Introduction
Syndecan (SDC)4 is a cell surface heparan sulfate
proteoglycan (HSPG). Its heparan sulfate (HS) chains and core
protein could control the stability, movement and reception of
diffusible heparin-binding growth factors (1). In addition, it can form physical
connections between the extracellular matrix (ECM) and
intracellular signaling to affect the growth and differentiation of
a number of tissues and organs (2,3).
SDC4 is highly complex by virtue of its external
side chains, and thus interacts with a variety of ligands, such as
vascular endothelial growth factors (VEGFs), platelet-derived
growth factors (PDGFs) and fibroblast growth factors (FGFs)
(4). At the cell membrane, SDC4
stabilizes the interactions between ligands and receptors by
forming a ternary complex, which has been demonstrated in several
signaling pathways (5,6). Proteolytic cleavage and shedding of
its extracellular domain can spread FGF signaling to adjacent
cells, thus regulating the local reception of FGF signaling
activity (7). Its cytoplasmic
domains can also initiate FGF-induced signaling independently of
FGFRs through the activation of Rho GTPases, such as Rac1, which
plays an essential role in the dental epithelium, involving
cell-matrix interactions and matrix biomineralization (8,9).
FGF signaling plays critical roles in tooth
development (10). In molar tooth
development, FGF4, 8 and 9 act as epithelial signals, mediating
inductive interactions between the dental epithelium and the
mesenchyme during the initiation of tooth development and the
regulation of tooth shape (11).
FGF10 signaling is also important for the continuous growth of
murine incisors as it maintains dental epithelial stem cells and
regulates enamel formation. FGF10 knockout mice lack cervical loop
structure and have enamel defects (12,13).
In the present study, to determine the roles of SDC4
in rodent tooth development, particularly in dental epithelial cell
differentiation, the expression patterns of SDC4 during the late
bell stage of development of molar tooth germs and its localization
in postnatal mice incisors were examined. The rat dental epithelial
cell line, HAT-7, was used to examine the interactions between SDC4
and FGF10 and their effects on dental epithelial cell proliferation
and differentiation. Our results reveal the temporospatial
expression of SDC4 in molars and incisors during amelogenesis,
providing some evidence that FGF10 signaling interacts with SDC4 to
affect dental epithelial cell behavior.
Materials and methods
Animals and tissue preparation
The animal experimental protocol was approved by the
Ethics Committee of West China College of Stomatology, Sichuan
University, Chengdu, China. C57BL/6J mice were purchased from the
experimental Animal Laboratory of Sichuan University. The
appearance of a vaginal plug was designated as day 0. Heads were
dissected from embryonic mice on embryonic day 18 (E18) and from
newborn mice on postnatal days (P)2, 4 and 7. These heads were
fixed in freshly prepared 4% paraformaldehyde overnight at 4°C, cut
in half along the midline, dehydrated, embedded in paraffin wax,
serially sectioned at 6 μm and either stained with hematoxylin and
eosin (H&E) or used in immunohistochemistry.
Immunohistochemistry
Tooth germs (incisors and molars), sectioned as
described above, were incubated with rabbit polyclonal anti-SDC4
antibody (1:500; Abcam, Cambridge, MA, USA). The sections were then
stained using a 3,3′-diaminobenzidine DAB kit (Dako, Carpinteria,
CA, USA). Immunohistochemical control was performed by replacing
the primary antibody with phosphate-buffered saline (PBS). These
immunostained sections contained no specific immunoreactions and
were counterstained with hematoxylin. The immune reactions were
visualized under a light microscope (Olympus BX43F; JEOL, Tokyo,
Japan).
Cell culture and transfection with small
interfering RNA (siRNA)
HAT-7 cell line was graciously provided by Professor
Hidemitsu Harada, Department of Oral Anatomy and Developmental
Biology, Osaka University Graduate School of Dentistry, Osaka,
Japan. The HAT-7 cells were plated with Dulbecco’s modified Eagle’s
medium/F-12 (DMEM/F-12; Gibco-BRL, Grand Island, NY, USA)
supplemented with 10% fetal bovine serum (FBS) and
penicillin-streptomycin. The cells were grown in a humidified
atmosphere at 37°C with 5% CO2, and the medium was
changed every 3 days. For transient transfections, the cells were
cultured in medium without antibiotics 24 h prior to transfection.
Three different nucleotides (SDC4 siRNA 1, 2 and 3) targeting rat
SDC4 mRNA (GenBank accession no. NM_012649.2) were designed and
tested for silencing. Their sequences are shown in Table I. The cells were transfected with
the siRNAs (50 nM) and Lipofectamine® RNAiMAX
transfection reagent (Invitrogen, Carlsbad, CA, USA) (1:1 v/v).
Non-silencing siRNA with no homology to any known mammalian gene
was used as a negative control. The RNAi-mediated knockdown of SDC4
expression was verified by quantitative reverse
transcription-polymerase chain reaction (RT-qPCR) and western blot
analysis.
 | Table ISequences of the 3 different
nucleotides (SDC4 siRNA 1, 2, 3) targeting rat SDC4 mRNA expression
(GenBank accession no. NM_012649.2). |
Table I
Sequences of the 3 different
nucleotides (SDC4 siRNA 1, 2, 3) targeting rat SDC4 mRNA expression
(GenBank accession no. NM_012649.2).
| Nucleotide | Sequence (5′→3′) |
|---|
| siRNA 1 |
| Sense strand |
GGCAGAUACUUCUCUGGAGdTdT |
| Antisense
strand |
dTdTCCGUCUAUGAAGAGACCUC |
| siRNA 2 |
| Sense strand |
CCUUGGUGCCACUAGAUAAdTdT |
| Antisense
strand |
dTdTGGAACCACGGUGAUCUAUU |
| siRNA 3 |
| Sense strand |
GGUCUUGGCAGCUCUGAUUdTdT |
| Antisense
strand |
dTdTCCAGAACCGUCGAGACUAA |
Cell proliferation assay
Cells transfected with SDC4 siRNA were used 24 h
after transfection, and recombinant mouse FGF10 at 10 ng/ml
(R&D Systems, Minneapolis, MN, USA) was added for a further 48
h. Cell proliferation was assayed with a BrdU incorporation
experiment. Briefly, cells on the coverglass were incubated with
BrdU (10 μM; Sigma-Aldrich, St. Louis, MO, USA) for 1 h and fixed
by immersing the cells in 4% paraformaldehyde for 10 min. DNA
denaturation was performed in 2N HCI for 30 min at 37°C prior to
incubation with 6 μg/ml anti-BrdU antibody (Millipore, Billerica,
CA, USA) overnight at 4°C. Alexa-Fluor 488-labeled goat anti-mouse
IgGs (1:1,000; Invitrogen) was used as a secondary antibody. After
counterstaining with 4′,6-diamidino-2-phenylindole (DAPI) (1:1,000;
Sigma-Aldrich), the number of BrdU-positive cells was quantified
under a fluorescence microscope (Leica, Wetzlar, Germany) and the
results are presented as a ratio.
RT-qPCR
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen). cDNA was prepared from 5 μg total RNA using a
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham,
MA, USA). After the cells were transfected with SDC4 siRNAs, the
expression of SDC4 mRNA was quantified by quantitative PCR using a
SYBR-Green probe and used to screen the most effective siRNA. After
the cells were transfected with siRNA 1 for 48 h, the expression of
SDC1, 2, 3 and 4 was also assayed. Amelogenesis-related gene
expression was assessed by RT-qPCR following transfection and/or
FGF10 treatment. The target gene primers used are listed in
Table II. The genes of interest
were normalized to the levels of glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) and presented relative to the control
levels.
 | Table IIOligonucleotide primer sequences
utilized in RT-qPCR. |
Table II
Oligonucleotide primer sequences
utilized in RT-qPCR.
| Gene name | Primer sequence
(5′→3′) | Product size
(bp) |
|---|
| SDC1 | F:
TTCTCATTGTGGGGAGGTCTA
R: CTGCTGGGGCTCTAAAACAG | 82 |
| SDC2 | F:
CACCGAGAAACATTCAGACAA
R: GGCAAAGAGAAAGCCAATCA | 88 |
| SDC3 | F:
TGCGGTTCATTCCTGACATA
R: GAGTTCCTCAAACGGGGTATC | 98 |
| SDC4 | F:
GGGCAAGAAACCCATCTACA
R: TGAAGTCCAAGCAGCACTCA | 100 |
| AMBN | F:
GAGAAAGGAGAGGGTCCAGAAG
R: GTCATTGGGGAAAGCAAGAAGT | 126 |
| AMGN | F:
ACCTCTGCCTCCACTGTTCTC
R: ACTTCTTCCCGCTTGGTCTT | 102 |
| ENAM | F:
GGTGTCTTCCCTCTCCCTAAA
R: AGTGGTTTGCCATTGTCTTTCT | 141 |
| KLK4 | F:
CCGAACTACAATGACCCTTCTT
R: TCAGATGCTACCGAGAGATTCA | 209 |
| MMP20 | F:
GCCTTGCTGTCCTTGTCAC
R: GAGGTGGTAGTTGCTCCTGAAG | 95 |
| RUNX2 | F:
GAAATGCCTCTGCTGTTATGAA
R: CCGTTATGGTCAAAGTGAAACTC | 102 |
| GAPDH | F:
ATCATCCCTGCCTCTACTGG
R: CTGCTTCACCACCTTCTTGA | 177 |
Western blot analysis
Cell lysates were collected from siRNA-and/or
FGF10-treated HAT-7 cells to screen the effective siRNA and to
evaluate the effects of siRNA and FGF10 on the protein expression
of SDC4, amelogenin (AMGN), ameloblastin (AMBN) and kallikrein 4
(KLK4). The samples were boiled in Laemmli sample buffer and loaded
onto a sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE)
followed by transfer onto nitrocellulose membranes. The primary
antibodies used were rabbit polyclonal anti-SDC4 (1:500; Abcam),
anti-AMGN (1:500), anti-AMBN (1:500) and anti-KLK4 (1:500)
antibodies (all from Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA). Anti-β-actin (1:1,000; Abcam) antibody was used as an
internal standard.
Statistical analysis
All experiments were performed independently at
least 3 times. All data are presented as the means ± standard
deviation (SD). Statistical significance was assessed using
two-tailed Student’s t-tests for 2 groups or the analysis of
variance Tukey’s test for more than 2 groups. Values of P<0.05
and P<0.001 were considered to indicate statistically signficant
differences.
Results
SDC4 expression in mouse molars during
the late bell stage of development
SDC4 expression was mainly detected in the oral
epithelium and dental epithelial cells, including the inner and
outer enamel epithelium and stellate reticulum (SR) cells (Fig. 1B). It was also found in
mesenchymal cells adjacent to the lateral sides of the epithelial
bud at E18 (Fig. 1A). On P2, SDC4
was expressed in the pre-ameloblasts (Fig. 1C and D), but its expression was
almost undetectable in the ameloblasts on P4 and P7 (Fig. 1E–H). Immunostaining in the SR also
decreased along with amelogenesis (Fig. 1).
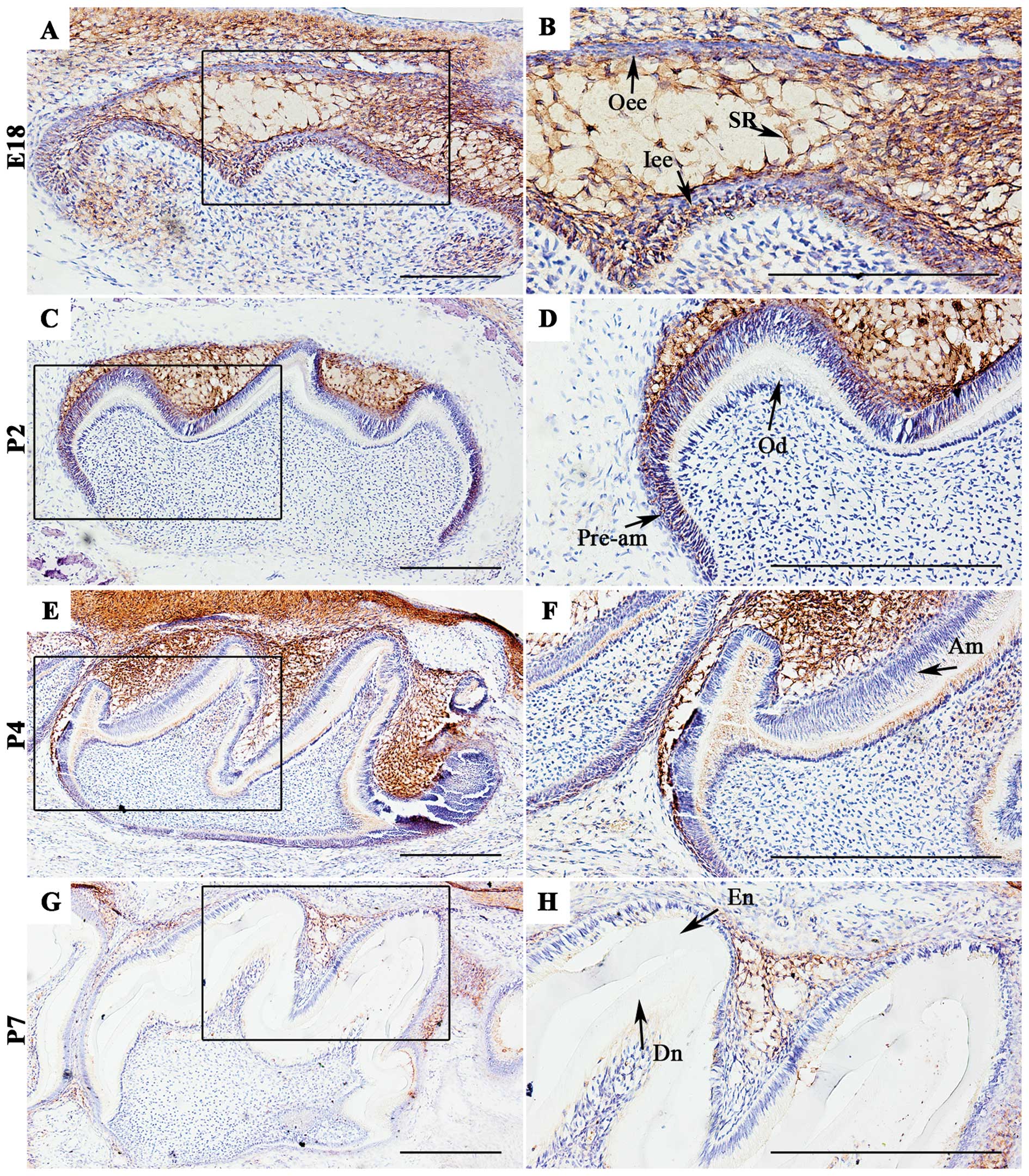 | Figure 1Localization of syndecan-4 (SDC4) in
first molar tooth germs of mice at embryonic day 18 (E18) and
newborn mice at postnatal day(P)2, P4 and P7. (A and B) At the late
bell stage of development (E18), SDC4 was intensely expressed in
the oral epithelium, stellate reticulum (SR) cells, and inner and
outer enamel epithelium. It was also observed in mesenchymal cells
adjacent to the cervical loop structure. (C and D) At P2, SDC4 was
observed among the pre-ameloblasts. (D–F) At P4, SDC4 protein was
faintly detected in newly differentiated ameloblasts and
odontoblasts. (G and H) At P7, SDC4 was almost undetectable in
ameloblasts and odontoblasts. The expression of SR was also
decreased. (B, D, F and H) Magnifications of the dark boxed areas
in (A, C, E and G). Oee, outer enamel epithelium; Iee, inner enamel
epithelium; Pre-am, pre-ameloblasts; Od, odontoblasts; Am,
ameloblasts; De, dentin; En, enamel. Scale bar, 100 μm. |
SDC4 expression in mouse postnatal
incisors
SDC4 expression was detected in the labial cervical
loop, lingual cervical loop and surrounding mesenchymal cells
(Fig. 2B–D). From cervical loops
to the distal part of incisors, SDC4 protein expression was not
detectable in pre-ameloblasts, pre-odontoblasts, ameloblasts or
odontoblasts. It was restricted to the stratum intermedium (SI)
cells, and the staining in this area gradually declined along with
cell differentiation (Fig.
2E–G).
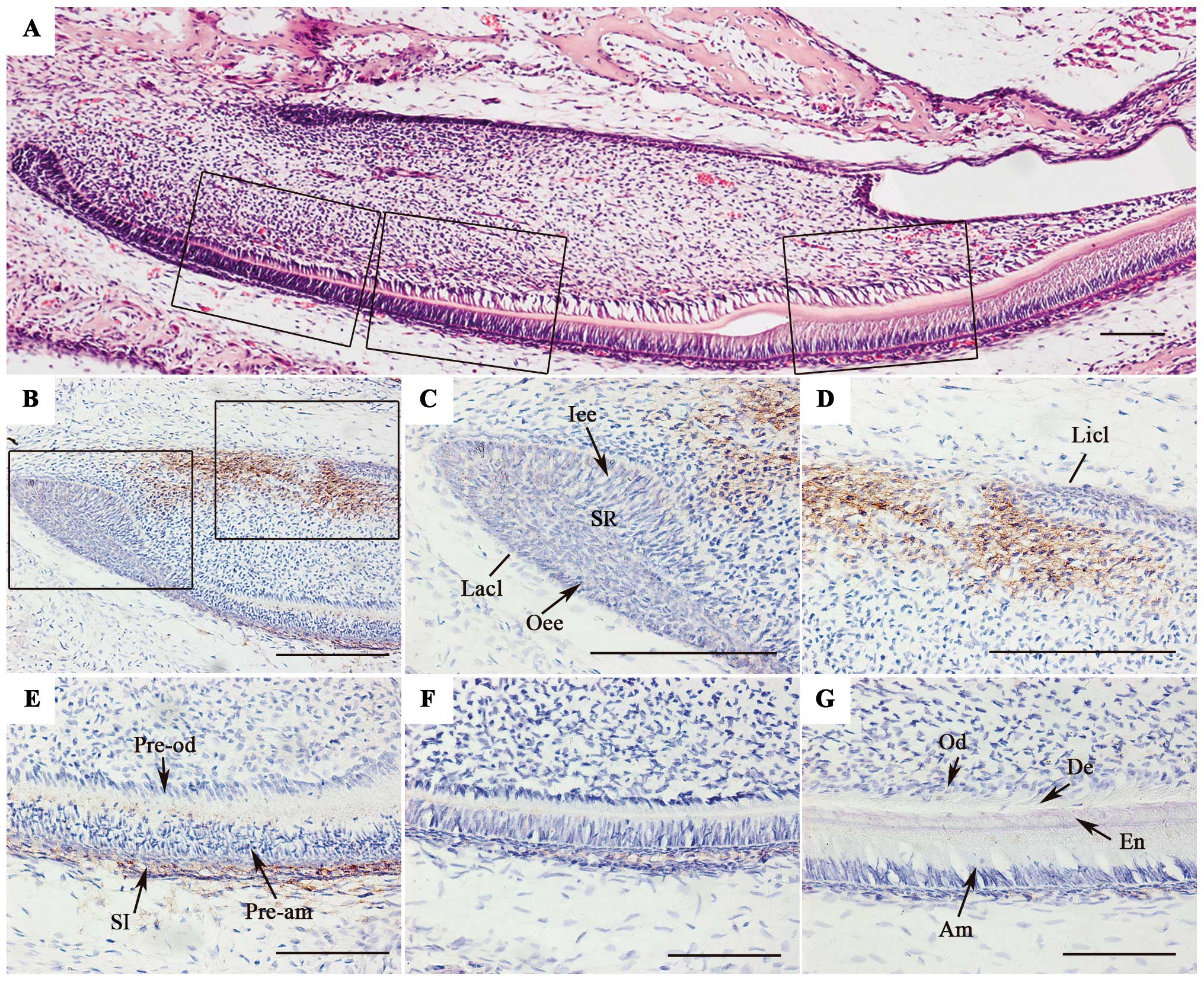 | Figure 2Localization of syndecan-4 (SDC4) in
incisors of postnatal mice. (A) Hematoxylin and eosin (H&E)
staining of postnatal mouse incisors. (B–D) SDC4 protein expression
was detected in labial and lingual cervical loops and surrounding
mesenchymal cells. (E–G) From the proximal to distal parts of the
incisor, SDC4 protein expression was not detectable in
pre-ameloblasts, pre-odontoblasts, ameloblasts or odontoblasts. It
was restricted to stratum intermedium (SI) cells, and the staining
in this area gradually declined along with cell differentiation. (C
and D) Magnifications of the boxed areas in (B), and (E–G) are
magnifications of the dark boxed areas in (A). Lacl, labial
cervical loop; Licl, lingual cervical loop; Oee, outer enamel
epithelium; SR, stellate reticulum; Iee, inner enamel epithelium;
Pre-od, pre-odontoblasts; Pre-am, ameloblasts; Od, odontoblasts;
Am, ameloblasts; De, dentin; En, enamel. Scale bar, 100 μm. |
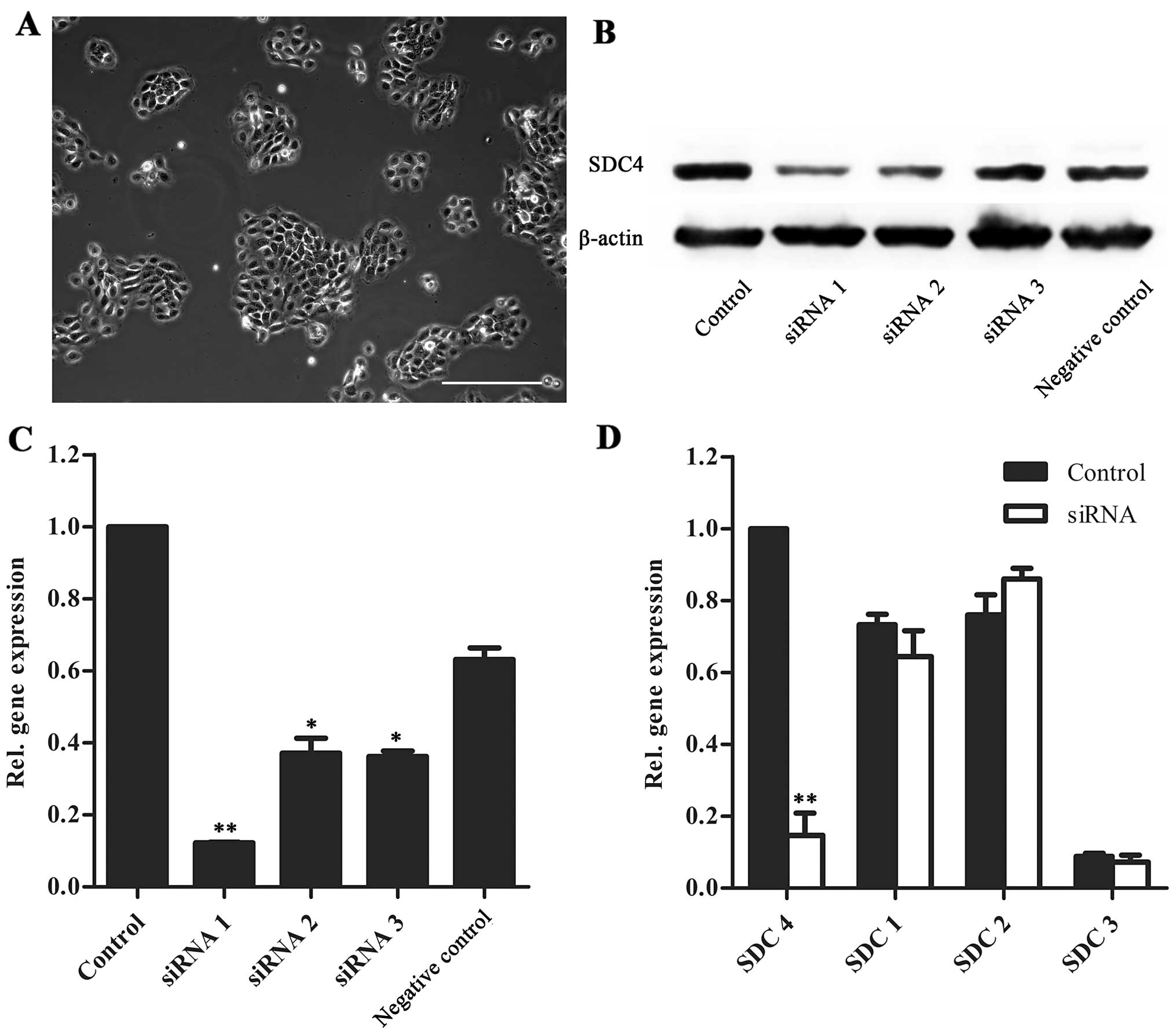
SDC4 silencing in HAT-7 cells
For the analysis of molecular function, SDC4 was
knocked down in HAT-7 cells, an immortalized dental epithelial cell
line derived from the cervical stem cell population of 6-day-old
rat mandibular incisors [provided by Professor Hidemitsu Harada
(14); (Fig. 3A)]. RT-qPCR and western blot
analysis revealed that SDC4 was successfully knocked down using
siRNA. siRNA 1 showed the highest inhibition efficiency and had no
effect on the gene expression of SDC1, 2 or 3 (Fig. 3B–D).
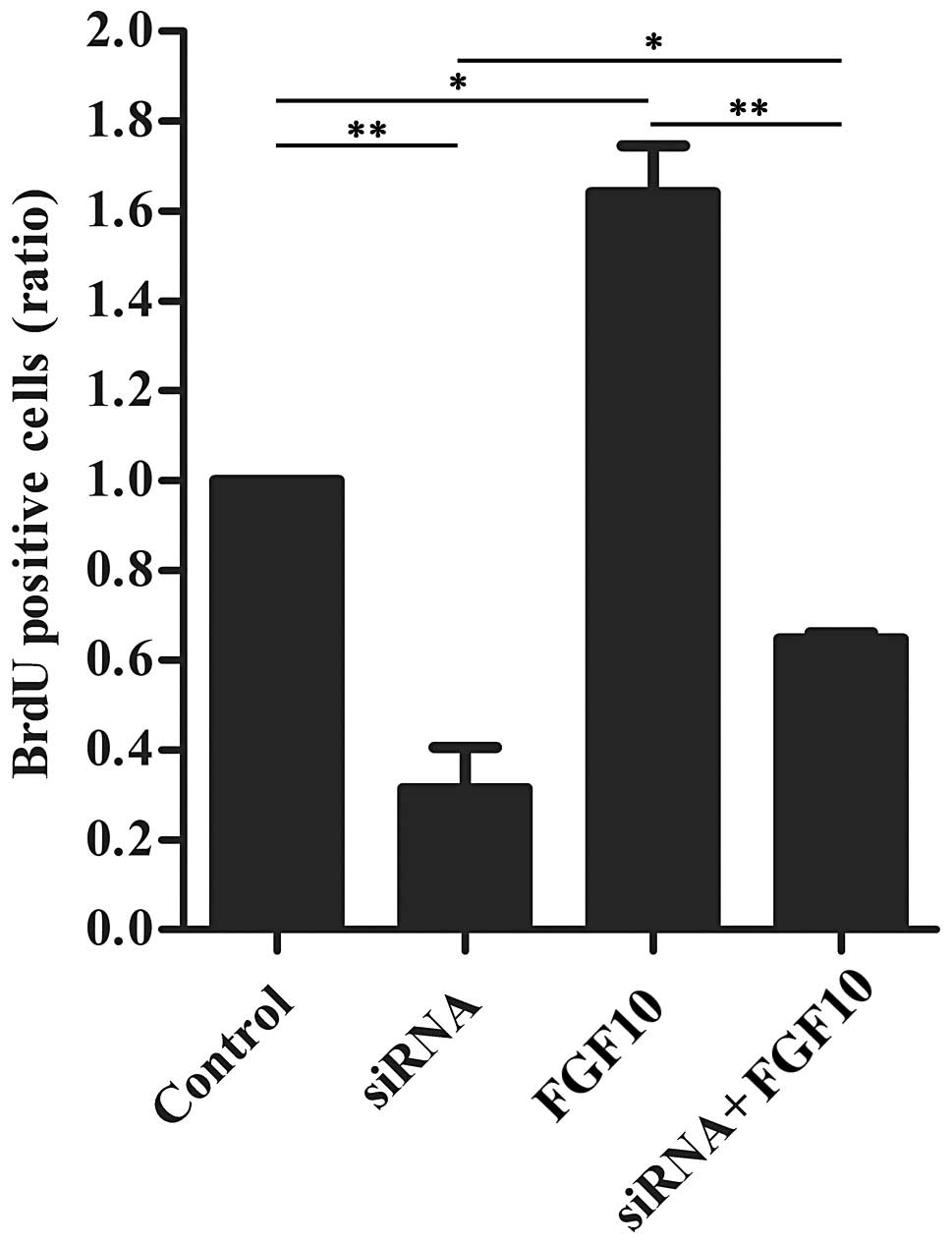
SDC4 and FGF10 co-regulate HAT-7 cell
proliferation
To determine the effects of SDC4 on cell
proliferation and the response of these siRNA-treated cells to
FGF10, recombinant mouse FGF10 was added to the HAT-7 cell culture
medium following transfection, and a BrdU incorporation analysis
was then performed (Fig. 4). The
cells treated solely with FGF10 exhibited an approximately 1.6-fold
greater number of BrdU-positive cells than the controls, and the
cells transfected with siRNA exhibited a marked decrease in
proliferation. Exogenous FGF10 partially reversed the inhibitory
effect exerted by siRNA targeting SDC4.
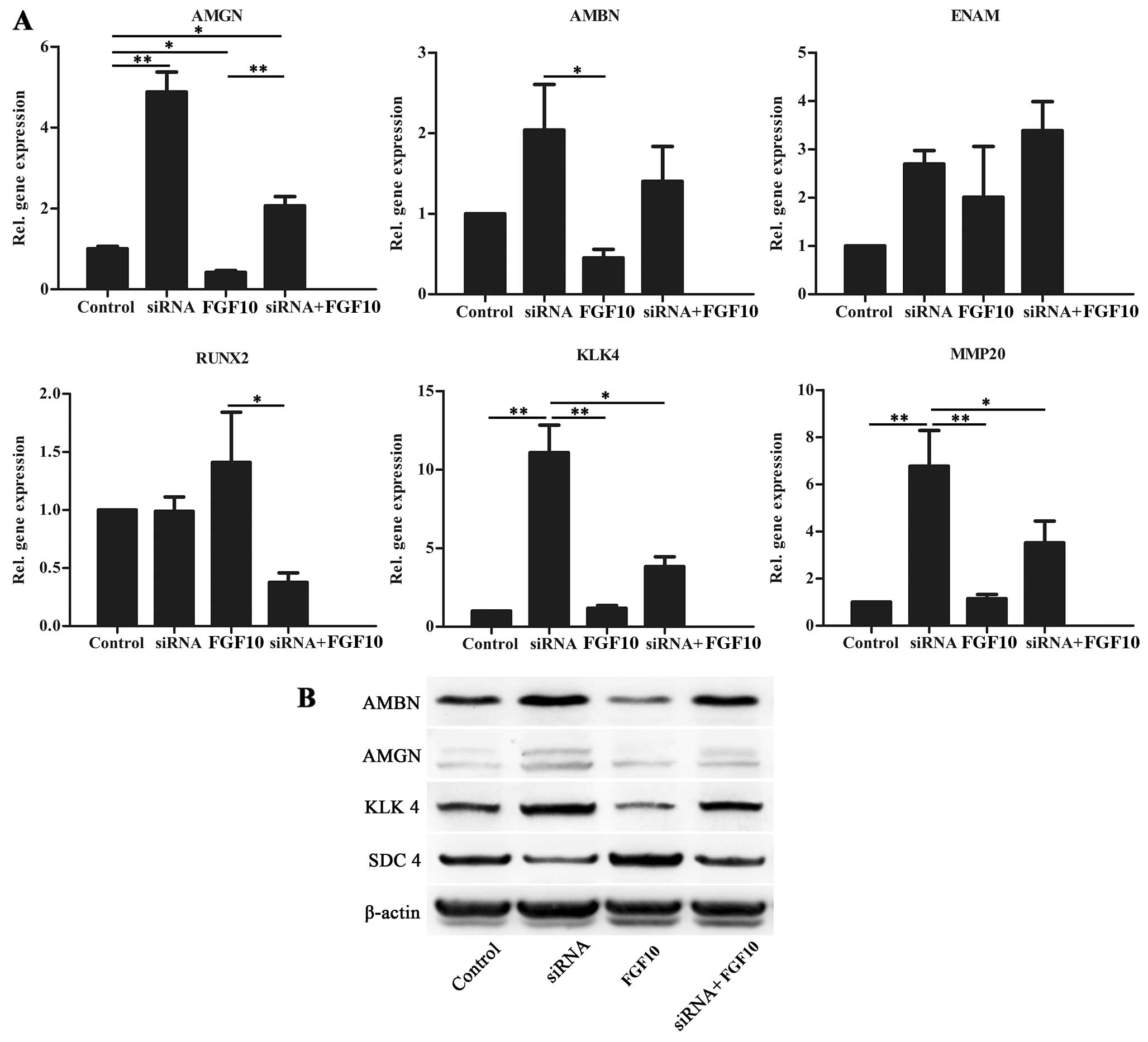
SDC4 affects HAT-7 cell differentiation
and is regulated by FGF10 signaling
Following treatment with siRNA and/or recombinant
protein FGF10, the expression of amelogenesis-related genes were
evaluated by RT-qPCR and western blot analysis (Fig. 5). When SDC4 expression was reduced
by siRNA, the expression of AMGN, AMBN, KLK4 and matrix
metalloproteinase (MMP)20 increased. Enamelin (ENAM) was unaffected
and RUNX2 expression slightly decreased when SDC4 expression
decreased (Fig. 5A). FGF10
downregulatex the expression of AMGN, AMBN and KLK4. The addition
of exogenous FGF10 weakened the effect exerted by siRNA targeting
SDC4 and upregulated SDC4 protein expression (Fig. 5B).
Discussion
Tooth formation is regulated by interactions between
the epithelium and the underlying neural crest-derived mesenchyme
(15). The earliest morphological
sign of tooth formation in mammals is the appearance of the dental
lamina, forming as a thickening of oral epithelium. The epithelium
of the lamina thickens at sites where teeth will grow, forming
dental placodes at these sites. During this bud stage, the dental
epithelium segregates into peripheral basal cells (inner and outer
enamel epithelium) and centrally located loosely arranged cells,
termed SR cells. The size and shape of the tooth crown becomes
apparent during the cap and bell stages of development. Beginning
from late bell stage (E18), the inner enamel epithelium
differentiates into pre-ameloblasts and then ameloblasts, which
secrete enamel matrix (16,17).
To the best of our knowledge, this study is the
first to evaluate SDC4 expression in molar germs at the late bell
stage of development. It was detected primarily in the oral
epithelium and the dental epithelial cells of the enamel organ in
molar germs, including SR cells, the inner and outer enamel
epithelium, and pre-ameloblasts, but not in ameloblasts. The
intense immunostaining in the SR was decreased along with
amelogenesis, possibly due to the retraction of the SR and the
decrease in the number of cells (18,19). These results indicate that SDC4
expression patterns may be critically controlled during enamel
organ development.
The expression of SDC4 was also analyzed in mouse
incisors. Rodent incisors continue to grow throughout adult life
due to the presence of dental epithelial stem cells, which give
rise to enamel-secreting ameloblasts. Incisors are an excellent
model for investigating the molecular underpinnings of amelogenesis
as epithelial cells at different stages of differentiation can be
easily detected in postnatal incisors. In the present study, SDC4
protein expression was detected in the cervical loops. A loss of
SDC4 expression was also evident in the epithelial cells of the
postnatal incisors when the inner epithelium gave rise to
ameloblasts. In addition, the amount staining in the SI cells
gradually decreased along with amelogenesis. Previous studies have
demonstrated that epithelial cell behaviors are regulated by an
integrated gene regulatory network, among which FGF10 signaling
regulates epithelial stem cell proliferation and SI cell
differentiation (12,13). FGF10, detected in the mesenchyme
surrounding the labial cervical loop, binds to FGFR2b, which is
expressed in the epithelium of the cervical loop (20). In this study, the distribution of
SDC4 in incisors suggests it is closely related to dental
epithelial cell differentiation and may be involved in FGF10
signaling.
However, this localization of SDC4 in incisors
contradicts earlier findings presented in the study by Muto et
al (21), who synthesized
rabbit antibody to the ectodomain of the mouse SDC4 core protein
and found that SDC4 was expressed throughout the tooth cellular
compartments, including ameloblasts. In this study, the anti-SDC4
antibody was the synthetic peptide surrounding the intracellular
domain of SDC4 (22). Due to the
fact that the cytoplasmic domains of the core protein are
conserved, and the extracellular domain of SDC4 can be
proteolytically cleaved, a process mediated by a variety of
proteases of the MMP family, it can thus be suggested that the SDC4
ectodomain may be cleaved to the ameloblasts that synthesize MMP20
at the secretory stage (23).
SDC4 was also expressed in dental mesenchymal cells
adjacent to cervical loops both in molars (E18) and postnatal
incisors. In molar germs, the dental mesenchyme is not segregated
into the dental papilla and the peripheral dental follicle at the
beginning of the bell stage (24). It has recently been suggested that
there is a population of putative mesenchymal stem cells between
cervical loops in incisors (25).
This intriguing similarity between molar and incisor germs suggests
that SDC4 may also have a function in regulating dental mesenchymal
cells.
Coincidentally, the suggestion of SDC4 functionality
was replicated in our in vitro studies. The HAT-7 cells were
used here to assess the mechanisms through which SDC4 affects the
proliferation and differentiation of dental epithelial cells. The
HAT-7 cell line is an immortalized dental epithelial cell line
derived from the cervical stem cell population of rat mandibular
incisors. It can shift from the transient amplification stage to
ameloblast lineage cells (26).
SDC4 expression was downregulated by siRNA, which had no effect on
the gene expression of SDC1, 2 or 3, the other three known members
of the SDC family (27).
Our results revealed that SDC4 expression was
associated with low levels of AMGN, AMBN, KLK4 and MMP20, while the
specific siRNA knockdown of SDC4 led to a highly significant
increase in the expression of these 4 genes; the gene expression of
ENAM and RUNX2 was unaffected. As regards the amelogenesis-related
genes, AMGN, AMBN and ENAM are scaffold proteins required to
support rod formation and hydroxyapatite (HA) crystallization
during enamel formation (16).
RUNX2 is a key regulatory transcription factor that suppresses
genes that are expressed during the secretory stage, such as AMGN
and ENAM, and thus regulates enamel formation (28). MMP20 and KLK4 are proteases that
break down enamel proteins to form a mineralized layer of enamel
(23). Among the genes
investigated in this study, they were the most profoundly affected
genes when SDC4 expression was inhibited.
In addition, the present study demonstrated that
FGF10 promoted cell proliferation and inhibited
amelogenesis-related genes expression. Previous studies have
demonstrated that FGF10 regulates dental epithelial stem cell
proliferation and maintenance. Arrest in FGF10 signaling
contributes to the terminal differentiation of the cells (20). Therefore, it is possible that
exogenous FGF10 kept the cells in a less differentiated state.
Furthermore, the addition of exogenous FGF10 weakened the effects
of siRNA through the upregulation of SDC4 protein expression,
indicating that SDC4 is under the control of FGF10 signaling.
SDC4 is thought to be involved in the formation of
the receptor-ligand complex (29,30). It is possible that the ternary
combination of FGF10, its receptor and SDC4 decrease when SDC4
expression decreases. In this study, we demonstrate that SDC4 may
regulate epithelial cell behaviors through FGF10 signaling.
However, several heparin-binding growth factors interact with SDC4
and may also regulate cell proliferation and differentiation during
tooth development (31). The
specific mechanisms through which SDC4 affects amelogenesis remain
to be elucidated in future sutdies.
Acknowledgements
This study was supported by the National Basic
Research Program (China, 2010CB944800), the National
High-Technology Research and Development Program (China,
2011AA030107), the Nature Science Foundation of China (China,
81271095, 81271119 and 81200792), the International Cooperation
Program of China (China, 2013DFG32770 and 2011DFA51970), the China
Postdoctoral Science Foundation (China, 2012M511934), the Key
Technology R&D Program of Sichuan Province (2012SZ0013,
12ZC0493, 13ZC0971, 2013GZX0158 and 13ZC0979) and the Basic
Research Program of Sichuan Province (12JC0212 and 2013JY0019).
References
|
1
|
Choi Y, Chung H, Jung H, Couchman JR and
Oh ES: Syndecans as cell surface receptors: Unique structure
equates with functional diversity. Matrix Biol. 30:93–99. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bellin RM, Kubicek JD, Frigault MJ, et al:
Defining the role of syndecan-4 in mechanotransduction using
surface-modification approaches. Proc Natl Acad Sci USA.
106:22102–22107. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elfenbein A and Simons M: Syndecan-4
signaling at a glance. J Cell Sci. 126:3799–3804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tkachenko E, Rhodes JM and Simons M:
Syndecans: new kids on the signaling block. Circ Res. 96:488–500.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuriyama S and Mayor R: A role for
Syndecan-4 in neural induction involving ERK- and PKC-dependent
pathways. Development. 136:575–584. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iwabuchi T and Goetinck PF: Syndecan-4
dependent FGF stimulation of mouse vibrissae growth. Mech Dev.
123:831–841. 2006. View Article : Google Scholar
|
|
7
|
Shimokawa K, Kimura-Yoshida C, Nagai N, et
al: Cell surface heparan sulfate chains regulate local reception of
FGF signaling in the mouse embryo. Dev Cell. 21:257–272. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Goodwin AF, Tidyman WE, Jheon AH, et al:
Abnormal Ras signaling in Costello syndrome (CS) negatively
regulates enamel formation. Hum Mol Genet. 23:682–692. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang Z, Kim J, Lacruz RS, et al:
Epithelial-specific knockout of the Rac1 gene leads to enamel
defects. Eur J Oral Sci. 119(Suppl 1): S168–S176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Porntaveetus T, Otsuka-Tanaka Y, Basson
MA, Moon AM, Sharpe PT and Ohazama A: Expression of fibroblast
growth factors (Fgfs) in murine tooth development. J Anat.
218:534–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kettunen P and Thesleff I: Expression and
function of FGFs-4, -8, and -9 suggest functional redundancy and
repetitive use as epithelial signals during tooth morphogenesis.
Dev Dyn. 211:256–268. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harada H, Toyono T, Toyoshima K, et al:
FGF10 maintains stem cell compartment in developing mouse incisors.
Development. 129:1533–1541. 2002.PubMed/NCBI
|
|
13
|
Wang XP, Suomalainen M, Felszeghy S, et
al: An integrated gene regulatory network controls stem cell
proliferation in teeth. PLoS Biol. 5:e1592007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kawano S, Morotomi T, Toyono T, Nakamura
N, Uchida T, Ohishi M, Toyoshima K and Harada H: Establishment of
dental epithelial cell line (HAT-7) and the cell differentiation
dependent on Notch signaling pathway. Connect Tissue Res.
43:402–412. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blanpain C, Horsley V and Fuchs E:
Epithelial stem cells: turning over new leaves. Cell. 128:445–458.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu JC, Sun X, Zhang C and Simmer JP: A
comparison of enamelin and amelogenin expression in developing
mouse molars. Eur J Oral Sci. 109:125–132. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thesleff I and Tummers M: Tooth
organogenesis and regeneration. StemBook. Harvard Stem Cell
Institute; Cambridge, MA: 2008, View Article : Google Scholar
|
|
18
|
Ida-Yonemochi H, Satokata I, Ohshima H, et
al: Morphogenetic roles of perlecan in the tooth enamel organ: an
analysis of overexpression using transgenic mice. Matrix Biol.
30:379–388. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baratella L, Arana-Chavez VE and
Katchburian E: Apoptosis in the early involuting stellate reticulum
of rat molar tooth germs. Anat Embryol (Berl). 200:49–54. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harada H, Kettunen P, Jung HS, Mustonen T,
Wang YA and Thesleff I: Localization of putative stem cells in
dental epithelium and their association with notch and FGF
signaling. J Cell Biol. 147:105–120. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Muto T, Miyoshi K, Munesue S, et al:
Differential expression of syndecan isoforms during mouse incisor
amelogenesis. J Med Invest. 54:331–339. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Corti F, Finetti F, Ziche M and Simons M:
The syndecan-4/protein kinase Cα pathway mediates prostaglandin
E2-induced extracellular regulated kinase (ERK) activation in
endothelial cells and angiogenesis in vivo. J Biol Chem.
288:12712–12721. 2013.PubMed/NCBI
|
|
23
|
Bartlett JD, Yamakoshi Y, Simmer JP, Nanci
A and Smith CE: MMP20 cleaves E-cadherin and influences ameloblast
development. Cells Tissues Organs. 194:222–226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rothová M, Peterková R and Tucker AS: Fate
map of the dental mesenchyme: dynamic development of the dental
papilla and follicle. Dev Biol. 366:244–254. 2012.PubMed/NCBI
|
|
25
|
Seidel K, Ahn CP, Lyons D, et al: Hedgehog
signaling regulates the generation of ameloblast progenitors in the
continuously growing mouse incisor. Development. 137:3753–3761.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsumoto A, Harada H, Saito M and
Taniguchi A: Induction of enamel matrix protein expression in an
ameloblast cell line co-cultured with a mesenchymal cell line in
vitro. In Vitro Cell Dev Biol Anim. 47:39–44. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xian X, Gopal S and Couchman JR: Syndecans
as receptors and organizers of the extracellular matrix. Cell
Tissue Res. 339:31–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Athanassiou-Papaefthymiou M, Kim D,
Harbron L, et al: Molecular and circadian controls of ameloblasts.
Eur J Oral Sci. 119(Suppl 1): S35–S40. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Elfenbein A, Lanahan A, Zhou TX, et al:
Syndecan 4 regulates FGFR1 signaling in endothelial cells by
directing macropinocytosis. Sci Signal. 5:ra362012.PubMed/NCBI
|
|
30
|
Horowitz A, Tkachenko E and Simons M:
Fibroblast growth factor-specific modulation of cellular response
by syndecan-4. J Cell Biol. 157:715–725. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thesleff I, Vaahtokari A and Partanen AM:
Regulation of organogenesis. Common molecular mechanisms regulating
the development of teeth and other organs. Int J Dev Biol.
39:35–50. 1995.PubMed/NCBI
|