Introduction
In response to various stimuli under conditions of
physiological stress, heat shock transcription factors (HSFs)
regulate the dynamic expression of various heat shock proteins
(HSPs), which are responsible for the subsequent downstream
effects, including stress-related cytoprotective events, the
folding and assembly of nascent polypeptides and the intracellular
transport of proteins (1–4).
Heat shock factor 2 (HSF2), which belongs to the HSF
family, has been proven to play a key role in regulating the
ubiquitin proteasome pathway and differentiation (1–2,5).
HSF2 is abundantly expressed and is activated in stem cells and
embryonic carcinoma cells and also during embryogenesis and
spermatogenesis (1–6,7).
The transcription of HSF2 is complicated by the existence of two
isoforms, HSF2-α and HSF2-β, which are generated by alternative
splicing events. The ratio of HSF2-α and HSF2-β isoforms varies
significantly between different adult tissues, such as the brain,
heart and testes, suggesting that these two proteins are
functionally distinct (1,4). Similar to heat shock factor 1
(HSF1), HSF2 was recently found to be activated during heat shock
and its expression is induced upon exposure to proteasome
inhibitors; it was also reported that its deficiency increased the
sensitivity of vertebrate cells to heat shock (8–11).
Hsf2−/− mice have a
male hypofertile phenotype that is characterized by reduced testis
size and brain abnormalities associated with enlarged ventricles
(6,12). Although HSF2 typically functions
as a transcription factor, it also induces gene bookmarking, such
as for the hsp70i gene, as demonstrated in mitotic cells (13). Additionally, HSF2 modulates the
expression of heat shock genes by interacting directly with HSF1 or
heat shock factor 4 (HSF4) (14–17). However, little is known concerning
the exact transcriptional regulation of HSF2 during cellular
processes.
To the very best of our knowledge, in the present
study, we provide the first direct evidence that HSF2 transcription
is inhibited and regulated by an autoregulatory mechanism through
regions of its own promoter, thus providing a novel mechanism
responsible for the regulation of HSF2 through various cellular
signals.
Materials and methods
Cell culture and reagents
Human K562 erythroleukemia cells [from the Americal
Type Culture Collection (ATCC), Manassas, VA, USA] were cultured in
a 5% CO2 atmosphere at 37°C in RPMI-1640 (Sigma-Aldrich,
Co. Wicklow, Ireland) supplemented with 10% fetal bovine serum
(FBS; Life Technologies, Carlsbad, CA, USA) and antibiotics
(penicillin 10,000 U/ml, streptomycin 10,000 µg/ml; WelGENE
Inc, Daegu, Korea). HEK293 embryonic kidney cells (ATCC) were
cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma, St.
Louis, MO, USA) supplemented with 10% (v/v) FBS. Hemin was
purchased from Sigma. Wild-type,
Hsf1−/− and
Hsf2−/− mouse
embryonic fibroblasts (MEFs) were maintained in DMEM with 10% FBS.
Wild-type MEFs and
Hsf1−/− MEFs were a
gift from Ivor Benjamin (University of Texas Southwestern Medical
Center, Dallas, TX, USA).
Hsf2−/− MEFs were
kindly provided by Dr Valérie Lallemand-Mezger (Paris Diderot
University, Paris, France).
Plasmid constructs
Human HSF1 and HSF2 coding regions were generated by
PCR amplification and subcloned into the pcDNA3 plasmid
(Invitrogen™, Carlsbad, CA, USA). The HSF2 promoter
(pGL3-HSF2-luc-P1, -2.68 kb) was constructed by PCR, with human
genomic DNA as the template. The HSF2 promoter was PCR-amplified
using the following primers: forward, 5′-CTAGCT
AGCGCCAGTAGCATCTGCGTCATCT-3′, and reverse,
5′-AGCTCATTAGCCAAATGCATGAGCCTC-3′. The deleted HSF2 promoters were
PCR- amplified using the following forward primers: pGL3-HSF2-P2,
5′-GGAAAGGGCACATACTTTTGAG CTC-3′; pGL3-HSF2-P3,
5′-CTAGCTAGCACTCTCCCATTTAC TTGCTGTGACTG-3′; and pGL3-HSF2-P4,
5′-CTAGCTAGCCTAGTTCATTGGGTTGTTGTGAGGATTC-3′. The reverse primer was
5′-AGCTCATTAGCCAAATGCATGAGCCTC-3′. pGL3-HSF2-P5 was created by the
HindIII digestion of pGL3-HSF2-P1. Luciferase reporter
assays were performed as previously described (16). Briefly, the wild-type,
Hsf1−/− and Hsf2−/− MEFs were grown in
12-well plates and then co-transfected with HSF2
promoter-luciferase plasmid DNA and either with pcDNA3-HSF1 or
-HSF2. After 48 h of transfection, cell lysates were analyzed for
luciferase activity. The luciferase reporter assays were performed
using the Dual-Luciferase Reporter assay system (Promega, Madison,
WI, USA) following the manufacturer's instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was prepared using TRIzol reagent
(Invitrogen). RNA (1 µg) was treated with DNase (Promega)
and reverse transcribed using the Maxime RT PreMix (iNtRON
Biotechnology, Seongnam-si, Korea). The following primers were used
for RT-PCR: hHSF2-ORF-forward, 5′-TAGAGAACCCACTGCTTACTGG-3′ and
hHSF2-ORF-reverse, 5′-GTTGCTCATCCAAGACCAGAA-3′; hHSF2-endo-forward,
5′-CCCCAGGAAGTGGACTTTACATGTA-3′ and hHSF2-endo-reverse,
5′-TATGGAGCTGGAACCCTATCAGACA-3′. The GAPDH RT-PCR primers were:
forward, 5′-AGCCAAAAGGGTCATCATCTCTGC-3′ and reverse,
5′-GCATTGCTGATGATCTTGAGGCTG-3′. GADPH was used as an internal
control. The following primers were used for quantitative
(real-time) PCR: hHSF2-ORF-forward, 5′-ATTCAGAGTGGAGAGCAGAATG-3′
and hHSF2-ORF-reverse, 5′-CTG GACAGCACTAGACATGAGA-3′;
hHSF2-endo-forward, 5′-CCGCGTTAACAATGAAGCAG-3′ and
hHSF2-endo-reverse, 5′-CATTCTGGCTCCAGGTG ATG-3′. After the reaction
mixture was loaded into a glass capillary tube, the following
cycling conditions were used: initial denaturation at 95°C for 10
min, followed by 40 cycles of denaturation at 95°C for 10 sec,
annealing at 55°C for 30 sec and a final extension at 72°C for 10
sec. In the final cycle, the melting curve was obtained by
initially heating to 95°C and subsequently cooling to 40°C for 30
sec. Our method was optimized for the relative quantification
module of LightCycler software, version 4.0.
Western blot analysis
The cells were treated with 30 µM hemin for
24 h or transiently transfected with the pcDNA3 plasmids containing
HSF1 or HSF2. The cells were then washed with PBS and harvested in
lysis buffer. Samples containing equal amounts of protein were
loaded into each lane of an SDS-polyacrylamide gel for
electrophoresis and subsequently transferred onto polyvinylidene
difluoride membranes. The membranes were blocked and then incubated
with the following antibodies: antibodies against HSF1 (sc-17757)
and HSF2 (sc-13517) obtained from Santa Cruz Biotechnology (Santa
Cruz, CA, USA).
Chromatin immunoprecipitation (ChIP)
assay
The K562 cells were grown to almost 80% confluence
and cross-linked with formaldehyde (Sigma) at room temperature for
10 min. The cross-linked chromatin was prepared with a commercial
ChIP assay kit (EZ-Magna ChIP; Millipore, Billerica, MA, USA) and
immunoprecipitated using 4 µg of normal rabbit anti-IgG
(Santa Cruz Biotechnology) or 4 µg of anti-HSF2 antibody
(Santa Cruz Biotechnology). The HSF2 binding site was PCR-amplified
using the input DNA or DNA isolated from the precipitated chromatin
as the template, in combination with primers flanking the putative
HSF binding sites in the HSF2 promoter. The primer sequences were
as follows: forward, 5′-CTCTCCCATTTACTTGCTGTGACTGAAG-3′ and
reverse, 5′-GAGCCCTTATATATGCCAAGGGCTTTAC-3′.
Purification of TAT fusion proteins
The TAT-Hsp40 expression vector was constructed as
previously described (18). The
TAT-HSF2 protein was expressed in E. coli BL21(DE3) pLysS
cells (Invitrogen) and purified using the urea-denaturing protein
purification method, as previously described (18,19). The cells were lysed by sonication
in lysis buffer (1 mM imidazole, 100 mM NaCl, 20 mM HEPES, pH 8.0)
containing 8 M urea. The cell lysates were centrifuged at 12,000 ×
g for 30 min at 4°C, and 1 ml Ni2+-NTA agarose was added
to the cleared supernatant. Following 2 h of gentle mixing at 4°C,
the resin was transferred to a column and subsequently washed 3
times with 10 ml washing buffer (20 mM imidazole, 300 mM NaCl, 50
mM phosphate buffer, pH 8.0). The proteins were eluted 4 times with
1 ml elution buffer (500 mM imidazole, 300 mM NaCl, 50 mM phosphate
buffer, pH 8.0). The urea denaturant was removed with a Mono Q ion
exchange column and desalinated with a PD-10 Sephadex size
exclusion column (both from GE Healthcare Bio-Sciences Corp.,
Piscataway, NJ, USA). The protein concentration was quantified
using the Bradford assay and confirmed by SDS-PAGE.
Statistical analysis
All data are expressed as the means ± SD of at least
3 independent experiments. One-way analysis of variance (ANOVA),
followed by the Student's t-test, was used for statistical
evaluations. Values significantly different from the relative
control are indicated with an asterisk, and values significantly
different from another group are indicated with a hash symbol. A
P-value <0.05 was considered to indicate a statistically
significant difference.
Results
HSF2 transcription is downregulated by
HSF2 overexpression
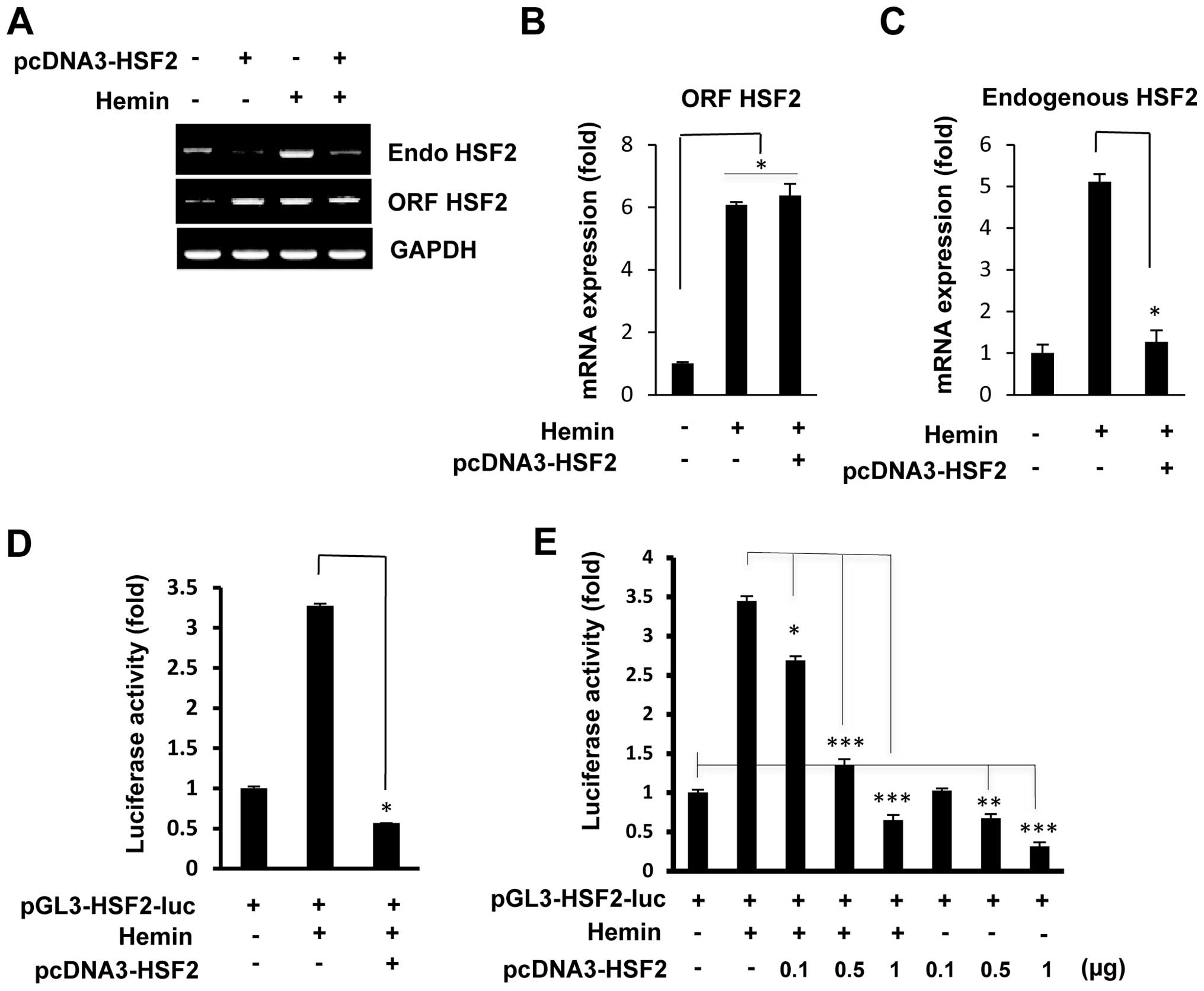
To examine whether HSF2 directly regulates its own
expression, the K562 cells were transiently transfected with an
HSF2 expression plasmid, and the mRNA expression levels of
endogenous HSF2 were measured by RT-qPCR. The primer set used for
endogenous HSF2 mRNA was designed to detect the 5′-untranslated
region (5′-UTR) of HSF2 mRNA. We also analyzed total HSF2 RNA using
open reading frame (ORF)-specific primers. As shown in Fig. 1A, the overexpression of HSF2
markedly inhibited the mRNA expression of endogenous HSF2. Hemin is
a well-established inducer of HSF2 in K562 cells, and in accordance
with our previous study (16),
hemin treatment induced the mRNA expression of endogenous HSF2.
However, even after treatment with hemin, the induction of the
overexpression of HSF2 in the K562 cells by transfection with the
HSF2 expression vector markedly inhibited the levels of endogenous
HSF2 mRNA. The total expression levels of HSF2 (ORF HSF2) were
similar between the HSF2-overexpressing and hemin-treated cells
(Fig. 1A). RT-qPCR confirmed that
the increased levels of endogenous HSF2 mRNA induced by hemin were
significantly decreased in the cells which overexpressed HSF2 and
that the total expression levels of HSF2 (ORF HSF2) were similar
between the HSF2-overexpressing and hemin-treated cells (Fig. 1B and C).
To investigate the effects of HSF2 on its own
promoter, a plasmid expressing HSF2 was co-transfected with the
human HSF2 promoter (2.68 kb/+19)-luciferase construct into the
K562 cells. As shown in Fig. 1D,
the overexpression of HSF2 markedly reduced the hemin-induced HSF2
promoter activity, indicating the specific repression by HSF2 of
its own promoter. This repression was shown to be
concentration-dependent by co-transfection with a fixed amount of
pGL3-HSF2-luc and increasing amounts of the plasmid pcDNA3-HSF2.
Both with and without hemin treatment, transfection with increasing
amounts of the expression plasmid, pcDNA3-HSF2, led to a marked
reduction in HSF2 promoter activity (Fig. 1E). These results strongly suggest
that the promoter of HSF2 (at position 2.68 kb/+19) contains an
HSF2-responsive region.
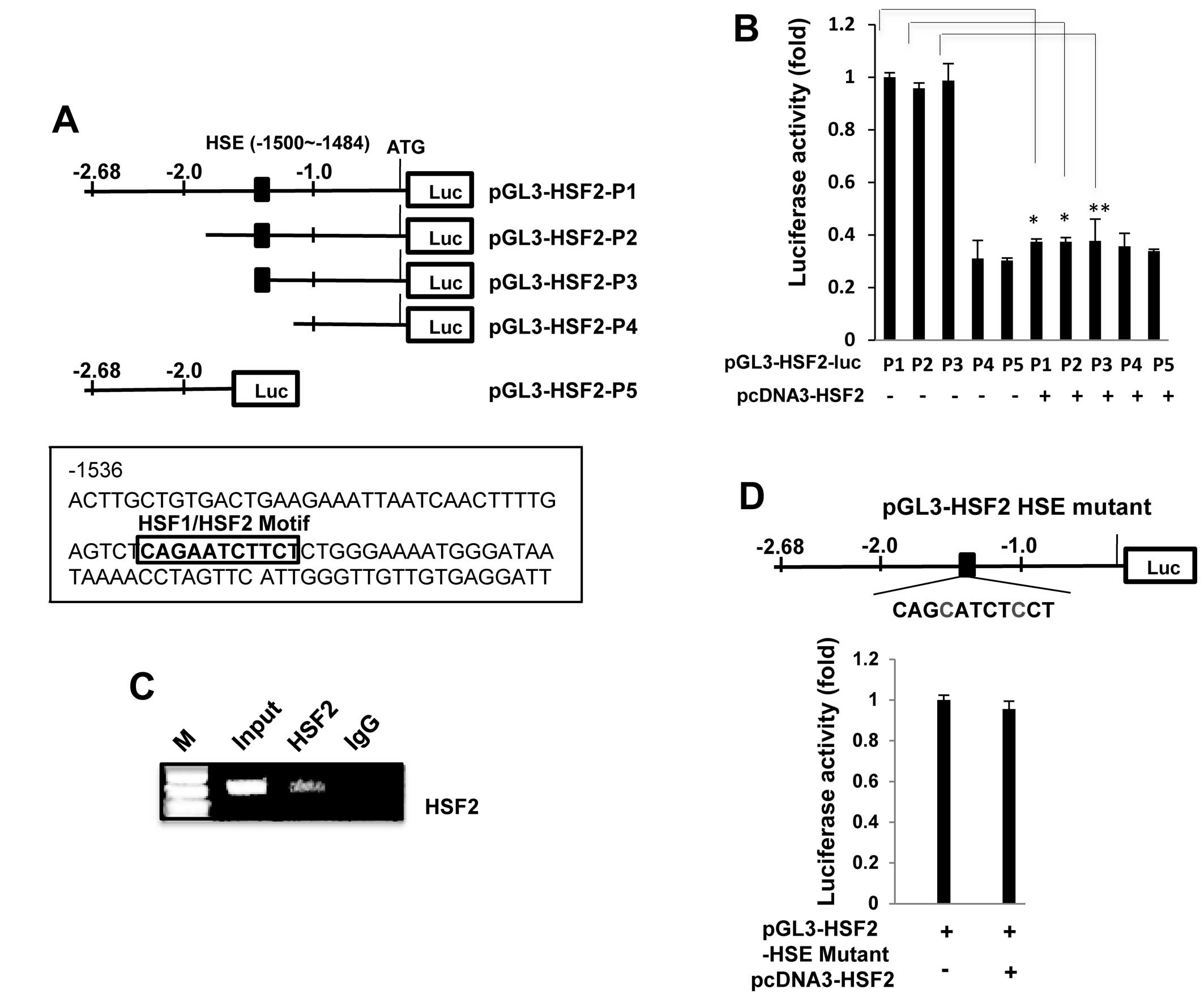
HSF2 binds to heat shock element (HSE)
sites in its own promoter
As HSF2 is known to be a transcription factor
involved in DNA-binding activity (20), we examined whether the HSF2
promoter contains typical HSF-binding sites. A HSF binding site
sequence analysis (http://molbioltools.ca/Transcriptional_factors.htm) of
the HSF2 promoter revealed one potential HSE site at −1500/−1484,
and therefore we examined whether this HSE motif plays a role in
the negative regulation of the HSF2 promoter. Deleted HSF2
promoters were constructed (Fig.
2A) and transfected into the K562 cells in order to analyze
promoter activity. As shown in Fig.
2B, deletion of the HSE motif reduced promoter activity by up
to 60% relative to the non-deleted promoter (pGL3-HSF2-luc-P1),
indicating that this HSE site is a critical region. The
overexpression of HSF2 significantly decreased the luciferase
activity of the wild-type promoter (pGL3-HSF2-luc-P1), but not the
HSE truncated-promoter (pGL3-HSF2-P4 and pGL3-HSF2-P5), indicating
that the HSE site contributes to the observed responsiveness to
HSF2-mediated repression.
To further confirm these results, we performed a
ChIP assay using cross-linked genomic DNA prepared from
HSF2-transfected K562 cells. As clearly shown in Fig. 2C, the PCR product containing the
putative HSE region was specifically and markedly amplified,
indicating that exogenous HSF2 directly binds to the HSE site; IgG
was employed as a negative control for this experiment.
Additionally, an HSF2 promoter (pGL3-HSF2-HSE mutant-P1) plasmid
containing a mutation at the HSE site was constructed (Fig. 2D) and transfected into the K562
cells in order that we could analyze promoter activity. Notably,
the pGL3-HSF2 HSE mutant did not significantly reduce the promoter
activity induced by HSF2 overexpression, indicating that the HSE
site contributes to the observed responsiveness to HSF2-mediated
repression.
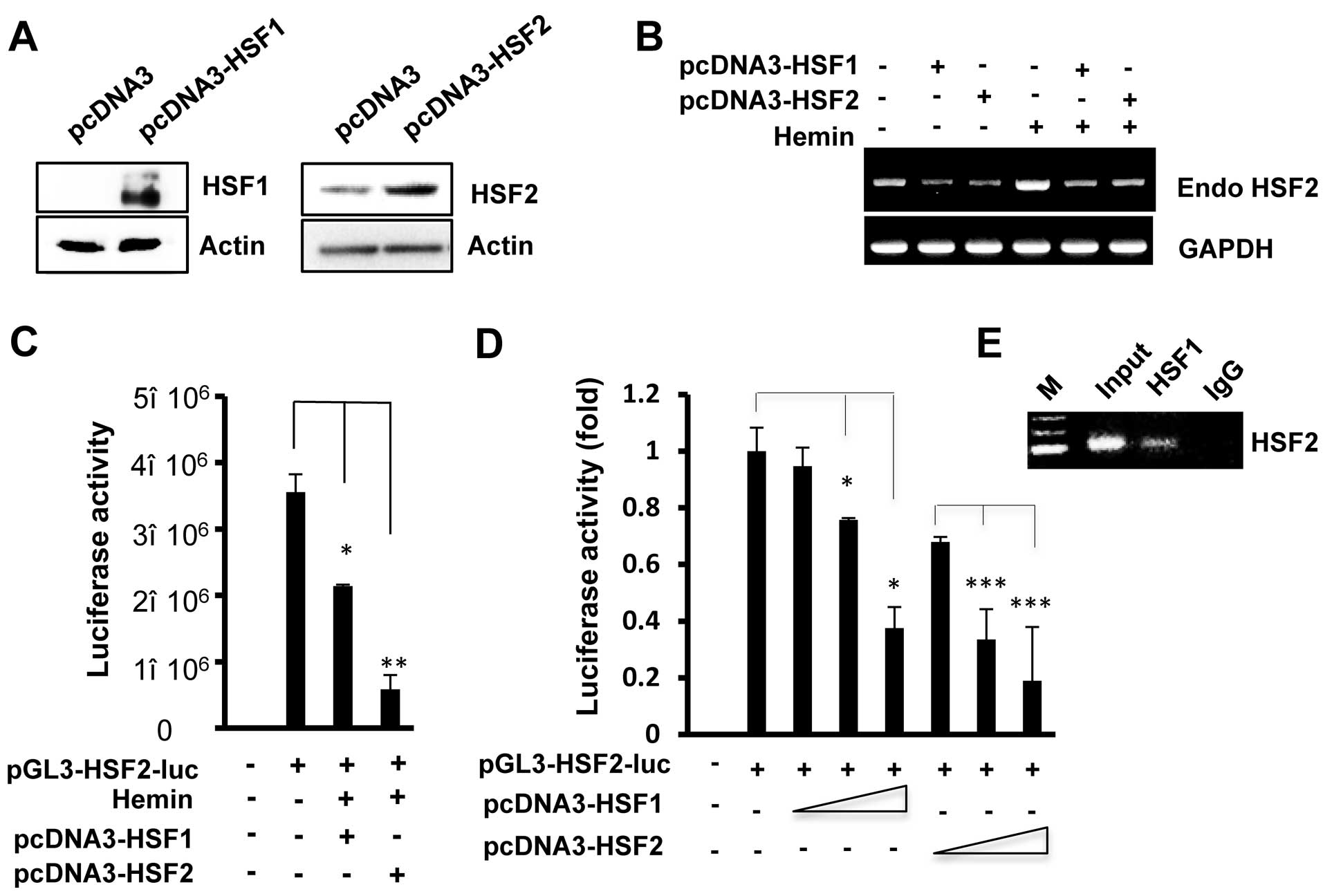
HSF1 is partially involved in the
regulation of HSF2 promoter activity
HSF1 is a transcription factor that contains a
DNA-binding domain and exhibits DNA-binding activity at the same
DNA sequences (HSE) (20). To
further analyze the transcriptional regulation of the HSF2 promoter
by HSF1, the cells were co-transfected with the pcDNA3-HSF1 and
HSF2 promoter. The protein levels of HSF1 or HSF2 increased in the
HSF1- or HSF2-transfected cells compared to the empty vector
(pcDNA2)-transfected cells (Fig.
3A). As shown in Fig. 3B–D,
as observed with HSF2, HSF1 overexpression also led to reduced the
levels of hemin-induced endogenous HSF2 mRNA and HSF2 promoter
activity. In addition, increasing the amounts of the expression
plasmid, pcDNA3-HSF1, inhibited HSF2 promoter activity in a
concentration-dependent manner (Fig.
3D). However, HSF2 resulted in a stronger inhibition of its own
promoter compared to when HSF1 was overexpressed.
To examine whether the sequence containing the HSE
site in the HSF2 promoter is recognized by HSF1, we performed a
ChIP assay. HSF1 antibody was used to immunoprecipitate the
chromatin from HSF1-transfected cells, and the associated DNA
fragments were amplified using primers flanking the HSE region in
the HSF2 promoter. As shown in Fig.
3E, the PCR product containing the putative HSE region was
specifically amplified, indicating that exogenous HSF1 also binds
to the HSE region. In addition, similar results were observed with
the HEK293 cells (data not shown). Thus, we speculate that the
binding of HSF1 and/or HSF2 on the HSE motif mediates the
repressive effect of the HSF2 promoter.
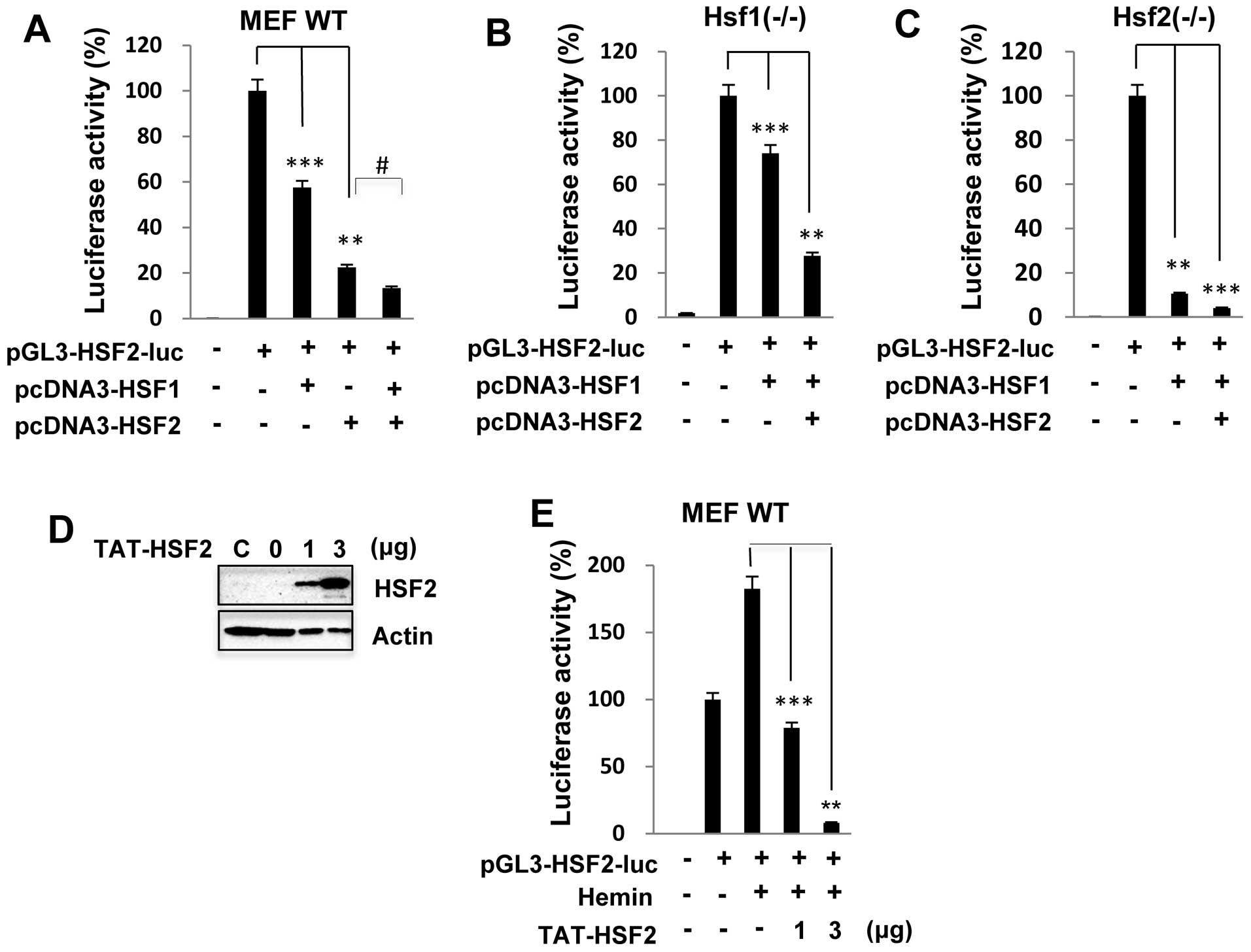
HSF1/HSF2 is recruited to the HSF2
promoter to regulate promoter activity
In a previous study, it was shown that HSF1 and HSF2
can form heterotrimers and bind to DNA following proteasome
inhibitor treatment (14,15). We thus examined whether HSF2
transcriptional activity is synergistically inhibited by complexes
of HSF1 and HSF2 proteins. We co-transfected the expression
plasmids HSF1 and/or HSF2 with the HSF2 reporter construct into the
MEFs. HSF2 promoter activity in the MEF wild-type cells was
significantly inhibited by up to 40 and 80% when HSF1 and HSF2 were
overexpressed, respectively; however, following co-transfection
with HSF1 and HSF2, promoter activity was decreased to an event
greater extent than following transfection with HSF1 or HSF2 alone
(Fig. 4A). Similar results were
also observed with the
Hsf1−/− or
Hsf2−/− MEFs
transfected with the HSF2 promoter luciferase construct (data not
shown). These results clearly suggest that HSF2 regulates its own
promoter activity through interplay with HSF1. As expected, HSF2
promoter activity was markedly inhibited in the
Hsf1−/− or
Hsf2−/− MEFs
transfected with HSF1 or HSF2 (Fig.
4B and C).
To determine whether HSF2 transcriptional activity
is functionally inhibited by the HSF2 protein, we assessed the
effects of a purified TAT-tagged HSF2 protein on HSF2 promoter
activity. Previous studies have demonstrated the potential ability
of the HIV-1 TAT protein transduction domain to modulate the
biology of living organisms through the direct cellular delivery of
proteins and peptides (18,19). In the present study, the purified
TAT-HSF2 fusion protein was directly added to wild-type MEFs for 24
h, and the level of transduced HSF2 was determined by a western
blot analysis. As shown in Fig.
4D, TAT-HSF2 was delivered successfully into the MEFs in a
dose-dependent manner. Consistent with the results shown in
Fig. 1, treatment with hemin
alone induced HSF2 promoter activity. However, the
TAT-HSF2-transduced cells exhibited lower levels of promoter
activity following treatment with hemin (Fig. 4E).
Discussion
HSF2 is a transcription factor that displays tightly
regulated gene expression. Its expression can be stimulated by
physiological signals triggered by differentiation or development
(1–21,22) and also by environmental stress
conditions, such as heat shock or proteasome inhibition (10,11). Although the HSF2 gene promoter
contains many putative responsive elements (23), the precise transcription factors
involved in the regulation of HSF2 transcription with various
stimuli remain unknown.
The majority of studies on HSF2 have focused on
protein misfolding diseases, delaying aging, and the development of
the embryo and sperm. It has been proposed that HSF2 is an upstream
regulator of oncogenic mechanisms relevant for tumor progression
and invasion, which are attractive therapeutic targets.
Understanding the mechanisms underlying the variable expression of
HSF2 is crucial to understanding the possible role of HSF2 under
both physiological and pathophysiological conditions. Previously, a
molecular characterization of the human HSF2 promoter was published
by Lee et al (24), who
observed that the several transcription factors play a critical
role in determining the levels of Hsf2 expression. However, no
information is available concerning the role of HSF2 in the
activity of its own promoter.
In the present study, we demonstrated that HSF2
transcription is regulated by its overexpression in a negative
manner. Promoter activity analysis revealed that HSF2 regulates its
own promoter, thus providing evidence for the hypothesis that an
autoregulatory mechanism exists at the transcriptional level, and
ChIP assays confirmed that the promoter binding of HSF2 is mediated
by a putative HSE motif. Our results also suggest that HSF2
transcription is partially repressed by HSF1.
In our previous study, we demonstrated that HSF4a
was able to inhibit hemin-induced HSF2 mRNA and protein expression
(16). Based on the results of
this study, and other previous studies, we suggest that HSF2
expression is regulated by the HSF family and by the
transcriptional and/or functional association between HSFs. It is
also possible that overexpressed HSFs regulate HSF2 expression by
preventing the induction of HSF2 or through the expression of other
factors controlled by HSF-mediated signaling. An alternative
explanation involves the presence of post-translational
modifications, such as phosphorylation and/or sumoylation, which
may stabilize the binding of HSF2 to the promoter (25).
It has been reported that the heterotrimerization
HSF1-HSF2 provides a transcriptional switch in response to stress
and developmental stimuli (14),
and previous studies have shown that HSF2 is associated with HSF1
and activates the hsp70 promoter in vitro and in
vivo (15,17). Furthermore, it is possible that
with various stimuli, HSF1 and HSF2 interact and form
heterocomplexes that can be recruited to specific promoters
(16). For example, in
endothelial cells, the arseniteinducible RNA-associated protein
(AIRAP) transcriptional level is regulated by HSF1-HSF2
heterotrimeric complexes following treatment with the anticancer
drug, bortezomib, suggesting that these two factors have a close
functional association and also that HSF2 alone can negatively
regulate bortezomib-induced AIRAP expression (26). Treatment with the proteasome
inhibitor MG132 or the amino acid analog L-azetidine-2-carboxylic
acid (AZC) has been shown to induce the formation of a HSF1/HSF2
heterocomplex that binds to the clusterin element and increases
both clusterin protein and mRNA levels in the human glial cell
line, U-251 MG (27).
In the present study, we also demonstrated that both
HSF1 and HSF2 are recruited to the HSF2 promoter under
overexpression conditions. The activity of the HSF2 promoter was
decreased to a greater extent by the overexpression of both HSF1
and HSF2 than by HSF1 or HSF2 alone, which suggests that HSF1 and
HSF2 interacts directly and/or form heterocomplexes to bind to the
HSF2 promoter. Notably, HSF1 is able to inhibit HSF2 promoter
activity in Hsf2−/−
MEF cells, indicating that HSF1 plays an important role in HSF2
transcription. Although the potential impact of HSF1 on
stress-regulated HSF2 transcriptional expression is not yet well
defined, at the transcriptional level, we do know that an HSF can
positively and/or negatively modulate the expression of other HSF
genes as well as that of its own gene.
In conclusion, in the present study, we provide
molecular evidence for an autoregulatory mechanism that allows HSF2
to control its own expression. We believe that these findings
provide new insight into the pathogenetic mechanisms of human
HSF2-related diseases.
Abbreviations:
|
HSF2
|
heat shock factor 2
|
|
HSF1
|
heat shock factor 1
|
|
HSE
|
heat shock element
|
|
ChIP
|
chromatin immunoprecipitation
|
|
TAT PDT
|
protein transduction domain of Tat
|
Acknowledgments
The present study was supported by the National
Research Foundation of Korea (NRF) grant funded by the Korea
government MSIP (No. 2008–0062283). This study was also supported
by the Basic Science Research Program through the National Research
Foundation of Korea (NRF) funded by the Ministry of Education,
Science and Technology (2010–0023366).
References
|
1
|
Akerfelt M, Trouillet D, Mezger V and
Sistonen L: Heat shock factors at a crossroad between stress and
development. Ann NY Acad Sci. 1113:15–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fujimoto M and Nakai A: The heat shock
factor family and adaptation to proteotoxic stress. FEBS J.
277:4112–4125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akerfelt M, Morimoto RI and Sistonen L:
Heat shock factors: integrators of cell stress, development and
lifespan. Nat Rev Mol Cell Biol. 11:545–555. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pirkkala L, Nykänen P and Sistonen L:
Roles of the heat shock transcription factors in regulation of the
heat shock response and beyond. FASEB J. 15:1118–1131. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pirkkala L, Alastalo TP, Zuo X, Benjamin
IJ and Sistonen L: Disruption of heat shock factor 1 reveals an
essential role in the ubiquitin proteolytic pathway. Mol Cell Biol.
20:2670–2675. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang G, Zhang J, Moskophidis D and Mivechi
NF: Targeted disruption of the heat shock transcription factor
(hsf)-2 gene results in increased embryonic lethality, neuronal
defects, and reduced spermatogenesis. Genesis. 36:48–61. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Choi MR, Jung KH, Park JH, Das ND, Chung
MK, Choi IG, Lee BC, Park KS and Chai YG: Ethanol-induced small
heat shock protein genes in the differentiation of mouse embryonic
neural stem cells. Arch Toxicol. 85:293–304. 2011. View Article : Google Scholar
|
|
8
|
Shinkawa T, Tan K, Fujimoto M, Hayashida
N, Yamamoto K, Takaki E, Takii R, Prakasam R, Inouye S, Mezger V
and Nakai A: Heat shock factor 2 is required for maintaining
proteostasis against febrile-range thermal stress and polyglutamine
aggregation. Mol Biol Cell. 22:3571–3583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Trinklein ND, Chen WC, Kingston RE and
Myers RM: Transcriptional regulation and binding of heat shock
factor 1 and heat shock factor 2 to 32 human heat shock genes
during thermal stress and differentiation. Cell Stress Chaperones.
9:21–28. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mathew A, Mathur SK and Morimoto RI: Heat
shock response and protein degradation: regulation of HSF2 by the
ubiquitin-proteasome pathway. Mol Cell Biol. 18:5091–5098.
1998.PubMed/NCBI
|
|
11
|
Sistonen L, Sarge KD, Phillips B, Abravaya
K and Morimoto RI: Activation of heat shock factor 2 during
hemin-induced differentiation of human erythroleukemia cells. Mol
Cell Biol. 12:4104–4111. 1992.PubMed/NCBI
|
|
12
|
Kallio M, Chang Y, Manuel M, Alastalo TP,
Rallu M, Gitton Y, Pirkkala L, Loones MT, Paslaru L, Larney S, et
al: Brain abnormalities, defective meiotic chromosome synapsis and
female subfertility in HSF2 null mice. EMBO J. 21:2591–2601. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xing H, Wilkerson DC, Mayhew CN, Lubert
EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge OK and Sarge KD:
Mechanism of hsp70i gene bookmarking. Science. 307:421–423. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sandqvist A, Björk JK, Akerfelt M,
Chitikova Z, Grichine A, Vourc'h C, Jolly C, Salminen TA, Nymalm Y
and Sistonen L: Heterotrimerization of heat-shock factors 1 and 2
provides a transcriptional switch in response to distinct stimuli.
Mol Biol Cell. 20:1340–1347. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ostling P, Björk JK, Roos-Mattjus P,
Mezger V and Sistonen L: Heat shock factor 2 (HSF2) contributes to
inducible expression of hsp genes through interplay with HSF1. J
Biol Chem. 282:7077–7086. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim SA, Yoon JH and Ahn SG: Heat shock
factor 4a (HSF4a) represses HSF2 expression and HSF2-mediated
transcriptional activity. J Cell Physiol. 227:1–6. 2012. View Article : Google Scholar
|
|
17
|
He H, Soncin F, Grammatikakis N, Li Y,
Siganou A, Gong J, Brown SA, Kingston RE and Calderwood SK:
Elevated expression of heat shock factor (HSF) 2A stimulates
HSF1-induced transcription during stress. J Biol Chem.
278:35465–35475. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Becker-Hapak M, McAllister SS and Dowdy
SF: TAT-mediated protein transduction into mammalian cells.
Methods. 24:247–256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim SA, Chang S, Yoon JH and Ahn SG:
TAT-Hsp40 inhibits oxidative stress-mediated cytotoxicity via the
inhibition of Hsp70 ubiquitination. FEBS Lett. 582:734–740. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ahn SG, Liu PC, Klyachko K, Morimoto RI
and Thiele DJ: The loop domain of heat shock transcription factor 1
dictates DNA-binding specificity and responses to heat stress.
Genes Dev. 15:2134–2145. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morange M: HSFs in development. Handb Exp
Pharmacol. 172:153–169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goodson ML, Park-Sarge OK and Sarge KD:
Tissue-dependent expression of heat shock factor 2 isoforms with
distinct transcriptional activities. Mol Cell Biol. 15:5288–5293.
1995.PubMed/NCBI
|
|
23
|
Nykänen P, Alastalo TP, Ahlskog J,
Horelli-Kuitunen N, Pirkkala L and Sistonen L: Genomic organization
and promoter analysis of the human heat shock factor 2 gene. Cell
Stress Chaperones. 6:377–385. 2001. View Article : Google Scholar
|
|
24
|
Lee SS, Kwon SH, Sung JS, Han MY and Park
YM: Cloning and characterization of the rat Hsf2 promoter: a
critical role of proximal E-box element and USF protein in Hsf2
regulation in different compartments of the brain. Biochim Biophys
Acta. 1625:52–63. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu YM, Huang DY, Chiu JF and Lau ATY:
Post-translational modification of human heat shock factors and
their functions: a recent update by proteomic approach. J Proteome
Res. 11:2625–2634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rossi A, Riccio A, Coccia M, Trotta E,
Frazia SL and Santoro MG: The proteasome inhibitor bortezomib is a
potent inducer of zinc-finger AN1-type domain 2a gene expression:
Role of HSF1/HSF2 heterocomplexes. J Biol Chem. 289:12705–12715.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Loison F, Debure L, Nizard P, le Goff P,
Michel D and le Dréan Y: Up-regulation of the clusterin gene after
proteotoxic stress: Implication of HSF1-HSF2 heterocomplexes.
Biochem J. 395:223–231. 2006. View Article : Google Scholar :
|