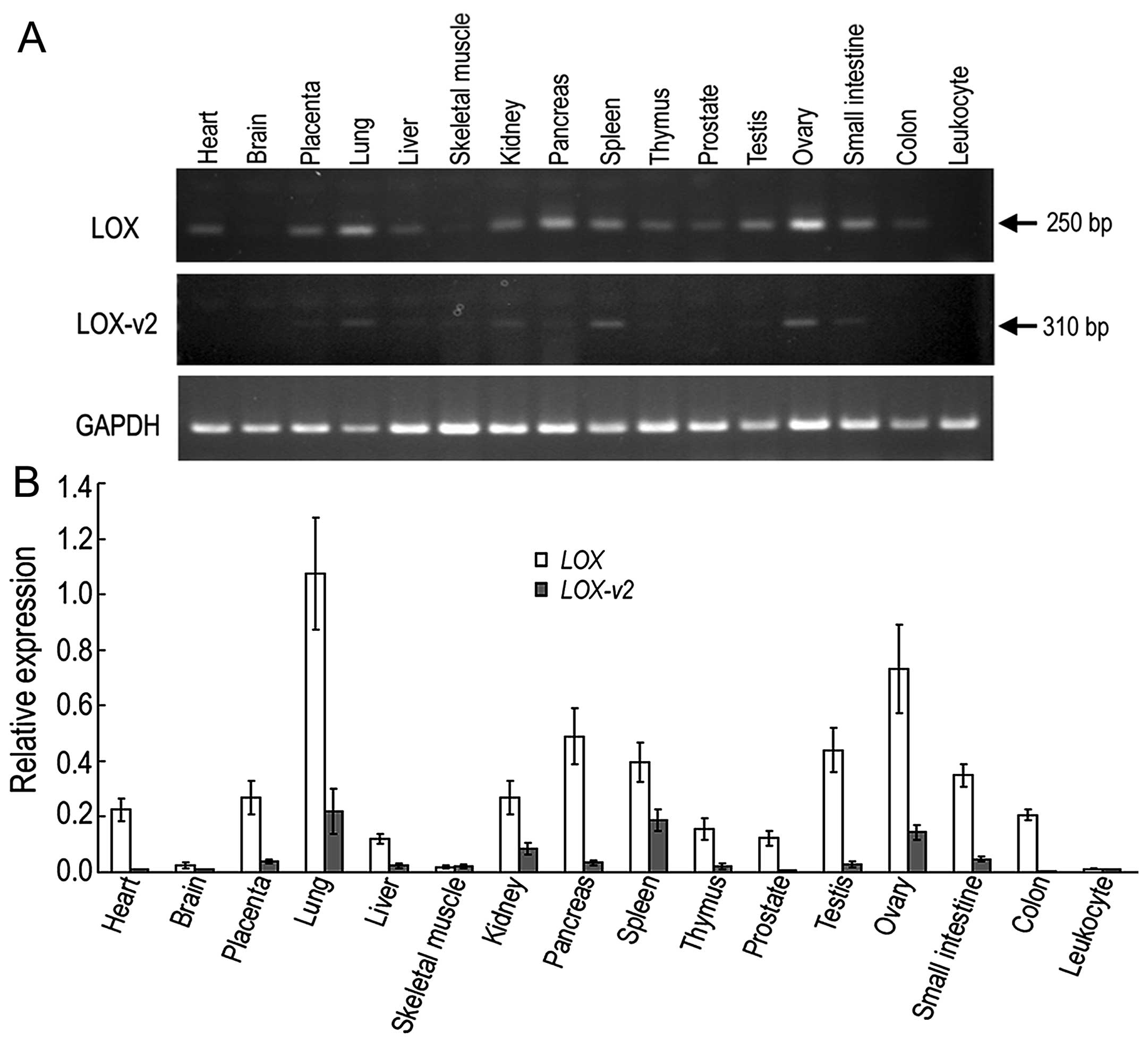
Different tissue-specificity of LOX and
LOX-v2 in human tissue
To compare the expression of LOX and
LOX-v2 in human tissue, RT-PCR was performed with
poly(A)+ RNA isolated from 16 different human tissues.
The forward primer for the LOX-specific amplicon was derived
from exon 1 of LOX, which was deleted in the LOX-v2
mRNA. By contrast, the forward primer for the
LOX-v2-specific amplicon was derived from the 5′-flanking
intronic region of exon 2 of LOX, which was included in the
LOX-v2 mRNA, but not in the LOX mRNA. A reverse
primer from exon 3 of LOX was used for both LOX- and
LOX-v2-specific PCR amplicons. In general, the expression of
LOX-v2 was lower than that of LOX in most tissues
tested, but there were some differences in expression patterns. For
instance, the LOX-specific band was more ubiquitously
detected in the tissues tested apart from the brain and leukocytes,
whereas the LOX-v2-specific band was observed in a more
tissue-specific pattern, showing relatively higher levels in the
lungs, kidneys, spleen, ovaries and small intestine (Fig. 2).
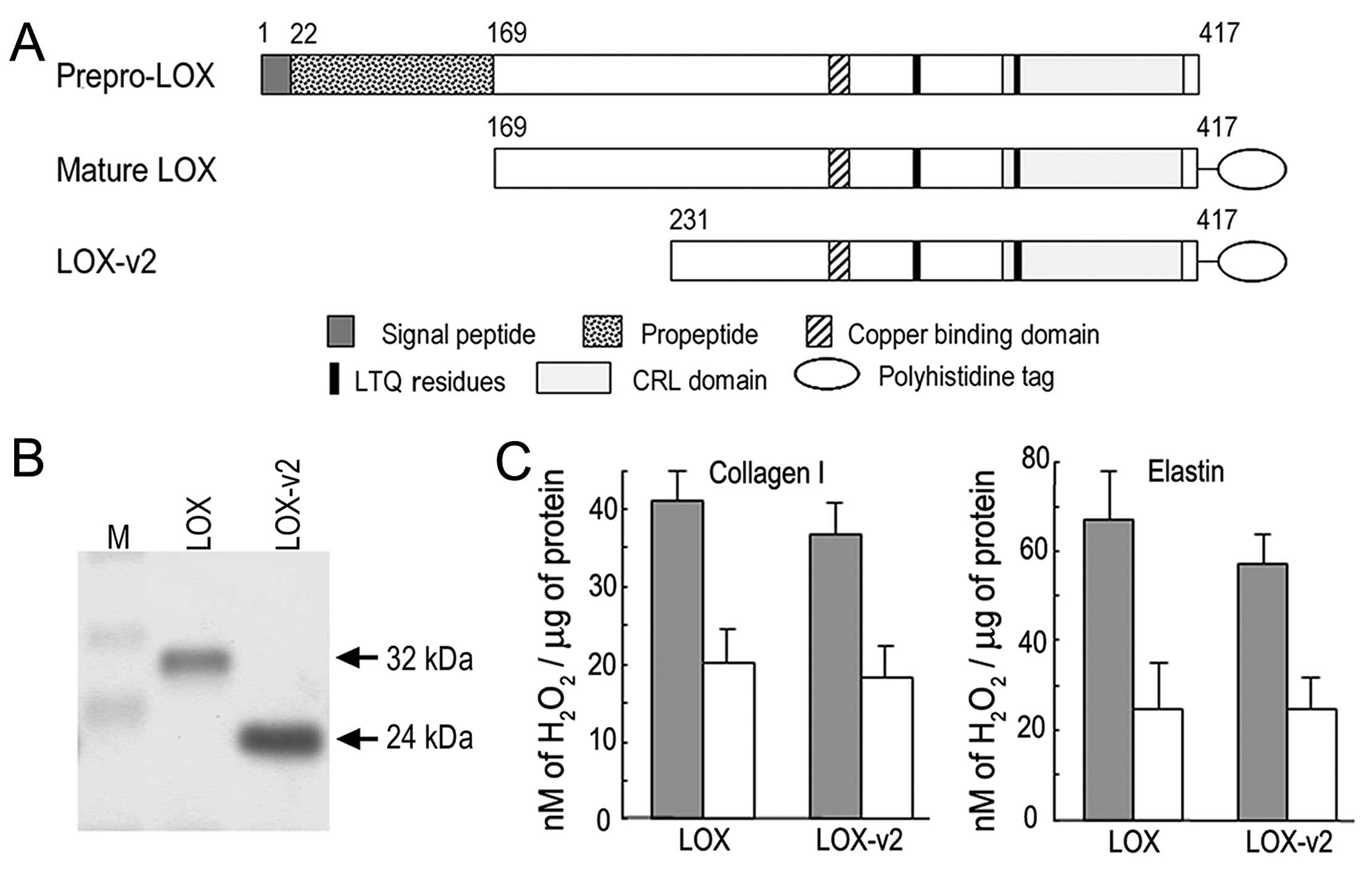
Amine oxidase activity of LOX-v2
To determine whether LOX-v2 functions as an amine
oxidase, we expressed and purified the LOX-v2 protein in a
hexa-histidine recombinant form as previously described for LOX and
other LOX family proteins (17,19,20) (Fig.
3A). For comparison with LOX for amine oxidase activity, the
LOX protein was also expressed and purified in a hexa-histidine
recombinant form as previously reported (17). The apparent sizes of the purified
recombinant proteins were in agreement with the deduced molecular
mass; 32 and 24 kDa for the recombinant LOX and LOX-v2 proteins,
respectively (Fig. 3B).
The recombinant LOX and LOX-v2 proteins were
assessed for amine oxidase activity toward collagen type I and
elastin using a peroxidase-coupled fluorometric assay. Both the
recombinant LOX and LOX-v2 proteins showed significant levels of
amine oxidase activity toward both collagen type I and elastin. The
amine oxidase activity of LOX and LOX-v2 was sensitive to BAPN, an
irreversible inhibitor of LOX (Fig.
3C). The LOX protein showed a slightly higher amine oxidase
activity than the LOX-v2 protein; however, the difference was not
statistically significant (Fig.
3C). These results indicate that LOX-v2 also functions as an
amine oxidase toward physiological substrates of LOX, such as
collagen type I and elastin.
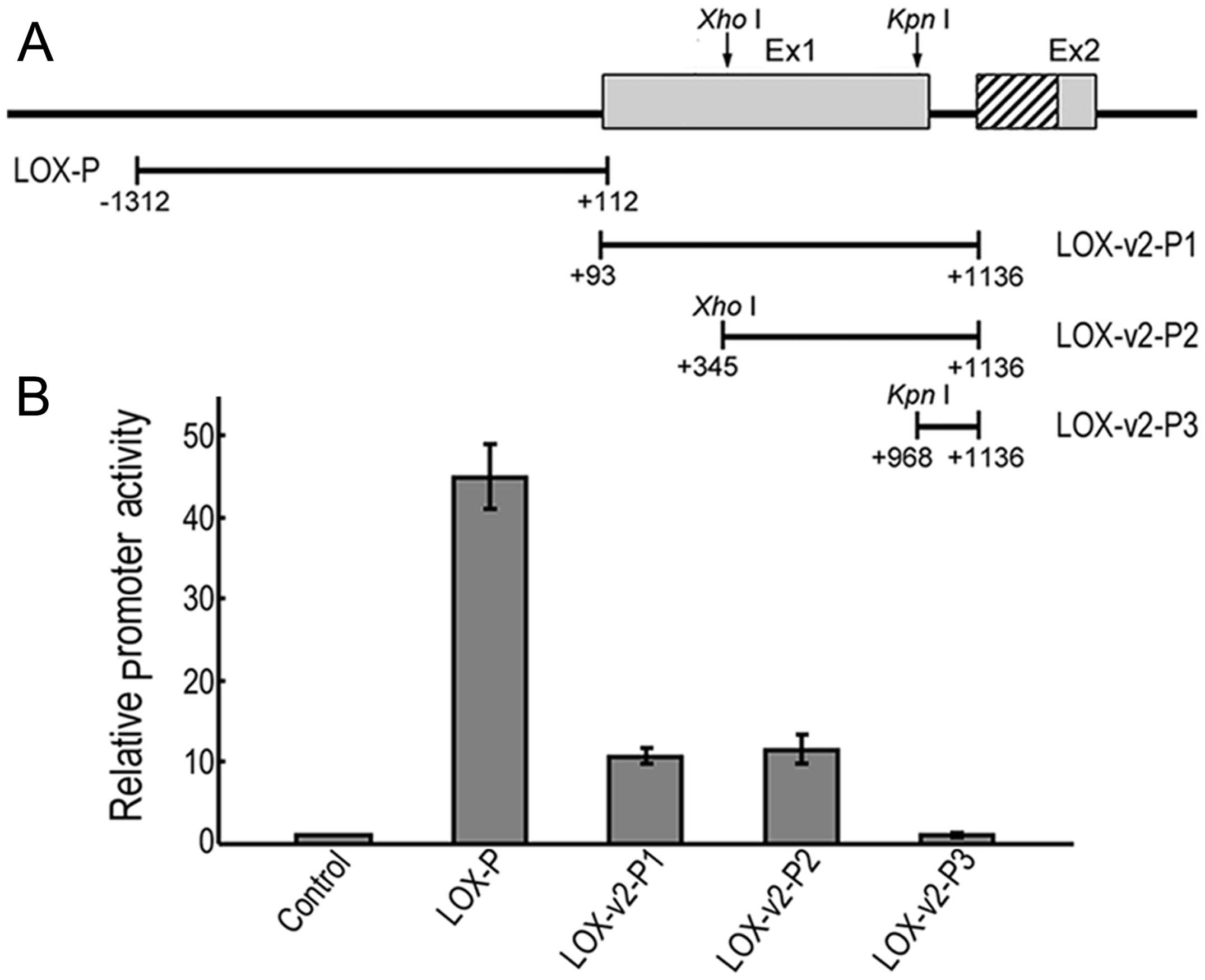
An alternative promoter for the
expression of LOX-v2
To establish whether the 5′-flanking region of exon
2 of LOX is sufficient for transcriptional initiation of
LOX-v2, a series of promoter reporter constructs was
generated, each containing a different portion of the 5′-proximal
genomic region of exon 2 (Fig.
4A). The 5′-flanking genomic region of exon 1 of LOX was
also examined for comparison. In the HEK 293 cells, the LOX
promoter construct containing a 1,424 bp fragment of the
5′-proximity of exon 1 showed approximately 45-fold higher promoter
activity than the promoterless negative control (Fig. 4B). The LOX-v2-promoter
constructs 1 and 2, each containing a 1,044 or 792 bp fragment of
the 5′-proximal region of exon 2 of LOX, respectively,
showed 11 to 12-fold higher promoter activities than the negative
control (Fig. 4B). By contrast,
the LOX-v2-promoter construct 3 containing a 169 bp fragment
of the 5′-proximal region of exon 2 of LOX showed no
detectable promoter activity (Fig.
4B). These results indicate that an alternative promoter
element required for the transcriptional activation of
LOX-v2 may reside in the region corresponding to exon 1 of
LOX, although the promoter activity of this alternative
promoter is significantly weaker than that of the promoter required
for the transcriptional activation of LOX. Both LOX
and LOX-v2 promoter regions contained no typical canonical
elements, TATA and CAAT boxes, in the proximity of the
transcription initiation site, but displayed high CpG-rich
sequences, as previously obsered in other LOX family genes
(4,5,19).
Discussion
From the time of the discovery of LOX in 1968
(21), its molecular weight has
remained uncertain as this enzyme tends to extensively aggregate
during purification. Utilization of urea extraction buffers has
allowed for the isolation of chromatographically distinct variants
of LOX with a molecular weight varying from 28 to 32 kDa, which
showed subtle differences in amino acid compositions (22–24). Four LOX-like genes (LOXL1–4) have
been identified, each encoding the characteristic C-terminal
domains of LOX; however, the expected molecular weights of these
LOX-like proteins were significantly larger (63–87 kDa) than the
molecular weight of the secreted mature LOX (4–7).
These findings suggested that the previously identified LOX
variants were neither derived from post-translational modifications
of the LOX precursor nor encoded by different genes. Furthermore,
smaller sizes of LOX ranging from 24 to 28 kDa have been observed
in various cultured cells and tissues. A 24 kDa band was observed
in chromatographically purified fractions of bovine aorta (23) and a 25–28 kDa band was detected in
the nuclear extract of rat aorta smooth muscle cells (25), adding even more puzzling
complexity on the size of the mature form of LOX. Thus, the
identification of LOX-v2 may suggest that the smaller sizes of LOX
previously identified may be derived, at least in part, from LOX-v2
that is expressed by an alternative promoter located in the
LOX gene.
The human LOX precursor is synthesized as a 48 kDa
prepro-protein of 417 amino acids and following cleavage of the
signal peptide composed of 21 amino acids, the proprotein undergoes
extensive post-translational modifications throughout the secretion
pathway (26). After being
secreted into the extracellular space, the propeptide region of 147
amino acids is cleaved off by bone morphogenic protein-1 (BMP-1)
between residues Gly168 and Asp169, resulting
in an enzymatically active 32 kDa protein of 249 amino acids
(26). Previous studies have
reported that LOX is not only secreted in the extracellular matrix,
but is also present in intranuclear locations, suggesting a novel
function for LOX in the alteration of chromatin structure and the
modification of histone proteins (25,27,28). The absence of the propeptide
region in LOX-v2 suggests that this novel variant of LOX may be
subject to different cellular processing and post-translational
modification from LOX, playing a distinct functional role in
different cellular compartments.
Since the discovery that LOX functions as the
ras recision gene in the phenotypic reversion of ras-transformed
cells (8), perplexing expression
patterns of LOX have been observed in various neoplastic
tissues and cells. LOX upregulation has been detected in
breast (29) and renal cell
carcinomas (30), while
LOX downregulation has been observed in prostate (31), bronchogenic (32) and gastric carcinomas (33). Furthermore, the upregulation of
LOX is more evident in invasive cells of breast carcinomas
than in non-metastatic cells (34). These results suggest that
LOX plays paradoxical roles in the suppression or
progression of tumorigenesis, depending on tumor status and type.
In its role as a tumor suppressor, the propeptide domain of LOX,
not the C-terminal domains, has been shown to inhibit ras-dependent
cell transformation (35).
Furthermore, the propeptide domain has been reported to reduce the
invasive phenotype of Her-2/neu-driven breast cancer cells
(36), to inhibit the
fibronectin-mediated activation of focal adhesion kinase in breast
cancer cells (37), and to
interfere with fibroblast growth factor-2-induced proliferation in
prostate cancer cells (38). By
contrast, the proteolytically processed 32 kDa LOX that does not
contain the propeptide region has been shown to play a pivotal role
in the hypoxia-induced metastasis of breast and head and neck
carcinomas (10). Thus, the
non-existence of the propeptide domain in LOX-v2 may lead to a
plausible hypothesis that the tumor progression activity currently
assigned to LOX may be attributed, at least to a certain extent, to
LOX-v2, while LOX with the propeptide domain may be more associated
with tumor suppression. Depending on the cell status and signals,
the LOX gene may be subject to alternative promoter
activation, resulting in either LOX or LOX-v2 mRNA,
which may function as a regulatory mechanism for determining the
functional role of LOX in tumorigenesis. Studies on cellular
processing and the compartmentalization of LOX-v2 along with
detailed expression analysis in various tumor types are required in
order to determine the specific functional roles of LOX-v2, which
may help broaden our understanding of the perplexing functional
roles of LOX in tumorigenesis.
Acknowledgements
This study was supported by a grant from the
National Research Foundation (NRF) of Korea funded by the Ministry
of Education, Science and Technology (2011-0030130).
References
|
1
|
Kagan HM and Trackman PC: Properties and
function of lysyl oxidase. Am J Respir Cell Mol Biol. 5:206–210.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith-Mungo LI and Kagan HM: Lysyl
oxidase: Properties, regulation and multiple functions in biology.
Matrix Biol. 16:387–398. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hämäläinen ER, Jones TA, Sheer D, Taskinen
K, Pihlajaniemi T and Kivirikko KI: Molecular cloning of human
lysyl oxidase and assignment of the gene to chromosome 5q23.3–31.2.
Genomics. 11:508–516. 1991.PubMed/NCBI
|
|
4
|
Kim Y, Boyd CD and Csiszar K: A new gene
with sequence and structural similarity to the gene encoding human
lysyl oxidase. J Biol Chem. 270:7176–7182. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jourdan-Le Saux C, Tronecker H, Bogic L,
Bryant-Greenwood GD, Boyd CD and Csiszar K: The LOXL2 gene encodes
a new lysyl oxidase-like protein and is expressed at high levels in
reproductive tissues. J Biol Chem. 274:12939–12944. 1999.PubMed/NCBI
|
|
6
|
Mäki JM and Kivirikko KI: Cloning and
characterization of a fourth human lysyl oxidase isoenzyme. Biochem
J. 355:381–387. 2001.PubMed/NCBI
|
|
7
|
Asuncion L, Fogelgren B, Fong KS, Fong SF,
Kim Y and Csiszar K: A novel human lysyl oxidase-like gene (LOXL4)
on chromosome 10q24 has an altered scavenger receptor cysteine rich
domain. Matrix Biol. 20:487–491. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kenyon K, Contente S, Trackman PC, Tang J,
Kagan HM and Friedman RM: Lysyl oxidase and rrg messenger RNA.
Science. 253:8021991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Giampuzzi M, Botti G, Cilli M, Gusmano R,
Borel A, Sommer P and Di Donato A: Down-regulation of lysyl
oxidase-induced tumorigenic transformation in NRK-49F cells
characterized by constitutive activation of ras proto-oncogene. J
Biol Chem. 276:29226–29232. 2001. View Article : Google Scholar
|
|
10
|
Erler JT, Bennewith KL, Nicolau M,
Dornhöfer N, Kong C, Le QT, Chi JT, Jeffrey SS and Giaccia AJ:
Lysyl oxidase is essential for hypoxia-induced metastasis. Nature.
440:1222–1226. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schietke R, Warnecke C, Wacker I, Schödel
J, Mole DR, Campean V, Amann K, Goppelt-Struebe M, Behrens J,
Eckardt KU and Wiesener MS: The lysyl oxidases LOX and LOXL2 are
necessary and sufficient to repress E-cadherin in hypoxia: insights
into cellular transformation processes mediated by HIF-1. J Biol
Chem. 285:6658–6669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peinado H, Del Carmen Iglesias-de la Cruz
M, Olmeda D, Csiszar K, Fong KS, Vega S, Nieto MA, Cano A and
Portillo F: A molecular role for lysyl oxidase-like 2 enzyme in
snail regulation and tumor progression. EMBO J. 24:3446–3458. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lucero HA, Ravid K, Grimsby JL, Rich CB,
DiCamillo SJ, Mäki JM, Myllyharju J and Kagan HM: Lysyl oxidase
oxidizes cell membrane proteins and enhances the chemotactic
response of vascular smooth muscle cells. J Biol Chem.
283:24103–24117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kagan HM, Williams MA, Calaman SD and
Berkowitz EM: Histone H1 is a substrate for lysyl oxidase and
contains endogenous sodium borotritide-reducible residues. Biochem
Biophys Res Commun. 115:186–192. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Giampuzzi M, Oleggini R and Di Donato A:
Demonstration of in vitro interaction between tumor suppressor
lysyl oxidase and histones H1 and H2: definition of the regions
involved. Biochim Biophys Acta. 1647:245–251. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Herranz N, Dave N, Millanes-Romero A,
Morey L, Díaz VM, Lórenz-Fonfría V, Gutierrez-Gallego R, Jerónimo
C, Di Croce L, García de Herreros A and Peiró S: Lysyl oxidase-like
2 deaminates lysine 4 in histone H3. Mol Cell. 46:369–376. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jung ST, Kim MS, Seo JY, Kim HC and Kim Y:
Purification of enzymatically active human lysyl oxidase and lysyl
oxidase-like protein from Escherichia coli inclusion bodies.
Protein Expr Purif. 31:240–246. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Palamakumbura AH and Trackman PC: A
fluorometric assay for detection of lysyl oxidase enzyme activity
in biological samples. Anal Biochem. 300:245–251. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee JE and Kim Y: A tissue-specific
variant of the human lysyl oxidase-like protein 3 (LOXL3) functions
as an amine oxidase with substrate specificity. J Biol Chem.
281:37282–37290. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim MS, Kim SS, Jung ST, Park JY, Yoo HW,
Ko J, Csiszar K, Choi SY and Kim Y: Expression and purification of
enzymatically active forms of the human lysyl oxidase-like protein
4. J Biol Chem. 278:52071–52074. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pinnell SR and Martin G: The cross-linking
of collagen and elastin: enzymatic conversion of lysine in peptide
linkage to alpha-aminoadipic-delta-semialdehyde (allysine) by an
extract from bone. Proc Natl Acad Sci USA. 61:708–716. 1968.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kagan HM, Sullivan KA, Olsson TA III and
Cronlund AL: Purification and properties of four species of lysyl
oxidase from bovine aorta. Biochem J. 177:203–214. 1979.PubMed/NCBI
|
|
23
|
Sullivan KA and Kagan HM: Evidence for
structural similarities in the multiple forms of aortic and
cartilage lysyl oxidase and a catalytically quiescent aortic
protein. J Biol Chem. 257:13520–13526. 1982.PubMed/NCBI
|
|
24
|
Kuivaniemi H, Savolainen ER and Kivirikko
KI: Human placental lysyl oxidase. Purification, partial
characterization, and preparation of two specific antisera to the
enzyme. J Biol Chem. 259:6996–7002. 1984.PubMed/NCBI
|
|
25
|
Li W, Nellaiappan K, Strassmaier T, Graham
L, Thomas KM and Kagan HM: Localization and activity of lysyl
oxidase within nuclei of fibrogenic cells. Proc Natl Acad Sci USA.
94:12817–12822. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cronshaw AD, Fothergill-Gilmore LA and
Hulmes DJS: The proteolytic processing site of the precursor of
lysyl oxidase. Biochem J. 306:279–284. 1995.PubMed/NCBI
|
|
27
|
Hayashi K, Fong KS, Mercier F, Boyd CD,
Csiszar K and Hayashi M: Comparative immunocytochemical
localization of lysyl oxidase (LOX) and the lysyl oxidase-like
(LOXL) proteins: changes in the expression of LOXL during
development and growth of mouse tissues. J Mol Histol. 35:845–855.
2004. View Article : Google Scholar
|
|
28
|
Mello ML, Contente S, Vidal BC, Planding W
and Schenck U: Modulation of ras transformation affecting chromatin
supraorganization as assessed by image analysis. Exp Cell Res.
220:374–382. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kirschmann DA, Seftor EA, Nieva DR,
Mariano EA and Hendrix MJ: Differentially expressed genes
associated with the metastatic phenotype in breast cancer. Breast
Cancer Res Treat. 55:127–136. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stassar MJ, Devitt G, Brosius M, Rinnab L,
Prang J, Schradin T, Simon J, Petersen S, Kopp-Schneider A and
Zoller M: Identification of human renal cell carcinoma associated
genes by suppression subtractive hybridization. Br J Cancer.
85:1372–1382. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ren C, Yang G, Timme TL, Wheeler TM and
Thompson TC: Reduced lysyl oxidase messenger RNA levels in
experimental and human prostate cancer. Cancer Res. 58:1285–1290.
1998.PubMed/NCBI
|
|
32
|
Woznick AR, Braddock AL, Dulai M, Seymour
ML, Callahan RE, Welsh RJ, Chmielewski GW, Zelenock GB and Shanley
CJ: Lysyl oxidase expression in bronchogenic carcinoma. Am J Surg.
189:297–301. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kaneda A, Wakazono K, Tsukamoto T,
Watanabe N, Yagi Y, Tatematsu M, Kaminishi M, Sugimura T and
Ushijima T: Lysyl oxidase is a tumor suppressor gene inactivated by
methylation and loss of heterozygosity in human gastric cancers.
Cancer Res. 64:6410–6415. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kirschmann DA, Seftor EA, Fong SF, Nieva
DR, Sullivan CM, Edwards EM, Sommer P, Csiszar K and Hendrix MJ: A
molecular role for lysyl oxidase in breast cancer invasion. Cancer
Res. 62:4478–4483. 2002.PubMed/NCBI
|
|
35
|
Palamakumbura AH, Jeay S, Guo Y, Pischon
N, Sommer P, Sonenshein GE and Trackman PC: The propeptide domain
of lysyl oxidase induces phenotypic reversion of ras-transformed
cells. J Biol Chem. 279:40593–40600. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Min C, Yu Z, Kirsch KH, Zhao Y, Vora SR,
Trackman PC, Spicer DB, Rosenberg L, Palmer JR and Sonenshein GE: A
loss-of-function polymorphism in the propeptide domain of the LOX
gene and breast cancer. Cancer Res. 69:6685–6693. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao Y, Min C, Vora SR, Trackman PC,
Sonenshein GE and Kirsch KH: The lysyl oxidase pro-peptide
attenuates fibronectin-mediated activation of focal adhesion kinase
and p130Cas in breast cancer cells. J Biol Chem. 284:1385–1393.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Palamakumbura AH, Vora SR, Nugent MA,
Kirsch KH, Sonenshein GE and Trackman PC: Lysyl oxidase propeptide
inhibits prostate cancer cell growth by mechanisms that target
FGF-2-cell binding and signaling. Oncogene. 28:3390–3400. 2009.
View Article : Google Scholar : PubMed/NCBI
|