1. Introduction
The kidneys play a physiologic role in the
regulation of urine formation and nutrient reabsorption in the
proximal tubule epithelial cells. Kidney development has been shown
to be regulated through calcium (Ca2+) signaling
processes that are present through numerous steps of tubulogenesis
and nephron induction during embryonic development of the kidneys
(1). Ca2+-binding
proteins, such as calbindin-D28k and regucalcin have been shown to
be important proteins that are commonly used as biomarkers in
pronephric tubules, and the ureteric bud and metanephric mesenchyme
(1). Thus, regucalcin has been
focused to be Ca2+ sensors that are involved in renal
organogenesis in the link between Ca2+-dependent signals
and polycystins. Moreover, there is accumulating evidence that
regucalcin plays a multifunctional role in kidney cell regulation
(2).
Regucalcin, which was discovered in 1978 as a
Ca2+-binding protein (3), is known to play a pivotal role as a
suppressor protein in intracellular Ca2+ signaling in
various types of cells and tissues (4–6).
The regucalcin gene, which is localized on the X chromosome, is
identified in over 15 species consisting of the regucalcin family
and is highly conserved in vertebrate species throughout evolution
(7). The rat regucalcin gene
consists of 7 exons and 6 introns (7). Various transcription factors
[including activator protein (AP)-1, nuclear factor I-A1 (NF1-A1),
regucalcin gene promoter region-related protein 117 (RGPR-p117),
β-catenin, SP1 and others] have been identified as enhancers and
suppressors of regucalcin gene expression (7,8).
Regucalcin gene expression has been shown to be pronounced in the
liver and kidney proximal tubule epithelial cells in rats and is
regulated by various transcription factors (1,7,9).
Regucalcin plays a role in the regulation of
Ca2+ in kidney proximal tubule epithelial cells. It is
involved in the regulation of intracellular Ca2+
homeostasis and thus in the transcellular transport and
reabsorption of Ca2+ from filtrated urinary
Ca2+, in the suppression of cell proliferation and
apoptotic cell death that are mediated through various signaling
factors, and in the regulation of the gene expression of various
proteins related to mineral transport-related proteins,
transcription factors, cell proliferation and apoptosis-related
proteins (4–8). This review discusses the recent
findings concerning the potential role of regucalcin in the
regulation of the kidney proximal tubule epithelial cells.
2. Various factors regulate regucalcin gene
expression
Regucalcin in rat kidney tissues is estimated to be
present in the range of 1.74-3.50×10−6 moles/g tissues
in male or female rats, as measured using an enzyme-linked
immunoadsorbent assay (9). This
expression does not decrease with aging (9). Regucalcin mRNA expression is
predominant in the kidney cortex, but not in the medulla of rats
(10). Kidney cortex is comprised
of nephrons which include the glomerulus and renal tubule. The
transcription factors, NF1-A1 and RGPR-p117, which were identified
as hepatic nuclear factors that bind to the TTGGC(N)6CC
sequence of the rat regucalcin gene promoter region, have been
shown to enhance regucalcin gene expression (6,7,11).
RGPR-p117 was found to be a novel transcription factor (8). The regucalcin gene has been shown to
be expressed in kidney proximal tubule epithelial NRK52E cells
derived from normal rat kidney cortexes (12). NF1-A1 and RGPR-p117, which are
localized in the nuclei of NRK52E cells, have been shown to
increase regucalcin promoter activity (13–15). The enhanced regucalcin gene
promoter activity with the overexpression of NF1-A1 or RGPR-p117
has been shown to be mediated through protein phosphorylation and
dephosphorylation, which are regulated by Ca2+-dependent
protein kinases, mitogen-activated protein kinase (MAPK) and
protein phosphatases in NRK52E cells (13–15). Thus, NF1-A1 or RGPR-p117 may play
an essential role as transcription factors (enhancers) in
regucalcin promoter activity in rat kidney cells. The involvement
of other transcription factors remains to be elucidated.
Regucalcin gene expression has been shown to be
regulated through various hormones. Regucalcin mRNA expression in
the kidney cortex was shown to be markedly stimulated after a
single intraperitoneal administration of calcium chloride in normal
and thyroparathyroidectomized rats with calcitonin and parathyroid
hormone (PTH) deficiencies in vivo (10), which is known to regulate calcium
reabsorption in kidney proximal tubule cells (16,17). The stimulatory effect of calcium
administration on regucalcin mRNA expression in the kidneys was
blocked by treatment with trifluoperazine (TFP), an inhibitor of
Ca2+/calmodulin, suggesting that its expression is
mediated through Ca2+/calmodulin, which is involved in
the activation of protein kinases (10,18). PTH, 1,25-dihydroxyvitamin
D3 and calcitonin play a role in the regulation of
calcium transport in NRK52E cells (16,17). Among these hormones, PTH was found
to stimulate regucalcin mRNA expression and its protein levels in
NRK52E cells (12) in
vitro. PTH exerts a stimulatory effect on the reabsorption of
calcium in the kidney proximal tubule (16,17). The effects of PTH are known to be
mediated by cyclic AMP (cAMP) or inositol 1,4,5-trisphosphate
(IP3)-released Ca2+ and protein kinase C in
cells (19). In a previous study,
regucalcin mRNA expression was enhanced by dibutyryl cAMP or
phorbol-12-myristate 13-acetate (PMA), an activator of protein
kinase C, in NRK52E cells (12),
suggesting that it is partly mediated through signaling pathways
related to cAMP or protein kinase C in NRK52E cells.
PD98059 is an inhibitor of the extracellular
signal-related kinase (ERK) pathway (20). Regucalcin mRNA expression in
NRK52E cells is not altered in the presence of PD98059 in
vitro (12), suggesting that
its expression is not mediated through a MAPK that is related to
the ERK pathway. Regucalcin mRNA expression has been shown to be
suppressed following culture with staurosporine, an inhibitor of
protein kinase C in NRK52E cells, supporting the hypothesis that
its expression is mediated through a cell signaling pathway related
to protein kinase C in kidney cells (12). Regucalcin mRNA expression is not
altered by culture with vanadate, which is an inhibitor of protein
tyrosine phosphatase (21), in
NRK52E cells (12). Of note,
regucalcin mRNA expression has been found to be suppressed
following culture with tumor necrosis factor-α (TNF-α) or
transforming growth factor-β (TGF-β) in NRK52E cells (22). The effects of TNF-α are mediated
through the nuclear factor-κB (NF-κB) signaling pathways. TGF-β is
mediated through the Smads signaling pathways. Regucalcin mRNA
expression may be suppressed through transcription factors that are
related to NF-κB and Smads.
Steroid hormones have been shown to regulate
regucalcin gene expression in kidney cells. Regucalcin mRNA
expression was shown to be enhanced following culture with
dexamethasone in NRK52E cells in vitro (12). Regucalcin mRNA expression was
shown to be stimulated after a single subcutaneous administration
of dexamethasone in rats, whereas it was suppressed after a single
subcutaneous administration of aldosterone or estrogen in
vivo (23). The
administration of hydrocortisone in rats did not alter regucalcin
mRNA expression in the kidney cortex in vivo (23). It has been suggested that the
regucalcin gene promoter region is the location of the response
elements for glucocorticoid, aldosterone or estrogen receptors.
Regucalcin mRNA expression has been shown to be suppressed in the
kidney cortex of adrenalectomized (ADX) rats, which diminishes the
secretion of endogenous steroid hormones from adrenal glands in
vivo (24,25). Thus, the adrenal glands may
participate in the regulation of regucalcin mRNA expression in the
kidney cortex of rats.
3. Regucalcin regulates intracellular
calcium transport
The kidneys play a physiological role in the
regulation of Ca2+ homeostasis in the blood through the
reabsorption of urinary Ca2+ (24,25). Renal cortex cells, which
constitute the proximal tubule epithelial cells, may play a role in
the reabsorption of urinary Ca2+. Active Ca2+
reabsorption involves transcellularlly transportation, and
Ca2+ pumps in the basolateral membranes are involved to
overcome the energy barriers at the peritubular cell side (24,25). The regulation of intracellular
Ca2+ homeostasis is important in the promotion of
intracellular Ca2+ transport. Intracellular
Ca2+ homeostasis is regulated through the plasma
membrane (Ca2+-Mg2+)-adenosine
5′-triphosphatase (ATPase), microsomal Ca2+-ATPase,
mitochondrial Ca2+ uptake and nuclear Ca2+
transport in the cells. The Ca2+-ATPase system exceeds
the capacity of the Na+/Ca2+ exchanger and
plays a primary role in Ca2+ homeostasis of in rat
kidney cortex cells (24).
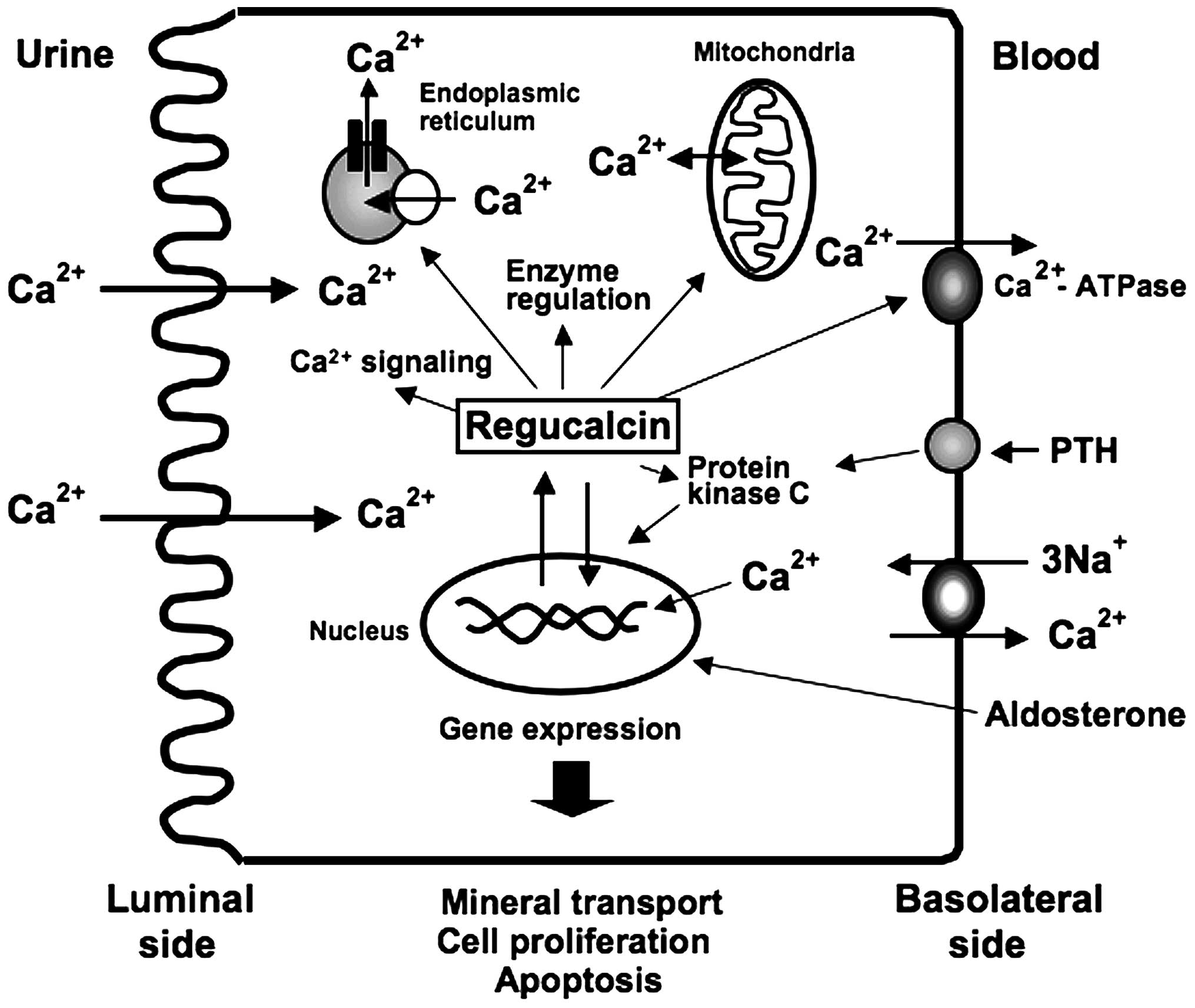
Regucalcin has been found to play a role as an activator of the
adenosine-5′-triphosphate (ATP)-dependent Ca2+ pumps
(Ca2+-ATPase) in the basolateral membranes isolated from
the rat kidney cortex; regucalcin (100 and 1,000 nM) increased
Ca2+-ATPase activity and stimulated
45Ca2+ uptake by the basolateral membranes
in vitro (26). The effect
of regucalcin on Ca2+ pump enzyme activity was inhibited
in the presence of vanadate, an inhibitor of phosphorylation of
ATPase, N-ethylmaleimide (SH-group modifying reagent) and
DcAMP, but not IP3 (26). It was suggested that regucalcin
binds to the lipids at the site of the Ca2+ pump enzyme
in the basolateral membranes of the rat kidney cortex and may
activate the enzyme by acting on the SH-group of the enzyme
(26).
Regucalcin has been shown to increase
Ca2+-ATPase activity and ATP-dependent calcium uptake in
the microsomes isolated from the rat kidney cortex in vitro
(27). This increase resulted
from the binding of regucalcin to the SH-group of active sites of
the enzyme and the stimulation of the phosphorylation of the enzyme
in the microsomes of the rat kidney cortex (27). Kidney cortex microsomal
Ca2+-ATPase activity and ATP-dependent Ca2+
uptake were inhibited by DcAMP or IP3 (27). Calmodulin increased microsomal
Ca2+-ATPase activity, and this increase was lower than
that effected by regucalcin; both proteins may be important as
activators in the microsomal ATP-dependent Ca2+
sequestration (27).
An ATP-dependent Ca2+ uptake system
(Ca2+ uniporter) exists in the mitochondria of the
kidney cortex of rats (28).
Regucalcin has also been shown to stimulate
Ca2+-ATPase-related Ca2+ uniporter activity
in the mitochondria (28). This
effect was completely blocked by ruthenium red or lanthanum
chloride, which is a specific inhibitor of Ca2+
uniporter in the mitochondria (28), suggesting that regucalcin
stimulates Ca2+-ATPase-related Ca2+ uniporter
activity in renal cortex mitochondria. Regucalcin has been
suggested to bind to the membranous lipids of renal cortex
mitochondria and act on the SH-groups, which are active sites of
Ca2+-ATPase (28).
It has been observed that reabsorption of urinary
calcium is promoted through transcellular Ca2+ transport
in renal proximal tubule epithelial cells (24,25). Thus, regucalcin may play a
physiological role in the regulation of intracellular
Ca2+ homeostasis in the renal proximal tubular
epithelial cells, due to its activation of ATP-dependent
Ca2+-transport systems in the basolateral membranes,
microsomes and mitochondria. Regucalcin may promote Ca2+
reabsorption, which is based on the ATP-dependent transcellular
transport of Ca2+, in the proximal tubular epithelial
cells of the nephron tubule of the kidney cortex. Thus, regucalcin
may play a physiological role in the regulation of Ca2+
homeostasis in the blood through reabsorption of urinary
Ca2+ in the kidneys.
Regucalcin has been demonstrated to regulate the
gene expression of various proteins related to mineral ion
transport in kidney cells. To determine the role of endogenous
regucalcin, NRK52E cells (transfectants) overexpressing regucalcin
were generated in a previous study (29). In regucalcin/pCXN2-transfected
cells regucalcin overexpression was increased 21-fold as compared
with that of the parental wild-type NRK52E cells (29). The overexpression of endogenous
regucalcin was found to increase rat outer medullary K+
channel (ROMK) mRNA expression in NRK52E cells, although it did not
alter the expression of type II NaPi cotransporter (NaPi-IIa),
Na+, K+-ATPase, epithelial sodium channel
(ENaC) or angiotensinogen mRNAs (30). Culture with aldosterone caused an
increase in ENaC, Na+, K+-ATPase and ROMK
mRNA expression in wild-type NRK52E cells (30). The effect of aldosterone in
increasing ENaC and Na+, K+-ATPase mRNA
expression was not observed in the transfectants (30). Aldosterone was shown to upregulate
regucalcin mRNA expression in NRK52E cells (12). The stimulatory effect of
aldosterone on ENaC and Na+, K+-ATPase mRNA
expressions may thus be partly mediated through endogenous
regucalcin in NRK52E cells.
The overexpression of endogenous regucalcin was
found to exert suppressive effects on the mRNA expression of the
L-type Ca2+ channel and calcium-sensing receptor (CaR),
which regulate intracellular Ca2+ signaling and renal
Ca2+ transport in NRK52E cells (30,31). The entry of Ca2+
through L-type Ca2+ channels induces mitochondrial
disruption and cell death (31).
The blockade of Ca2+ influx through L-type
Ca2+ channels has been shown to attenuate mitochondrial
injury and apoptosis in hypoxia of renal tubular cells (31). Regucalcin may regulate the
intracellular Ca2+ signaling pathway, which is mediated
through its suppressive effect on L-type Ca2+ channel or
CaR mRNA expressions in the kidney proximal tubular epithelial
cells.
Taking the above discussion into account, it is
suggested that regucalcin plays a pivotal role in the regulation of
intracellular Ca2+ homeostasis and mineral transport in
kidney proximal tubule epithelial cells, as summarized in Fig. 1.
4. Regucalcin regulates cell
signaling-related enzyme activity
Regucalcin has been demonstrated to exert
suppressive effects on the activity of various enzymes that
regulate cell-signaling pathways in kidney cells. Multifunctional
Ca2+/calmodulin-dependent protein kinases play an
important role in the Ca2+ signaling response of many
cells (32). Regucalcin (10-1,000
nM) was found to exert suppressive effects on the activation of
Ca2+/calmodulin-dependent protein kinase in the cytosol
of rat kidney cortex in vitro (33). This effect was also noted when
higher concentrations of calcium chloride (100-1,000 µM) and
calmodulin (4-20 µg/ml) were added to the enzyme reaction
mixture (33), suggesting that
regucalin has a direct inhibitory effect on the protein kinase in
renal cortex cytosol.
Protein kinase C is a diacylglycerol-activated
Ca2+- and phospholipid-dependent protein kinase that is
related to Ca2+ signaling in many cell types. Regucalcin
has been found to inhibit protein kinase C activity in the cytosol
of rat kidney cortex in vitro (34). The inhibitory effect of regucalcin
on protein kinase C activity was not observed in a reaction mixture
without Ca2+, but was noted in the presence of
Ca2+, phosphatidylserine and dioctanoylglycerol
(34). Moreover, regucalcin
suppressed protein kinase C activity caused by the addition of
diacylglycerol or PMA, which directly activated the enzyme, in the
presence of both Ca2+ and phosphatidylserine (34). Thus, it is suggested that
regucalcin directly binds to protein kinase C.
Protein phosphorylation-dephosphorylation is a
universal mechanism through which numerous cellular events are
regulated (21). Protein
phosphatases play an important role in intracellular signal
transduction due to hormonal stimulation. Calcineurin, a
calmodulin-binding protein, possesses a Ca2+-dependent
and calmodulin-stimulated protein phosphatase that is a protein
serine/threonine phosphatase (35). Regucalcin was found to inhibit
calcineurin activity in rat renal cortex cytosol in vitro
(36). This inhibitory effect was
independent of Ca2+, suggesting that regucalcin acted
directly on the enzyme (36).
Regucalcin has been shown to bind to calmodulin in a study using
calmodulin-agarose beads in vitro (37). The inhibitory effect of regucalcin
on enzyme activity may be related to its binding to calmodulin.
Regucalcin has also been shown to suppress the activities of
Ca2+/calmodulin-independent protein phosphatases for
tyrosine, phosphoserine and phosphothreonine in rat renal cortex
cytosol (38). The presence of
anti-regucalcin monoclonal antibody in an enzyme reaction mixture
caused an increase in protein phosphatase activity toward three
phosphoamino acids in the renal cortex cytosol (38).
Endogenous regucalcin may suppress the activity of
various protein phosphatases in the kidney cortex cells. Of note,
nuclear regucalcin was found to suppress calcineurin,
serine/threonine phosphatase and tyrosine phosphatase that are
present in the nucleus of rat kidney cortex cells (39). The administration of calcium in
rats was shown to increase the levels of regucalcin and cause a
corresponding rise in protein phosphatase activity in the cytoplasm
and nucleus of the kidney cortex in vivo (40). The presence of anti-regucalcin
monoclonal antibody in the enzyme reaction mixture caused an
increase in protein phosphatase in the cytoplasm and nucleus of
normal rat kidney cortex (40).
The increased levels of endogenous regucalcin were suggested to
suppress protein phosphatase activity enhanced in the cytoplasm and
nucleus of the kidney cortex of calcium-administered rats. The
dephosphorylation of many phosphorylated proteins is regulated by
protein phosphatase, which is implicated in nuclear transcriptional
regulation in various cell types (21). Thus, it is posited in this study
that regucalcin may play a role in the regulation of nuclear
signaling-related gene expression, through the inhibition of
nuclear protein phosphatase activity.
cAMP, which is generated through the activation of
the plasma membrane adenylate cyclase due to hormonal stimulation,
activates the cAMP-dependent protein kinase, which plays an
important role in the cAMP signaling pathway. cAMP is degraded by
cAMP phosphodiestrase, which is activated by
Ca2+/calmodulin (41).
Regucalcin was shown to inhibit
Ca2+/calmodulin-dependent cAMP phosphodiesterase
activity in the cytosol of rat renal cortex (41). Regucalcin plays a role in the
regulation of both cAMP-dependent and Ca2+-dependent
signaling pathways, which are modulated through hormonal
stimulation in the renal cortex cells.
Nitric oxide (NO) acts as a messenger or modulator
molecule in many biological systems (42). NO has an unpaired electron that
reacts with proteins and targets primarily through their thiol or
heme groups, and it acts as a messenger or modulator molecule in
many biological systems. NO is produced from L-arginine with
L-citrulline as a co-product in a reaction catalysed by NO synthase
that requires Ca2+/calmodulin (42). Regucalcin was found to inhibit
Ca2+/calmodulin-dependent NO synthase activity in the
renal cortex cytoplasm of rats (43). NO acts as a messenger or modulator
in kidney cortex cells. Regucalcin may play a role as a suppressor
protein in NO production in the kidney cells and may regulate many
cellular events that are involved in NO signaling.
Of note, regucalcin was found to stimulate
proteolytic activity in the cytoplasm of rat kidneys (44,45). Regucalcin uniquely activated thiol
proteases (including calpines) independently of Ca2+ in
the cytoplasm of rat kidney cortex, although it did not have an
effect on serine proteases and metalloproteases (44,45). Regucalcin was shown to directly
act on the SH-groups of protease in the kidney cortex cytoplasm.
Regucalcin activated protease at concentrations of 10-250 nM in
in vitro experiments (44). The concentration of regucalcin in
the cytosol of rat kidney tissues may be ~5.3 µM (9). Regucalcin may play a physiological
role in the activation of thiol proteases in renal cortex cells.
Calpains, which are thiol (SH) proteases, are ubiquitous,
non-lysosomal and Ca2+-dependent proteases (46). The ability of calpains to alter
limited proteolysis, the activity or function of numerous
cytoskeletal proteins, protein kinases, receptors and transcription
factors is related to Ca2+-regulated cellular functions
(45). Regucalcin may play a
pivotal role in the degradation of proteins that are involved in
the regulation of signaling pathways.
As described above, regucalcin may play a pivotal
role as a regulatory protein involved in the activation of many
enzymes that are related to signaling pathways, which are mediated
through Ca2+, cAMP, NO, Ca2+-dependent
protein kinases, protein phosphatases and proteases in the kidney
cortex cells.
5. Regucalcin regulates nuclear
function
Regucalcin in the cytoplasm of kidney cells is
demonstrated to localize into the nucleus, and it has been found to
regulate nuclear function; regucalcin was found to localize in the
nucleus of HA-regucalcin/phCMV2-transfected NRK52E cells using
immunocytochemical and western blot analysis (39). The nuclear localization of
regucalcin was enhanced after culture with FBS, PTH, Bay K 8644 or
PMA (47). This enhancement was
remarkable following culture with PMA, which is an activator of
protein kinase C, and it was suppressed by staurosporine, an
inhibitor of protein kinase C (47). Thus, the nuclear localization of
regucalcin was enhanced through Ca2+-signaling factors
including protein kinase C in NRK52E cells (47). The stimulatory effect of PTH on
regucalcin mRNA expression in NRK52E cells was mediated through
cAMP or IP3-released Ca2+ and protein kinase
C (12). Thus, it has been
demonstrated that protein kinase C enhances both regucalcin mRNA
expression and nuclear localization of regucalcin protein in NRK52E
cells.
Regucalcin was found to inhibit the activity of
various protein phosphatases, including the
Ca2+/calmodulin-dependent enzyme in the nuclei of rat
kidney cortex in vitro (39,40).
Regucalcin has been shown to exert suppressive
effects on DNA synthesis activity in the nuclei isolated from rat
renal cortiex in vitro (48). Such an effect was induced by
adding of regucalcin (0.1-0.5 µM) to the reaction mixture
(48). Also, the suppressive
effects of regucalcin were noted in the presence of calcium
chloride (50 µM) in the reaction mixture, and this
suppression was potentiated in the presence of EGTA (1 mM)
(48). The suppressive effects of
regucalcin on nuclear DNA synthesis were thus mediated through a
mechanism unrelated to Ca2+. The presence of
anti-regucalcin monoclonal antibody (10-50 ng/ml) in the reaction
mixture caused an increase in nuclear DNA synthesis activity
(48). This increase was
completely abolished by the addition of exogenous regucalcin (0.5
µM) (48). Regucalcin can
directly bind to nuclear DNA and inhibit DNA synthesis (49). Nuclear regucalcin may play a role
in the suppression of nuclear DNA synthesis in kidney proximal
tubular epithelial cells (50).
Moreover, regucalcin, which was localized in the
nucleus, has been found to regulate the gene expression of various
proteins involved in ion transport (31), proliferation and apoptosis in
kidney cells.
6. Suppressive effect of regucalcin on the
proliferation of kidney cells
The overexpression of endogenous regucalcin was
found to suppress the proliferation of NRK52E cells in vitro
(30). The suppressive effects of
regucalcin on cell proliferation were prevented following culture
with butyrate, roscovitine and sulforaphane, which induce cell
cycle arrest (30). Butyrate
induces the inhibition of G1 progression (50). Roscovitine is a potent and
selective inhibitor of the cyclin-dependent kinase cdc2, cdk2 and
cdk5 (51) and can lead to cell
cycle arrest in G1 and accumulation in the G2 phase of the cell
cycle. Sulforaphane can induce G2/M phase cell cycle arrest
(52). The effect of butyrate,
roscovitin or sulforaphan, which inhibit the proliferation of
wild-type NRK52E cells, was not noted in the transfectants
overexpressing regucalcin (29).
These findings support the view that endogenous regucalcin induces
G1 and G2/M phase cell cycle arrest in NRK52E cells.
The proliferation of NRK52E cells was also
suppressed following culture with PD98059, staurosporine or
dibucaine, which are an inhibitors of various protein kinases
(29). Such suppression was not
observed in the transfectants that overexpressed endogenous
regucalcin (29). The suppressive
effects of endogenous regucalcin on cell proliferation were
suggested to result from inhibitory effects of regucalcin on
various protein kinases that are involved in the stimulation of
cell proliferation. The suppressive effects of endogenous
regucalcin on cell proliferation were shown to be mediated through
the inhibition of PI3-kinase, using its inhibitor wortmannin
(53). Moreover, the
overexpression of regucalcin was shown to prevent the suppression
of cell proliferation induced by Bay K 8644, an agonist of calcium
entry into cells (29),
supporting the view that endogenous regucalcin maintains
intracellular Ca2+ homeostasis.
The overexpression of regucalcin was found to
regulate the gene expression of proteins that are involved in cell
proliferation and cell cycle. The expression of c-jun and
checkpoint kinase 2 (chk2) mRNAs was suppressed by overexpression
of regucalcin (29). The
expression of p53 mRNA was enhanced by overexpression of
regucalcin, while the expression of c-myc, c-fos, cdc2 and p21
mRNAs was not changed (29). The
suppression of c-jun and chk2 mRNA expression may lead to
retardation of cell proliferation. Regucalcin may exert suppressive
effects on cell proliferation by regulating the gene expression of
many proteins related to cell proliferation in NRK52E cells.
Thus, the suppressive effects of regucalcin on cell
proliferation may be mediated through a decrease in various
Ca2+ signaling-dependent protein kinases and PI3-kinase
activities, the suppression of c-jun and chk2 mRNA expression, or
the enhancement of p53 mRNA expression in kidney NRK52E cells.
7. Suppressive effect of regucalcin on
apoptotic cell death
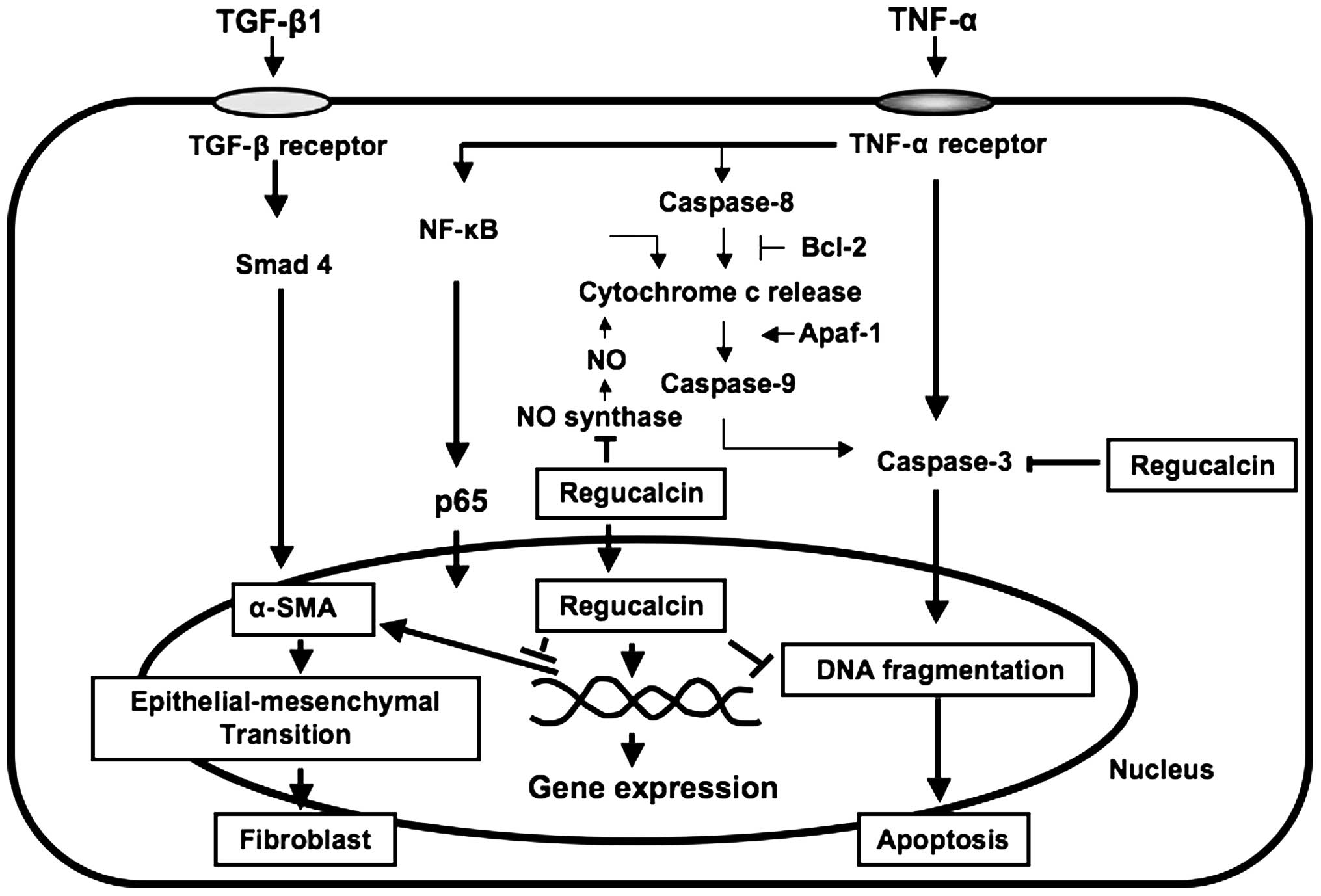
The role of endogenous regucalcin in apoptotic cell
death has been demonstrated in NRK52E cells overexpressing
regucalcin (22,54). The overexpression of regucalcin
was found to prevent the apoptotic cell death of NRK52E cells
induced by culture with TNF-α, TGF-β1, lipopolysaccharide (LPS),
Bay K 8644, or thapsigargin, suggesting the enhancement of nuclear
DNA fragmentation (54). It has
been suggested that the suppressive effect of regucalcin on
apoptotic cell death is mediated through the suppression of various
intracellular signaling pathways, which was induced by caspase-3,
NO and Ca2+ in NRK52E cells (54).
Bcl-2 is a suppressor of apoptotic cell death
(55). Apaf-1 participates in the
activation of caspase-3 (56).
Akt-1 is involved in survival signaling for cell death (56). The overexpression of regucalcin
was found to cause a marked elevation of Bcl-2 mRNA expression in
NRK52E cells, and it slightly stimulated Akt-1 mRNA expression in
the cells (54). The
overexpression of endogenous regucalcin prevented LPS-suppressed
Bcl-2 mRNA expression and LPS-stimulated Apaf-1 mRNA expression,
and it suppressed LPS-induced cell death in NRK52E cells (54). Caspase-3 mRNA expression in NRK52E
cells was enhanced following culture with TNF-α (54). This enhancement was prevented by
the overexpression of regucalcin (54). Culture with Bay K 8644 or
thapsigargin increased caspase-3 mRNA expression in wild-type
NRK52E cells, and these increases were prevented by regucalcin
overexpression (54). Thus,
overexpression of endogenous regucalcin enhances the expression of
Bcl-2 and Akt-1 mRNAs, and it suppresses the expression of
caspase-3 and Apaf-1 mRNAs in NRK52E cells, thereby inducing
suppression of apoptotic cell death. Many toxic factors have been
reported to induce renal failure by stimulating apoptotic cell
death (57). Thus, endogenous
regucalcin may play an important role as a suppressor protein in
the promotion of apoptotic cell death and renal failure in kidney
proximal tubule epithelial cells.
Smads are involved in the signal transduction of
TGF-β1 (58). NF-κB is involved
in the intracellular signaling of TNF-α (59). The expression of Smad 2 and NF-κB
mRNAs in wild-type NRK52E cells was enhanced by the overexpression
of regucalcin (22), suggesting
that regucalcin stimulates the gene expression of Smad 2 or NF-κB.
Of note, the overexpression of regucalcin was found to suppress
α-smooth muscle actin expression in NRK52E cells (22). Culture with TNF-α or TGF-β1 caused
a marked increase in α-smooth muscle actin levels in NRK52E cells
(22). This increase was
suppressed by the overexpression of regucalcin (22). These findings suggest that
endogenous regucalcin suppresses α-smooth muscle actin production
in NRK52E cells cultured with TNF-α or TGF-β1. TGF-β1 is a key
mediator that regulates transdifferentiation of NRK52E cells into
myofibroblasts, which express α-smooth muscle actin (60). This is known to lead to renal
fibrosis associated with overexpression of TGF-β1 that is seen in
the diseased kidney (60).
As summarized in Fig.
2, suppressed regucalcin gene expression may lead to apoptotic
cell death and renal fibrosis, suggesting that regucalcin plays a
pathophysiological role in kidney proximal tubule epithelial cells.
Regucalcin exerts suppressive effects on proliferation and
apoptotic cell death induced through various stimuli in kidney
proximal tubule epithelial cells. Thus, it can be suggested that
regucalcin may play a physiological role in maintaining cell
homeostasis in kidney proximal tubular epithelial cells.
8. Involvement of regucalcin in kidney
failure
Regucalcin gene expression has been shown to
suppress kidney failure induced in various pathophysiologic states.
Kidney regucalcin gene expression has been found to be suppressed
in hypertensive states. Regucalcin expression has been shown to be
suppressed in spontaneously hypertensive rats, suggesting the
involvement of regucalcin in hypertensive states (61,62). Regucalcin mRNA expression was also
suppressed in the kidney cortex of rats following saline
administration for 7 days (61–63). This intake caused an increase in
the serum calcium and blood urea nitrogen (BUN) concentrations,
which are an index of kidney disorder (61,62), and it also caused an increase in
Ca2+-ATPase activity in the basolateral membranes of the
kidney cortex and a corresponding increase in renal cortex calcium
content (62). It is therefore
suggested that suppressed regucalcin expression may lead to a
disturbance of kidney calcium metabolism, which is related to renal
hypertension.
Several drugs are known to cause nephrotoxicity.
Cisplatin, a nephrotoxic antitumor drug (64), or cephaloridine, a nephrotoxic
cephalosporin antibiotic (65),
is known to change the thiol status in the renal cortex, and this
takes place before significant morphological changes occur.
Regucalcin mRNA expression and its protein levels were markedly
reduced in the kidney cortex of rats which received a single
intraperitoneal administration of cisplatin or cephaloridine, and
this also induced a marked accumulation of calcium in the kidney
cortex and a corresponding elevation of BUN (66,67). Moreover, regucalcin was
downregulated by the administration of hexachloro-1:3-butadiene
(HCBD) in low doses (68).
Glutamine synthase activity in the kidney cortex was also
downregulated, whereas BUN and creatinine levels increased after a
high dose of HCBD (68).
Regucalcin gene expression appears to be a sensitive genomic marker
which can be used to evaluate the renal impairment caused by
chemicals, and its downregulation seems to be related to damage of
the proximal tubule (68).
Ochratoxin A (OTA), a naturally occurring
mycotoxin, is nephrotoxic in all animal species investigated thus
far and is considered a potent renal carcinogen, although the
mechanism of its toxicity remains unknown (69). The gene expression profiles in
target and non-target organs were analyzed after oral
administration of OTA in rats (69). The number of differentially
expressed genes in the kidney was much higher than those in the
liver (541 vs. 11 at both time-points) (69). Downregulation was predominant in
the genes involved in the oxidative stress response pathway, and
metabolism and transport (69).
Regucalcin was strongly suppressed at both time points, while the
genes implicated in cell survival and proliferation were
upregulated at day 21, and translation factors and Annexin were
upregulated at both time points (69).
The prolonged intake of aristolochic acid (AA) has
been shown to be associated with the development of certain renal
disorders in rats (70). Renal
tubular atrophy and interstitial fibrosis are the early symptoms of
AA nephropathy. Differentiated proteins have been identified in the
kidney tissues through proteomics investigations (70). Upregulated proteins identified
included ornithine aminotransferase, sorbitol dehydrogenase, actin,
aspartoacylase, 3-hydroxyisobutyrate dehydrogenase and
peroxiredoxin-1 (70).
Downregulated proteins included regucalcin, ATP synthase subunit β,
glutamate dehydrogenase 1, glutamate-cysteine ligase regulatory
subunit, dihydropteridine reductase, hydroxyacyl-coenzyme A
dehydrogenase, voltage-dependent anion-selective channel protein 1,
prohibitin and adenylate kinase isoenzyme 4 (70). Thus, these identified protein
markers are suggested to have biological and medical
significance.
The basic mechanism underlying calcineurin
inhibitor (cyclosporine) nephrotoxicity with enhancement by
sirolimus is still largely unknown (71). The effects of cyclosporine, alone
and in combination with sirolimus, on the renal proteome have been
investigated (71).
Twenty-four-hour urine was collected for nuclear magnetic resonance
(NMR)-based metabolic analysis, and the kidneys were harvested for
two dimensional-gel electrophoresis and histology (71). Cyclosporine affected the following
groups of proteins: calcium homeostasis (regucalcin and calbindin),
cytoskeleton (vimentin and caldesmon), response to hypoxia and
mitochondrial function [prolyl 4-hydroxylase, proteasome and
reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase],
and cell metabolism (kidney aminoacylase, pyruvate dehydrogenase
and fructose-1, 6-bisphosphate) (71). It was noted that regucalcin was
found to be increased in the urine of rats with kidney disorder,
indicating that regucalcin is a useful potential biomarker of
kidney disorders (71).
As described above, the suppression of regucalcin
gene expression and its association with nephrotoxicity may play a
pathophysiological role in the development of renal disorders.
9. Prospects
Ca2+, which plays a multifunctional role
in many biological systems, plays a pivotal role during embryonic
development of kidneys, and it is present throughout numerous steps
of tubulogenesis and nephron induction, from the formation of a
simple kidney in amphibian larvae (pronephros) to the formation of
the more complex mammalian kidney (metanephrons) (2). Regucalcin, a regulatory protein of
Ca2+ signaling in kidney cells, plays a pivotal role in
the development of the kidneys, as it is involved in tubulogenesis
and also in nephron induction.
Regucalcin has been demonstrated to play a
multifunctional role in the regulation of intracellular
Ca2+ transport, the activity of various cell
signaling-related enzymes, nuclear DNA synthesis, gene expression,
proliferation and apoptotic cell death in kidney cells. Regucalcin
gene expression was found to be suppressed in various
pathophysiological conditions including hypertensive states, and
nephrotoxicants are associated with kidney failure. An analysis for
proteome and differential gene expression demonstrated a potential
suppression of regucalcin expression among many proteins. Thus, it
can be suggested that suppressed regucalcin gene expression may
lead to development of renal failure. The pathophysiological role
of regucalcin in kidney diseases in human subjects is poorly
understood. Of note, regucalcin gene expression and its protein
have been shown to be suppressed in kidney tumor tissues as
compared with kidney tissues of healthy humans (72), suggesting that regucalcin plays a
suppressive role in the development of carcinogenesis in the
kidneys of human subjects. Clinical studies on regucalcin are thus
necessary in order to examine its role further.
Acknowledgments
The author was partly supported by the Foundation
for Biomedical Research on Regucalcin.
References
|
1
|
Gilbert T, Leclerc C and Moreau M: Control
of kidney development by calcium ions. Biochimie. 93:2126–2131.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yamaguchi M: Regucalcin, cell
signaling-related protein: its multifunctional role in kidney cell
regulation. OA Biotechnology. 1:22012. View Article : Google Scholar
|
|
3
|
Yamaguchi M: Role of regucalcin in calcium
signaling. Life Sci. 66:1769–1780. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamaguchi M: Role of regucalcin in
maintaining cell homeostasis and function (Review). Int J Mol Med.
15:371–389. 2005.PubMed/NCBI
|
|
5
|
Yamaguchi M: Regucalcin and cell
regulation: role as a suppressor protein in signal transduction.
Mol Cell Biochem. 353:101–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamaguchi M: The transcriptional
regulation of regucalcin gene expression. Mol Cell Biochem.
346:147–171. 2011. View Article : Google Scholar
|
|
7
|
Yamaguchi M: Novel protein RGPR-p117: its
role as the regucalcin gene transcription factor. Mol Cell Biochem.
327:53–63. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shimokawa N and Yamaguchi M: Calcium
administration stimulates the expression of calcium-binding protein
regucalcin mRNA in rat liver. FEBS Lett. 305:151–154. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamaguchi M and Isogai M: Tissue
concentration of calcium-binding protein regucalcin in rats by
enzyme-linked immunoadsorbent assay. Mol Cell Biochem. 122:65–68.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamaguchi M and Kurota H: Expression of
calcium-binding protein regucalcin mRNA in the kidney cortex of
rats: the stimulation by calcium administration. Mol Cell Biochem.
146:71–77. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamaguchi M: Role of regucalcin in cell
nuclear regulation: involvement as a transcription factor. Cell
Tissue Res. 354:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakagawa T and Yamaguchi M: Hormonal
regulation on regucalcin mRNA expression in cloned normal rat
kidney proximal tubular epithelial NRK52E cells. J Cell Biochem.
95:589–597. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sawada N and Yamaguchi M: Involvement of
nuclear factor I-A1 in the regulation of regucalcin gene promoter
activity in cloned normal rat kidney proximal tubular epithelial
cells. Int J Mol Med. 18:665–671. 2006.PubMed/NCBI
|
|
14
|
Sawada N and Yamaguchi M: Overexpression
of RGPR-p117 enhances regucalcin gene expression in cloned normal
rat kidney proximal tubular epithelial cells. Int J Mol Med.
16:1049–1055. 2005.PubMed/NCBI
|
|
15
|
Sawada N and Yamaguchi M: Overexpression
of RGPR-p117 enhances regucalcin gene promoter activity in cloned
normal rat kidney proximal tubular epithelial cells: involvement of
TTGGC motif. J Cell Biochem. 99:589–597. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van Os CH: Transcellular calcium transport
in intestinal and renal epithelial cells. Biochim Biophys Acta.
906:195–222. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Taylor CW: Calcium regulation in
vertebrates: an overview. Comp Biochem Physiol. 249–255. 1985.
View Article : Google Scholar
|
|
18
|
Murata T and Yamaguchi M: Binding of
kidney nuclear proteins to the 5′-flanking region of the rat gene
for Ca2+-binding protein regucalcin: involvement of
Ca2+/calmodulin signaling. Mol Cell Biochem. 199:35–40.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Verheijen MH and Defize LHK: Parathyroid
hormone activates mitogen-activated protein kinase via a
cAMP-mediated pathway independent of Ras. J Biol Chem.
272:3423–3429. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Yang Y, Ye YC, Shi QF, Chai K,
Tashiro S, Onodera S and Ikejima T: Activation of ERK-p53 and
ERK-mediated phosphorylation of Bcl-2 are involved in autophagic
cell death induced by the c-Met inhibitor SU11274 in human lung
cancer A549 cells. J Pharmacol Sci. 118:423–432. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hunter T: Protein kinases and
phosphatases: the yin and yang of protein phosphorylation and
signaling. Cell. 80:225–236. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakagawa T and Yamaguchi M: Overexpression
of regucalcin suppresses cell response for tumor necrosis factor-α
or transforming growth factor-β1 in cloned normal rat kidney
proximal tubular epithelial NRK52E cells. J Cell Biochem.
100:1178–1190. 2007. View Article : Google Scholar
|
|
23
|
Kurota H and Yamaguchi M: Steroid hormonal
regulation of calcium-binding protein regucalcin mRNA expression in
the kidney cortex of rats. Mol Cell Biochem. 155:105–111. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
van Heeswijk MPE, Geertsen JAM and van Os
CH: Kinetic properties of the ATP-dependent Ca2+ pump
and the Na+/Ca2+ exchange system in
basolateral membranes from rat kidney cortex. J Membr Biol.
79:19–31. 1984. View Article : Google Scholar
|
|
25
|
Agus ZS, Chiu PJS and Goldberg M:
Regulation of urinary calcium excretion in the rat. Am J Physiol.
232:F545–F549. 1977.PubMed/NCBI
|
|
26
|
Kurota H and Yamaguchi M: Activatory
effect of calcium-binding protein regucalcin on ATP-dependent
calcium transport in the basolateral membranes of rat kidney
cortex. Mol Cell Biochem. 169:149–156. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kurota H and Yamaguchi M: Regucalcin
increases Ca2+-ATPase activity and ATP-dependent calcium
uptake in the microsomes of rat kidney cortex. Mol Cell Biochem.
177:201–207. 1997. View Article : Google Scholar
|
|
28
|
Xue JH, Takahashi H and Yamaguchi M:
Stimulatory effect of regucalcin on mitochondrial ATP-dependent
calcium uptake activity in rat kidney cortex. J Cell Biochem.
80:285–292. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakagawa T, Sawada N and Yamaguchi M:
Overexpression of regucalcin suppresses cell proliferation of
cloned normal rat kidney proximal tubular epithelial NRK52E cells.
Int J Mol Med. 16:637–643. 2005.PubMed/NCBI
|
|
30
|
Nakagawa T and Yamaguchi M: Overexpression
of regucalcin enhances its nuclear localization and suppresses
L-type Ca2+ channel and calcium-sensing receptor mRNA
expressions in cloned normal rat kidney proximal tubular epithelial
NRK52E cells. J Cell Biochem. 99:1064–1077. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Magno AL, Ward BK and Ratajczak T: The
calcium-sensing receptor: a molecular perspective. Endocr Rev.
32:3–30. 2011. View Article : Google Scholar
|
|
32
|
Kennedy MB, Bennett MK, Erondu NE and
Miller SG: Calcium/calmodulin-dependent protein kinases. Calcium
and Cell function. Cheung WY: 7. Academic Press Inc; New York: pp.
61–107. 1987, View Article : Google Scholar
|
|
33
|
Kurota H and Yamaguchi M: Inhibitory
effect of regucalcin on Ca2+/calmodulin-dependent
protein kinase activity in rat renal cortex cytosol. Mol Cell
Biochem. 177:239–243. 1997. View Article : Google Scholar
|
|
34
|
Kurota H and Yamaguchi M: Inhibitory
effect of calcium-binding protein regucalcin on protein kinase C
activity in rat renal cortex cytosol. Biol Pharm Bull. 21:315–318.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pallen CJ and Wang JH:
Calmodulin-stimulated dephosphorylation of p-nitrophenyl phosphate
and free phosphotyrosine by calcineurin. J Biol Chem.
258:8550–8553. 1983.PubMed/NCBI
|
|
36
|
Omura M, Kurota H and Yamaguchi M:
Inhibitory effect of regucalcin on
Ca2+/calmodulin-dependent phosphatase activity in rat
renal cortex cytosol. Biol Pharm Bull. 21:440–443. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Omura M and Yamaguchi M: Inhibition of
Ca2+/calmodulin-dependent phosphatase activity by
regucalcin in rat liver cytosol: involvement of calmodulin binding.
J Cell Biochem. 71:140–148. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Morooka Y and Yamaguchi M: Suppressive
role of endogenous regucalcin in the regulation of protein
phosphatase activity in rat renal cortex cytosol. J Cell Biochem.
81:639–646. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Morooka Y and Yamaguchi M: Inhibitory
effect of regucalcin on protein phosphatase activity in the nuclei
of rat kidney cortex. J Cell Biochem. 83:111–120. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Morooka Y and Yamaguchi M: Endogenous
regucalcin suppresses the enhancement of protein phosphatase
activity in the cytosol and nucleus of kidney cortex in
calcium-administered rats. J Cell Biochem. 85:553–560. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamaguchi M and Kurota H: Inhibitory
effect of regucalcin on Ca2+/calmodulin-dependent cyclic
AMP phosphodiesterase activity in rat kidney cytosol. Mol Cell
Biochem. 177:209–214. 1997. View Article : Google Scholar
|
|
42
|
Lowenstein CJ, Dinerman JL and Snyder SH:
Nitric oxide: a physiologic messenger. Ann Intern Med. 120:227–237.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma ZJ and Yamaguchi M: Regulatory effect
of regucalcin on nitric oxide synthase activity in rat kidney
cortex cytosol: Role of endogenous regucalcin in transgenic rats.
Int J Mol Med. 12:201–206. 2003.PubMed/NCBI
|
|
44
|
Baba T and Yamaguchi M: Stimulatory effect
of regucalcin on proteolytic activity in rat renal cortex cytosol:
involvement of thiol proteases. Mol Cell Biochem. 195:87–92. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Baba T and Yamaguchi M: Stimulatory effect
of regucalcin on proteolytic activity is impaired in the kidney
cortex cytosol of rats with saline ingestion. Mol Cell Biochem.
206:1–6. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Croall DE and DeMartino GN:
Calcium-activated neutral protease (calpain) system: structure,
function, and regulation. Physiol Rev. 71:813–847. 1991.PubMed/NCBI
|
|
47
|
Nakagawa T and Yamaguchi M: Nuclear
localization of regucalcin is enhanced in culture with protein
kinase C activation in cloned normal rat kidney proximal tubular
epithelial NRK52E cells. Int J Mol Med. 21:605–610. 2008.PubMed/NCBI
|
|
48
|
Morooka Y and Yamaguchi M: Suppressive
effect of endogenous regucalcin on deoxyribonuclic acid synthesis
in the nuclei of rat renal cortex. Mol Cell Biochem. 229:157–162.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tsurusaki Y and Yamaguchi M: Role of
regucalcin in liver nuclear function: Binding of regucalcin to
nuclear protein or DNA and modulation of tumor-related gene
expression. Int J Mol Med. 14:277–281. 2004.PubMed/NCBI
|
|
50
|
Charollais RH, Buquet C and Mester J:
Butyrate blocks the accumulation of CDC2 mRNA in late G1 phase but
inhibits both the early and late G1 progression in chemically
transformed mouse fibroblasts BP-A31. J Cell Physiol. 145:46–52.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Meijer L, Borgne A, Mulner O, Chong JP,
Blow JJ, Inagaki N, Inagaki M, Delcros JG and Moulinoux JP:
Biochemical and cellular effects of roscovitine, a potent and
selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and
cdk5. Eur J Biochem. 243:527–536. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Singh SV, Herman-Antosiewicz A, Singh AV,
Lew KL, Srivastava SK, Kamath R, Brown KD, Zhang L and Baskaran R:
Sulforaphane-induced G2/M phase cell cycle arrest involves
checkpoint kinase 2-mediated phosphorylation of cell division cycle
25C. J Biol Chem. 279:25813–25822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Park YC, Lee CH, Kang HS, Chung HT and Kim
HD: Wortmannin, a specific inhibitor of
phosphatidylinositol-3-kinase, enhances LPS-induced NO production
from murine peritoneal macrophages. Biochem Biophys Res Commun.
240:692–696. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nakagawa T and Yamaguchi M: Overexpression
of regucalcin suppresses apoptotic cell death in cloned normal rat
kidney proximal tubular epithelial NRK52E cells: change in
apoptosis-related gene expression. J Cell Biochem. 96:1274–1285.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Widmann C, Gibson S and Johnson GL:
Caspase-dependent cleavage of signaling proteins during apoptosis.
A turn-off mechanism for anti-apoptotic signals. J Biol Chem.
273:7141–7147. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dieguez-Acuña FJ, Polk WW, Ellis ME,
Simmonds PL, Kushleika JV and Woods JS: Nuclear factor kappaB
activity determines the sensitivity of kidney epithelial cells to
apoptosis: implications for mercury-induced renal failure. Toxicol
Sci. 82:114–123. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang YQ, Kanzaki M, Furukawa M, Shibata
H, Ozeki M and Kojima I: Involvement of Smad proteins in the
differentiation of pancreatic AR42J cells induced by activin A.
Diabetologia. 42:719–727. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hammar EB, Irminger JC, Rickenbach K,
Parnaud G, Ribaux P, Bosco D, Rouiller DG and Halban PA: Activation
of NF-kappaB by extracellular matrix is involved in spreading and
glucose-stimulated insulin secretion of pancreatic beta cells. J
Biol Chem. 280:30630–30637. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fan JM, Ng YY, Hill PA, Nikolic-Paterson
DJ, Mu W, Atkins RC and Lan HY: Transforming growth factor-β
regulates tubular epithelial-myofibroblast transdifferentiation in
vitro. Kidney Int. 56:1455–1467. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shinya N, Kurota H and Yamaguchi M:
Calcium-binding protein regucalcin mRNA expression in the kidney
cortex is suppressed by saline ingestion in rats. Mol Cell Biochem.
162:139–144. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shinya N and Yamaguchi M: Alterations in
Ca2+-ATPase activity and calcium-binding protein
regucalcin mRNA expression in the kidney cortex of rats with saline
ingestion. Mol Cell Biochem. 170:17–22. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Shinya N and Yamaguchi M: Stimulatory
effect of calcium administration on regucalcin mRNA expression is
attenuated in the kidney cortex of rats ingested with saline. Mol
Cell Biochem. 178:275–281. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Montine TJ and Borch RF: Role of
endogenous sulfur-containing nucleophiles in an in vitro model of
cis-diamminedichloroplatinum(II)-induced nephrotoxicity. Biochem
Pharmacol. 39:1751–1757. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Goldstein RS, Pasino DA, Hewitt WR and
Hook JB: Biochemical mechanisms of cephaloridine nephrotoxicity:
time and concentration dependence of peroxidative injury. Toxicol
Appl Pharmacol. 83:261–270. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kurota H and Yamaguchi M: Suppressed
expression of calcium-binding protein regucalcin mRNA in the renal
cortex of rats with chemically induced kidney damage. Mol Cell
Biochem. 151:55–60. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Misawa H and Yamaguchi M: Involvement of
nuclear factor-1 (NF1) binding motif in the regucalcin gene
expression of rat kidney cortex: the expression is suppressed by
cisplatin administration. Mol Cell Biochem. 219:29–37. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chiusolo A, Defazio R, Casartelli A,
Bocchini N, Mongillo M, Zanetti E, Cristofori P and Trevisan A:
Regucalcin down-regulation in rat kidney tissue after treatment
with nephrotoxicants. Toxicol Lett. 182:84–90. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Arbillaga L, Vettorazzi A, Gil AG, van
Delft JH, García-Jalón JA and López de Cerain A: Gene expression
changes induced by ochratoxin A in renal and hepatic tissues of
male F344 rat after oral repeated administration. Toxicol Appl
Pharmacol. 230:197–207. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wu HZ, Guo L, Mak YF, Liu N, Poon WT, Chan
YW and Cai Z: Proteomics investigation on aristolochic acid
nephropathy: a case study on rat kidney tissues. Anal Bioanal Chem.
399:3431–3439. 2011. View Article : Google Scholar
|
|
71
|
Klawitter J, Klawitter J, Kushner E,
Jonscher K, Bendrick-Peart J, Leibfritz D, Christians U and Schmitz
V: Association of immunosuppressant-induced protein changes in the
rat kidney with changes in urine metabolite patterns: a
proteo-metabonomic study. J Proteome Res. 9:865–875. 2010.
View Article : Google Scholar
|
|
72
|
Murata T and Yamaguchi M: Alternatively
spliced variants of the regucalcin gene in various human normal and
tumor tissues. Int J Mol Med. 34:1141–1146. 2014.PubMed/NCBI
|