Introduction
Male infertility is a major factor in the inability
of a couple to conceive. The most common cause of male infertility
is disorders affecting spermatogenesis, which is a complex process
strictly regulated by the cooperation of genetic factors, hormones
and cytokines. Testicular cytokines and growth factors, such as
interleukin (IL)-1, tumor necrosis factor (TNF), leukemia
inhibitory factor (LIF), stem cell factor (SCF) and transforming
growth factor (TGF) have been shown to affect both germ cell
proliferation and testicular function (1,7).
Under pathological conditions, the levels of cytokines are altered
and negatively affect spermatogenesis. Thus, the expression levels
and the regulatory pathway of cytokines in the testis should be
taken into consideration in the development of therapeutic
strategies for male infertility (1–3).
TGF-β1 is a member of the TGF-β superfamily that
controls proliferation, differentiation, embryonic development,
angiogenesis and other functions in various cell types (4). It has been found that the expression
levels of TGF-βs vary at different stages of testicular
development. They induce and maintain the specification of the germ
cell lineage in the fetal testis, and regulate spermatogonial
differentiation in the adult testis (5–7).
Although studies on the TGF-β signaling pathway have mainly focused
on the ligand-receptor interaction and the specific biological
effects at a cellular and/or molecular level (8), there also exist unknown cross-talks
between TGF-βs and other different signaling molecules, such as
microRNAs (miRNAs or miRs), which are small non-coding RNAs that
post-transcriptionally regulate gene expression by targeting mRNAs
for translational repression and degradation (9).
Increasing evidence indicates that miRNAs are likely
to be critically involved in the majority of biological processes,
including mammalian germ-cell development (10). For instance, in Dicer-deleted
testis, spermatogenesis is retarded at an early stage of
proliferation and/or early differentiation (11). Recent data have indicated that the
expression levels of miR-141, miR-200a, miR-200c and miR-323 in
mice are downregulated gradually in both male and female germ cells
throughout their development, suggesting that miRNAs are involved
in translational repression during spermatogenesis (12). On the other hand, miRNAs have been
found to target the TGF-β1 super-family receptors, Smads or
multiple components of the TGF-β1 signaling pathway, and thereby
also affect TGF-β1-regulated physiological or pathological
processes, indicating that miRNAs are involved in the cellular
response to TGF-β1 signaling (13). However, to the best of our
knowledge, no systemic studies on the changes that occur in the
expression levels of miRNAs induced by TGF-β1 signaling in mouse
male germ cells have been published to date. Thus, in the present
study, we investigated the effects of TGF-β1 on global miRNA and
protein expression profiles in the mouse GC-1 spg germ cell line.
In addition, we aimed to elucidate the association between these
miRNAs and proteins and TGF-β1 signaling.
Materials and methods
Cell line
Mouse GC-1 spg germ cells (CRL-2053; obtained from
ATCC, Manassas, VA, USA) were cultured in Dulbecco’s modified
Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum
(FBS), 100 μg/ml penicillin-streptomycin, and maintained in
an atmosphere of 5% CO2 and 95% humidified air at
37°C.
Treatment with TGF-β1
The cells (60% confluent) were maintained in
serum-free DMEM for 24 h prior to stimulation with TGF-β1
(PeproTech, Rocky Hill, NJ, USA). Subsequently, the cells were
stimulated with 5 or 10 ng/ml of TGF-β1 in DMEM containing 1% FBS
for 48 h.
RNA isolation
Total RNA was extracted using the RNAiso reagent
(Takara Bio, Inc., Shiga, Japan) according to the manufacturer’s
instructions, digested by RNase-free DNase (Fermentas, Burlington,
ON, Canada), dissolved in diethylpyro-carbonate-treated water, and
stored at −80°C until use. For quality control, RNA purity and
integrity were evaluated by agarose gel electrophoresis and the
OD260/OD280 ratio, respectively.
miRNA microarray and data analysis
The Affymetrix GeneChip miRNA 3.0 microarray
(CapitalBio Corp., Beijing, China) covering the miRNAs from Sanger
miRBase v17.0 (www.mirbase.org), snoRNAs and scaRNAs
from snoRNABase (www.snorna.biotoul.fr/coordinates.php) and the Ensembl
Archive (www.ensembl.org/biomart/martview) was applied to
investigate the possible changes in miRNA expression in the GC-1
spg cells treated with different doses of TGF-β1 in comparison to
the control (untreated) GC-1 spg cells. Total RNA was labeled with
the FlashTag™ Biotin RNA Labeling kit following the manufacturer’s
instructions. Subsequently, hybridization, scanning and data
extraction were carried out at CapitalBio Corp.. The number of
miRNAs affected by TGF-β1 was determined using the scatter plots of
the control GC-1 spg cells versus the GC-1 spg cells treated with
TGF-β1. Finally, the images were gridded and analyzed using ImaGene
7.0 software (BioDiscovery Inc., Hawthorne, CA, USA).
Reverse transcription-quantitative
(real-time) PCR (RT-qPCR)
For the analysis of miRNA expression, RT-qPCR was
performed. cDNA was made from enriched miRNA using miRNA
PrimeScript RT Enzyme Mix (Takara Bio, Inc.). The SYBR-Green
real-time PCR protocol (Takara Bio, Inc.) was then applied by using
a MX3000 (Stratagene, La Jolla, CA, USA) instrument. PCR was
performed in a 10-μl reaction volume containing 3 μl
of nuclease-free water, 5 μl of SYBR Premix Ex Taq II, 0.2
μl ROX Reference Dye II (both from Takara Bio, Inc.), 1
μl of cDNA and 0.4 μl each of the 10 μM
gene-specific primers. Following initial denaturation for 20 sec at
95°C, 39–45 cycles of PCR were performed. Each cycle consisted of a
denaturation period (5 sec at 95°C), an annealing phase (30 sec at
55°C) and an extension period (30 sec at 72°C). U6 RNA was used as
an endogenous control. miRNA primers were provided by Takara Bio,
Inc.
Preparation of protein samples and
two-dimensional gel electrophoresis (2-DE)
In brief, the harvested cells were extracted using
lysis buffer (9 M urea, 4% CHAPS, 40 mM Tris-HCl, 2% pharmylate, 40
mM DTT and 1 mM PMSF). The protein concentration in the
supernatants was determined using the Non-Interference Protein
Assay kit (Sangon Biotech, Shanghai, China) and then used for 2-DE.
The total proteins (250 μg) were loaded onto 24-cm
non-linear IPG strips at pH 3–10 (GE Healthcare Bio-Sciences,
Pittsburgh, PA, USA) for simultaneous hydration, then isoelectric
focusing (IEF) was performed using the Ettan™IPGphor™ Isoelectric
Focusing System (GE Healthcare Bio-Sciences) following the
voltage-time program of 30 V for 13 h, 500 V for 1 h, 1,000 V for 1
h, 8,000 V for 3 h, 8,000 V for 44,000 Vh with a total power of
69,890 Vh. Following IEF, the strips were equilibrated for 2
intervals of 15 min in an equilibration buffer containing 6 M urea,
0.375 M Tris-HCl, 2% SDS, and 30% glycerol. The strips were then
transferred onto the second-dimensional SDS-PAGE and run on 10%
polyacrylamide gels at the program of 2.5 w/gel for 30 min; 15
w/gel for 5 h. The gels were fixed for silver nitrate staining
prior to scanning.
In-gel digestion and matrix-assisted
laser desorption/ionization time-of-flight mass spectrometry
(MALDI-TOF/TOF MS)
ImageMaster 2D Platinum analysis software (GE
Healthcare Bio-Sciences) was used for spot detection, gel matching
and spot quantification. Protein spots showing significant changes
(protein spots with a ratio >1.5 or <0.5) following
stimulation with TGF-β1 were manually excised and subjected to
in-gel digestion. The digested samples were extracted and analyzed
by MALDI-TOF/TOF MS (Sangon Biotech). For peptide mass
fingerprinting (PMF), each mass spectrum was obtained from signals
generated from at least 500 laser shots. The mass data were used to
search the UniProt database (http://www.pir.uniprot.org) using the MS-Fit database
search engine (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msfitstandard).
Bioinformatics analysis
The miRNA sequences and annotation were accessible
at miRBase 2.0 (http://www.mirbase.org). The potential miRNA targets
for the selected miRNAs were screened using TargetScan 6.2
(http://www.targetscan.org/). The minimum
free energy hybridization of the miRNAs and mRNAs was analyzed
using the RNAhybrid tool (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid).
The secondary structure of single-stranded RNAs was predicted using
the RNAFold WebServer (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The
proteins identified by proteomics analysis were analyzed using the
Mascot database (http://www.matrixscience.com/).
Statistical analysis
The data are presented as the means ± SD (n=3).
Statistical analysis was conducted using the Student’s t-test.
Differences were considered statistically significant at p-values
≤0.05.
Results
Alterations in miRNA profiles induced by
TGF-β1 in GC-1 spg cells
Global miRNA expression levels induced by different
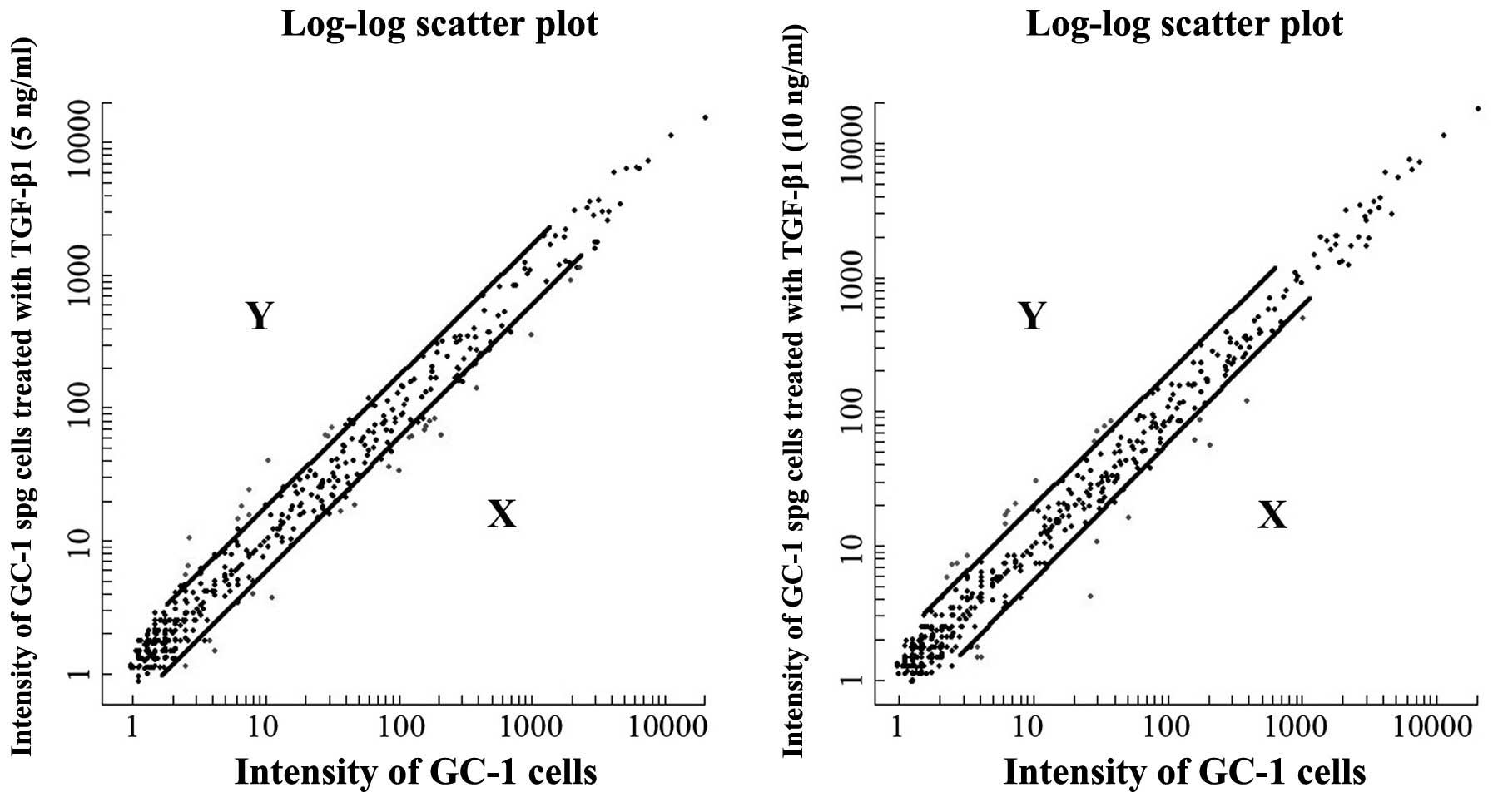
doses of exogenous TGF-β1 in the mouse GC-1 spg cells, as shown by
microarray analysis, are presented in Fig. 1. The differentially expressed
miRNAs with a fluorescence signal of ≥20 and fold changes of ≥2 or
≤0.5 (p≤0.05) were analyzed using the t-test. Our data demonstrated
that the expression of 24 out of approximately 924 miRNAs was
altered following stimulation with TGF-β1. Of these, 7 miRNAs were
upregulated and 17 miRNAs were downregulated (Table I). In addition, 4 miRNAs
(mmu-miR-196a, miR-497, miR-199a-3p and miR-199a-5p) were
upregulated following stimulation of the cells with 5 and 10 ng/ml
TGF-β1, and 5 miRNAs (mmu-miR-23a*, miR-1941-5p,
miR-5112, miR-423-5p and miR-700) were downregulated following
stimulation with the 2 different concentrations of TGF-β1. These
miRNAs were mapped onto almost all chromosomes and approximately
83% of these miRNAs were located in intronic or intergenic regions.
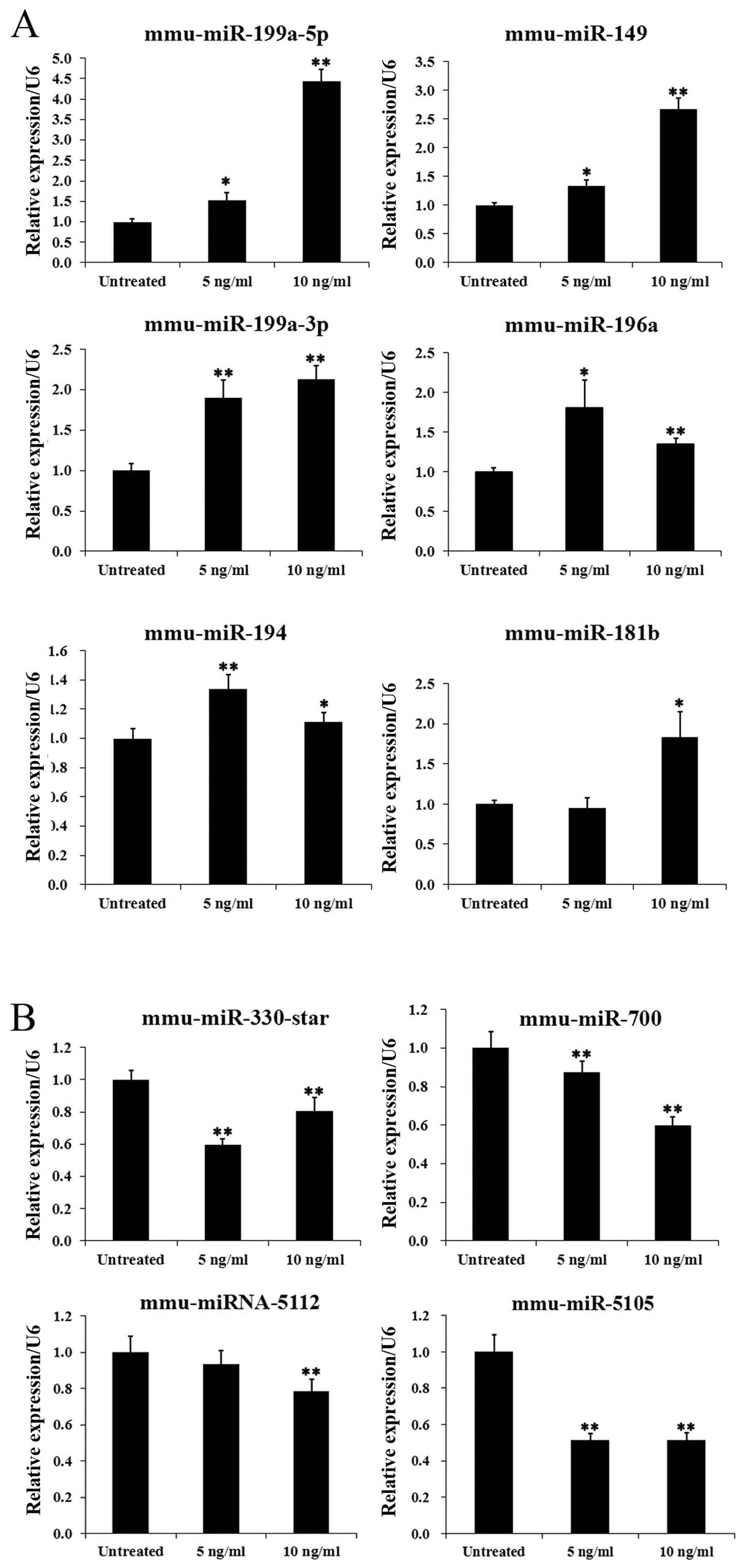
In order to test the reliability of the microarray data, the
expression patterns of 10 selected miRNAs, including 6 upregulated
miRNAs and 4 downregulated miRNAs, were examined by RT-qPCR. The
overall profile of miRNA expression by RT-qPCR analysis was similar
to that revealed by the microarray data for all the selected miRNAs
(Fig. 2).
 | Table IThe differentially expressed miRNAs in
GC-1 spg cells following stimulation with transforming growth
factor-(TGF)-β1. |
Table I
The differentially expressed miRNAs in
GC-1 spg cells following stimulation with transforming growth
factor-(TGF)-β1.
| Differential
expression | miRNA | Signal ratio (5 ng/ml
vs. con) | Signal ratio (10
ng/ml vs. con) | Chromosome | Overlapping
transcripts |
|---|
| Upregulation | mmu-miR-196a | 3.8771 | 2.95 | chr11 or chr15 | Intergenic or Hoxc5
Mir196a |
| mmu-miR-497 | 3.2642 | 2.7882 | chr11 | Intergenic |
| mmu-miR-194 | 2.2598 | – | chr1 or chr19 | Iars2 or
intergenic |
| mmu-miR-199a-3p | 2.2036 | 2.0962 | chr1 or chr9 | Dnm3os Dnm3 or
Dnm2 |
| mmu-miR-199a-5p | 2.0496 | 2.4190 | | – |
| mmu-miR-181b | – | 2.2239 | chr1 or chr2 | RP23 mmu-mir-181b or
Nr6a1 |
| mmu-miR-149 | – | 2.2931 | chr1 | Gpc1 |
| Downregulation |
mmu-miR-3102-5p.2 | 0.4955 | – | chr7 | Arhgef17 |
| mmu-miR-320 | 0.4955 | – | chr14 | Intergenic |
| mmu-miR-296-3p | 0.4864 | – | chr2 | Intergenic |
| mmu-miR-5105 | 0.4614 | – | – | – |
|
mmu-miR-23a* | 0.4604 | 0.4993 | chr8 | Intergenic |
|
mmu-miR-330* | 0.4597 | – | chr7 | Eml2 |
|
mmu-miR-351* | 0.4572 | – | chrX | Intergenic |
| mmu-miR-503 | 0.4353 | – | chrX | Intergenic |
| mmu-miR-1941-5p | 0.4295 | 0.3884 | chr15 | Krt80 |
| mmu-miR-877 | 0.4266 | – | chr17 | Abcf1 |
| mmu-miR-1943 | 0.3967 | – | chr15 | Tmem184b |
| mmu-miR-5112 | 0.3604 | 0.3048 | chr18 | Intergenic |
| mmu-miR-423-5p | 0.3520 | 0.4899 | chr11 | Ccdc55 |
| mmu-miR-714 | 0.3312 | – | – | – |
| mmu-miR-700 | 0.2998 | 0.2706 | chr4 | Rcan3 |
|
mmu-miR-3096-3p | – | 0.3208 | – | – |
|
mmu-miR-486* | – | 0.3563 | chr8 | Ank1 AC126445.2
Gm15816 |
Alterations in protein profiles induced
by TGF-β1 in GC-1 spg cells
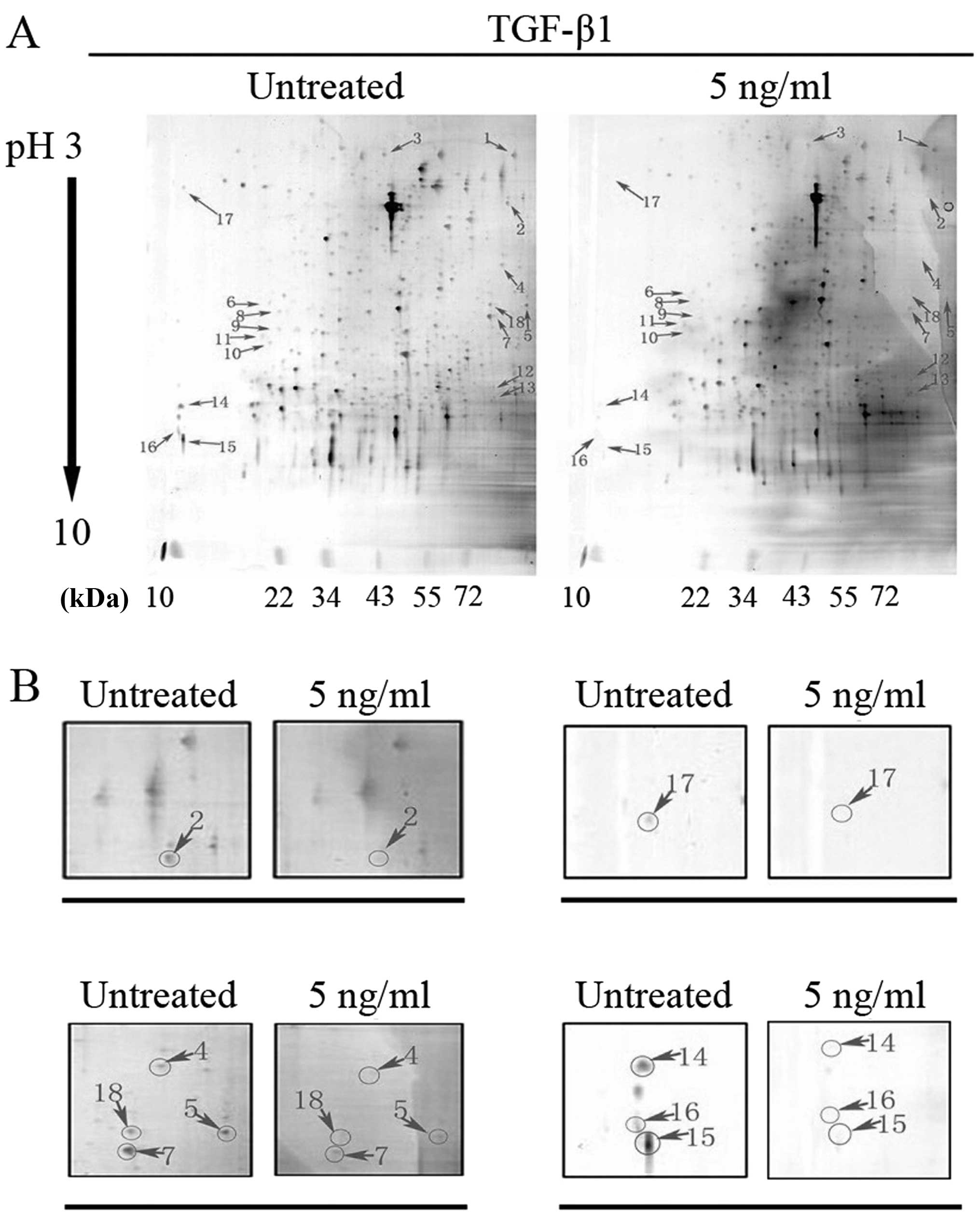
2-DE and MALDI-TOF/TOF MS-based proteomics analysis
were performed to explore the expression profiles of proteins
associated with TGF-β1 signaling in mouse GC-1 spg cells. In the
first dimension, the pH range of the IPG strips is from 3 to 10
with most of the proteins located between pH 4.0 and pH 8.0
(Fig. 3A). Along the second
dimension, proteins are mainly distributed within the range of
20–70 kDa according to their molecular weight. In this study,
protein spots with a ratio >1.5 or <0.5 were identified as
differentially expressed. Of the identified proteins, 11 proteins
were selected and subjected to MALDI-TOF/TOF MS of these
differentially expressed spots. Fig.
3 shows the total number of identified proteins (Fig. 3A) and the downregulated protein
spots (Fig. 3B). Table II shows the relevant information
of the downregulated polypeptides, including their annotation,
function, molecular weight (MW)/isoelectric point (pI), score and
sequence coverage. The function of these identified proteins
[gelsolin, vinculin, ezrin, nucleoside diphosphate kinase B
(NDKB)/Nme2, peptidyl-prolyl isomerase A (PPIA), cofilin-1,
transitional endoplasmic reticulum ATPase (TERA) and eukaryotic
translation initiation factor 5A-1 (IF5A1)] are mostly associated
with invasion, cell-cell adhesion, actin dynamics, signal
transduction and so on, suggesting that they may be involved in the
cellular process of cell polarity, morphology and locomotion.
 | Table IIThe characteristics of the
downregulated proteins. |
Table II
The characteristics of the
downregulated proteins.
| Spot No. | Protein/gene
name | Annotation | Proposed
functiona | MW/pI | Score | Sequence
coverage | Protein
alteration |
|---|
| 2 |
TERA/Vcp | Transitional
endoplasmic reticulum ATPase | Involved in the
formation of the tER, DNA damage, DNA repair, transport and Ubl
conjugation pathway. | 89950/5.14 | 622 | 17% | ↓ |
| 4 |
GELS/Gsn | Gelsolin | Plays a role in
ciliogenesis. | 86287/5.83 | 208 | 7% | ↓ |
| 5 |
VINC/Vcl | Vinculin | Involved in
cell-matrix adhesion and cell-cell adhesion, and may also play an
important role in cell morphology and locomotion. | 117215/5.77 | 359 | 13% | ↓ |
| 14 |
NDKB/Nme2 | Nucleoside
diphosphate kinase B | Major role in the
synthesis of nucleoside triphosphates other than ATP, negatively
regulates Rho activity. | 17466/6.97 | 409 | 47% | ↓ |
| 15 |
COF1/Cfl1 | Cofilin-1/p18 | Regulates actin
cytoskeleton dynamics, plays a role in the regulation of cell
morphology and cytoskeletal organization. | 18776/8.22 | 464 | 46% | ↓ |
| 16 |
PPIA/Ppia | Peptidyl-prolyl
cis-trans isomerase A | PPIases accelerate
the folding of proteins. | 18131/7.74 | 469 | 43% | ↓ |
| 17 |
IF5A1/Eif5a | Eukaryotic
translation initiation factor 5A-1 | mRNA-binding
protein, involved in translation elongation and actin dynamics and
cell cycle progression. | 17049/5.08 | 674 | 58% | ↓ |
| 18 |
EZRI/Ezr | Ezrin | Probably involved
in connections of major cytoskeletal structures to the plasma
membrane. In epithelial cells, required for the formation of
microvilli and membrane ruffles on the apical pole. Along with
PLEKHG6, required for normal macropinocytosis. | 69478/5.83 | 299 | 14% | ↓ |
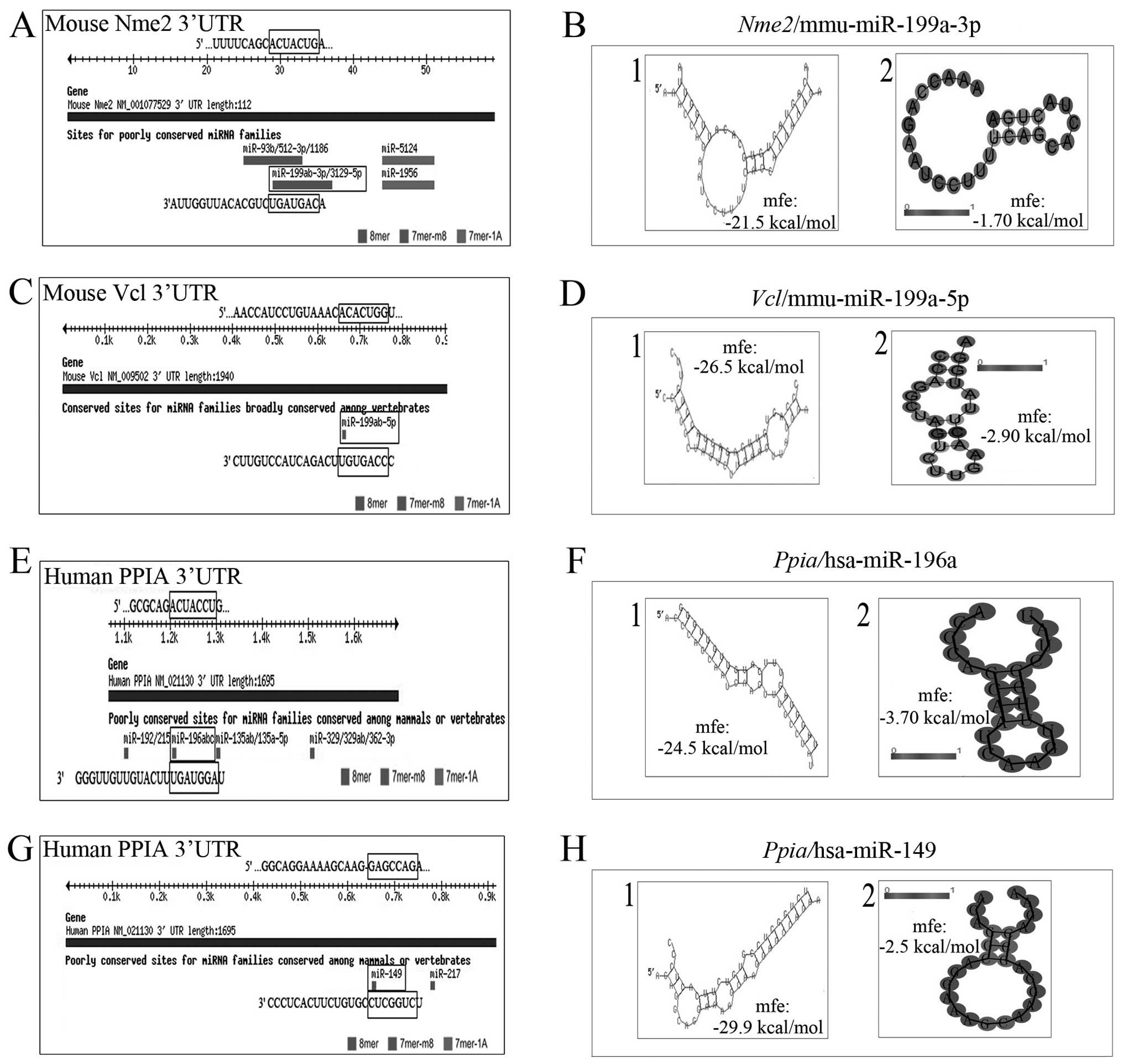
Predicted miRNA binding sites of the
selected proteins following stimulation with TGF-β1 in GC-1 spg
cells
Using miRBase 2.0 and TargetScan 6.2, the miRNA
targets following the induction of TGF-β1 were predicted. It was
predicted that miR-199a-3p would bind to target sequences in the
3′UTR of the mouse gene NDKΒ/Nme2 or Vcl mRNA, which was
downregulated in the GC-1 spg cells following stimulation with
TGF-β1 (Fig. 4A and C). Both
miR-149 and miR-196a bind to target sequences in the 3′UTR of the
downregulated gene, PPIA (Fig. 4E and
G). These 3 miRNAs were upregulated in response to TGF-β1, as
shown by miRNA microarray analysis. In addition, bioinformatics
analysis further revealed that the minimum free energy
hybridization of miR-199a-3p binding with the target gene NDKB/Nme2
(or Vcl) 3′UTR was markedly lower than that of the secondary
structure of single-stranded NDKB/Nme2 (or Vcl) mRNA, indicating
that NDKB/Nme2 and miR-199a-3p have a higher possibility for
binding (Fig. 4B and D). We also
analyzed this binding possibility between miR-149 and PPIA, or
miR-196a and PPIA (Fig. 4F and H)
and obtained similar results. These results suggest that there may
be a bio-functional link between these miRNAs and proteins.
Discussion
The TGF-β1 superfamily comprises a broad range of
signaling ligands, including TGF-β1, activin, nodal and bone
morphogenetic protein (BMP), which regulate a variety of cellular
processes, and its dysregulation often leads to cancer, male
infertility and other diseases (4). Recently, miRNAs have emerged as
major regulators of gene expression (14), and many more miRNAs have been
identified and proven to be involved in various physical and
pathological processes by regulating cell-fate decisions. In the
present study, we performed miRNA microarray analysis in mouse GC-1
spg cells and identified a total of 24 miRNAs (Table I) that were regulated in response
to stimulation of the cells with TGF-β1. Of these, 16 miRNAs are
molecules associated with TGF-β1 signaling, while miR-181, miR-23a,
miR-194, miR-199a and miR-196a family members have been previously
reported to respond to TGF-β1 treatment in different cells and
tissues (15–19). Taking the miR-199a family member
as an example, researchers found that TGF-β1 increased the
expression of miR-199a-3p in primary human adult pulmonary
fibroblasts and was associated with fibrotic remodeling (20). Furthermore, recent data suggest
that miR-199a suppresses cell growth, cancer migration, invasion
and metastasis in testicular cancer by regulating TGF-β1 signaling
through the regulation of Smad4 (13). As the activation of TGF-β
signaling plays a central role in the pathogenesis of development,
proliferation, differentiation, or apoptosis, the identification of
these miRNAs regulated by TGF-β1 in GC-1 spg cells provides a
possible link and a rationale for the hypothesis that they are
downstream mediators of this pathway and mediate the functions of
the pathway.
On the other hand, in order to identify candidate
downstream target proteins in the GC-1 spg cells following
stimulation with TGF-β1, we utilized comparative proteomics
analysis to identify a group of 8 proteins, including gelsolin,
vinculin, ezrin, NDKB/Nme2, PPIA, cofilin-1, TERA and IF5A1, which
were suppressed following stimulation with TGF-β1 (Fig. 3). Some of these 8 proteins which
were downregulated following stimulation with TGF-β1 have not been
previously linked with TGF-β1 signaling. Bioinformatics analysis of
these 8 candidate proteins using miRNA target prediction algorithms
revealed that all 8 proteins contained more than one predicted
miRNA binding site. Of note, some predicted miRNAs, which targeted
those 8 proteins were also identified by miRNA microarray analysis
and their expression levels showed an opposite trend in response to
stimulation with TGF-β1. More specifically, we noted that miR-149
and miR-199a-3p directly target PPIA and NDKB, respectively in
response to stimulation with TGF-β1 in the mouse GC-1 spg cells,
suggesting that these miRNAs play an important role in controlling
downstream gene expression following the activation of TGF-β1
signaling. We therefore hypothesized that these proteins may be
suppressed by the miRNAs in response to TGF-β1.
Nucleoside diphosphate (NDP) kinase B, also known as
NME2, NM23B and NM23-H2, is an isoform of multifunctional proteins
found to be responsible for the synthesis of nucleoside
triphosphates in eukaryotes and is involved in a variety of
cellular activities, including proliferation, development, adhesion
and differentiation (21,22). NDKB is strongly expressed in the
testis (23). High levels of NDKB
have been observed at specific locations in post-meiotic germ
cells, and its distribution is reminiscent of the microtubular
structure of the manchette. In mature spermatozoa, NDKB is present
at specific locations in the head and flagellar region. Normally,
during sperm differentiation, microtubules and motor proteins
(cytosolic dynein and kinesin) of the manchette and the centrosome
region require nucleotide synthesis for the spermatid nuclear
shaping and sperm tail assembly (24,25). Studies have demonstrated an
association of NDP kinases with microtubules and NDKB may thus have
specific functions in the phosphotransfer network involved in
spermiogenesis and flagellar movement (26,27). Our findings provide evidence of a
direct mechanistic link of NDKB protein in TGF-β1 signaling, in
which NDKB may be regulated by miR199a-3p, a member of an essential
family that controls the fate of cell survival and death.
Another downregulated protein, PPIA, also known as
cyclophilin A (CyPA), is an ubiquitously distributed protein
belonging to the immunophilin family and regulates protein folding
and trafficking (28). Although
PPIA/CyPA was initially believed to function primarily as an
intracellular protein, it has recently been shown that it can be
secreted by cells in response to inflammatory stimuli (28). Another study demonstrated that
PPIA/CyPA is present in rat germ cells, including pachytene
spermatocytes, spermatids, interstitial cells and sertoli cell
nuclei, and is associated with spermatocyte apoptosis (29). As TGF-β1 signaling plays a major
role throughout development and in adult tissue homeostasis, our
findings also provide evidence of a direct mechanistic link between
PPIA protein and miR-149 or miR-196a in TGF-β1 signaling. This
suggests the existence of some key mediators of the TGF-β1 pathway
in early germ cell development.
In conclusion, we identified several important
miRNAs and proteins as potential targets for TGF-β1 signaling. Of
these miRNAs, miR-199a-3p and miR-149 (or miR-196a) were shown to
directly target PPIA and NDKB and modulate the response to TGF-β1
signaling in the GC-1 spg cell line. The results obtained in this
study may prove to be useful for further research on the function
and mechanisms of action of miRNAs, as well as their targets which
are involved in the process of TGF-β1-mediated spermatogenesis. Our
data may lead to the development of novel therapeutic approaches
for the treatment of male infertility.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (no. 81270735) and project 111
of the Ministry of Education of China (B14033).
References
|
1
|
Huleihel M and Lunenfeld E: Regulation of
spermatogenesis by paracrine/autocrine testicular factors. Asian J
Androl. 6:259–268. 2004.PubMed/NCBI
|
|
2
|
Petersen C, Svechnikov K, Fröysa B and
Söder O: The p38 MAPK pathway mediates interleukin-1-induced
Sertoli cell proliferation. Cytokine. 32:51–59. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hedger MP and Meinhardt A: Cytokines and
the immune-testicular axis. J Reprod Immunol. 58:1–26. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gordon KJ and Blobe GC: Role of
transforming growth factor-beta superfamily signaling pathways in
human disease. Biochim Biophys Acta. 1782:197–228. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang YQ, He XZ, Zhang JS, et al:
Stage-specific localization of transforming growth factor beta1 and
beta3 and their receptors during spermatogenesis in men. Asian J
Androl. 6:105–109. 2004.PubMed/NCBI
|
|
6
|
Dobashi M, Fujisawa M, Yamazaki T, et al:
Distribution of intracellular and extracellular expression of
transforming growth factor-beta1 (TGF-beta1) in human testis and
their association with spermatogenesis. Asian J Androl. 4:105–109.
2002.PubMed/NCBI
|
|
7
|
Fan YS, Hu YJ and Yang WX: TGF-β
superfamily: how does it regulate testis development. Mol Biol Rep.
39:4727–4741. 2012. View Article : Google Scholar
|
|
8
|
Massagué J: TGF beta in cancer. Cell.
134:215–230. 2008. View Article : Google Scholar
|
|
9
|
Shukla GC, Singh J and Barik S: Micrornas:
processing, maturation, target recognition and regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
10
|
Ro S, Park C, Sanders KM, McCarrey JR and
Yan W: Cloning and expression profiling of testis-expressed
microRNAs. Dev Biol. 311:592–602. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hayashi K, Chuva de Sousa Lopes SM, Kaneda
M, et al: MicroRNA biogenesis is required for mouse primordial germ
cell development and spermatogenesis. PLOS One. 3:e17382008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McIver SC, Roman SD, Nixon B and
McLaughlin EA: miRNA and mammalian male germ cells. Hum Reprod
Update. 18:44–59. 2012. View Article : Google Scholar
|
|
13
|
Zhang Y, Fan KJ, Sun Q, et al: Functional
screening for miRNAs targeting Smad4 identified miR-199a as a
negative regulator of TGF-β signalling pathway. Nucleic Acids Res.
40:9286–9297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gargalionis AN and Basdra EK: Insights in
microRNAs biology. Curr Top Med Chem. 13:1493–1502. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang B, Hsu SH, Majumder S, et al:
TGFbeta-mediated upregulation of hepatic miR-181b promotes
hepatocarcinogenesis by targeting TIMP3. Oncogene. 29:1787–1797.
2010. View Article : Google Scholar :
|
|
16
|
Cao M, Seike M, Soeno C, et al: miR-23a
regulates TGF-β-induced epithelial-mesenchymal transition by
targeting E-cadherin in lung cancer cells. Int J Oncol. 41:869–875.
2012.PubMed/NCBI
|
|
17
|
Jenkins RH, Martin J, Phillips AO, et al:
Transforming growth factor β1 represses proximal tubular cell
microRNA-192 expression through decreased hepatocyte nuclear factor
DNA binding. Biochem J. 443:407–416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lino Cardenas CL, Henaoui IS, Courcot E,
et al: miR-199a-5p Is upregulated during fibrogenic response to
tissue injury and mediates TGFbeta-induced lung fibroblast
activation by targeting caveolin-1. PLoS Genet. 9:e10032912013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Honda N, Jinnin M, Kajihara I, et al:
TGF-β-mediated down-regulation of microRNA-196a contributes to the
constitutive upregulated type I collagen expression in scleroderma
dermal fibroblasts. J Immunol. 188:3323–3331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mungunsukh O and Day RM: Transforming
growth factor-β1 selectively inhibits hepatocyte growth factor
expression via a micro-RNA-199-dependent posttranscriptional
mechanism. Mol Biol Cell. 24:2088–2097. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kang Y, Lee DC, Han J, et al: NM23-H2
involves in negative regulation of Diva and Bcl2L10 in apoptosis
signaling. Biochem Biophys Res Commun. 359:76–82. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Polanski R, Maguire M, Nield PC, et al:
MDM2 interacts with NME2 (non-metastatic cells 2, protein) and
suppresses the ability of NME2 to negatively regulate cell
motility. Carcinogenesis. 32:1133–1142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Munier A, Serres C, Kann ML, et al:
Nm23/NDP kinases in human male germ cells: role in spermiogenesis
and sperm motility? Exp Cell Res. 289:295–306. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshida T, Ioshii SO, Imanaka-Yoshida K
and Izutsu K: Association of cytoplasmic dynein with manchette
microtubules and spermatid nuclear envelope during spermiogenesis
in rats. J Cell Sci. 107:625–633. 1994.PubMed/NCBI
|
|
25
|
Kierszenbaum AL: Intramanchette transport
(IMT): managing the making of the spermatid head, centrosome, and
tail. Mol Reprod Dev. 63:1–4. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nosaka K, Kawahara M, Masuda M, et al:
Association of nucleoside diphosphate kinase nm23-H2 with human
telomeres. Biochem Biophys Res Commun. 243:342–348. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boissan M and Lacombe ML: Learning about
the functions of NME/NM23: lessons from knockout mice to silencing
strategies. Naunyn Schmiedebergs Arch Pharmacol. 384:421–431. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nigro P, Pompilio G and Capogrossi MC:
Cyclophilin a: a key player for human disease. Cell Death Dis.
4:e8882013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wine RN, Ku WW, Li LH and Chapin RE:
Cyclophilin A is present in rat germ cells and is associated with
spermatocyte apoptosis. Reproductive Toxicology Group. Biol Reprod.
56:439–446. 1997. View Article : Google Scholar : PubMed/NCBI
|