Introduction
Epilepsy is one of the most common neurological
disorders and nearly a quarter of patients with seizures have
intractable epilepsy, which is also referred to as ‘medically
refractory/drug-resistant’ or ‘pharmacoresistant’ (1). Malformations of cortical development
(MCD) are often observed in patients with intractable epilepsy
(2). In our previous
observations, we collected 3 surgical specimens from patients with
intractable epilepsy and MCD. Using Illumina BeadChip technology,
we identified >400 abnormally expressed genes in the
epileptogenic zone (unpublished data). The RNA-binding Fox 1
(Rbfox1) gene is one of the intriguing abnormally expressed
genes.
Mutations in RBFOX1 [also known as ataxin
2-binding protein 1 (A2BP1)] have been observed in a growing number
of neurodevelopmental disorders, including epilepsy, mental
retardation (3,4) and autism spectrum disorder (5,6).
RBFOX1 can regulate both splicing and transcriptional networks in
human neuronal development, and can control neuronal excitation
(7-9). Alternative splicing is the process
of removing introns from pre-mRNA transcripts and joining exons in
different combinations (10).
There is increasing evidence indicating that alternative splicing
affects neuronal development, controls functions in the mature
brain and plays important roles in a number of neurological
disorders (11). Rbfox1 regulates
alternative splicing of neuronal transcripts by binding the
sequence (U) GCAUG in introns flanking alternative exons (12,13). Target transcripts of Rbfox1,
include those encoding N-methyl D-aspartate 1 [Grin1, also
known as N-methyl-D-aspartate receptor subunit NR1 (NMDAR1)],
synaptosomal-associated protein, 25 kDa (SNAP-25 or Snap25)
and sodium channel, voltage gated, type VIII, alpha subunit
(Scn8a, also known as Nav1.6) have been implicated in
epileptogenesis (9).
In 2011, Gehman et al found that the stimulus
intensities required to evoke field excitatory post-synaptic
potentials (fEPSPs) in the Rbfox1−/− brain were
lower than those required for the wild-type brain; thus, the
authors came to the conclusion that the deletion of Rbfox1
in mice was responsible for neuronal hyperexcitation (9). Clinical studies have revealed
mutations (3–5) or the downregulation (5) of RBFOX1 in the specimens
obtained from patients with epilepsy or MCD; specifically, the
specimens were not from the actual epileptogenic lesions. However,
the expression of RBFOX1 varies according to tissue type
(12,14). Thus, it is necessary to re-examine
the expression of RBFOX1 in cortical lesions from patients with MCD
and epilepsy.
It has been suggested that the RBFOX1 protein plays
a major role in the cellular response to hyperexcitation. To test
this hypothesis, we altered the expression of Rbfox1 in cultured
cortical neurons and measured their electrophysiological properties
using whole-cell patch clamp recordings. Some well known Rbfox1
target transcripts which have been directly linked to epilepsy were
also investigated.
Materials and methods
Subjects
We recruited 15 patients with MCD and intractable
epilepsy and documented their brain malformations using MRI. All
patients underwent pre-operative clinical evaluations, including
seizure charts, MRIs, a 24-h electroencephalogram or a video
electroencephalogram, sphenoidal electrode monitoring and
intraoperative electrocorticography. An intractable seizure was
defined as the ‘failure of adequate trials of 2 tolerated and
appropriately selected and used anti-epileptic drug schedules to
achieve sustained seizure freedom’. Surgical specimens were
obtained from these patients at the First Affiliated Hospital of
Chongqing Medical University and Xinqiao Hospital of the Third
Military Medical University (both in Chongqing, China). We detected
the expression of doublecortin (DCX) and tuberous sclerosis
(TSC)1/TSC2 in all the specimens by immunohistochemistry. Samples
without an abnormal expression of DCX and TSC1/TSC2 were included
in this study. A summary of the clinical characteristics of all the
patients is shown in Table I.
 | Table IClinical characteristics of patients
with MCD and intractable epilepsy. |
Table I
Clinical characteristics of patients
with MCD and intractable epilepsy.
| Patient no. | Gender | Age at surgery
(years) | Seizure duration
(year) | AEDs prior to
surgery | Resected regions of
cases | Classification |
|---|
| 1 | F | 24 | 18 | CBZ, PHT, VPA,
PB | TNr | FCD |
| 2 | M | 33 | 10 | VPA, CBZ, TPM | TNl | FCD |
| 3 | M | 28 | 18 | CBZ, VPA, TPM,
LEV | TNr | FCD |
| 4 | M | 36 | 12 | VPA, TPM, LEV, PB,
CZP | TNr | FCD |
| 5 | F | 18 | 10 | CBZ, PHT, TPM,
PB | TNr | FCD |
| 6 | F | 30 | 12 | CBZ, TPM, PB | TNr | FCD |
| 7 | M | 36 | 13 | CBZ, VPA, TPM | TNl | FCD |
| 8 | F | 18 | 17 | VPA, OCBZ, CBZ | TNl | TSC |
| 9 | M | 23 | 20 | CBZ, VPA, TPM,
PB | TNl | TSC |
| 10 | M | 11 | 9 | VPA, OCBZ, CBZ,
TPM | TNr | TSC |
| 11 | M | 34 | 28 | CBZ, PHT, PB | TNr | DC |
| 12 | F | 20 | 15 | TB, CBZ, VPA,
TPM | TNl | DC |
| 13 | F | 18 | 12 | CBZ, PB, TPM | TNr | DC |
| 14 | F | 15 | 6 | VPA, CBZ, PB | TNl | DC |
| 15 | F | 25 | 22 | CBZ, VPA, PHT | TNl | DC |
The control specimens were obtained from 7 patients
who underwent neurosurgical intervention due to brain trauma at the
Neurosurgical Department of the First Affiliated Hospital of
Chongqing Medical University and Xinqiao Hospital of the Third
Military Medical University. The surgical specimens were not from
the contusion area. The normal brain tissues were resected as these
patients had sustained severe craniocerebral injury (SCCI) and
internal decompression treatments were unavoidable. None of the
specimens were exposed prior to the neurosurgical intervention. All
the specimens were resected within 1 h from injury. None of the
patients had ever suffered from epilepsy until the neurosurgical
intervention. There were no abnormalities observed in the specimens
from the control patients. The clinical characteristics of the
controls are presented in Table
II.
 | Table IIClinical characteristics of the
controls. |
Table II
Clinical characteristics of the
controls.
| Patient no. | Gender | Age (years) | Etiology
diagnosis | Resection
tissue | Adjacent tissue
pathology |
|---|
| 1 | M | 33 | Trauma | TNl | n |
| 2 | M | 21 | Trauma | TNr | n |
| 3 | F | 31 | Trauma | TNr | n |
| 4 | F | 14 | Trauma | TNr | n |
| 5 | F | 25 | Trauma | TNl | n |
| 6 | M | 13 | Trauma | TNl | n |
| 7 | M | 26 | Trauma | TNl | n |
The resected tissues from the parietal lobe were
accepted for study. All the specimens were washed with cold 0.9%
saline to erase superficial blood. Subsequently, one section from
each specimen was fixed in 4% paraformaldehyde immediately after
being resected, and the other section was preserved in liquid
nitrogen.
Ethics statement
The present study was approved by the Ethics
Committee on Human Research at Chongqing Medical University and
written informed consent was obtained from the patients or their
relatives in accordance with the Helsinki Declaration.
Cortical neuronal cultures and lentiviral
transfection
All animal procedures were approved by the
Commission of Chongqing Medical University for the ethics of
experiments on animals and were carried out in accordance with the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals (NIH publications nos. 80–23) revised in
1996. The neocortex was dissected from Sprague-Dawley rat pups on
postnatal age (P)0 as previously described (15,16). The neocortex was dissected from
Sprague-Dawley rat pups on P0, transferred to D-Hank’s solution
(HyClone, Logan, UT, USA) and cut into small sections (~1
mm3) using eye scissors. The tissue sections were
trypsinized in 4 ml solution (0.125%, w/v) for 10 min at 37°C, and
filtered through a mesh screen (39 μm) to obtain a
single-cell suspension, followed by centrifugation at 1,000 × g for
5 min at 25°C. The isolated neurons were then seeded in
poly-L-Lysine-coated 24-well plates at 5–8×105
cells/well. The neurons were cultured in DMEM/F12 medium
supplemented with 10% fetal bovine serum and 1%
penicillin-streptomycin (all from Gibco, Carlsbad, CA, USA). After
6–8 h of incubation, the medium was changed to DMEM/F12
supplemented with 2% B27 (Gibco) and 1% penicillin-streptomycin.
The cells were cultured in an incubator at 37°C with circulating
air and 5% carbon dioxide and the medium was changed every
3–4 days. All efforts were made to minimize the number of
animals used and any possible suffering caused to them.
Lentiviral-mediated-α-Rbfox1 (Rbfox1) and the
control lentivirus (lentivirus) were synthesized at Shanghai
GeneChem Co., Ltd. (Shanghai, China; nos. V20130406004 and
V20130303005). Following 72 h of incubation, the
lentiviral-mediated-α-Rbfox1 and the control lentivirus media were
added to the cultured neurons with a multiplicity of infection
(MOI) of 5. Untransfected cells were used as controls. To
calclulate the amount of virus required for a certain MOI, the
following formula was used: total ml of virus required = cells ×
desired MOI/(plaque forming units/ml). All the media was changed 12
h later. Four days after transfection, RT-qPCR, western blot
analysis, immunofluorescence assay and whole cell patch-clamp
recordings were performed.
Immunohistochemistry
Immunohistochemical analyses were performed using a
previously published streptavidin-biotin-peroxidase complex method
(17). Paraffin sections were
dried at 60°C for 20 min, dewaxed in xylene, rehydrated through a
graded series of alcohol and immersed in 3% hydrogen peroxide for
15 min to block endogenous peroxidase activity. The sections were
then blocked with normal non-immune goat serum and incubated with
anti-Rbfox1 antibody (no. ab94581, 1:200; Abcam, Cambridge, UK) at
4°C overnight. Subsequently, the sections were incubated with
biotin-labeled secondary antibody at a 1:500 dilution (no. A0277;
Beyotime Institute of Biotechnology, Shanghai, China) for 1 h at
room temperature and stained with 3,3-diaminobenzidine (DAB;
Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China)
after washing with phosphate-buffered saline (PBS) again. Finally,
the sections were counterstained with haematoxylin, dehydrated and
mounted. PBS was used to replace the primary antibody as a negative
control. Three types of MCD, including focal cortical dysplasia
(FCD), TSC and double cortex (DC) were examined. The sections were
observed and images were acquired using an Olympus BX51 microscope
fitted with a digital camera and DP Controller software (Olympus,
Tokyo, Japan).
Immunofluorescence
Immunofluorescence assay was performed to examine
the expression of Rbfox1 and its target transcripts. The cells were
washed twice with ice-cold PBS and fixed in 4% paraformaldehyde for
15 min at room temperature. After washing with PBS 3 times, the
cells were incubated with 0.2% Triton X-100 for 15 min. To block
nonspecific binding, the cells incubated with 5% albumin from
bovine serum (BSA) at room temperature for 30 min, followed by
incubation with a primary antibody at 4°C overnight. The cells were
then incubated with a secondary antibody for 30 min at 37°C in a
humidified atmosphere. After washing 3 times with PBS, the nuclei
were stained with DAPI (no. C1005; Beyotime Institute of
Biotechnology). The following primary antibodies were used:
α-Rbfox1 (no. ab94581, 1:1,000; Abcam), microtubule-associated
protein 2 (MAP2; no. sc-74421, 1:100; Santa Cruz Biotechnology
Inc., Delaware, CA, USA), Nav1.6 (no. ab65166, 1:1,000), NMDAR1
(no. ab28669, 1:1,000) and SNAP25 (no. ab24732, 1:1,000) (all from
Abcam). DyLight 649-conjugated goat anti-rabbit IgG (1:100) and
DyLight 649-conjugated goat anti-mouse IgG (1:100) (both from
CWbiotech, Beijing, China), DyLight 649-conjugated donkey anti-goat
IgG (1:100; Jackson ImmunoResearch Laboratories, Inc., West Grove,
PA, USA), FITC-conjugated goat anti-rabbit IgG (1:100) and
FITC-conjugated rabbit anti-mouse IgG (1:100) (both from Beijing
DingGuo ChangSheng Biotechnology Co., Ltd., Beijing, China) were
used as the secondary antibodies. Neurons were observed under a
laser scanning confocal microscope (TCS-SP2; Leica Microsystems
GmbH, Wetzlar, Germany).
Whole-cell patch clamp recordings
Whole-cell patch clamp recordings were acquired from
cortical neurons isolated from the rat neocortex following a 4-day
culture using previously established protocols (18). Briefly, a neuronal culture plate
was mounted on the stage of an inverted microscope (IX-51; Olympus)
and continuously perfused with standard extracellular solution
containing 140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 2 mM
CaCl2, 10 mM HEPES, 10 mM D-glucose and 10 mM
tetraethyl-ammonium (TEA) Cl (pH 7.4 with CsOH). The experiments
were performed at room temperature (22–24°C). Recordings
were made using patch electrodes with a resistance of 5-10 MΩ. The
internal solution contained 130 mM KAsp, 5 mM ATP-Na2, 2 mM
MgCl2, 1 mM CaCl2 and 10 mM HEPES (pH 7.2
with CsOH). The osmolarity was adjusted to 290±10 mOsm with
sucrose. The intracellular recordings were carried out using an
EPC-10 amplifier (HEKA Elektronik Dr. Schulze GmbH,
Lambrecht/Pfalz, Germany) in current clamp mode. The pipette
resistance and capacitance were compensated electronically after
the establishment of a gigaseal. Following whole-cell capacitance
compensation, recordings were made only when the series resistance
was <20 MΩ. Cultured neurons with small dendritic arborizations,
long axon and soma diameters of 20-26 μm were selected for
the electrophysiological recordings to avoid space clamp artifacts.
Routinely, 60-80% series resistance compensation was employed,
continually monitored and adjusted as required. Whole-cell
resistance and resting membrane potential were also monitored
before and during the experiments and a cell was accepted for study
only if these parameters remained stable.
Spontaneous firing, voltage-gated Na+
currents, and evoked action potentials (APs) were measured. Under
current-clamp mode, the spontaneous firing of the cultured cortical
neurons was firstly recorded. In order to further investigate
whether Rbfox1 can influence the excitability of the cultured
cortical neurons, the cells were held at −80 mV, and a series of 20
msec depolarizing current pulses in 10 mV increments from −80 to 70
mV was delivered to elicit the cell generating voltage-gated
Na+ currents. The cells were then held at −70 mV, and a
series of 40 msec depolarizing current pulses in 10 pA increments
from 0 to 80 pA was delivered to elicit the cell generating evoked
APs. Voltage-gated Na+ currents were measured from
threshold to the Na+ currents peak. The AP amplitudes
were measured from the threshold to the AP peak. Data were
digitized and transferred to videotape using a Clamp-fit 10.0
device (from Axon Instruments, Sunnyvale, CA, USA). Fitting and
statistical analysis were performed using Igor4.03 (WaveMetrics,
Portland, OR, USA), SPSS 19 (SPSS Inc., Chicago, IL, USA) and
Origin7.5 sofware (OriginLab Corp., Northampton, MA, USA).
Western blot analysis
Protein levels in the human cortical tissue and
cultured cortical neurons were measured by western blot analysis.
We analyzed all the 15 specimens obtained from patients with both
MCD and intractable epilepsy and then analyzed the optical density
of 7 representative examples, including 2 patients with tuberous
sclerosis (lanes/bars 4 and 5), 2 with focal cortical dysplasia
(lanes/bars 6 and 7) and 3 with double cortex (lanes/bars 8, 9 and
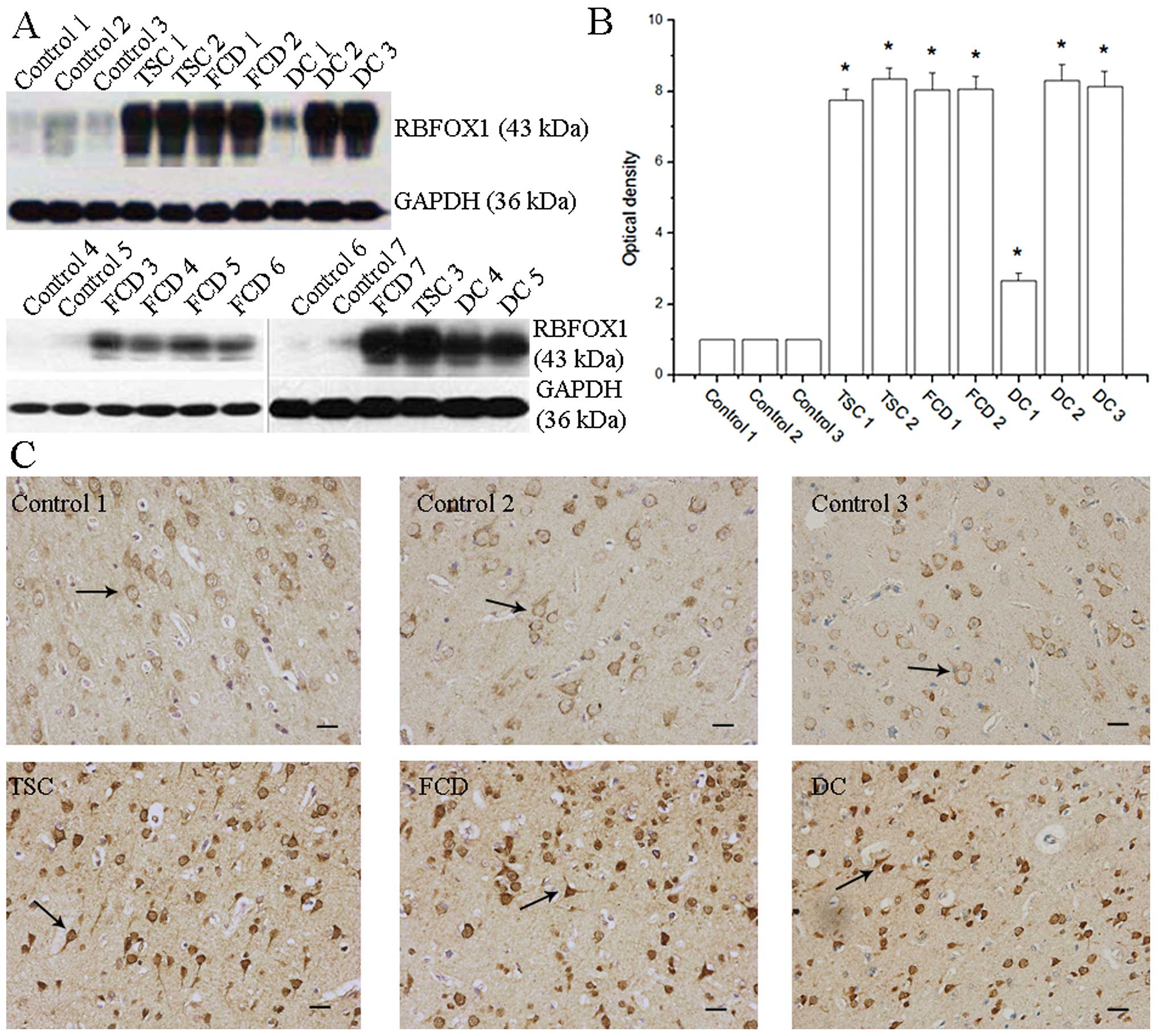
10) (Fig. 1).
Total protein which was derived from the human
cortical tissue and cultured rat cortical neurons was harvested
using RIPA lysis buffer containing a protease and phosphatase
inhibitor cocktail (KeyGen Biotech Co., Ltd., Nanjing, China). The
protein concentrations were measured using the Bradford method
(Beyotime Institute of Biotechnology). The protein samples (50
μg) were fractionated by 10% SDS-polyacrylamide gel
electrophoresis, electroblotted onto a polyvinylidene difluoride
(PVDF; Millipore, Billerica, MA, USA) membrane and immunoblotted
with a primary antibody and GAPDH (1:1,000; Beijing DingGuo
ChangSheng Biotechnology Co., Ltd.) as an internal control. The
following primary antibodies were used: α-Rbfox1 (no. ab94581,
1:1,000; Abcam), Nav1.6 (no. ASC-009, 1:1,000; Alomone, Jerusalem,
Israel), NMDAR1 (no. ab28669, 1:1,000) and SNAP25 (no. b24732,
1:1,000) (both from Abcam). Horseradish peroxidase-conjugated
secondary antibodies (nos. A0208, A0216 and A0181, 1:1,000;
Beyotime Institute of Biotechnology) were used. RBFOX1, Nav1.6,
NMDAR1 and SNAP25 proteins were identified as immunopositive bands
with molecular weights of 43, 220, 120 and 29 kDa, respectively.
The bands were detected using the Pierce ECL western blotting
substrate (Thermo Fisher Scientific, Inc., Rockford, IL, USA),
scanned with a Bio-rad GS-800™ calibrated densitometer and analyzed
using Quantity One software version 4.6.2 (Bio-Rad Laboratories,
Hercules, CA, USA).
RT-qPCR
RT-qPCR was used for Rbfox1 only. The
cultured rat cortical neurons were homogenized prior to RNA
extraction in 1 ml of TRIzol. All RNA was quantified using a
spectrophotometer (NanoDrop1000; Thermo Fisher Scientific, Inc.) to
determine the optical density at 260/280 nm ratios. Reverse
transcription was performed on 2 μg RNA using the K1622
RevertAid First Strand cDNA Synthesis kit (Fermentas Canada Co.,
Ltd., Burlington, ON, Canada) according to the manufacturer’s
instructions. The primers for rat α-Rbfox1 (forward,
5′-CTACAGTGACAGTTACGGACGAG-3′ and reverse,
5′-ATGAAGAAAGAACGAGACCC-3′) were purchased from BGI Tech (Shenzhen,
China). RT-qPCR was performed using 2X SYBR Mix SsoAdvanced
SYBR-Green Supermix (Bio-Rad Laboratories). Amplification was
conducted with an initial denature action step at 95°C for 5 min,
followed by 95°C for 5 sec, 60°C for 30 sec and 72°C for 15 sec, 40
cycles, 72°C for 10 min and a final melting curve. Gene analysis
was carried out in triplicate. Gapdh was used as a loading
control. The data were collected and analyzed using OneStep
Software (Applied Biosystems, Foster City, CA, USA). Relative
quantification was performed using the 2−ΔΔCt method, as
previously described (19).
Statistical analysis
All data are expressed as the means ± SD. An
independent samples t-test was used for 2 sample comparison.
One-way analysis of variance (ANOVA) followed by Dunnett t (2
sided) test or Dunnett’s T3 test was used for multiple comparisons
of treated/patient groups against the controls. Linear fits and
statistical analyses were performed using Igor 4.03 (WaveMetrics),
SPSS 19 (SPSS Inc.) and OriginPro 7.5 sofrware (OriginLab Corp.)
software. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Patient profiles
A total of 15 individuals (8 females and 7 males)
aged from 11 to 36 years with both MCD and intractable epilepsy
were recruited in this study. In total, 7 patients had focal
cortical dysplasia, 5 had a double cortex and 3 had tuberous
sclerosis. No altered expression of the DCX or
TSC1/TSC2 genes was observed in the 15 specimens obtained
from these patients.
Control samples were obtained from 7 brain trauma
patients who underwent neurosurgical intervention. There were no
histopathological abnormalities observed in these patients
(Table II). There was no
statistically significant difference in the mean age between the
controls and the patients with MCD/epilepsy [t0.05/2(14,4)=0.363, P=0.721, independent
samples t-test].
Protein expression of RBFOX1 in
patients
The expression of RBFOX1 protein in tissues from
both the controls and patients with MCD and intractable epilepsy
was analyzed by western blot analysis and immunohistochemistry
assays. The results from western blot analysis revealed that the
protein expression of RBFOX1 was markedly upregulated, not only in
the patients with double cortex, but also in patients with focal
cortical dysplasia and tuberous sclerosis (Fig. 1A). There was a significant
difference in the RBFOX1 protein level between the patients with
epilepsy/MCD and the controls [F(9,20)=199.855, P=0.000, ANOVA].
ANOVA followed by the Dunnett t (2 sided) test was used for
multiple comparisons of the protein levels in the patients with
epilepsy/MCD compared to the average levels of the controls (lanes
1–3). The results revealed a significant difference in the
protein levels in the epilepsy/MCD group (P=0.004, 0.004, 0.009,
0.005, 0.041, 0.008 and 0.007 for bars 4–10, respectively) against
the average of the controls (bars 1–3; Fig. 1B). Specimens from patients with
double cortex, focal cortical dysplasia and tuberous sclerosis were
analyzed by immunohistochemistry. A significantly stronger RBFOX1
staining signal was observed in the cortical lesions of the
patients with MCD/epilepsy compared to the controls (Fig. 1C). In summary, these data
demonstrate the overexpression of RBFOX1 protein in the cortical
lesions of patients with MCD and intractable epilepsy.
Assessment of transfection
efficiency
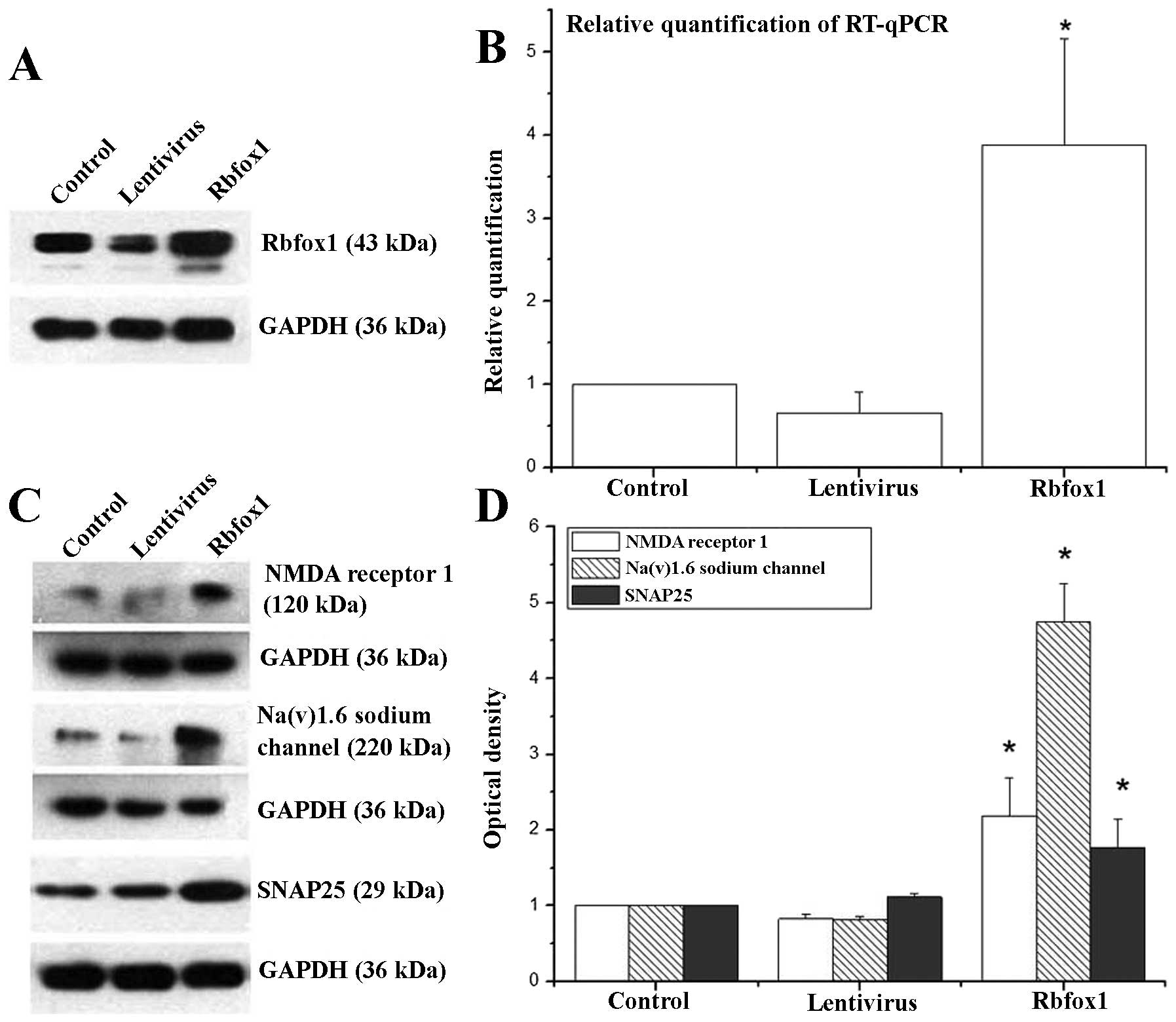
The transfection efficiency was assessed 4 days
following transfection by western blot analysis, RT-qPCR and
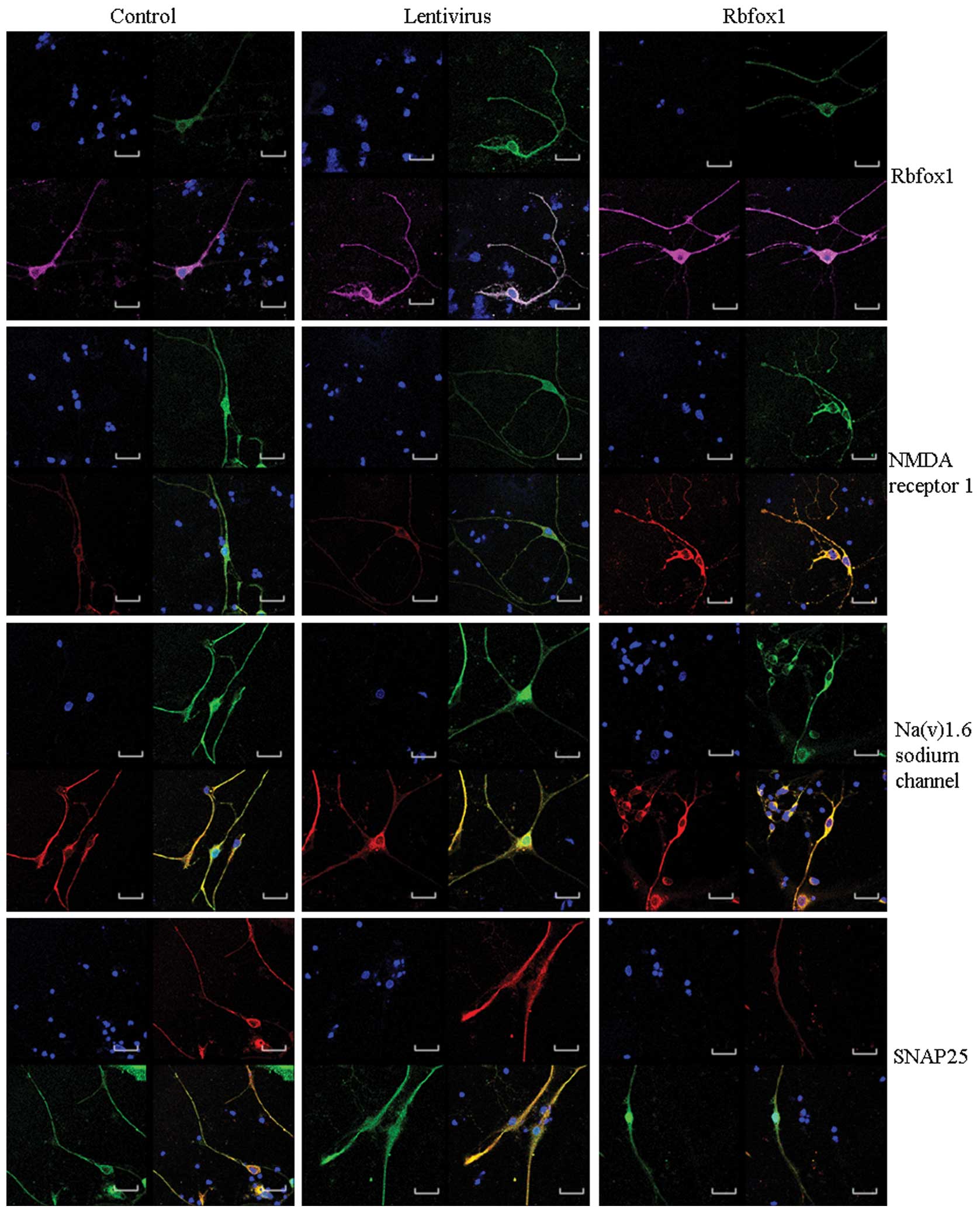
immunofluorescence staining. Western blot analysis (Fig. 2A) and immunofluorescence staining
(Fig. 4) revealed a significant
upregulation of Rbfox1 expression in the lentiviral-mediated-Rbfox1
transfected neuron group (Rbfox1 group). As shown by western blot
analysis, there was a significant difference in the Rbfox1 protein
level between the Rbfox1 group, the control-lenti-viral-vector
transfected neurons (lentivirus group) and the untransfected group
(control group) [F(2,6)=220.821,
P=0.000, ANOVA]. ANOVA followed by Dunnett’s T3 test was used for
multiple comparisons of the Rbfox1 protein level in the 3 groups.
Statistical analysis revealed a significant difference between the
Rbfox1 group (P=0.010) and the control group; however, no
significant difference was observed between the lentivirus group
(P=0.056) and the control group. RT-qPCR analysis (Fig. 2B) revealed that the Rbfox1
mRNA level was 387 and 63% of the controls in the Rbfox1 group and
lentivirus group, respectively. There was a significant difference
in the Rbfox1 mRNA level between the Rbfox1 and lentivirus
and the control groups [F(2,6)=16.242, P=0.004, ANOVA]. ANOVA
followed by the Dunnett t (2 sided) test was used for
multiple comparisons of the mRNA levels in the 3 groups. The
results revealed a significant difference in Rbfox1 mRNA
levels between the Rbfox1 group (P=0.006) and the control group;
however, no significant difference was observed between the
lentivirus group (P=0.812) and the control group.
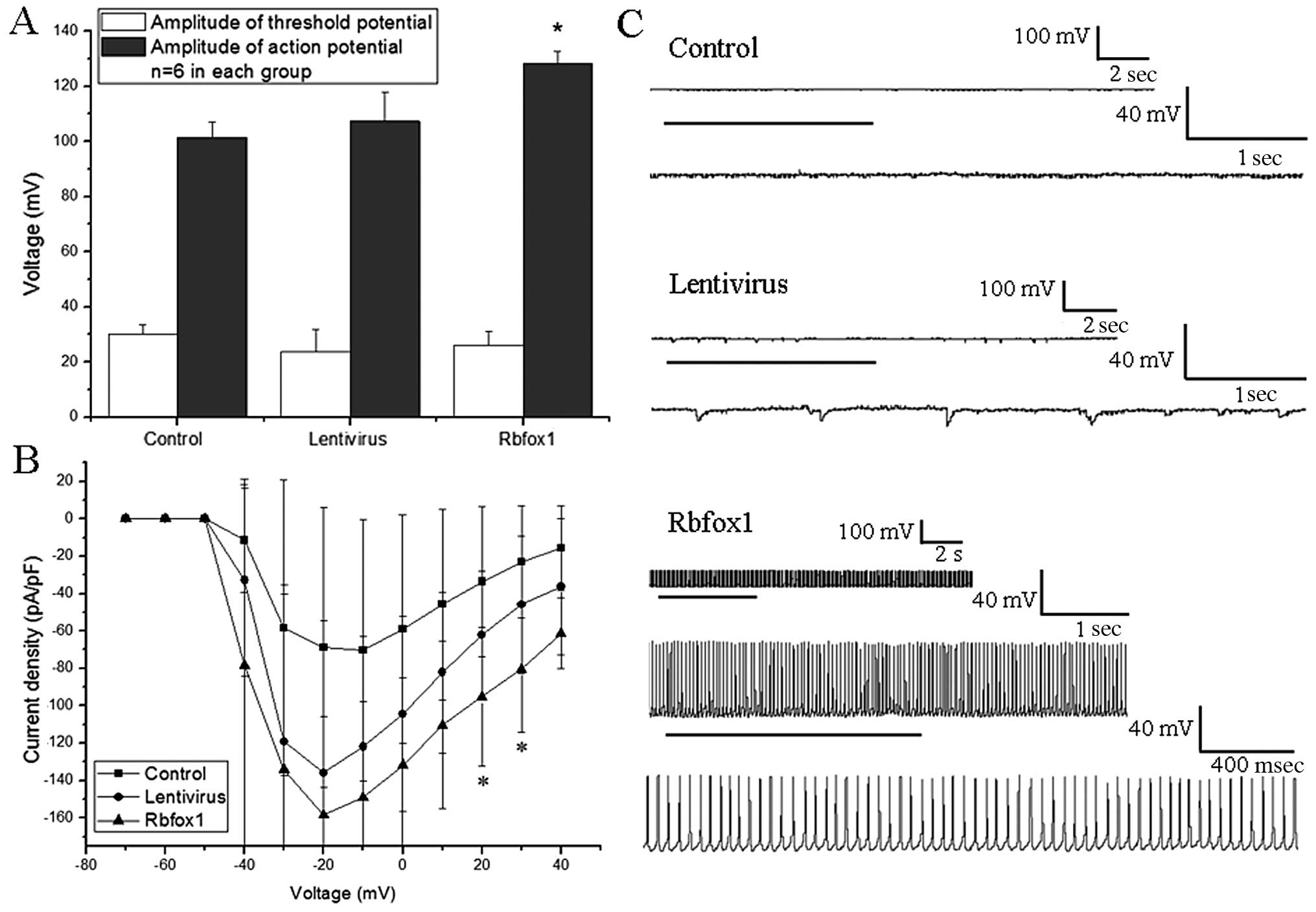
Whole-cell patch clamp recordings
We measured the absolute value of threshold
potential, AP and sodium currents in each group (n=6). Spontaneous
discharges were also recorded in each group. The lentivirus group
was used as a reference. There was no statistically significant
difference observed in membrane capacitance [F(2,15)=2.889, P=0.061, ANOVA] or
threshold potential between the Rbfox1, lentivirus and control
groups [F(2,15)=2.022, P=0.167, ANOVA].
Statistical analysis revealed a significant difference in AP
between the 3 groups [F(2,15)=22.115, P=0.000, ANOVA,
Fig. 3A]. ANOVA followed by the
Dunnett t (2-sided) test was used for multiple comparisons of the
AP between the 3 groups. The amplitude of APs in the Rbfox1 group
was markedly increased compared to the lentivirus group (P=0.000).
No significant difference was observed however, between the
untransfected (control) group and the lentivirus group
(P=0.307).
No statistical difference was observed in the
Na+ current density when the step voltage was from −40
to 10 mV [F(2,15)=1.578, 1.314, 2.626, 2.631,
2.813, 2.924, respectively, P=0.239, 0.298, 0.105, 0.105, 0.092,
0.085, respectively, ANOVA] and 40 mV [F(2,15)=3.399, P=0.061, ANOVA].
Statistical analysis revealed a significant difference in the
Na+ current density between the 3 groups when the step
voltage was 20 or 30 mV [F(2,15)=4.093 and 4.441, P=0.038 and
0.031, respectively, ANOVA, Fig.
3B]. ANOVA followed by the Dunnett t (2-sided) test was used
for multiple comparisons of the Na+ current density
between the Rbfox1 and untransfected groups against the lentivirus
group. Statistical analysis revealed a significant difference
between the Rbfox1 group and the lentivirus group at 20 and 30 mV
(P=0.022 and 0.018, respectively). No significant difference was
observed between the untransfected group and the lentivirus group
at 20 and 30 mV (P=0.246 and 0.165, respectively).
In the case of spontaneous discharges, there were no
spontaneous, recurrent, epileptiform discharges (SREDs) observed in
the untransfected and lentivirus groups. This epileptiform activity
was characterized by abruptly developing, repetitive high-frequency
burst spiking that overlaid a paroxysmal depolarization shift
(20,21). In the Rbfox1 group, 5 (26.32%) out
of 19 neurons displayed SREDs (Fig.
3C).
Overall, these data suggest that changes in the
Rbfox1 protein level result in changes in AP amplitude and SREDs,
and cause a generalized increase in the excitability of cultured
cortical neurons.
Identification of Rbfox1 target
transcripts involved in epilepsy
We examined the expression of NMDAR1 (Grin1),
SNAP-25 (Snap25) and Scn8a (or Nav1.6) in our
transfected cortical neurons by western blot analysis (Fig. 2C) and immunofluorescence staining
(Fig. 4). When the Rbfox1 protein
level was increased, all 3 proteins were upregulated.
Statistical analysis revealed a significant
difference in the levels of the NMDAR1, SNAP-25 and Nav1.6
transcripts between the 3 groups [F(2,6)=19.961, 8.33 and 162.779,
respectively, P=0.002, 0.008 and 0.000, respectively, ANOVA,
Fig. 2D]. ANOVA followed by the
Dunnett (2-sided) test was used for multiple comparisons between
the levels of NMDAR1 and SNAP-25 in the Rbfox1 and lentivirus group
and the control group. Statistical analysis revealed a significant
difference in the levels of these transcripts between the Rbfox1
group and the control group (P=0.004 and 0.010, respectively). No
significant difference was observed however, between the lentivirus
group and the control group (P=0.698 and 0.909, respectively).
ANOVA followed by the Dunnett’s T3 test was used for multiple
comparisons between the levels of Nav1.6 in the Rbfox1 and
lentivirus group and the control group. Statistical analysis
revealed a significant difference in the Nav1.6 expression level
between the Rbfox1 group (P=0.013) and the cotnrol group. No
significant difference was observed however, between the lentivirus
group (P=0.056) and the control group.
Discussion
Intractable epilepsy and neurodevelopmental delay
are the most common clinical manifestations of MCD (22). Modern advanced imaging techniques
have improved our ability to detect MCD in patients with
intractable epilepsy (23).
Neurosurgical intervention is a potential curative treatment for a
subset of patients with intractable epilepsy and MCD (22,24). This makes it possible to explore
abnormally expressed genes in the epileptogenic zone. Mutations in
RBFOX1 have been observed in epilepsy or mental retardation,
although brain tissues were not available in these studies
(3,4). In this study, we provided new
evidence to support the hypothesis that abnormally expressed RBFOX1
is related to epilepsy/MCD by analyzing the expression of RBFOX1 in
the epileptogenic zone. We demonstrated that RBFOX1 protein is
upregulated in cortical lesions obtained from patients with
epilepsy/MCD. By electrophysiological recordings analysis, we found
that cultured rat cortical neurons with an increased Rbfox1
expression were hyperexcitated, which provides direct evidence to
support the hypothesis that Rbfox1 may contribute to neuronal
hyperexcitation and epilepsy.
We analyzed the expression of RBFOX1 in brain
tissues obtained from patients with epilepsy/MCD and control
subjects. Although the patients had different types of MCD, they
all overexpressed RBFOX1. In fact, there is a notion that the
boundaries between disorders of neuronal proliferation, migration,
or cortical organisation are fading, as supported by a recent
report of mutations in WD repeat domain 62 (WDR62), dynein,
cytoplasmic 1, heavy chain 1 (DYNC1H1) and tubulin, gamma 1 (TUBG1)
with a broad range of MCD types (25). Previous studies have suggested
that mutations in RBFOX1 are related to epilepsy (3,4).
We suspected that the overexpression of RBFOX1 in the epileptogenic
zone may contribute to epilepsy. While the deletion of the
Rbfox1 gene has been shown to result in neuronal
hyperexcitation and seizures in mice (9), it thus surprised us that in the
cortical tissue obtained from patients with intractable epilepsy
and MCD, the protein expression of RBFOX1 was increased. However,
in their study, Gehman et al (9) did not use a transgenic mouse model
by increasing the Rbfox1 protein level to explore the effects on
neuronal hyperexcitation. Thus, the increased protein level of
RBFOX1 in the brain tissues of patients with epilepsy/MCD is a
significant finding and worthy of further study. A question which
remains unanswered however, is whether the increased RBFOX1 protein
expression caused the seizures, or whether the seizures themselves
resulted in the increased protein levels of RBFOX1. In order to
attempt to address this issue, we investigated neuronal excitation
in cultured cortical neurons with different levels of RBFOX1
protein.
We used the lentivirus-mediated transfection
technique to alter the Rbfox1 protein level in cultured cortical
neurons. The AP, Na+ current density and SREDs of these
neurons were measured by whole-cell patch clamp recordings. To
avoid the possible influence of the lentivirus vector on the
electrophysiological recordings, we selected the control lentivirus
group as a reference. Our results revealed that the AP amplitude
and Na+ current density increased as the Rbfox1 protein
level increased. In fact, some neurons even displayed SREDs. Based
on these data, we concluded that the increased Rbfox1 protein
expression may be the cause of hyperexcitation in cultured cortical
neurons, and the increased RBFOX1 protein epxression is likely the
cause of epilepsy in human patients rather than a symptom of the
epilepsy.
Once we established this causal relationship between
the altered Rbfox1 protein expression and neuronal hyperexcitation,
we then wished to determine whether this increase in excitability
results from changes in certain known Rbfox1 target transcripts
that have been directly linked to epilepsy. As mentioned above, the
expression levels of NMDAR1 (or Grin1), the Nav1.6 (or
Scn8a) and SNAP-25 (or Snap25) were elevated in the
neurons with an increased Rbfox1 protein expression.
NMDAR1 (or Grin1), one of the fundamental
excitatory neurotransmitter receptors for basic brain development
and function (26), was
upregulated in the cultured neurons with an increased Rbfox1
protein expression and this was expected. Both in animal models and
patients with epilepsy, the increased expression of NMDAR1 has been
reported to be associated with epilepsy (27,28). Nav1.6 is the most abundantly
expressed sodium channel in the adult central nervous system and is
critical for AP generation and propagation (29). Blumenfeld et al found that
kindling was associated with increased persistent sodium current
density in CA3 neurons and an increased mRNA and protein expression
of Nav1.6 in these neurons, suggesting that Nav1.6 may participate
in a self-reinforced cycle of kindling (30). APs are initiated by Nav1.6
channels in the axon initial segment based on their low threshold
and high channel density (29,31). No matter the excitability of other
neurons and neurotransmitters, neurons with an increased Nav1.6
protein level tend to display evoked epileptiform events. In this
study, we demonstrated that SREDs and an enhanced protein
expression of Nav1.6 may contribute to SREDs in the neurons with an
altered Rbfox1 protein expression. This may partially explain why
these neurons exist in an epileptogenic zone and are prone to the
initial ictal period. SNAP25 is a plasma membrane protein which
mediates exocytosis by participating in forming the soluble
N-ethylmaleimide factor attachment receptor (SNARE) complex
(32). It has been suggested that
changes in SNAP25 gene expression may also contribute to epilepsy
(33).
Unexpectedly, our findings are inconsistent with
those found in the study by Gehman et al (9) using Rbfox1 knockout mice. They found
that the deletion of the Rbfox1 gene resulted in increased
excitability in the dentate gyrus, and the decreased expression of
NMDAR1, Nav1.6 and SNAP-25 (9).
One possible explanation for this difference is that their
experiments were performed in vitro and may represent more
immediate changes that are not observed in vivo. The
increased expression of NMDAR1, Nav1.6 and SNAP-25 in cultured
cortical neurons with an increased Rbfox1 protein expression may be
involved in Rbfox1-related neuronal hyper-excitation, although
further studies are required to confirm these findings.
Acknowledgments
Whole-cell patch clamp recordings were supported by
Professor Yun Xiu, Institute of Life Sciences, Chongqing Medical
University, Chongqing 400016, P.R. China.
References
|
1
|
Kwan P, Schachter SC and Brodie MJ: Drug-
resistant epilepsy. N Engl J Med. 365:919–926. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Piao YS, Lu DH, Chen L, Liu J, Wang W, Liu
L, Yu T, Wang YP and Li YJ: Neuropathological findings in
intractable epilepsy: 435 Chinese cases. Brain Pathol. 20:902–908.
2010.PubMed/NCBI
|
|
3
|
Bhalla K, Phillips HA, Crawford J,
McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G and
Callen DF: The de novo chromosome 16 translocations of two patients
with abnormal phenotypes (mental retardation and epilepsy) disrupt
the A2BP1 gene. J Hum Genet. 49:308–311. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lal D, Trucks H, Møller RS, Hjalgrim H,
Koeleman BP, de Kovel CG, Visscher F, Weber YG, Lerche H, Becker F,
Schankin CJ, Neubauer BA, Surges R, Kunz WS, Zimprich F, Franke A,
Illig T, Ried JS, Leu C, Nürnberg P and Sander T; EMINet Consortium
and EPICURE Consortium: Rare exonic deletions of the RBFOX1 gene
increase risk of idiopathic generalized epilepsy. Epilepsia.
54:265–271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin CL, Duvall JA, Ilkin Y, Simon JS,
Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM,
Rao K, Cook E and Geschwind DH: Cytogenetic and molecular
characterization of A2BP1/FOX1 as a candidate gene for autism. Am J
Med Genet B Neuropsychiatr Genet. 144B:869–876. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Voineagu I, Wang X, Johnston P, Lowe JK,
Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ and Geschwind DH:
Transcriptomic analysis of autistic brain reveals convergent
molecular pathology. Nature. 474:380–384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Damianov A and Black DL: Autoregulation of
Fox protein expression to produce dominant negative splicing
factors. RNA. 16:405–416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fogel BL, Wexler E, Wahnich A, Friedrich
T, Vijayendran C, Gao F, Parikshak N, Konopka G and Geschwind DH:
RBFOX1 regulates both splicing and transcriptional networks in
human neuronal development. Hum Mol Genet. 21:4171–4186. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gehman LT, Stoilov P, Maguire J, Damianov
A, Lin CH, Shiue L, Ares M Jr, Mody I and Black DL: The splicing
regulator Rbfox1 (A2BP1) controls neuronal excitation in the
mammalian brain. Nat Genet. 43:706–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cartegni L, Chew SL and Krainer AR:
Listening to silence and understanding nonsense: exonic mutations
that affect splicing. Nat Rev Genet. 3:285–298. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng D and Xie J: Aberrant splicing in
neurological diseases. Wiley Interdiscip Rev RNA. 4:631–649.
2013.PubMed/NCBI
|
|
12
|
Underwood JG, Boutz PL, Dougherty JD,
Stoilov P and Black DL: Homologues of the Caenorhabditis elegans
Fox- 1 protein are neuronal splicing regulators in mammals. Mol
Cell Biol. 25:10005–10016. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Auweter SD, Fasan R, Reymond L, Underwood
JG, Black DL, Pitsch S and Allain FH: Molecular basis of RNA
recognition by the human alternative splicing factor Fox- 1. EMBO
J. 25:163–173. 2006. View Article : Google Scholar
|
|
14
|
Hammock EA and Levitt P: Developmental
expression mapping of a gene implicated in multiple
neurodevelopmental disorders, A2bp1 (Fox1). Dev Neurosci. 33:64–74.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gavazzo P, Tedesco M, Chiappalone M,
Zanardi I and Marchetti C: Nickel modulates the electrical activity
of cultured cortical neurons through a specific effect on N-
methyl- D- aspartate receptor channels. Neuroscience. 177:43–55.
2011. View Article : Google Scholar
|
|
16
|
Yan L, Zhou X, Zhou X, Zhang Z and Luo HM:
Neurotrophic effects of 7,8- dihydroxycoumarin in primary cultured
rat cortical neurons. Neurosci Bull. 28:493–498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasaki S, Shirata A, Yamane K and Iwata M:
Parkin- positive autosomal recessive juvenile Parkinsonism with
alpha- synuclein- positive inclusions. Neurology. 63:678–682. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deshpande LS, Blair RE, Ziobro JM, Sombati
S, Martin BR and DeLorenzo RJ: Endocannabinoids block status
epilepticus in cultured hippocampal neurons. Eur J Pharmacol.
558:52–59. 2007. View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real- time quantitative PCR and
the 2(- Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Wang YY, Qin J, Han Y, Cai J and Xing GG:
Hyperthermia induces epileptiform discharges in cultured rat
cortical neurons. Brain Res. 1417:87–102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deshpande LS and DeLorenzo RJ:
Acetaminophen inhibits status epilepticus in cultured hippocampal
neurons. Neuroreport. 22:15–18. 2011. View Article : Google Scholar :
|
|
22
|
Guerrini R and Barba C: Malformations of
cortical development and aberrant cortical networks:
epileptogenesis and functional organization. J Clin Neurophysiol.
27:372–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andrade CS and Leite Cda C: Malformations
of cortical development: current concepts and advanced neuroimaging
review. Arq Neuropsiquiatr. 69:130–138. 2011.PubMed/NCBI
|
|
24
|
Papayannis CE, Consalvo D, Kauffman MA,
Seifer G, Oddo S, D’Alessio L, Saidon P and Kochen S: Malformations
of cortical development and epilepsy in adult patients. Seizure.
21:377–384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guerrini R and Dobyns WB: Malformations of
cortical development: clinical features and genetic causes. Lancet
Neurol. 13:710–726. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Furukawa H, Singh SK, Mancusso R and
Gouaux E: Subunit arrangement and function in NMDA receptors.
Nature. 438:185–192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wasterlain CG, Naylor DE, Liu H, Niquet J
and Baldwin R: Trafficking of NMDA receptors during status
epilepticus: therapeutic implications. Epilepsia. 54(Suppl 6):
78–80. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
de Moura JC, Tirapelli DP, Neder L,
Saggioro FP, Sakamoto AC, Velasco TR, Panepucci RA, Leite JP,
Assirati Júnior JA, Colli BO and Carlotti Júnior CG: Amygdala gene
expression of NMDA and GABA(A) receptors in patients with mesial
temporal lobe epilepsy. Hippocampus. 22:92–97. 2012. View Article : Google Scholar
|
|
29
|
Dulla CG and Huguenard JR: Who let the
spikes out? . Nat Neurosci. 12:959–960. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Blumenfeld H, Lampert A, Klein JP, Mission
J, Chen MC, Rivera M, Dib-Hajj S, Brennan AR, Hains BC and Waxman
SG: Role of hippocampal sodium channel Nav1.6 in kindling
epileptogenesis. Epilepsia. 50:44–55. 2009. View Article : Google Scholar
|
|
31
|
Kole MH, Ilschner SU, Kampa BM, Williams
SR, Ruben PC and Stuart GJ: Action potential generation requires a
high sodium channel density in the axon initial segment. Nat
Neurosci. 11:178–186. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jahn R and Scheller RH: SNAREs- engines
for membrane fusion. Nat Rev Mol Cell Biol. 7:631–643. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Vilaythong AP, Yoshor D and
Noebels JL: Elevated thalamic low- voltage- activated currents
precede the onset of absence epilepsy in the SNAP25- deficient
mouse mutant coloboma. J Neurosci. 24:5239–5248. 2004. View Article : Google Scholar : PubMed/NCBI
|