Introduction
Thyroid cancer is a relatively common type of cancer
associated with the endocrine system, and has increased in
incidence during the past 10 years; with premenopausal women at
highest risk for papillary and follicular thyroid carcinoma,
implicating the role of estrogens in thyroid cancer (1) and suggesting a specific role for
17β-estradiol (E2) stimulation and estrogen receptor (ER)
expression in thyroid tumorigenesis. There are two different
isoforms of nuclear estrogen receptors in humans, ERα and ERβ,
together mediating the role of estrogens. They have several
similarities in structural and functional domains, however, they
differ in tissue distribution and function (2,3).
In general, ERα is involved in a series of cellular processes,
including cell proliferation, invasion, differentiation and
apoptosis (2), whereas E2 acts in
the target cells through binding to the ligand-binding domain of
ERα. Notably, dysfunction of the estrogenic system is often
associated with the progression of several types of lesion,
including cancer (4).
IQ-domain GTPase-activating proteins (IQGAPs) are an
evolutionary conserved family containing multi-domain proteins,
which have been identified in numerous organisms ranging from
yeasts to mammals. Of the IQGAP proteins, IQGAP1, IQGAP2 and
IQGAP3, have been confirmed to be ubiquitously expressed in humans
and involved in multiple cellular processes, including cell
adhesion, cell migration, extracellular signaling and cytokinesis
(5–7). The most extensively investigated
family member is the scaffold IQGAP1 protein, which is involved in
multiple essential biology processes by binding to numerous
interacting proteins and mediating their functions (8). Increasing evidence has emerged to
suggest that IQGAP1 may contribute to the progression of several
types of cancer, including thyroid cancer, lung cancer, ovarian
cancer gastric cancer, hepatocellular carcinoma, breast cancer and
melanoma (9–13). In addition, IQGAP1 knockdown
attenuates the transcription ability of estrogen-responsive genes,
induced by estrogen via binding to ERα (4), and the overexpression of IQGAP1 has
been associated with the invasiveness of thyroid cancer cells
(14).
In addition, the extracellular signal-regulated
kinase (ERK) pathway, a mitogen-activated protein kinase (MAPK)
pathway, is known to be activated by receptor tyrosine kinases,
cytokine receptors, and G-protein-coupled receptors (15,16). In particular, the phosphorylation
of certain target proteins by ERK1/2 is involved in transcriptional
activation by coordinating extracellular cues and intracellular
signals (15,16). Previous studies have shown that
IQGAP1 modulates the ERK pathway in certain cells and can also
function independently of this mechanism (2–4),
and that ERα is a major driver of growth in tamoxifen-resistant
breast cancer cells, supported by human epithelial growth factor
receptor (HER)/ERK signaling (17). This indicates that IQGAP1 and ERα
are associated with the ERK signaling pathway in cancers.
The present study aimed to investigate how the
overexpression or knockdown of IQGAP1 in thyroid cancer cells
alters cell proliferation, in particular cell invasion, as well as
the ERK pathway, thus identifying the physiological functions of
IQGAP1. In addition, the effect of IQGAP1 knockdown on the
transcriptional activity of ERα and the IQGAP1/ERα interaction were
evaluated in vitro using co-immunoprecipitation and
protein-binding assays, to investigate their role in the
proliferation and invasion of thyroid cancer cells.
Materials and methods
Tissue samples and cell culture
Follicular thyroid cancer tissue samples (n=10) and
adjacent non-neoplastic tissue samples (n=10) were originally
obtained from patients at the First Affiliated Hospital of
Zhengzhou University with informed consent at the time of surgery
in accordance with the Declaration of Helsinki. The 10 patients
were females with a mean age of 41.4 years (SD, ± 8.9 years) and
were diagnosed with poorly differentiated follicular cancer. The
tissue samples were then prepared for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blot analyses. This study was approved by the Ethics
Committee of Zhengzhou University (Zhengzhou, China). The FTC133
human follicular thyroid cancinoma cell line and the Nthy-ori 3-1
normal thyroid follicular epithelial cell line were obtained from
the European Collection of Cell Cultures (Wiltshire, UK). The
simian-derived COS-7 cell line was purchased from American Type
Culture Collection (Manassas, VA, USA). The FTC133 cell line was
maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco Life
Technologies, Rockville, MD, USA)/F12 medium. The Nthy-ori 3-1 cell
line was maintained in RPMI-1640 medium (Gibco Life Technologies).
The COS-7 cell line was maintained in DMEM. The three cell lines
were equally supplemented with 10% fetal bovine serum (FBS; Gibco
Life Technologies), 2 mM L-glutamine, 100 U/ml penicillin and 100
μg/ml streptomycin (Gibco Life Technologies), in a
humidified atmosphere of 5% CO2/95% air at 37°C.
RNA extraction and RT-qPCR analysis
Total RNA was extracted from the FTC133 cells using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA),
according to the manufacturer’s instructions. Reverse transcription
was performed with 1 μg total RNA using the high capacity
cDNA reverse transcription kit (Applied Biosystems, Foster City,
CA, USA). The gene sequences encoding the human IQGAP1 and ERα
full-length molecules were then amplified by qPCR. Amplification
was performed using a Takara Ex Taq PCR kit (Takara Biotechnology
Co., Ltd., Dalian, China) and conducted in a 50 μL reaction
volume containing 5 μl cDNA, 1 U Ex Taq polymerase, 50 pM of
each primer (1 μl), 2.5 mM of each dNTP (8 μl), and 5
μl 10X PCR buffer on a I-cycler thermal cycler PCR (Bio-Rad,
Hercules, CA). The following specific primers were used: IQGAP1,
sense 5′-CTAGCTAGCATGTCCGCCGCAGACGAGG-3′ (NheI site) and
antisense 5′-CCGCTCGAGCCTCGTCTGCGGCGGACAT-3′ (XhoI site);
and ERα, sense 5′-CTAGCTAGCATGACCATGACCCTCCACACCA-3′ (NheI
site) and antisense 5′-CCGCTCGAGTGGTGTGGAGGGTCATGGTCAT-3′
(XhoI site). The qPCR reactions were performed using the
following conditions: 95°C for 1 min, and 30 cycles of 95°C for 1
min, 60°C for 1 min, 72°C for 2 min, then 72°C for 10 min and 4°C
for 5 min. All primers specific to IQGAP1 and ERα were designed
using Premier 5.0 software (Premier Biosoft International, Palo
Alto, CA, USA). All other regents used in the assay were purchased
from Takara Biotechnology Co., Ltd.
Plasmid construction of IQGAP1- and
ERα-small interfering RNA (siRNA), and IQGAP1 and ERα
overexpression plasmids
For gene silencing experiments, IQGAP1- and
ERα-specific siRNA and negative siRNA were synthesized by Takara
Biotechnology Co., Ltd. and annealed into double-stranded molecules
using an siRNA construction kit (Ambion Life Technologies, Austin,
TX, USA), which were then cloned into the BamHI/ClaI
sites of pSUPER-neo vectors (OligoEngine, Seattle, WA, USA). Using
this method, pSUPER-neo-IQGAP1-siRNA plasmids and
pSUPER-neo-ERα-siRNA plasmids were obtained. The sequences of the
specific siRNA against IQGAP1 were as follows: Forward,
5′-AAGGCCGAACTAGTGAAACTGCCTGTCTC-3′ and reverse,
5′-AACAGTTTCACTAGTTCGGCCCCTGTCTC-3′. The sequences of the
nonspecific (NS) siRNA control were as follows: Forward,
5′-AAGTACCAAGGACGCGAATGTCCTGTCTC-3′ and reverse,
5′-AAACATTCGCGTCCTTGGTACCCTGTCTC-3′; the sequences of specific
siRNA against ERα were as follows: Foward,
5′-CCTCGGGCTGTGCTCTTTTTTCCTGTCTC-3′ and reverse,
5′-AAAAGAGCACAGCCCGAGGTTCCTGTCTC-3′; the sequences of the NS siRNA
control were as follows: Forward,
5′-AATTCTCCGAACGTGTCACGTCCTGTCTC-3′ and reverse,
5′-AAACGTGACACGTTCGGAGAACCTGTCTC-3′.
In addition, pcDNA3.1-IQGAP1 and pcDNA3.1-ERα were
constructed by inserting the full-length coding region of the
IQGAP1 and ERα cDNA into the NheI/XhoI sites of the
pcDNA3.1 (Invitrogen Life Technologies), respectively, which were
finally identified by enzyme digestion and sequencing. The
constructed plasmids were digested with the restriction enzymes,
NheI and XhoI (Takara Biotechnology Co., Ltd.), at
37°C for 4 h and then sequenced by Takara Biotechnology Co.,
Ltd.
Transfection with the expression
vector
The cultured cells were seeded in 6-well plates at a
density of 4×105 cells/well and transfected with a
constructed vector, as described above, using Lipofectamine 2000
(Invitrogen Life Technologies), according to the instructions of
the manufacturer. The pSUPER-neo-IQGAP1-siRNA or
pSUPER-neo-IQGAP1-siRNA-N plasmids, and the pSUPER-neo-ERα-siRNA or
pSUPER-neo-ERα-siRNA-N plasmids were transfected into the FTC133
cells for detection of the function of IQGAP1. The pcDNA3.1-IQGAP1
plasmids and pcDNA3.1-ERα plasmids were co-transfected into COS-7
cells or FTC133 cells cells at a density of 4×105
cells/well in 6-well plates for in vivo protein-protein
interaction assays. The transfected cells were then exposed to 500
μg/ml Geneticin (G418 sulfate; Invitrogen Life Technologies)
for ~3 weeks for the formation of G418-resistant colonies at 37°C.
To ascertain the specificity of the siRNAs and expression vector
used, the mRNA and protein levels, as well as the effects on the
functional targets, of the IQGAP1 and ERα proteins were monitored
by RT-qPCR, western blot analysis and reporter luciferase
assay.
Western blot analysis
As regards the fresh tissue samples, ~500 mg of
frozen thyroid tissue was sectioned into small sections,
homogenized and dissolved in 500 μl RIPA lysis buffer
(Beyotime Institute of Biotechnology, Shanghai, China). The
cultured FTC133, Nthy-ori 3-1 and COS-7 cells were extracted, as
indicated, following transfection and treatment. The protein
concentrations of the samples were determined using a Bradford
assay, and equal quantities of protein were loaded onto a sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel
(Shanghai Sangon Biotechnology Co. Ltd, Shanghai, China), prior to
being transferred onto polyvinylidene difluoride membranes (PVDF;
Millipore, Bedford, MA, USA). Subsequently, the membranes were
blocked with 5% non-fat milk in 0.1% Tris-buffered saline with
Tween-20 (TBST; Shanghai Sangon Biotechnology Co. Ltd.) for 1 h at
room temperature, followed by incubation with the primary
antibodies overnight at 4°C. The antibodies used were mouse
anti-human IQGAP1 monoclonal antibody (#610611) or mouse anti-human
E-cadherin monoclonal antibody (#610181) (dilution, 1:2000; BD
Biosciences, San Jose, CA, USA); mouse anti-human ERα monoclonal
antibody (sc73479), rabbit polyclonal IgG anti-p-ERK1/2 antibody
(sc16981-R), mouse monoclonal IgG anti-ERK1/2 antibody (sc-135900),
mouse anti-human matrix metalloproteinase-9 (MMP-9) monoclonal
antibody (sc21733), mouse anti-human cyclin D1 monoclonal antibody
(sc20044), rabbit anti-β-actin polyclonal antibody (sc130657)
(dilution, 1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Following incubation with the appropriate secondary antibody, the
bands were visualized using enhanced chemiluminescence (Beyotime
Institute of Biotechnology). The absorbance values of the target
proteins and data analysis were performed using Gel-Pro Analyzer
version 4.0 software (Media Cybernetics, Silver Spring, MD, USA).
The protein expression levels were normalized to β-actin.
Cell proliferation analysis
The FTC133 cells were seeded at a density of
5×103/200 μl into 96-well microplates following
pSUPER-neo-IQGAP1-siRNA transfection and G418 selection, and were
then serum-starved overnight at 37°C, and treated with 10 nM (E2;
Sigma-Aldrich, St. Louis, MO, USA) or ethanol as a vehicle
(Invitrogen Life Technologies). Following culture for 24, 48 and 72
h, the target cells were added to 20 μl MTT (5 mg/ml;
Sigma-Aldrich) solution and incubated at 37°C for 4 h. The
supernatant was removed and 150 μl dimethyl sulfoxide
(Sigma-Aldrich) was added to each well. Following dissolving of the
dark-blue MTT crystals, the absorbance was determined using a
Bio-Rad microplate reader (Bio-Rad) at a wavelength of 490 nm.
Reporter luciferase assay
Luciferase and β-galactosidase assays were
performed, according to the manufacturer’s instructions (Promega
Corporation, Madison, WI, USA). The cultured FTC133 cells were
seeded at a density of 2×105/well into 6-well plates and
co-transfection was performed using the pSUPER-neo-IQGAP1-siRNA or
pSUPER-neo-IQGAP1-siRNA-N expression vector (1 μg), a
reporter gene (PC3-LUC vector; 300 ng) and a pCMV-β galactosidase
internal control vector (150 ng; Clontech, Palo Alto, CA, USA), or
as indicated in the figure legends using Lipofectamine 2000.
Following 36 h of transfection at 37°C, the FTC133 cells were
pretreated with E2, as described above, for 12 h at 37°C and then
harvested. The reporter gene activity was obtained following
normalization of the luciferase activity with that of
β-galactosidase.
Cell invasion assay
The invasion assay was performed using
Matrigel-coated 24-well Transwell chambers with 8.0 μm
poly-carbonated filters (Corning Incorporated, Corning, NY, USA).
Following transfection with pSUPER-neo-IQGAP1-siRNA and G418
selection, the FTC133 cells were pretreated with E2, as described
above, for 12 h, and then seeded in serum-free DMEM medium at a
density of 2×105 cells/300 μl into the upper
compartment, while the DMEM medium in the lower compartment was
supplemented with 15% FBS. After 24 h, the non-invasive cells on
the upper surface of the membrane were removed using cotton swabs,
and the invasive cells that had penetrated through the pores and
migrated to the underside of the membrane were fixed using 4%
paraformaldehyde and stained with 0.1% crystal violet
(Sigma-Aldrich). The number of invasive cells was counted under a
microscope (Olympus BX50, Tokyo, Japan).
Cell lysates and co-immunoprecipitation
assay
The COS-7 and FTC133 cells were plated in 6-well
plates to obtain 85% confluence at 37°C. After 24 h, each plate was
transfected with 2 μg myc-tagged pcDNA3.1-IQGAP1 plasmids
and 2 μg myc-tagged pcDNA3.1-ERα plasmids. After 48 h, the
cells were pretreated with E2, as described above, for 12 h, lysed
with 500 μl buffer A, which contained 50 mM Tris HCl (pH
7.4), 150 mM NaCl, 1% Triton X-100, protease and phosphatase
inhibitors and 1 mM PMSF. The lysates were incubated on ice for 30
min, and insoluble material was precipitated by centrifugation at
20,000 × g for 10 min at 4°C. The supernatants were pre-cleared
using glutathione-sepharose beads (GE Healthcare Life Sciences,
Piscataway, NJ, USA) for 1 h at 4°C. Equal quantities of protein
lysate were incubated with protein A-Sepharose beads (GE Healthcare
Life Sciences) and either anti-rabbit IgG (#43413; Sigma-Aldrich)
or mouse anti-human IQGAP1 monoclonal antibody (BD Biosciences), or
mouse anti-human ERα monoclonal antibody (Santa Cruz Biotechnology)
overnight at 4°C. The samples were then washed 5 times with Buffer
B (Shanghai Sangon Biotechnology Co. Ltd.), containing 50 mM Tris
HCl (pH 7.4), 150 mM NaCl and 1% Triton X-100, were resolved by
SDS-PAGE, and were processed by SDS-PAGE and western blot analysis
as described above.
Statistical analysis
The results are expressed as the mean ± standard
deviation of three individual experiments performed in triplicate.
All analyses were conducted using SPSS software (SPSS Inc.,
Chicago, IL, USA). Statistical analysis was performed using
Student’s t-test and analysis of variance. *P<0.05
was considered to indicate a statistically significant
difference.
Results
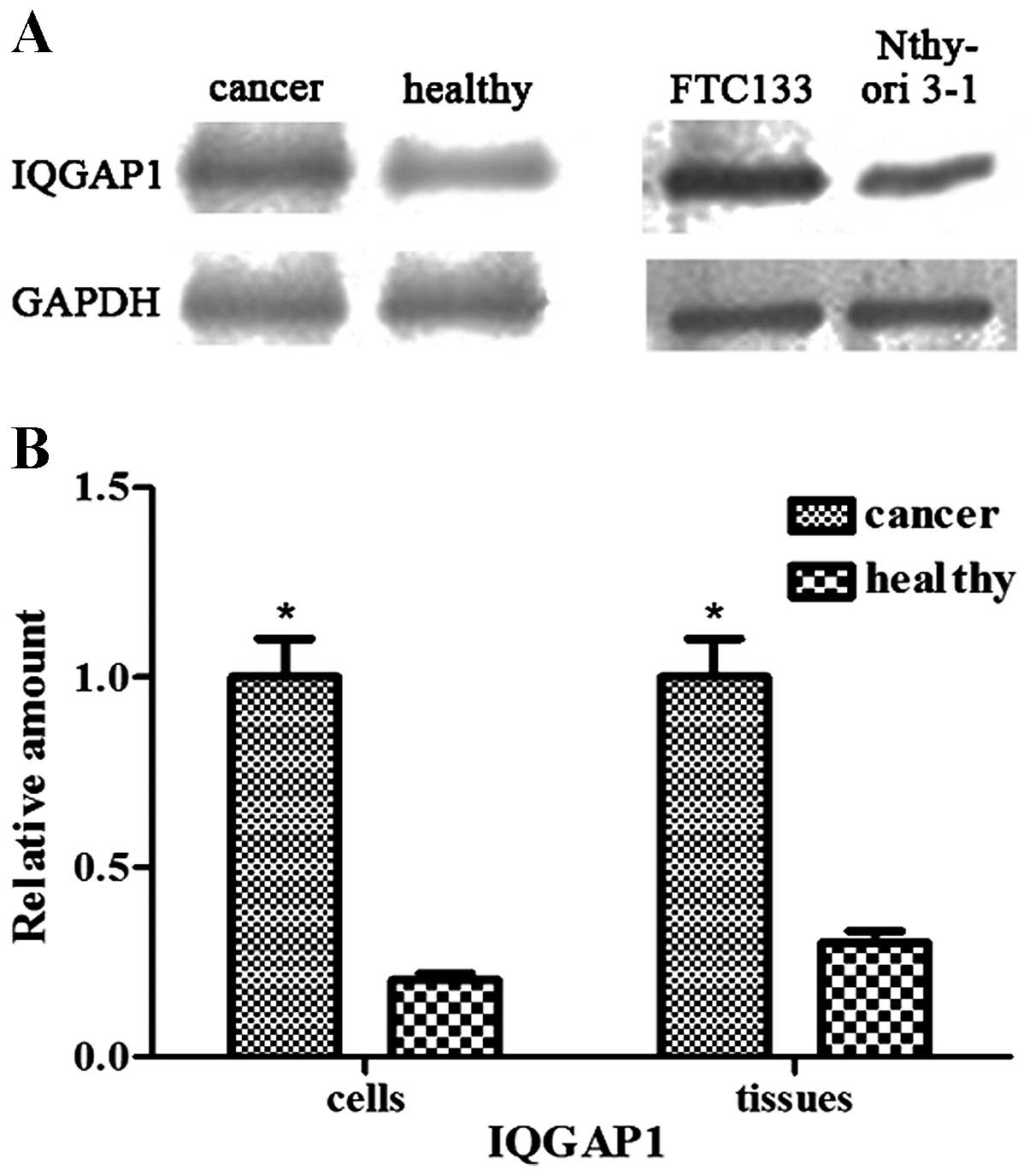
IQGAP1 is overexpressed in follicular
thyroid cancer and FTC133 cells
The results of the western blotting demonstrated
that the protein levels of IQGAP1 were significantly increased in
the follicular thyroid cancer tissues and FTC133 cells, compared
with the weak levels of expression observed in the adjacent
non-neoplastic tissues and Nthy-ori 3-1 cells (P<0.05; Fig. 1).
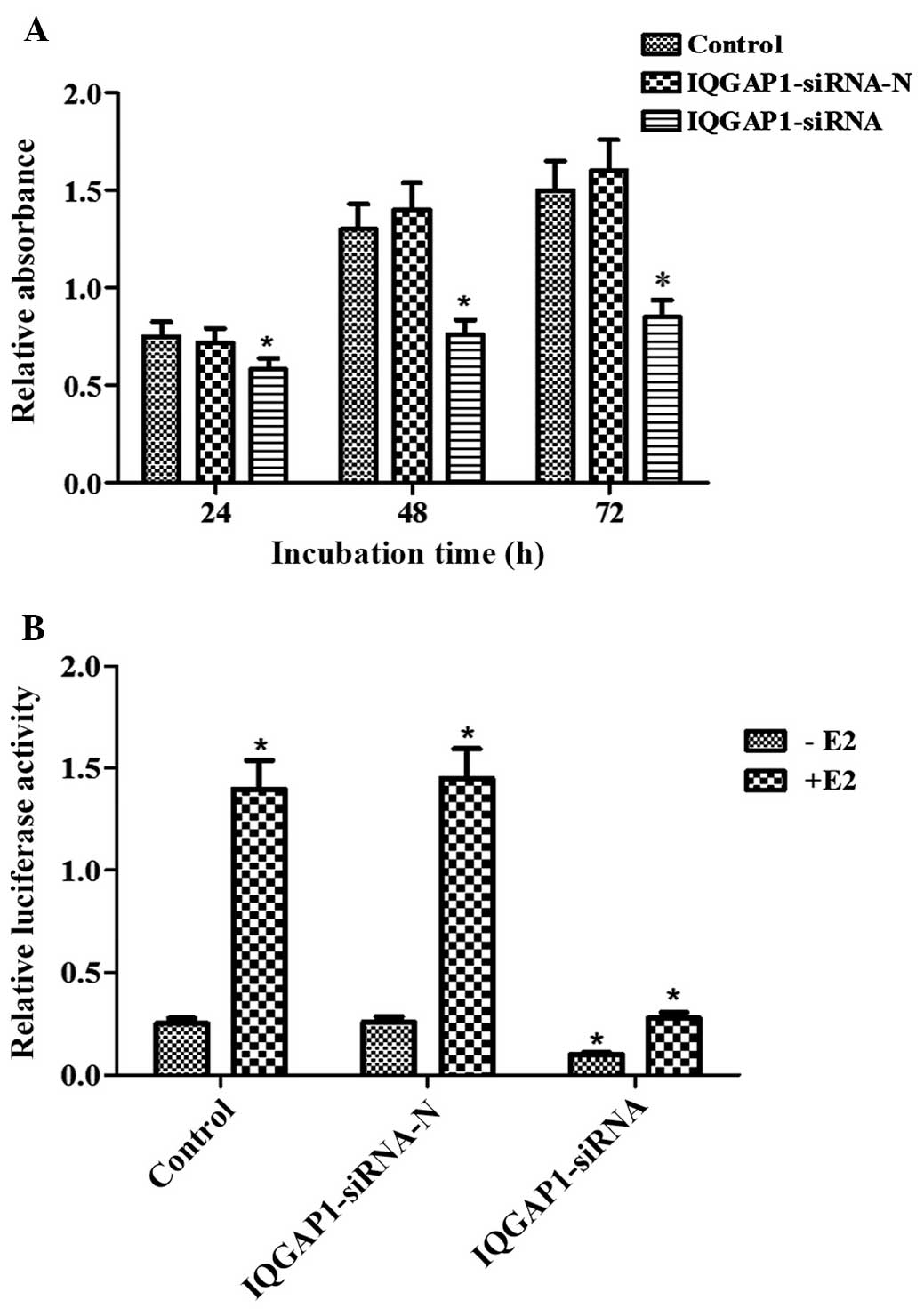
IQGAP1 knockdown represses cell
proliferation and ERα transcriptional activity
In the present study, an MTT assay was used to
investigate whether ectopic IQGAP1 affects cell proliferation in
the FTC133 thyroid cancer cell line. Compared with the FTC133 cells
transfected with pSUPER-neo-IQGAP1-siRNA-N, the proliferation of
the FTC133 cells transfected with pSUPER-neo-IQGAP1-siRNA was
significantly reduced to 79.27, 57.49 and 55.14% at 24, 48 and 72
h, respectively (P<0.05; Fig.
2A). No significant difference was observed between the control
cells and the cells transfected with pSUPER-neo-IQGAP1-siRNA-N
(P>0.05).
In addition, the present study used the
well-characterized ERE-reporter assay to examine the effect of
endogenous IQGAP1 on ERα transcriptional activity following E2
pretreatment in FTC133 cells. The expression of IQGAP1 was
effectively knocked down by pSUPER-neo-IQGAP1-siRNA plasmid
transfection, which led to a significant reduction of ERE reporter
activity, regardless of whether E2 treatment had been performed or
not (Fig. 2B). Therefore,
knockdown of the expression of IQGAP1 in FTC133 cells contributed
to decreased cell proliferation and inhibited the transcriptional
activity of ERα.
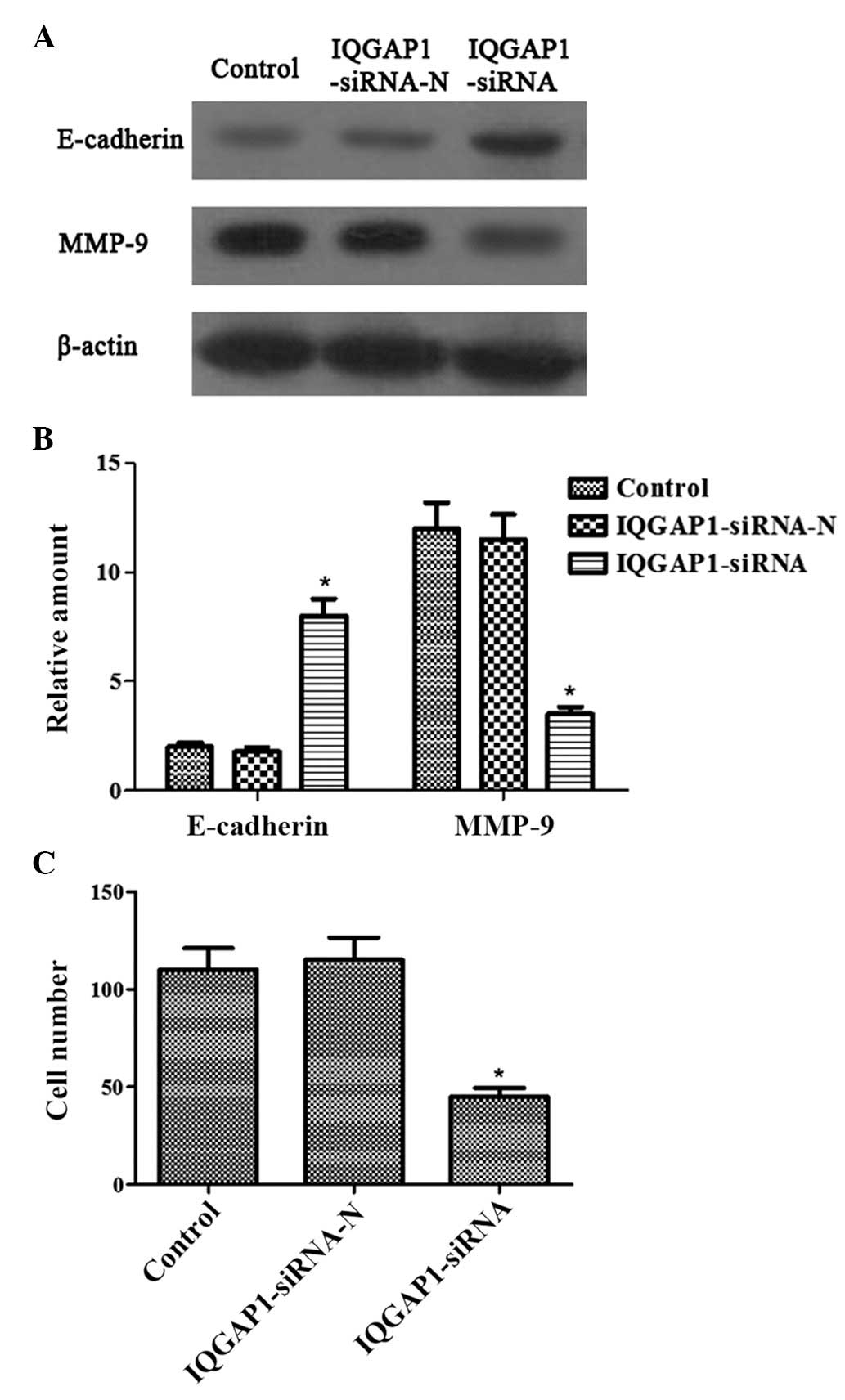
IQGAP1 knockdown represses the invasion
of FTC133 cells
In the present study, the effect of
pSUPER-neo-IQGAP1-siRNA on the protein expression levels of
E-cadherin and MMP-9 were examined, which are known to be involved
in cancer cell adhesion, invasion and progression. The protein
levels of E-cadherin were significantly upregulated, while those of
MMP-9 were significantly downregulated following transfection with
pSUPER-neo-IQGAP1-siRNA, compared with the control or the cells
transfected with pSUPER-neo-IQGAP1-si RNA-N (Fig. 3A and 3B).
Matrigel invasion assays were also used for
functional investigation of whether IQGAP1 is involved in thyroid
cancer cell invasion. pSUPER-neo-IQGAP1-siRNA transfection markedly
inhibited the invasion of FTC133 cells (Fig. 3C). Therefore, these data all
confirmed the specific and important role of IQGAP1 in the
invasiveness of thyroid cancer.
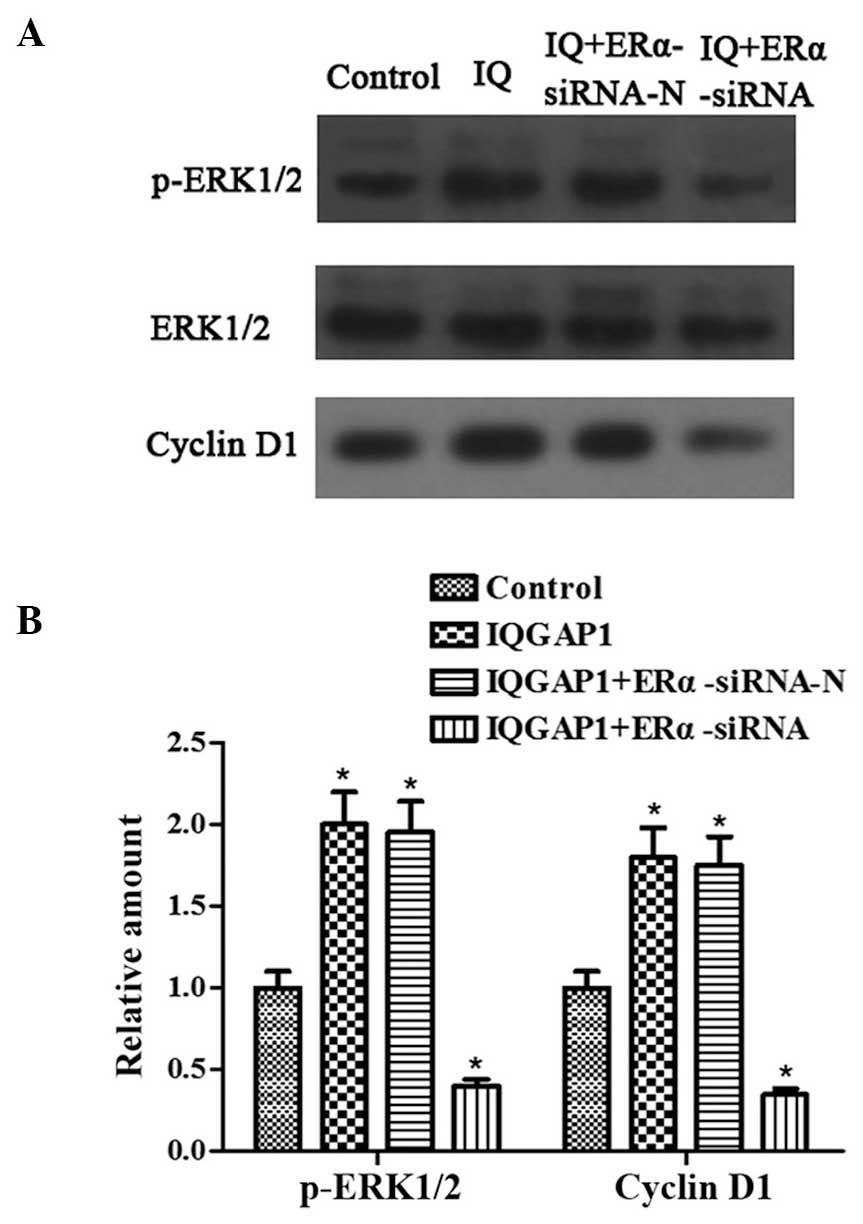
ERα knockdown inhibits the increases in
the expression levels of p-ERK1/2 and cyclin D1, induced by IQGAP1
overexpression
Overexpression of IQGAP1 and the ERK pathway are
critical in promoting thyroid cancer cell proliferation and
invasion. The present study examined whether ERα was involved in
ERK1/2 activation and cell proliferation in the FTC133 cells. The
expression levels of p-ERK1/2 and cyclin D1 were upregulated
following transfection with the pcDNA3.1-IQGAP1 and
pSUPER-neo-ERα-siRNA-N plasmids into the FTC133 cells (Fig. 4). By contrast, significant
decreases in the protein expression of p-ERK1/2 and cyclin D1 were
observed in the FTC133 cells co-transfected with pcDNA3.1-IQGAP1
and pSUPER-neo-ERα-siRNA. These data indicated that IQGAP1 promoted
activation of the ERK1/2 signaling pathway and cell proliferation,
which may be inhibited by the absence of ERα.
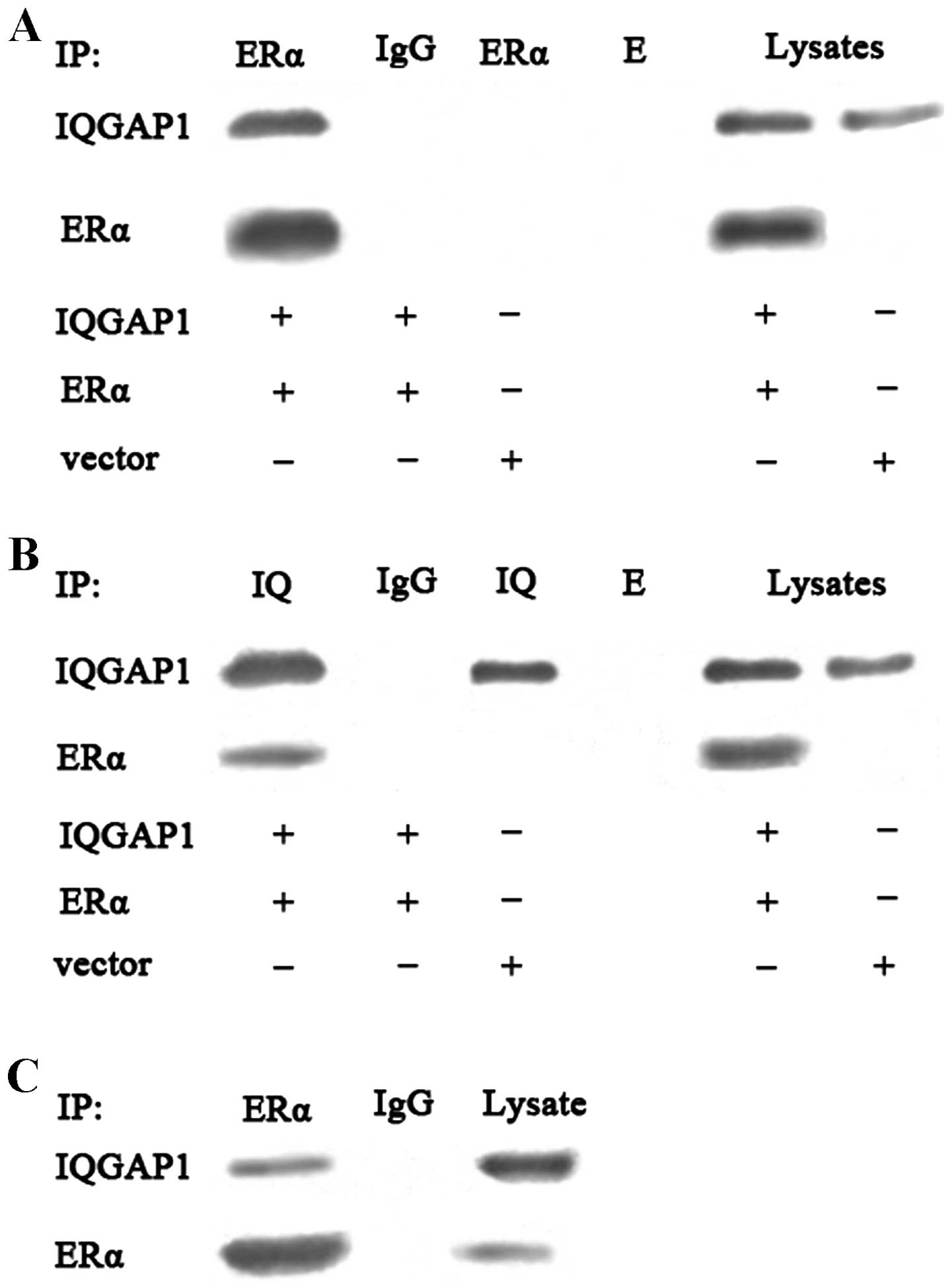
IQGAP1 directly interacts with ERα in
intact cells
The binding function between IQGAP1 and ERα has been
reported in breast cancer cells (4), thus the present study used
ERα-negative COS-7 cells to detect the potential interaction
between IQGAP1 and ERα in intact cells. The co-expression of IQGAP1
and ERα in COS-7 cells was evaluated using an immunoprecipitation
assay followed by western blot analysis. Immunoprecipitation with
the anti-ERα or anti-IQGAP1 antibody revealed that binding of
IQGAP1 to ERα in the COS-7 cells, with rabbit IgG considered a
negative control, validating the specificity of the interaction
(Fig. 5A and 5B). The interaction between IQGAP1 and
endogenous ERα in the FTC133 cells was further examined. The cells
were cross-linked using paraformaldehyde, followed by lysis.
Immunoprecipitation with the anti-ERα antibody revealed that IQGAP1
interacted with theERα from the FTC133 cells, suggesting that
endogenous IQGAP1 interacted directly with ERα in theFTC133 cells
(Fig. 5C).
Discussion
The present study aimed to investigate the
association between IQGAP1 and ERα with E2 pretreatment in the
FTC133 thyroid cancer cell line. ERα acts as a nuclear hormone
receptor, which is known to modulate the expression of genes that
are implicated in different cellular processes, including
tumorigenesis (18,19). IQGAP1 acts as a key mediator of
numerous cellular processes and signaling pathways in a broad range
of interacting partners (9).
Western blotting revealed that IQGAP1 protein was
consistently upregulated in thyroid cancer samples and FTC133
cells, compared with adjacent healthy non-neoplastic tissues and
Nthy-ori 3-1 cells, as has been confirmed in other types of cancer
(9,20). Therefore, the selected
ERα-positive FTC133 cell line expressed high levels of IQGAP1
protein, thus enabling evaluation of the role of IQGAP1 and ERα in
thyroid cancer cells (21).
Subsequently, the effects of IQGAP1 expression and E2 stimulation
on ERα transcriptional activity, cell proliferation, cell invasion
and IQGAP1/ERα protein-binding were examined in the FTC133
follicular thyroid cancer cell line.
Increased evidence has indicated the connection
between the expression of IQGAP1 and tumorigenesis (9), in particular the association with
hormone-associated tumorigenesis, including the development of
breast cancer (4). IQGAP1 can
bind to ERα and modulate its function under the stimulation of E2
in breast cancer cells; therefore, it may be considered a
therapeutic target for patients with breast carcinoma (14). However, whether the IQGAP1/ERα
interaction is involved in thyroid cancer processes remains to be
elucidated. To validate this hypothesis, a series of assays were
performed in the present study by expressing knockdown, transfected
or endogenous human IQGAP1 or ERα in the FTC133 cells, along with
pilot investigations on tissue protein detection. This study
demonstrated the IQGAP1/ERα interaction and its role in promoting
the proliferation and invasion of human follicular thyroid cancer
cells.
The present study also observedthat the knockdown of
IQGAP1 by two distinct IQGAP1 siRNAs significantly inhibited
proliferation of the FTC133 cells in a time-dependent manner, as is
also observed in certain other types of cancer cells (22–24). Furthermore, the present study
examined the possible role of IQGAP1 in E2-induced or
non-E2-induced ERα transcriptional activation using a reporter
luciferase assay (25,26). The results suggested that ERα
transcriptional activation was significantly increased by E2
stimulation in the FTC133 cells and was significantly decreased in
the cells, in which IQGAP1 was specifically knocked down by siRNA,
in the E2 group and non-E2 group. Therefore, the possible mechanism
may be that the interaction between IQGAP1 and ERα in FTC133 cells
is regulated, to a certain extent, by E2 stimulation, and that the
ability of E2 to induce transcriptional activity is impaired in
cells following IQGAP1 knockdown. It was hypothesized that
knockdown of IQGAP1 results in a loss of cell proliferation and the
E2-induced activation of ERα transcriptional activity in FTC133
cells.
The expression of MMP-9, which is known to promote
and enhance metastasis, has been reported to correlate with E2-ER
signaling (27,28). Furthermore, MMP-9 secretion is
increased in ER-positive thyroid cancer cells following E2
treatment (29). The abnormal
expression of E-cadherin has been reported in the majority of types
of human cancer, including thyroid cancer (30), and is found to correlate with the
invasion and metastasis of epithelial tumor cells (31). IQGAP1 is a multifunctional protein
involved in actin cytoskeleton assembly, which may be involved in
cancer invasion (32–34) and in regulating
E-cadherin-mediated cell-cell adhesion (35), particualrly in thyroid cancer
cells (14). In the present
study, IQGAP1 knockdown significantly affected the expression
levels of these two adhesion-associated proteins, decreasing MMP-9
and increasing E-cadherin, and inhibited the invasion of the E2
pretreated FTC133 cells.
Previous studies have demonstrated that activation
of ERK1/2 signaling is coupled with the upregulation of specific
target transcription factors under E2 stimulation, for example,
cyclic AMP response element-binding protein (36), a nuclear hormone receptor/estrogen
receptor (37) that modulates the
expression of B-cell lymphoma-2 or cyclin D1 and may be involved in
cell cycle progression and cell proliferation (36,37). Notably, the present study found
that ERα knockdown negatively acted on the connection between the
ERK signaling pathway and expression of cyclin D1 in the FTC133
cells though the overexpression of IQGAP1. These results indicated
that IQGAP1 and ERα were involved in the ERK pathway and regulated
the expression of cyclin D1. In addition, ER-dependent cyclin D1
transcription and DNA synthesis are considered to be downstream
targets of E2-induced ERK activation (38,39). These data suggested that the
interaction between the IQGAP1/ERα proteins enhanced the
ERα-mediated transactivation activity of cyclin D1 and p-ERK in the
E2-mediating signaling pathway, and that IQGAP1/ERα proteins may be
essential in the ERK pathway and in cell proliferation in thyroid
cancer.
Further in vitro analysis is required to
confirm the protein interactions between IQGAP1 and nuclear
receptors. In the present study this protein interaction was
identified in whole structural domains in a complex from cell
lysates using co-immunoprecipitation to validate a direct
association in COS-7 cells. The results demonstrated that IQGAP1
was able to bind to ERα distinctly, whether endogenous or
expressed, with E2 pretreatment from FTC133 cells and
co-transfected COS-7 cells, respectively. In addition, the binding
between intracellular IQGAP1 and ERα was enhanced by
co-immunoprecipitation.
Using the above approaches, the present study
successfully demonstrated that IQGAP1 was able to bind to
endogenous or expressed ERα and that IQGAP1 knockdown affected ERα
transcriptional activity, cell proliferation and invasion in the
FTC133 cells. In addition, the interaction of IQGAP1 and ERα
promoted the association of ERK1/2 with the IQGAP1 molecule, which
stimulated ERK1/2 phosphorylation and cyclin D1 activation. These
data offer novel insight into the involvement of IQGAP1 and ERα in
the progression of thyroid cancer. Further analyses are required to
determine whether the IQGAP1/ERα interaction identified in the
present study is involved in other cancer processes, which may
provide an effective target for cancer prevention and therapy.
References
|
1
|
Kumar A, Klinge CM and Goldstein RE:
Estradiol-induced proliferation of papillary and follicular thyroid
cancer cells is mediated by estrogen receptors α and β. Int J
Oncol. 36:1067–1080. 2010.PubMed/NCBI
|
|
2
|
Thomas C and Gustafsson JA: The different
roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer.
11:597–608. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heldring N, Pike A, Andersson S, et al:
Estrogen receptors: how do they signal and what are their targets.
Physiol Rev. 87:905–931. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Erdemir HH, Li Z and Sacks DB: IQGAP1
binds to estrogen receptor-α and modulates its function. J Biol
Chem. 289:9100–9112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brown MD and Sacks DB: IQGAP1 in cellular
signaling: bridging the GAP. Trends Cell Biol. 16:242–249. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Machesky LM: Cytokinesis: IQGAPs find a
function. Curr Biol. 8:R202–R205. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Noritake J, Watanabe T, Sato K, Wang S and
Kaibuchi K: IQGAP1: a key regulator of adhesion and migration. J
Cell Sci. 118:2085–2092. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
White CD, Erdemir HH and Sacks DB: IQGAP1
and its binding proteins control diverse biological functions. Cell
Signal. 24:826–834. 2012. View Article : Google Scholar :
|
|
9
|
Johnson M, Sharma M and Henderson BR:
IQGAP1 regulation and roles in cancer. Cell Signal. 21:1471–1478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
White CD, Brown MD and Sacks DB: IQGAPs in
cancer: a family of scaffold proteins underlying tumorigenesis.
FEBS Lett. 583:1817–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sugimoto N, Imoto I, Fukuda Y, et al:
IQGAP1, a negative regulator of cell-cell adhesion, is upregulated
by gene amplification at 15q26 in gastric cancer cell lines HSC39
and 40A. J Hum Genet. 46:21–25. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nabeshima K, Shimao Y, Inoue T and Koono
M: Immunohistochemical analysis of IQGAP1 expression in human
colorectal carcinomas: its overexpression in carcinomas and
association with invasion fronts. Cancer Lett. 176:101–109. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong P, Nabeshima K, Nishimura N, et al:
Overexpression and diffuse expression pattern of IQGAP1 at invasion
fronts are independent prognostic parameters in ovarian carcinomas.
Cancer Lett. 243:120–127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Liu D, Bojdani E, El-Naggar AK,
Vasko V and Xing M: IQGAP1 plays an important role in the
invasiveness of thyroid cancer. Clin Cancer Res. 16:6009–6018.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Z, Gibson TB, Robinson F, et al: MAP
kinases. Chem Rev. 101:2449–2476. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schaeffer HJ and Weber MJ:
Mitogen-activated protein kinases: specific messages from
ubiquitous messengers. Mol Cell Biol. 19:2435–2444. 1999.PubMed/NCBI
|
|
17
|
Thrane S, Lykkesfeldt AE, Larsen MS,
Sorensen BS and Yde CW: Estrogen receptor α is the major driving
factor for growth in tamoxifen-resistant breast cancer and
supported by HER/ERK signaling. Breast Cancer Res Treat. 139:71–80.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Walter P, Green S, Greene G, et al:
Cloning of the human estrogen receptor cDNA. Proc Natl Acad Sci
USA. 82:7889–7893. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kuiper GG, Enmark E, Pelto-Huikko M,
Nilsson S and Gustafsson JA: Cloning of a novel receptor expressed
in rat prostate and ovary. Proc Natl Acad Sci USA. 93:5925–5930.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang XX, Li XZ, Zhai LQ, Liu ZR, Chen XJ
and Pei Y: Overexpression of IQGAP1 in human pancreatic cancer.
Hepatobiliary Pancreat Dis Int. 12:540–545. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mirebeau-Prunier D, Le Pennec S, Jacques
C, et al: Estrogen-related receptor alpha and PGC-1-related
coactivator constitute a novel complex mediating the biogenesis of
functional mitochondria. FEBS J. 277:713–725. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jadeski L, Mataraza JM, Jeong HW, Li Z and
Sacks DB: IQGAP1 stimulates proliferation and enhances
tumorigenesis of human breast epithelial cells. J Biol Chem.
283:1008–1017. 2008. View Article : Google Scholar
|
|
23
|
Ma Y, Jin Z, Huang J, et al: IQGAP1 plays
an important role in the cell proliferation of multiple myeloma via
the MAP kinase (ERK) pathway. Oncol Rep. 30:3032–3038.
2013.PubMed/NCBI
|
|
24
|
Wang XX, Wang K, Li XZ, et al: Targeted
knockdown of IQGAP1 inhibits the progression of esophageal squamous
cell carcinoma in vitro and in vivo. PLoS One. 9:e965012014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Safe S: Transcriptional activation of
genes by 17 beta-estradiol through estrogen receptor-Sp1
interactions. Vitam Horm. 62:231–252. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cavarretta IT, Mukopadhyay R, Lonard DM,
et al: Reduction of coactivator expression by antisense
oligodeoxynucleotides inhibits ERalpha transcriptional activity and
MCF-7 proliferation. Mol Endocrinol. 16:253–270. 2002.PubMed/NCBI
|
|
27
|
Nilsson UW, Garvin S and Dabrosin C: MMP-2
and MMP-9 activity is regulated by estradiol and tamoxifen in
cultured human breast cancer cells. Breast Cancer Res Treat.
102:253–261. 2007. View Article : Google Scholar
|
|
28
|
Kousidou OC, Berdiaki A, Kletsas D, et al:
Estradiol-estrogen receptor: a key interplay of the expression of
syndecan-2 and metalloproteinase-9 in breast cancer cells. Mol
Oncol. 2:223–232. 2008. View Article : Google Scholar
|
|
29
|
Rajoria S, Suriano R, George A, et al:
Estrogen induced metastatic modulators MMP-2 and MMP-9 are targets
of 3,3′-diindolyl-methane in thyroid cancer. PLoS One.
6:e158792011. View Article : Google Scholar
|
|
30
|
Graff JR, Greenberg VE, Herman JG, et al:
Distinct patterns of E-cadherin CpG island methylation in
papillary, follicular, Hurthle’s cell, and poorly differentiated
human thyroid carcinoma. Cancer Res. 58:2063–2066. 1998.PubMed/NCBI
|
|
31
|
Shiozaki H, Oka H, Inoue M, Tamura S and
Monden M: E-cadherin mediated adhesion system in cancer cells.
Cancer. 77:1605–1613. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mataraza JM, Briggs MW, Li Z, Entwistle A,
Ridley AJ and Sacks DB: IQGAP1 promotes cell motility and invasion.
J Biol Chem. 278:41237–41245. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Watanabe T, Wang S, Noritake J, et al:
Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin
filaments during cell polarization and migration. Dev Cell.
7:871–883. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L,
et al: IQGAP1, a novel vascular endothelial growth factor receptor
binding protein, is involved in reactive oxygen species - dependent
endo-thelial migration and proliferation. Circ Res. 95:276–283.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fukata M, Kuroda S, Nakagawa M, et al:
Cdc42 and Rac1 regulate the interaction of IQGAP1 with
beta-catenin. J Biol Chem. 274:26044–26050. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu TW, Wang JM, Chen S and Brinton RD:
17Beta-estradiol induced Ca2+ influx via L-type calcium
channels activates the Src/ERK/cyclic-AMP response element binding
protein signal pathway and BCL-2 expression in rat hippocampal
neurons: a potential initiation mechanism for estrogen-induced
neuroprotection. Neuroscience. 135:59–72. 2005. View Article : Google Scholar
|
|
37
|
Acconcia F, Ascenzi P, Bocedi A, et al:
Palmitoylation-dependent estrogen receptor alpha membrane
localization: regulation by 17beta-estradiol. Mol Biol Cell.
16:231–237. 2005. View Article : Google Scholar :
|
|
38
|
Marino M, Acconcia F, Bresciani F, Weisz A
and Trentalance A: Distinct nongenomic signal transduction pathways
controlled by 17beta-estradiol regulate DNA synthesis and cyclin
D(1) gene transcription in HepG2 cells. Mol Biol Cell.
13:3720–3729. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Marino M, Acconcia F and Trentalance A:
Biphasic estradiol-induced AKT phosphorylation is modulated by PTEN
via MAP kinase in HepG2 cells. Mol Biol Cell. 14:2583–2591. 2003.
View Article : Google Scholar : PubMed/NCBI
|