Introduction
The hard tissue of teeth consists of enamel and
dentin. Enamel, which covers the crown, is formed by ameloblasts
derived from the dental epithelium, which never regenerates after
tooth eruption. Dentin, which is a major component of teeth, is
formed by odontoblasts derived from neural crest cells and includes
connective tissue, termed dental pulp (DP), in the center. The DP
is a non-hematopoietic connective tissue that is almost completely
surrounded by dentin (1,2). Odontoblasts are located peripherally
on the inside wall of the DP chamber, and they secrete dentin.
After tooth maturation, the DP is protected only by reparative or
secondary dentin, which is formed by odontoblasts in response to
general mechanical stress or disruption and dentinal degradation
caused by bacterial infections. In addition, dental pulp
progenitor/stem cells (DPSCs) exist in dental pulp and become
activated in response to odontoblast injury. DPSCs are multipotent
stem cells that are capable of differentiating into odontoblasts,
osteoblasts, adipocytes and neural cells in vivo (3). DPSCs are thought to be useful for
dentin regeneration. However, there are very few DPSCs in pulp
tissue (4). Mesenchymal stem
cells (MSCs) are also multipotent stem cells that can differentiate
not only into osteoblastic and chondroblastic cell lineages, but
also into adipocytes, tendon and muscle (3,5).
It has been demonstrated that MSCs can differentiate into
dentin-forming odontoblasts and participate in dentin regeneration
(3). However, the mechanisms
through which MSCs migrate into the DP have not yet been
elucidated.
Stromal cell-derived factor 1α (SDF1, also known as
CXCL12) is an α-chemokine that attracts MSCs and endothelial
progenitor cells (EPCs) through interaction with its unique
receptor, C-X-C chemokine receptor type 4 (CXCR4) (6–9).
In adults, tissue repair and regeneration, complex processes
involving the proliferation and migration of tissue stem
cell-dependent progenitor cells, are thought to involve the
selective recruitment of circulating or resident stem cell
populations. The expression of SDF1 in injured tissue correlates
with the recruitment of adult stem cells and tissue regeneration,
highlighting the importance of SDF1 in stem and progenitor cell
recruitment (6–9). Thus, SDF1 may play an important role
in coordinating tissue injury and repair in DP. However, the exact
which SDF1 plays in relation to dental pulp cells (DPCs) derived
from human teeth is not known.
Therefore, in the present study, we sought to
determine the functions of SDF1 and CXCR4 in DPC cultures and human
MSCs. To the best of our knowledge, this is the first report
describing the expression of SDF1 and CXCR4 in DPCs derived from
human deciduous teeth.
Materials and methods
Cell culture
The UE7T-13 cells, a human bone marrow-derived MSC
line infected with retroviruses expressing papillomavirus E7 and
hTERT in order to prolong the life span of the cells (10–13), were purchased from the Health
Science Research Resources Bank (JCRB1154; Japan Health Sciences
Foundation, Tokyo, Japan) and cultured in α-modified minimum
essential medium (α-MEM; Gibco-BRL, Gaithersburg, MD, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco-BRL). The
MG-63 cells derived from human osteosarcoma (14) were purchased from the American
Type Culture Collection (CRL-1427; ATCC, Manassas, VA, USA). The
HuO9 human osteosarcoma cells (15) were purchased from the Japan Health
Sciences Foundation (JCRB0427) and cultured in α-MEM containing 10%
FBS. All cultures were maintained at 37°C in a humidified
atmosphere of 5% CO2.
Isolation of DPCs
The DP tissues were obtained from the crown and root
of healthy human deciduous teeth (obtained from 3 donors, aged 6–8
years). Informed consent was obtained from the donors’ parents
prior to tooth extraction, which was carried out at Tokushima
University Hospital (Tokushima, Tokyo) during the course of
orthodontic treatment. The study protocol was approved by the
Ethics Committee of Iwate Medical University, School of Dentistry,
Morkioka Japan (no. 01101) and Tokushima University Hospital,
Tokushima, Japan.
The DP tissues were cut into sections using a
surgical blade and were digested with collagenase (2 mg/ml) at 37°C
for 30 min. The tissues were then washed with Dulbecco’s
phosphate-buffered saline (DPBS), placed in culture dishes and
maintained in α-MEM supplemented with 10% FBS. Fibroblastic cells
that grew from the DP tissues were used as DPCs. When the cells
reached confluence, they were detached with 0.2% trypsin and 0.02%
ethylenediaminetetraacetic acid tetrasodium salt (EDTA-4Na) in DPBS
(Gibco-BRL) and subcultured at a split ratio of 1:4, as previously
described (1).
Transfection of DPCs with the hTERT
gene
The DPCs were transfected with the
pBABE-neo-hTERT plasmid, which harbored a
neomycin-resistance gene (Addgene Inc., Cambridge, MA, USA) using
Lipofectamine LTX (Invitrogen Life Technologies, Carlsbad, CA, USA)
according to the manufacturer’s instructions. The cells were
exposed to α-MEM containing 10% FBS and 200 µg/ml G418
(Gibco-BRL) for 12–15 days. The surviving cells were trypsinized
and cultured further in 100-mm culture dishes, as previously
described (16).
Single-cell cloning
Single-cell clones were obtained using the limited
dilution method as previously described (16). Clones obtained after single-cell
cloning were named single cell clones derived from human deciduous
tooth pulp cells (termed SDPCs). All the experiments were performed
using clone no. 11 (SDP11 cells) cultured in α-MEM supplemented
with 10% FBS in the absence or presence 10 ng/ml fibroblast growth
factor (FGF)-2 (R&D Systems, Minneapolis, MN, USA) for 2 days.
All cultures were maintained at 37°C in a humidified atmosphere of
5% CO2 in air.
Isolation of total RNA
Total RNA was extracted from the cultured cells
using TRIzol reagent (Invitrogen Life Technologies) as previously
described (16). The total RNA
pellet was washed briefly with 75% ethanol, resuspended in 30
µl diethylpyrocarbonate (DEPC)-treated water (Invitrogen
Life Technologies) and stored at −80°C. The concentration of total
RNA was determined spectrophotometrically by measuring the optical
density at 260 nm, using a BioPhotometer (Eppendorf, Hamburg,
Germany).
Reverse transcription polymerase chain
reaction (RT-PCR)
RNA (1 µg) was reverse transcribed into
first-strand cDNA using a PrimeScript RT reagent kit (Takara Bio,
Kyoto, Japan), according to the manufacturer’s instructions. cDNA
samples were then amplified with specific primer pairs (listed in
Table I) and as previously
described: for SDF1 (30 cycles) (16), CXCR4 (35 cycles) (17) and glyceraldehyde 3-phosphate
dehydrogenase (GAPDH, 28 cycles) (16). PCR was performed for the given
number of cycles at 94°C for 1 min, 56°C for 1 min and 72°C for 1
min. Subsequently, 8 µl of the PCR product were resolved on
a 4.0% agarose gel and stained with ethidium bromide. After
staining, the bands were visualized using an imaging system
(AE-6932GXCF; ATTO, Tokyo, Japan).
 | Table IPrimers used for PCR in the present
study. |
Table I
Primers used for PCR in the present
study.
| Gene name | Primer | Oligonucleotide
sequence (5′→3′) | Predicted size
(bp) |
|---|
| SDF1 | Forward
Reverse |
GAGCCAACGTCAAGCATCTCAA
TTTAGCTTCGGGTCAATGCACA | 109 |
| CXCR4 | Forward
Reverse |
AGCTGTTGGCTGAAAAGGTGGTCTATG
GCGTTCTGGTGGCCCTTGGAGTGTG | 260 |
| GAPDH | Forward
Reverse |
GCACCGTCAAGGCTGAGAAC
TGGTGAAGACGCCAGTGGA | 267 |
Real-time PCR
One microgram of the RNA sample was reverse
transcribed into first-strand cDNA using a PrimeScript RT reagent
kit (Takara Shuzo Co., Ltd., Kyoto, Japan), according to the
manufacturer’s instructions and as previously described (1,16).
A Thermal Cycler Dice real-time system (Takara Shuzo Co., Ltd.) was
used for the two-step RT-PCR. The cDNA was amplified with SYBR
Premix Ex Taq and specific oligonucleotide primers for the target
sequences encoding parts of SDF1 and CXCR4. The primers (listed in
Table I and as previously
described) were designed based on the cDNA sequences of human mRNA
for SDF1 (16),
CXCR4 (17) and
GAPDH (16). The
amplification conditions were as follows: 10 sec at 95°C, followed
by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, with a final 15
sec at 95°C and 30 sec at 60°C in the Thermal Cycler Dice real-time
system.
Western blot analysis of SDF1 protein
expression in SDP11 cells
The cells were plated at a density of
1.5×105 cells/dish (60 mm in diameter) and cultured for
2 days. The cells were then washed twice with DPBS and treated with
lysis buffer (10 mM HEPES-KOH (pH 7.5), 100 mM KCl and 0.1% NP-40].
Conditioned medium (CM) was also collected. The protein
concentrations in the cell lysates (CLs) and CM were measured using
a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Equal
amounts of protein (20 µg) from each sample were separated
by sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) on 10% gels and then transferred onto polyvinylidene
difluoride membranes (Millipore, Bedford, MA, USA). After blocking
with 5% skim milk in Tris-buffered saline containing 0.1% Tween-20
(TBST), the membranes were incubated with rabbit anti-human SDF1
antibody (Cat. no. 500-P87A; PeproTech, Rocky Hill, NJ, USA) and
subsequently with anti-mouse secondary antibody (product no. 31432;
Zymed Laboratories Inc., San Francisco, CA, USA). Specific protein
bands on the membrane were detected using an enhanced horseradish
peroxidase (HRP) conjugate substrate kit (Bio-Rad) as previously
described (1).
Immunocytochemistry
The cells were fixed with cold 4% para-formaldehyde
in 0.1 M DPBS (4) for 15 min. The
cells were then rinsed in 1X Tris-buffered saline (TBS), blocked
with 10% normal goat serum (NGS) in 1X Triton X-100 in TBS for 1 h,
and incubated with primary antibodies overnight. The following
primary antibodies were used: antibodies against SDF1 (1:1,000;
PeproTech) and CXCR4 (1:1,000; Cat. no. MAB172-SP; R&D
Systems). After washing in TBS, the cells were incubated for 1 h at
room temperature with the appropriate secondary antibodies
[anti-mouse Alexa 594 (Cat. no. R37115) at a dilution of 1:1,000 or
anti-rabbit Alexa 488 (Cat. no. A-11034) at a dilution of 1:1,000
(both from Invitrogen Life Technologies)]. Finally, the nuclei were
stained with 4′,6-diamidino-2-phenylindole (DAPI). The cells were
then mounted on coverslips in ProLong anti-fade reagent (Invitrogen
Life Technologies) and examined under an epifluorescence microscope
(Nikon ECLIPSE TE2000-U; Nikon Corp., Tokyo, Japan).
Transmembrane migration assay
Cell migration was evaluated using a Transwell
system as previously described (10). Chambers with a 8-µm pore
size and 6.4 mm in diameter were used (Corning Cell Culture Insert
24-well System; Sigma-Aldrich, St. Louis, MO, USA). The UE7T-13
cells were treated with DPBS or 0.1 µM AMD3100 (a specific
antagonist of CXCR4; Millipore, Darmstadt, Germany) for 3 h prior
to the assay. The cells were then dissociated and re-seeded into
the upper chamber at a density of 1×105 cells/ml.
Subsequently, 700 µl of CM collected from the SDP11 cell
culture were added to the lower well. Following a 6-h incubation at
37°C, the cells migrating through the membrane were fixed in 4%
paraformaldehyde for 15 min, while the cells on the upper surface
of the inserts were removed using cotton-tipped swabs. The
Transwell chamber was immersed in 1 g/ml hematoxylin
(Sigma-Aldrich) for 15 min. To quantify the migrating cells, 5
random microscopic fields per membrane were photographed using a
Nikon inverted microscope system at ×40 magnification. All assays
were performed independently 3 times.
Effect of FGF-2 on SDF1 expression
In order to determine whether SDF1 expression
can be modulated by FGF-2 (Cat. no. 233-FB-01M; R&D Systems),
the SDP11 cells were cultured in the presence or absence of 20
ng/ml FGF-2 for 2 days. To confirm whether FGF-2 acts through the
FGF receptor (FGFR), 1 µM AZD4547 (Cat. no. sc-364421, Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) was also used as a
specific inhibitor of FGFR. Following culture, RNA was extracted
and real-time PCR was performed as described above.
Statistical analysis
The results are expressed as the means ± SEM.
Statistical significance was determined using one-way analysis of
variance and the Bonferri correction method between pairs of
groups. Differences with P-values of <0.01 were considered
statistically significant.
Results
Expression of SDF1 and CXCR4 in SDP11
cells
We first confirmed the mRNA expression of
SDF1 and CXCR4 in the SDP11, MG-63, HuO9 and UE7T-13
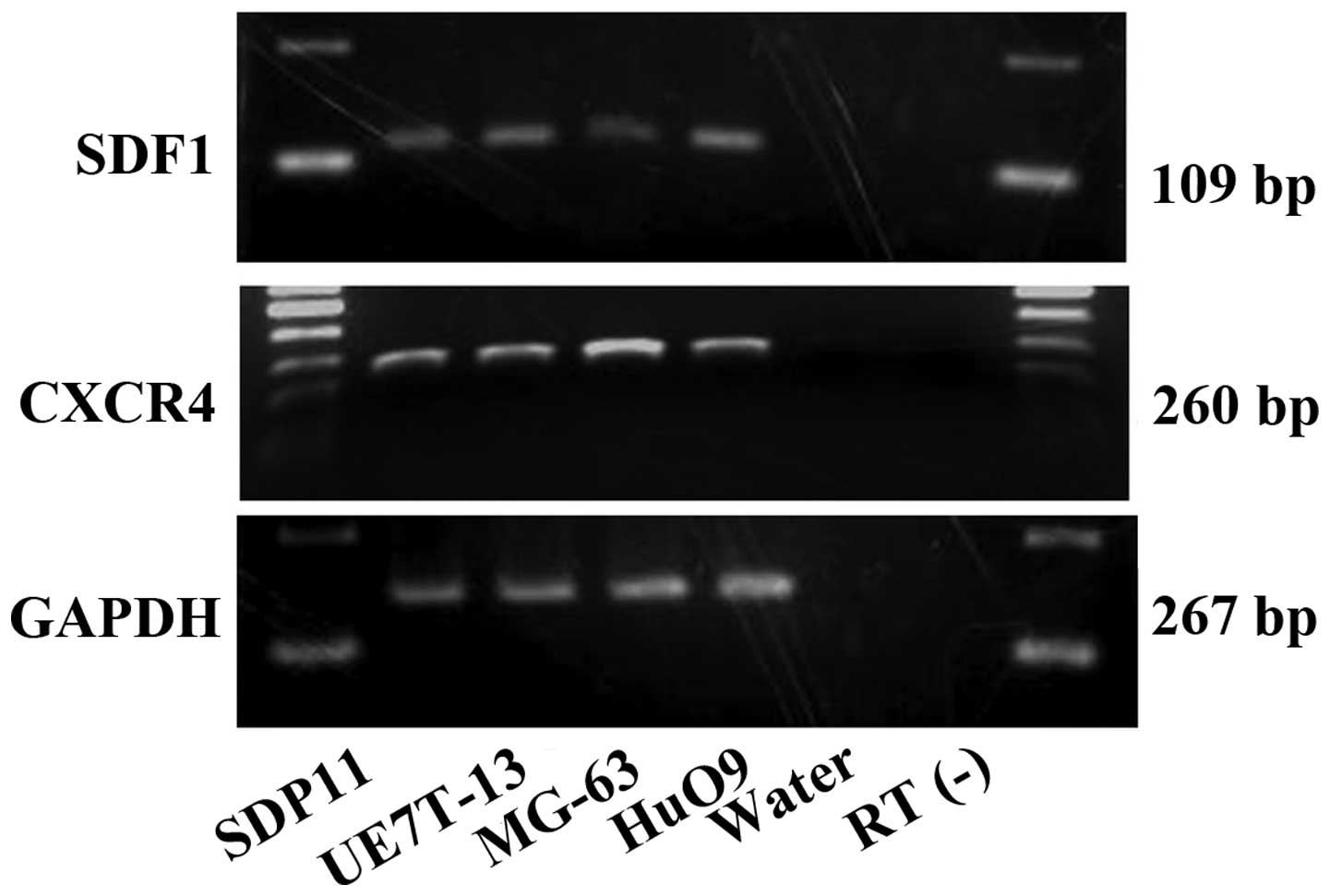
cells using RT-PCR (Fig. 1).
SDF1 and CXCR4 mRNA was observed in all the cell
lines examined in the present study (Fig. 1). No bands were obserbed for the
water and RT(−) negative controls.
Immunocytochemical analysis of SDF1 and
CXCR4
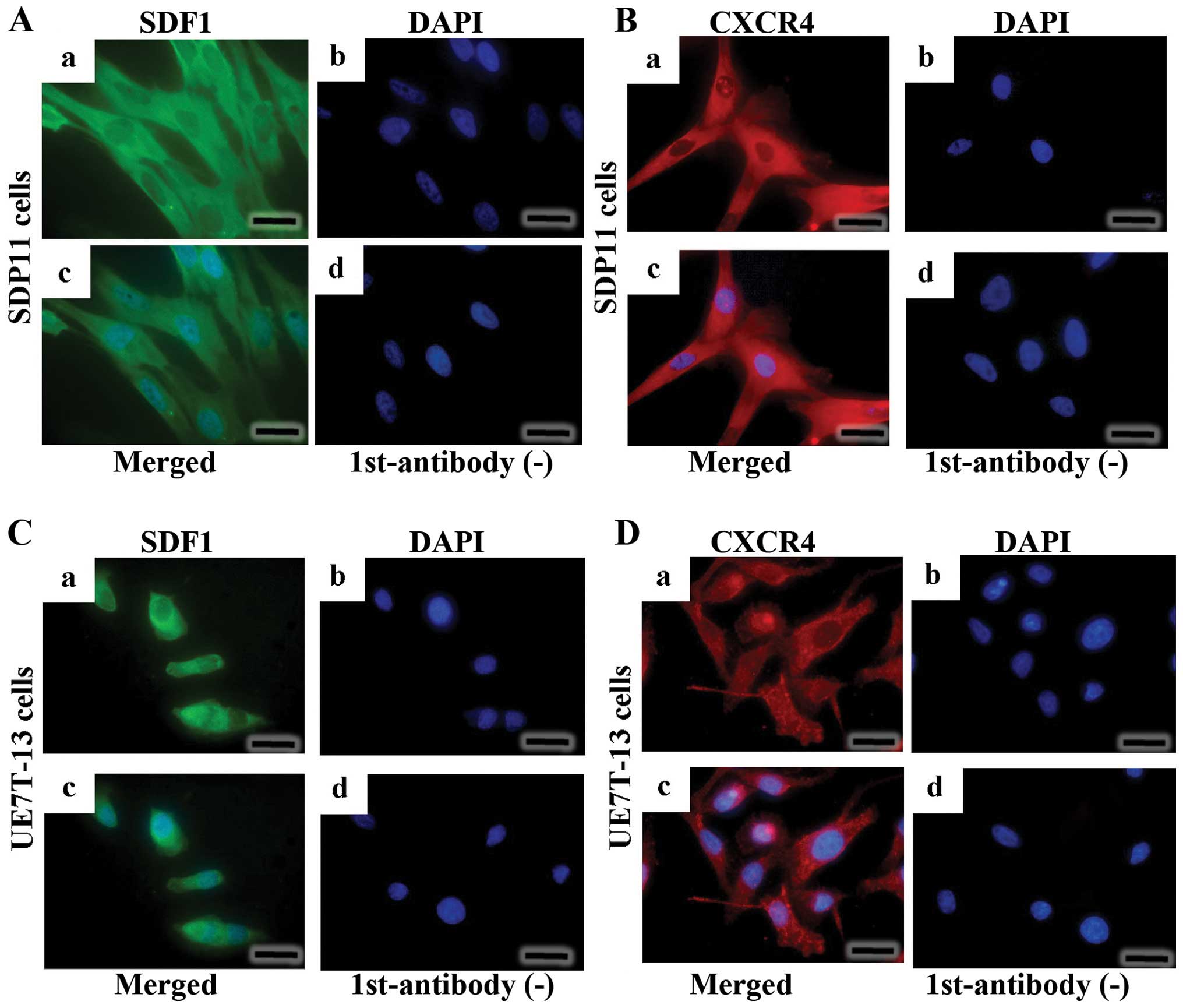
The SDP11 and UE7T-13 cells were stained using
anti-SDF1 antibody (Fig. 2A–a and
C–a). SDF1 presented as diffuse cytoplasmic fluorescent green
signals, with no staining in the nuclei (Fig. 2A–a and C–a). No signals were
observed when the primary antibody was omitted (Fig. 2A–d and C–d). CXCR4 was detected in
both the cytoplasmic and cell surface compartments (Fig. 2B–a and D–a). No signal was
detected when the primary antibody for CXCR4 was omitted (Fig. 2B–d and D–d).
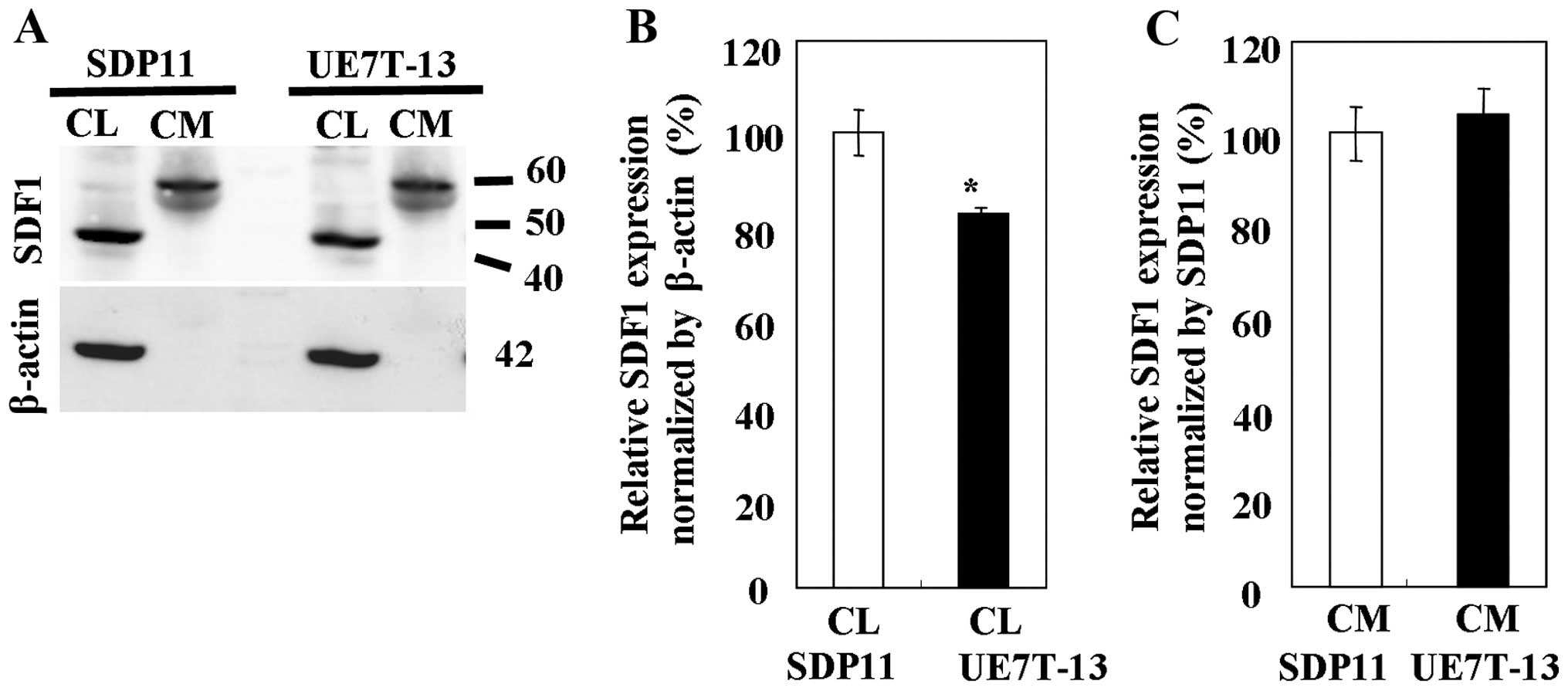
Western blot analysis of SDF1
To determine the protein expression of SDF1, whole
CLs and CM were prepared from the SDP11 and UE7T-13 cells. The
expression of SDF1 was detected in the CLs and CM from both cell
lines using anti-SDF1 antibody (Fig.
3). The band representing SDF1 appeared at a higher molecular
weight than the theoretical molecular weight, possibly due to
either an association of the chemokine with other molecules or the
aggregation of the chemokine, as SDF1 is able to form oligomers
spontaneously (18). The highest
levels of SDF1 protein in all samples tested were observed in the
CLs from the SDP11 cells (Fig.
3B). SDF1 expression was significantly higher in the CLs from
the SDP11 cells than in the CLs from the UE7T-13 cells (Fig. 3B); however, no significant
differences (very slight difference) in SDF1 expression were
observed between the CM of the SDP11 and UE7T-13 cells (Fig. 3C).
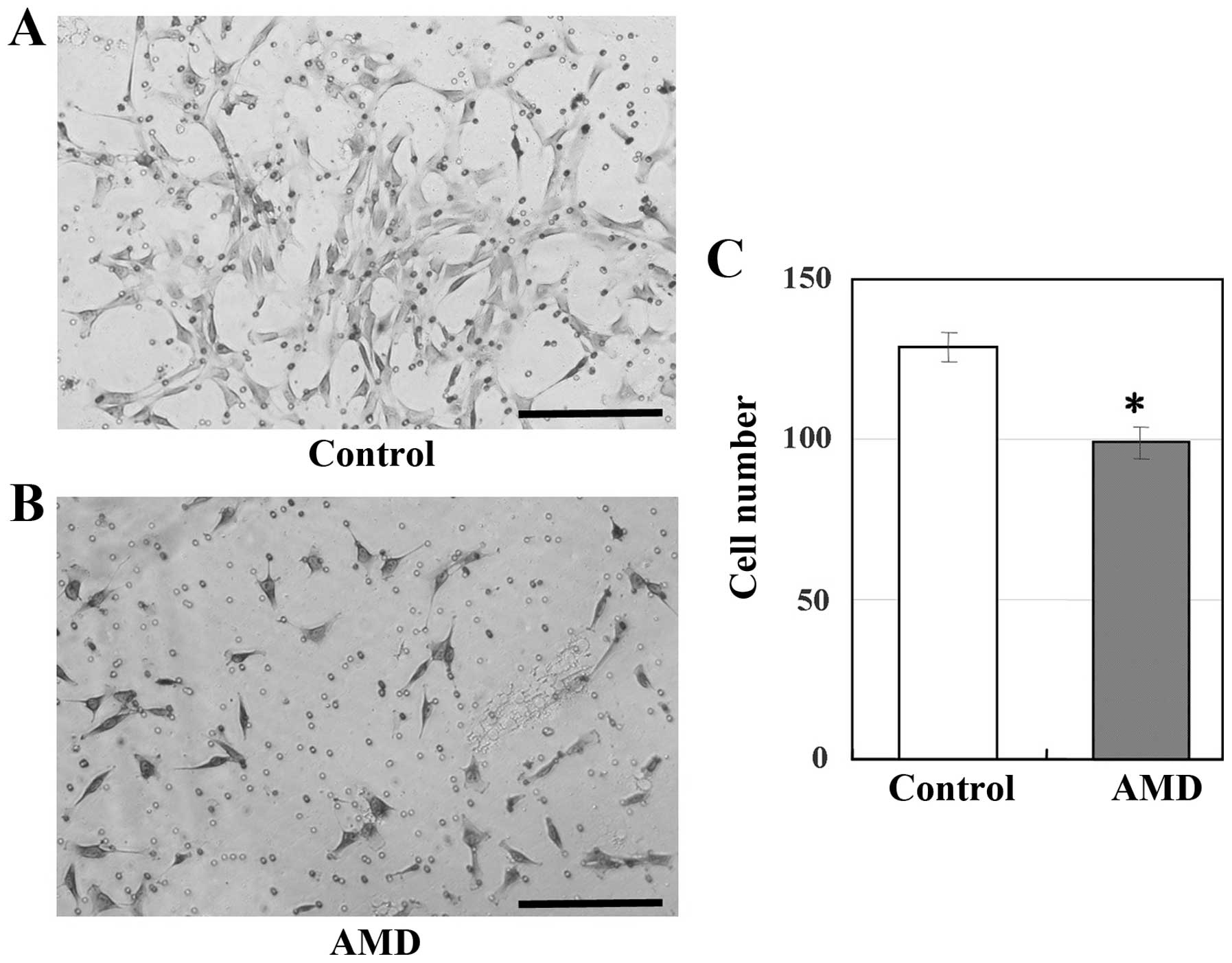
Transmembrane migration assay
To determine whether the inhibition of CXCR4 in
UE7T-13 cells inhibits cell migration, the cell cultures were
treated with the CXCR4-specific inhibitor, AMD3100. As shown in
Fig. 4, the migration of the
UE7T-13 cells was significantly inhibited by treatment with
AMD3100.
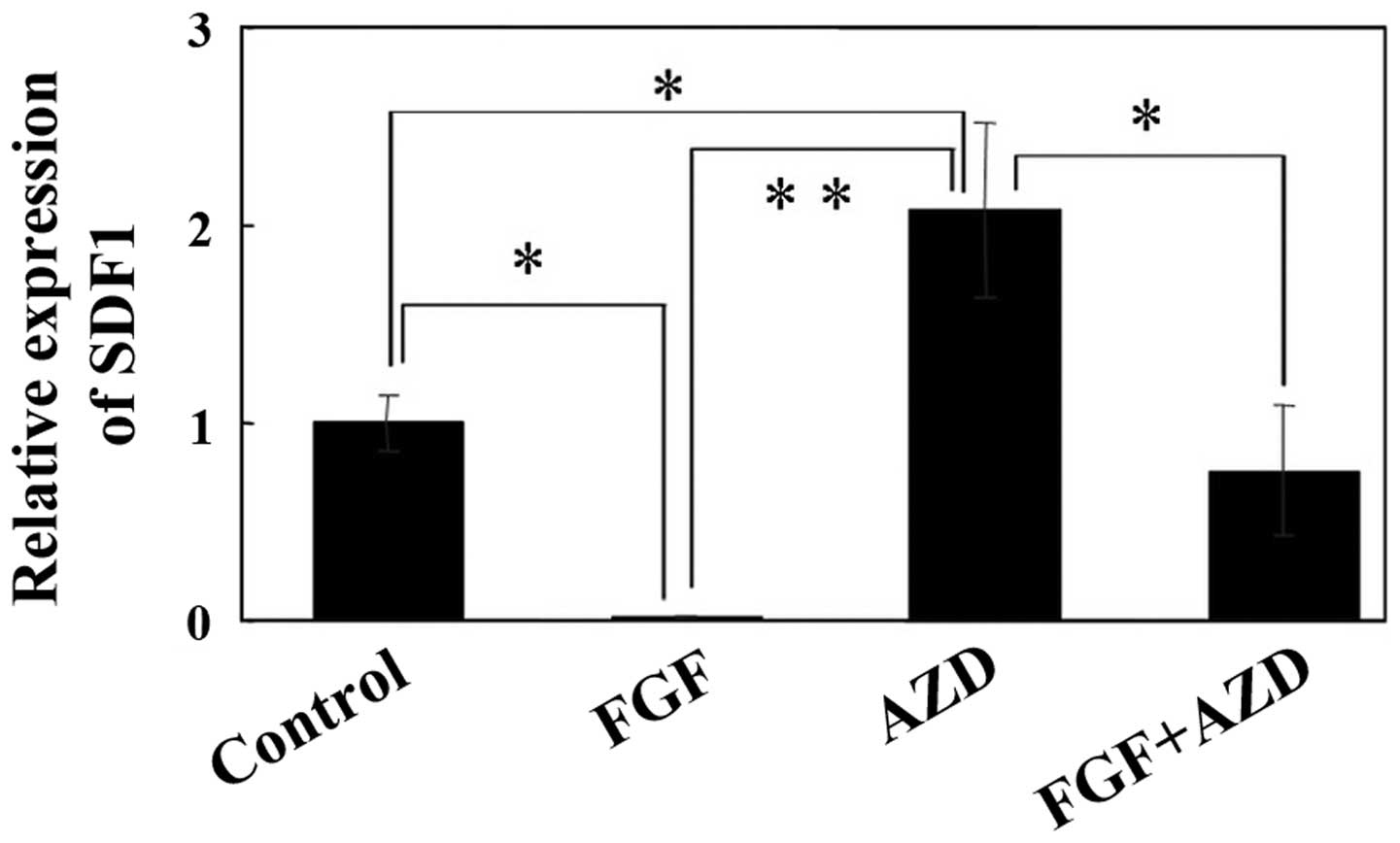
SDF1 expression is regulated by
FGF-2
Subsquently, the effects of FGF-2 on SDF1
expression were evaluated by real-time PCR. The results of
real-time PCR revealed that SDF1 expression was
substantially inhibited by treatment with 20 ng/ml FGF-2 for 48 h
in the SDP11 cells (Fig. 5). By
contrast, treatment with AZD4547 (an inhibitor of the FGF receptor)
alone significantly increased SDF1 expression, while
combined treatment with FGF-2 and 1 µM AZD4547 did not
significantly alter SDF1 expression (Fig. 5).
Discussion
In the present study, we examined the expression of
the chemokine, SDF1, and its receptor, CXCR4, in human DPCs and
MSCs. We found that both proteins were expressed in the CLs and CM
from the SDP11 and UE7T-13 cells. Moreover, the inhibition of the
CXCR4 using the specific inhibitor, AMD3100, in the UE7T-13 cells
markedly inhibited cell migration. These data highlight the
potential roles of SDF1 and CXCR4 in a variety of biological
processes in DP.
SDF1 is a chemokine that was first characterized as
a growth-stimulating factor in B cell precursors (19,20). CXCR4, the physiological receptor
of SDF1, is a seven-transmembrane receptor coupled to
heterotrimeric guanosine triphosphate(GTP)-binding proteins
(21). In a previous study, the
conditional knockout of CXCR4 in mice demonstrated that SDF1-CXCR4
signaling is essential to the maintenance of the hematopoietic stem
cell pool in the bone marrow stromal cell niche (22). Furthermore, SDF1 is upregulated in
response to injury from certain pathological conditions, such as
diabetes, liver cirrhosis and myocardial infarction, and promotes
the migration and differentiation of residential stem cells
(23–26). Therefore, SDF1 plays a critical
role in a variety of cellular functions, including embryogenesis,
tissue homeostasis and inflammation response.
In healthy human DP, 17 chemokine genes are
expressed, including SDF1, suggesting that locally produced
chemokines are involved in the migration of immune cells during
bacterial infections (27). Zhou
et al demonstrated that the engraftment of bone
marrow-derived cells (BMDCs) leads to their migration into DP
tissue, indicating that BMDCs may participate in the regeneration
of dentin, thereby serving as a source of stem cells which can
replace odontoblasts (28,29).
As SDF1 expression is significantly higher in dental tissue than in
other tissues (e.g., adipose, lung and liver tissue), the
SDF1-CXCR4 pathway is likely involved in the engraftment of BMDCs
(28). Hence, these residential
stem cells are capable of homing in on inflammatory sites in
response to injury signals, such as dental decay, and
differentiating into odontoblasts which repair dentin. In the
present study, we demonstrated DPCs from deciduous teeth (i.e.,
SDP11 cells) expressed SDF1 and CXCR4. Indeed, the migration of the
UE7T-13 cells was observed in the presence of CM. When the UE7T-13
cells were treated with AMD3100, a CXCR4 inhibitor, the cell
migration was inhibited. These results showed that the activity of
SDF1 in CM was inhibited by the CXCR4 inhibitor, AMD3100. From the
above results, it is demonstrated that SDF1 produced by pulp cells
regulates the migration of MSCs/DPSCs in DP.
It has been reported that healthy tissues basally
express SDF1 (30). In addition,
SDF1 is widely expressed in various tissues and cell lines, with
the exception of blood (31). Our
data are consistent with those of previous reports (30,31). Moreover, SDP11 cells from DP
constitutively expressed SDF1 mRNA at a level similar to
that of GAPDH (data not shown). Thus, these data suggest
that pulp cells attract and maintain DSPCs/MSCs in the pulp chamber
under normal conditions and that they participate in the response
to abnormal conditions, such as infection. SDF1 also promotes EPC
migration in a concentration-dependent manner (8). Therefore, it is reasonable to
speculate that DPSCs/MSCs and EPCs may be induced to migrate to
damaged sites, where they can then participate in tissue repair.
Thus, since there are both MSCs and EPCs in bone marrow,
circulatory system and areas surrounding DP as residential stem
cells, these cells may be involved in a coordinating mechanism for
pulpal tissue regeneration, which operates when pulp tissues are
damaged.
It has been demonstrated in previous studies that a
number of cytokines and bioactive materials have clinical uses
(32,33). One such cytokine molecule, FGF-2,
is a multifunctional growth factor that exerts various effects,
including the induction of proliferation and differentiation of a
wide range of mesodermal and neuroectodermal cells (10,32,33). FGF-2 is also a crucial factor in
wound healing and is involved in the induction of angiogenesis,
cell proliferation and the accumulation and modulation of the
extracellular matrix (34). Under
pathological conditions, such as moderate caries, growth factors
such as FGF-2 may be released from the dentin and travel to
pre-existing odontoblasts in order to secrete reparative dentin
(35). Moreover, compared with
erythropoietin (EPO), interleukin (IL)-6, SDF1β and vascular
endothelial growth factor (VEGF), FGF-2 is the most potent effector
of MSC migration (36). In light
of the above findings, we hypothesized that FGF-2 may affect
SDF1 expression in pulp cells and modulate DPSC/MSC
migration and odontoblast differentiation, and may thus have
potential therapeutic applications. Surprisingly, our results
indicated that treatment with FGF-2 for 48 h decreased SDF1
expression to <10% that of control pulp cells. FGF-2 transmits
signals via receptor-type tyrosine kinases. Following the
activation of a tyrosine kinase, various signaling pathways, such
as mitogen-activated protein kinase (MAPK), protein kinase C (PKC)
and phosphoinositol 3-kinase (PI3K) are triggered (32). In line with previous reports
(10,32), our data demonstrated that the FGF
receptor antagonist, AZD4547, abolished the effects of FGF-2 in
pulp cells. Since AZD4547 is an FGFR-specific tyrosine kinase
inhibitor, the decreased mRNA expression of SDF1 in the
SDP11 cells treated with FGF-2 was thought to result from the
modulation of FGF-2 via FGFR. However, SB203580 (a p38 inhibitor),
PD98059 [an extracellular signal-regulated kinase (ERK) inhibitor],
SP600125 [a c-Jun N-terminal kinase (JNK) inhibitor] and LY294002
(a PI3K inhibitor) did not inhibit the effects of FGF-2 (data not
shown). Thus, the intracellular signaling pathways dependent on the
mRNA expression of SDF1 in SDP11 cells and MSCs remain
unclear, and further studies are required to examine these effects.
The elucidation of the FGF-2-dependent signaling pathways in pulp
cells may be important for identifying therapeutic applications for
FGF-2 in the repair of dentin.
In conclusion, in the present study, we demonstrate
that DPCs may be important for maintaining the homeostasis of DP
tissue by controlling the migration of postnatal stem cells, e.g.,
MSCs and EPCs, to the required sites via SDF1-CXCR4 expression.
Moreover, as SDF1 expression may be regulated by FGF-2 in
DPCs, several other cytokines may be crucial for SDF1-CXCR4
expression and thus also for cell-based tissue regeneration. These
findings may facilitate our understanding of the mechanisms of
homeostasis in the DP via SDF1 expression.
Acknowledgments
The present study was supported in part by
Grants-in-Aid for Scientific Research (C) (no. 25463182 to T.H.)
and (B) (no. 26293435 to T.I.), Grants-in Aid for Exploratory
Research (no. 25670869 to T.I.), and Grants-in-Aid for Young
Scientists (B) (no. 26861790 to Y.A.) from the Ministry of
Education, Culture, Sports, Science and Technology (MEXT); the
Japan Society for the Promotion Science (JSPS); and the Futokukai
Foundation (to Y.A., 2013).
References
|
1
|
Hasegawa T, Chosa N, Asakawa T, Yoshimura
Y, Asakawa A, Ishisaki A and Tanaka M: Effect of fibroblast growth
factor-2 on dental pulp cells derived from human deciduous teeth in
vitro. Exp Ther Med. 1:477–480. 2010.
|
|
2
|
Orchardson R and Cadden SW: An update on
the physiology of the dentine-pulp complex. Dent Update.
28:200–206. 208–209. 2001.PubMed/NCBI
|
|
3
|
Miura M, Gronthos S, Zhao M, Lu B, Fisher
LW, Robey PG and Shi S: SHED: Sstem cells from human exfoliated
deciduous teeth. Proc Natl Acad Sci USA. 100:5807–5812. 2003.
View Article : Google Scholar
|
|
4
|
Arakaki M, Ishikawa M, Nakamura T, Iwamoto
T, Yamada A, Fukumoto E, Saito M, Otsu K, Harada H, Yamada Y and
Fukumoto S: Role of epithelial-stem cell interactions during dental
cell differentiation. J Biol Chem. 287:10590–10601. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fakhry M, Hamade E, Badran B, Buchet R and
Magne D: Molecular mechanisms of mesenchymal stem cell
differentiation towards osteoblasts. World J Stem Cells. 5:136–148.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Loberg R and Taichman RS: The
pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis.
Cancer Metastasis Rev. 25:573–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zernecke A, Schober A, Bot I, von
Hundelshausen P, Liehn EA, Möpps B, Mericskay M, Gierschik P,
Biessen EA and Weber C: SDF-1α/CXCR4 axis is instrumental in
neointimal hyperplasia and recruitment of smooth muscle progenitor
cells. Circ Res. 96:784–791. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kucia M, Ratajczak J, Reca R,
Janowska-Wieczorek A and Ratajczak MZ: Tissue-specific muscle,
neural and liver stem/progenitor cells reside in the bone marrow,
respond to an SDF-1 gradient and are mobilized into peripheral
blood during stress and tissue injury. Blood Cells Mol Dis.
32:52–57. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ratajczak MZ, Kucia M, Reca R, Majka M,
Janowska-Wieczorek A and Ratajczak J: Stem cell plasticity
revisited: CXCR4-positive cells expressing mRNA for early muscle,
liver and neural cells ‘hide out’ in the bone marrow. Leukemia.
18:29–40. 2004. View Article : Google Scholar
|
|
10
|
Hasegawa T, Chosa N, Asakawa T, Yoshimura
Y, Fujihara Y, Kitamura T, Tanaka M, Ishisaki A and Mitome M:
Differential effects of TGF-β1 and FGF-2 on SDF-1α expression in
human periodontal ligament cells derived from deciduous teeth in
vitro. Int J Mol Med. 30:35–40. 2012.PubMed/NCBI
|
|
11
|
Mori T, Kiyono T, Imabayashi H, Takeda Y,
Tsuchiya K, Miyoshi S, Makino H, Matsumoto K, Saito H, Ogawa S, et
al: Combination of hTERT and bmi-1, E6, or E7 induces prolongation
of the life span of bone marrow stromal cells from an elderly donor
without affecting their neurogenic potential. Mol Cell Biol.
25:5183–5195. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shimomura T, Yoshida Y, Sakabe T, Ishii K,
Gonda K, Murai R, Takubo K, Tsuchiya H, Hoshikawa Y, Kurimasa A, et
al: Hepatic differentiation of human bone marrow-derived UE7T-13
cells: Effects of cytokines and CCN family gene expression. Hepatol
Res. 37:1068–1079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aomatsu E, Takahashi N, Sawada S, Okubo N,
Hasegawa T, Taira M, Miura H, Ishisaki A and Chosa N: Novel
SCRG1/BST1 axis regulates self-renewal, migration, and osteogenic
differentiation potential in mesenchymal stem cells. Sci Rep.
4:36522014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lv Z, Yang D, Li J, Hu M, Luo M, Zhan X,
Song P, Liu C, Bai H, Li B, et al: Bone morphogenetic protein 9
overexpression reduces osteosarcoma cell migration and invasion.
Mol Cells. 36:119–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kimura K, Nakano T, Park YB, Tani M, Tsuda
H, Beppu Y, Moriya H and Yokota J: Establishment of human
osteosarcoma cell lines with high metastatic potential to lungs and
their utilities for therapeutic studies on metastatic osteosarcoma.
Clin Exp Metastasis. 19:477–485. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hasegawa T, Chosa N, Asakawa T, Yoshimura
Y, Ishisaki A and Tanaka M: Establishment of immortalized human
periodontal ligament cells derived from deciduous teeth. Int J Mol
Med. 26:701–705. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trubiani O, Isgro A, Zini N, Antonucci I,
Aiuti F, Di Primio R, Nanci A, Caputi S and Paganelli R: Functional
interleukin-7/interleukin-7Ralpha, and SDF-1alpha/CXCR4 are
expressed by human periodontal ligament derived mesenchymal stem
cells. J Cell Physiol. 214:706–713. 2008. View Article : Google Scholar
|
|
18
|
Vergote D, Butler GS, Ooms M, Cox JH,
Silva C, Hollenberg MD, Jhamandas JH, Overall CM and Power C:
Proteolytic processing of SDF-1alpha reveals a change in receptor
specificity mediating HIV-associated neurodegeneration. Proc Natl
Acad Sci USA. 103:19182–19187. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagasawa T, Kikutani H and Kishimoto T:
Molecular cloning and structure of a pre-B-cell growth-stimulating
factor. Proc Natl Acad Sci USA. 91:2305–2309. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nagasawa T, Hirota S, Tachibana K,
Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H and
Kishimoto T: Defects of B-cell lymphopoiesis and bone-marrow
myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature.
382:635–638. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tachibana K, Hirota S, Iizasa H, Yoshida
H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N,
Nishikawa S, et al: The chemokine receptor CXCR4 is essential for
vascularization of the gastrointestinal tract. Nature. 393:591–594.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sugiyama T, Kohara H, Noda M and Nagasawa
T: Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4
chemokine signaling in bone marrow stromal cell niches. Immunity.
25:977–988. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yano T, Liu Z, Donovan J, Thomas MK and
Habener JF: Stromal cell derived factor-1 (SDF-1)/CXCL12 attenuates
diabetes in mice and promotes pancreatic beta-cell survival by
activation of the prosurvival kinase Akt. Diabetes. 56:2946–2957.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsuchiya A, Imai M, Kamimura H, Takamura
M, Yamagiwa S, Sugiyama T, Nomoto M, Heike T, Nagasawa T, Nakahata
T, et al: Increased susceptibility to severe chronic liver damage
in CXCR4 conditional knock-out mice. Dig Dis Sci. 57:2892–2900.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Asano Y, Iimuro Y, Son G, Hirano T and
Fujimoto J: Hepatocyte growth factor promotes remodeling of murine
liver fibrosis, accelerating recruitment of bone marrow-derived
cells into the liver. Hepatol Res. 37:1080–1094. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saxena A, Fish JE, White MD, Yu S, Smyth
JW, Shaw RM, DiMaio JM and Srivastava D: Stromal cell-derived
factor-1alpha is cardioprotective after myocardial infarction.
Circulation. 117:2224–2231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Farges JC, Keller JF, Carrouel F, Durand
SH, Romeas A, Bleicher F, Lebecque S and Staquet MJ: Odontoblasts
in the dental pulp immune response. J Exp Zoolog B Mol Dev Evol.
312B:425–436. 2009. View Article : Google Scholar
|
|
28
|
Zhou J, Shi S, Shi Y, Xie H, Chen L, He Y,
Guo W, Wen L and Jin Y: Role of bone marrow-derived progenitor
cells in the maintenance and regeneration of dental mesenchymal
tissues. J Cell Physiol. 226:2081–2090. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kimura Y, Komaki M, Iwasaki K, Sata M,
Izumi Y and Morita I: Recruitment of bone marrow-derived cells to
periodontal tissue defects. Front Cell Dev Biol. 2:192014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luster AD: Chemokines - chemotactic
cytokines that mediate inflammation. N Engl J Med. 338:436–445.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ratajczak MZ, Zuba-Surma E, Kucia M, Reca
R, Wojakowski W and Ratajczak J: The pleiotropic effects of the
SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis.
Leukemia. 20:1915–1924. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Murakami S: Periodontal tissue
regeneration by signaling molecule(s): What role does basic
fibroblast growth factor (FGF-2) have in periodontal therapy?
Periodontol. 56:188–208. 2011. View Article : Google Scholar
|
|
33
|
Fujii S, Maeda H, Tomokiyo A, Monnouchi S,
Hori K, Wada N and Akamine A: Effects of TGF-β1 on the
proliferation and differentiation of human periodontal ligament
cells and a human periodontal ligament stem/progenitor cell line.
Cell Tissue Res. 342:233–242. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim YS, Min KS, Jeong DH, Jang JH, Kim HW
and Kim EC: Effects of fibroblast growth factor-2 on the expression
and regulation of chemokines in human dental pulp cells. J Endod.
36:1824–1830. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goldberg M and Smith AJ: Cells and
extracellular matrices of dentin and pulp: A biological basis for
repair and tissue engineering. Crit Rev Oral Biol Med. 15:13–27.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schmidt A, Ladage D, Schinköthe T,
Klausmann U, Ulrichs C, Klinz FJ, Brixius K, Arnhold S, Desai B,
Mehlhorn U, et al: Basic fibroblast growth factor controls
migration in human mesenchymal stem cells. Stem Cells.
24:1750–1758. 2006. View Article : Google Scholar : PubMed/NCBI
|