Introduction
Stroke is one of the most common causes of mortality
and disability worldwide (1–3).
Stroke is a clinical syndrome with pathological types including
ischemic, intracerebral hemorrhage, and sub-arachnoid hemorrhage
types (4,5). Stroke risk increases with age
(6). Ischemic stroke, which
accounts for 87% of all stroke subtypes presented clinically
(7), is prevalent in populations
of European origin and Chinese populations (5,8,9).
Acute ischemic stroke is the major cause of high morbidity and
mortality in the aging population worldwide (10). The prognosis is worse for stroke,
with up to 50% of the individuals worldwide confirmed to have
suffered a stroke succumbing or being dependent on carers 6 months
after the event (5). Current
therapeutic strategies for acute ischemic stroke are limited
(3). Intravenous thrombolysis
with aspirin improves the outcome for only a small proportion of
patients presenting early after symptom onset with acute ischemic
stroke (11). Thus, seeking
clinical diagnosis, prognostic biomarkers and therapeutic targets
for ischemic stroke is indispensable. Approximately 50% of ischemic
strokes are attributed to large-artery atherothrombotic disease,
25% to disease of the small intracranial arteries, 20% to cardiac
emboli, and 5% to various rare causes (e.g., extracranial artery
dissection) (4,5). The most common characteristic of
acute ischemic stroke is an occlusion of the artery. Plaque
composition is thought to be an independent risk factor for acute
ischemic stroke (12).
The thickness of the fibrous cap, the size of the
necrotic core and intraplaque hemorrhage, and the extent of
inflammatory activity within the plaque are key factors in advanced
plaque that are most likely to lead to complications (13). However, it was not possible to
demonstrate how the complicated plaques associate with symptoms,
since it appears that complicated plaques may occur at any time,
irrespective of symptoms (14).
Additionally, the exact mechanisms causing plaque rupture remain to
be determined (15). Detection of
vulnerable plaque is important for preventing future clinical
events and remains to be resolved.
MicroRNAs (miRNAs or miRs) are non-coding RNAs that
regulate gene expression by translation inhibition or messenger RNA
degradation (16). miRNAs that
are highly stable in serum (17)
and urine (18) are potential
biomarkers of pathology, such as stroke (19,20) and age-associated pathologies
(21). The differential
expression of miRNAs in peripheral blood of young ischemic stroke
patients in Malaysia has been previously examined (22). However, the miRNAs involved in the
formation and progression of atherosclerotic plaque have yet to be
thoroughly investigated.
An intra-patient comparison on the transcript
profile of atheromatous plaque and intact tissue samples was
performed by Ayari and Bricca (23). However, those authors only
analyzed the expression of CD163 and heme oxygenase (decycling) 1
(HMOX1), which are involved in the homeostasis of iron and heme in
atherosclerotic plaques (23).
In the present study, we aimed to identify which
miRNAs were associated with the formation and progression of
plaque, to investigate the potential mechanism of plaque
vulnerability to rupture, and to examine potential biomarkers and
therapeutic targets for acute ischemic stroke. The same microarray
data as that used by Ayari and Bricca (23) were downloaded, and differentially
expressed genes (DEGs) in paired atheromatous plaque and intact
tissue samples were identified using the Multtest and Limma
packages of R language. Enriched miRNAs that regulate the DEGs were
then predicted using WebGestalt software. The expression of those
predicted miRNAs in serum was confirmed using reverse transcription
quantitative PCR (RT-qPCR). Protein-protein interactions (PPIs) of
DEGs were constructed from STRING databases (http://string-db.org/), and confirmed miRNA-targeted
DEGs interactions were visualized using Cytoscape software
(http://www.cytoscape.org/). Gene
ontology (GO) function and pathway analysis of confirmed
miRNA-targeted DEGs were performed using the Bingo plug-in of
Cytoscape software.
Materials and methods
Microarray data and data
preprocessing
The gene microarray data GSE43292 (23) was downloaded from the Gene
Expression Omnibus (GEO), which includes paired atheromatous plaque
and intact tissue samples collected from 32 patients.
Endarterectomy specimens were characterized histologically
according to the classification proposed by the Nomenclature
Committee of the American Heart Association (24). The sections consisted of the
intima and a majority of tunica media, although the tunica
adventitia was excluded. Stage IV and later were considered as
atheromatous plaques, and intact tissues were almost exclusively
composed of stage I and II lesions (23). The platform of GSE43292 was
GPL6244 (HuGene-1_0-st) Affymetrix Human Gene 1.0 ST Array
[transcript (gene) version].
The mean expression of a total of 33,297 probes was
derived from GEO, and accurate annotations were made. Probes
matching >1 gene ID were discarded. All the probes were then
mapped to Entrez gene IDs.
Screening of the DEGs
The log2 transformation of the expression value was
performed for each probe (25).
Up- and downregulated probes were determined according to their
fold differences (26). DEGs were
identified using the Multtest and Limma packages of R language
(27). The false discovery rate
(FDR) correction was calculated using the Benjamini- Hochberg (BH)
method (FDR < 0.05) (28). A
two-fold change method was used to identify DEGs (|log FC (Fold
change)|> 1).
Prediction of enriched miRNAs in carotid
atheroma
The WebGestal, a web-based gene set analysis
toolkit, was used to predict miRNA associated with DEGs (29,30). The hyper-geometric test was widely
used to investigate the significance of functional terms enrichment
within a list of genes. The 10 most significant categories (Top 10)
and a minimum of two genes for a category were selected as the
threshold.
Patient samples and RNA isolation
Serum specimens were collected from 10 patients with
acute ischemia stroke (average age of 69±10) and 10 healthy
individuals (no disease) who received a health check-up. There were
no significant differences in age and gender distributions between
the patients with acute ischemic stroke and healthy controls
(P>0.05).
The acute ischemic stroke patients were associated
with significant infract lesions. Serum samples prior to miRNA
analysis were stored at −80°C within 4 h following collection. The
present study adhered to the tenets of the Declaration of Helsinki
and was approved by the Medical Ethics Committee of Sichuan
University (Sichuan, China). Informed consent was obtained from
participants or their close relatives for the use of their blood in
this study.
RT-qPCR
Total RNA was extracted from serum samples according
to a method described previously (31). Synthetic C. elegans miRNA
(cel-miR-39; Qiagen, Hilden, Germany) was spiked in each extracted
RNA at a final concentration of 4 fmol as an internal control. The
concentration and quality of RNA were measured by UV absorbance at
260 and 280 nm (A260/280 ratio).
Ten nanograms of total RNA was reverse transcribed
and qPCR was performed using an All-in-One™ miRNA qRT-PCR Detection
kit (GeneCopoeia, Rockville, MD, USA) according to the
manufacturer's instructions. The validated All-in-One miRNA forward
primers were purchased from GeneCopoeia. Forward primers specific
for cel-miR-39 were designed based on a sequence of mature miRNA
obtained from the miRbase (32).
The levels of miRNAs were normalized with cel-miR-39. The
2−ΔΔCT method, described by Livak et al (33), was used to analyze data. miR-9 in
each serum sample and no-template controls were run in
triplicate.
Statistical analysis
Statistical analysis was performed using the
two-tailed paired Student's t-test using IBM SPSS 21.0 (SPSS, Inc.,
Chicago, IL, USA). Differences were considered statistically
significant at P<0.05.
Confirmed miRNA-targeted gene interaction
network
PPIs of DEGs were constructed from the STRING
databases (34), and interactions
associated with confirmed miRNAs were retained. miRNA-targeted DEGs
interaction networks were visualized using the Cytoscape software
version 3.1.1 (35).
Hierarchical clustering of expressional
values of DEGs
Hierarchical clustering of expression data of the
signature DEGs and the confirmed miRNA-targeted DEGs that were
altered in atheromatous plaque was performed in a dChip Analyzer
(36). The clustering algorithms
with Euclidean distance (37)
metric and centroid linkage rule were used (36).
GO function and pathway analysis of
confirmed miRNA- targeted DEGs interaction
The Cytoscape software with plug-in Bingo was run to
determine which biological processes are statistically
overrepresented in the set of genes corresponding to the predicted
miRNA-targeted DEGs interaction networks (38). The hypergeometric test was
employed with a BH-FDR-based multiple testing correction (corr
P<0.01).
Results
Screening of the DEGs
The expression value was derived for each probe, and
an accurate annotation was made. DEGs were identified using the
Multtest and Limma packages of R language. In the present study, 56
and 69 genes that were significantly differentially down- and
upregulated, respectively, were identified (data not shown).
Prediction of enriched miRNAs
The top 10 miRNAs significantly associated with the
screened up- and downregulated DEGs were predicted using the
WebGestalt toolkit (Table I). Six
miRNAs were connected with the upregulated DEGs including miR-9,
-22, -23, -27, -125 and -524, and 10 miRNAs were connected with the
downregulated DEGs including miR-9, -30, -33, -124, -135, -181,
-197, -218, -330 and -452. A list of target DEGs associated with
these miRNAs is provided in Table
I. In the DEGs provided in Table
II, the expression of vav 3 guanine nucleotide exchange factor
(VAV3) and PR domain containing 1 (PRDM1) was 2.0-fold higher than
that in the intact tissue, and the expression of β-thyroid hormone
receptor (THRB) and contactin 4 (CNTN4) in atheromatous plaque was
a notable 2.1- and 3.5-fold lower than that in the intact tissue,
respectively. Notably, miR-9 was associated with both down- and
upregulated DEGs.
 | Table ITop 10 enriched miRNA with target
DEGs. |
Table I
Top 10 enriched miRNA with target
DEGs.
|
Up-/Downregulated | Enriched
miRNAs | Targets DEGs | Adj.P |
|---|
| Downregulated | hsa_TGTTTAC,
miR-30 |
LGI1|ACTC1|PRUNE2|NLGN1|GRIA2|FRK|TTLL7|LPHN3|PCDH20 | 0.0006 |
| hsa_GTGGTGA,
miR-197 |
NLGN1|GRIA2|TTLL7|LPHN3 | 0.0006 |
| hsa_AAGCCAT,
miR-135 |
THRB|LRRN1|FRK|PRUNE2|PDE8B|NEGR1 | 0.0029 |
| hsa_TGCCTTA,
miR-124 |
RYR2|THRB|CNTN3|GRIA2|CNTN1|NEGR1 | 0.0261 |
| hsa_TGAATGT,
miR-181 |
THRB|LRRN1|CNTN4|GRIA2|NEGR1 | 0.0563 |
| hsa_TGCTTTG,
miR-330 |
PDZRN3|PCDH11Y|FRK|PRUNE2 | 0.0609 |
| hsa_CAATGCA,
miR-33 | LGI1|CNTN4 | 0.0864 |
| hsa_TAGCTTT,
miR-9 |
THRB|CNTN4|NEGR1 | 0.0864 |
| hsa_AAGCACA,
miR-218 |
PLD5|GRIA2|FREM1|NPY1R | 0.0864 |
| hsa_TGCAAAC,
miR-452 | PLCB4|THRB | 0.0915 |
| Upregulated | hsa_AATGTGA,
miR-23 | PRDM1|CD163 | 0.1423 |
| hsa_ACCAAAG,
miR-9 | VAV3|PRDM1 | 0.1423 |
| hsa_GGCAGCT,
miR-22 | DPP4|AQP9 | 0.1423 |
| hsa_ACTGTGA,
miR-27 | VAV3|RGS1 | 0.1423 |
| hsa_CTTTGTA,
miR-524 | VAV3|PRDM1 | 0.1423 |
| hsa_CTCAGGG,
miR-125 | PRDM1|ANPEP | 0.1423 |
 | Table IIDEGs in the confirmed miRNA-target
DEGs interaction networks. |
Table II
DEGs in the confirmed miRNA-target
DEGs interaction networks.
| Gene symbol | Entrez ID | Adj.P | logFC | Gene name |
|---|
| CNTN1 | 1272 | 2.00E-05 | −1.91103 | Contactin 1 |
| CNTN4 | 152330 | 1.70E-05 | −1.79233 | Contactin 4 |
| CASQ2 | 845 | 1.91E-05 | −1.66766 | Calsequestrin 2
(cardiac muscle) |
| PCDH11Y | 83259 | 2.55E-05 | −1.38541 | Protocadherin 11
Y-linked |
| CNTN3 | 5067 | 3.89E-05 | −1.36174 | Contactin 3
(plasmacytoma associated) |
| NPY1R | 4886 | 8.70E-05 | −1.27438 | Neuropeptide Y
receptor Y1 |
| LRRN1 | 57633 | 2.85E-05 | −1.27233 | Leucine rich repeat
neuronal 1 |
| RYR2 | 6262 | 5.41E-05 | −1.24144 | Ryanodine receptor
2 (cardiac) |
| LGI1 | 9211 | 6.74E-05 | −1.15644 | Leucine-rich,
glioma inactivated 1 |
| PRUNE2 | 158471 | 3.53E-05 | −1.11997 | Prune homolog 2
(Drosophila) |
| PCDH20 | 64881 | 2.81E-05 | −1.11425 | Protocadherin
20 |
| PLD5 | 200150 | 3.62E-05 | −1.11138 | Phospholipase D
family, member 5 |
| NEGR1 | 257194 | 1.97E-05 | −1.10062 | Neuronal growth
regulator 1 |
| GRIA2 | 2891 | 1.42E-03 | −1.08966 | Glutamate receptor,
ionotropic, AMPA 2 |
| NPR1 | 4881 | 8.88E-05 | −1.07677 | Natriuretic peptide
receptor A/guanylate cyclase A |
| ACTC1 | 70 | 2.93E-04 | −1.07237 | Actin, α, cardiac
muscle 1 |
| PDZRN3 | 23024 | 2.55E-05 | −1.07182 | PDZ
domain-containing ring finger 3 |
| LPHN3 | 23284 | 3.67E-04 | −1.06682 | Latrophilin 3 |
| PLN | 5350 | 9.84E-05 | −1.06111 | Phospholamban |
| NLGN1 | 22871 | 6.43E-05 | −1.03802 | Neuroligin 1 |
| THRB | 7068 | 1.16E-04 | −1.0368 | Thyroid hormone
receptor, β |
| TTLL7 | 79739 | 7.12E-05 | −1.02607 | Tubulin tyrosine
ligase-like family, member 7 |
| GRIA1 | 2890 | 3.08E-04 | −1.02562 | Glutamate receptor,
ionotropic, AMPA 1 |
| FRK | 2444 | 4.00E-04 | −1.01427 | Fyn-related
kinase |
| FREM1 | 158326 | 1.57E-04 | −1.00531 | FRAS1 related
extracellular matrix 1 |
| ACP5 | 54 | 4.63E-04 | 1.00142 | Acid phosphatase 5,
tartrate resistant |
| PRDM1 | 639 | 3.75E-05 | 1.005106 | PR domain
containing 1, with ZNF domain |
| CR1 | 1378 | 7.47E-04 | 1.011272 | Complement
component (3b/4b) receptor 1 (Knops blood group) |
| VAV3 | 10451 | 3.72E-05 | 1.026726 | Vav 3 guanine
nucleotide exchange factor |
| CXCL10 | 3627 | 5.23E-04 | 1.051734 | Chemokine (C-X-C
motif) ligand 10 |
| NPL | 80896 | 5.61E-05 | 1.066364 |
N-acetylneuraminate-pyruvate lyase |
| ITGAX | 3687 | 1.91E-04 | 1.070598 | Integrin, α X
(complement component 3 receptor 4 subunit) |
| RGS1 | 5996 | 5.42E-03 | 1.094815 | Regulator of
G-protein signaling 1 |
| CYTIP | 9595 | 1.22E-04 | 1.099515 | Cytohesin 1
interacting protein |
| PLEK | 5341 | 4.59E-05 | 1.104737 | Pleckstrin |
| CD163 | 9332 | 3.84E-05 | 1.130551 | CD163 molecule |
| ADAMDEC1 | 27299 | 2.95E-03 | 1.138718 | ADAM-like, decysin
1 |
| SELE | 6401 | 2.36E-04 | 1.157189 | Selectin E |
| ANPEP | 290 | 8.68E-05 | 1.173833 | Alanyl (membrane)
aminopeptidase |
| CCR1 | 1230 | 3.94E-05 | 1.179454 | Chemokine (C-C
motif) receptor 1 |
| PLIN2 | 123 | 1.37E-04 | 1.18298 | Perilipin 2 |
| AQP9 | 366 | 1.82E-03 | 1.252998 | Aquaporin 9 |
| CD52 | 1043 | 1.35E-04 | 1.263187 | CD52 molecule |
| CHI3L1 | 1116 | 4.86E-03 | 1.289639 | Chitinase 3-like 1
(cartilage glycoprotein-39) |
| IL1RN | 3557 | 5.61E-05 | 1.394489 | Interleukin 1
receptor antagonist |
| HMOX1 | 3162 | 4.75E-05 | 1.419502 | Heme oxygenase
(decycling) 1 |
| MME | 4311 | 8.26E-05 | 1.485 | Membrane
metallo-endopeptidase |
| MMP12 | 4321 | 3.59E-03 | 1.574106 | Matrix
metallopeptidase 12 |
| DPP4 | 1803 | 3.69E-05 | 1.610873 |
Dipeptidyl-peptidase 4 |
| CD36 | 948 | 1.99E-04 | 1.802205 | CD36 molecule
(thrombospondin receptor) |
| MMP9 | 4318 | 1.86E-04 | 1.817804 | Matrix
metallopeptidase 9 |
| MMP7 | 4316 | 5.72E-04 | 1.840231 | Matrix
metallopeptidase 7 |
| FABP4 | 2167 | 2.39E-05 | 2.454461 | Fatty acid binding
protein 4, adipocyte |
Validation of predicted serum miRNA in
acute ischemic stroke
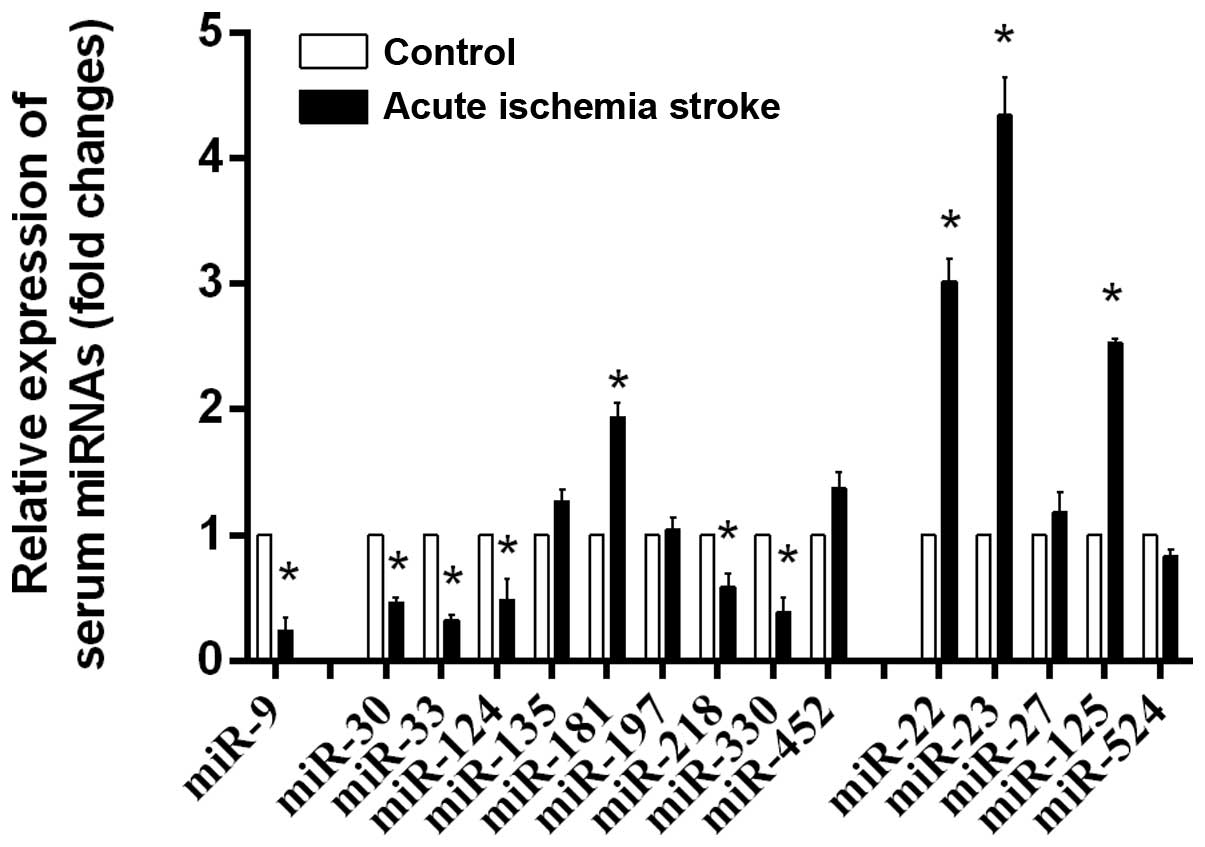
The expression of predicted serum miRNAs in acute
ischemic stroke was validated by RT-qPCR (Fig. 1). It was confirmed that the
expression of serum miR-9, -30, -33, -124, -181, -218, -330, -22,
-23 and -125 was significantly altered in ischemic stroke,
indicating these miRNAs are important in acute ischemic stroke.
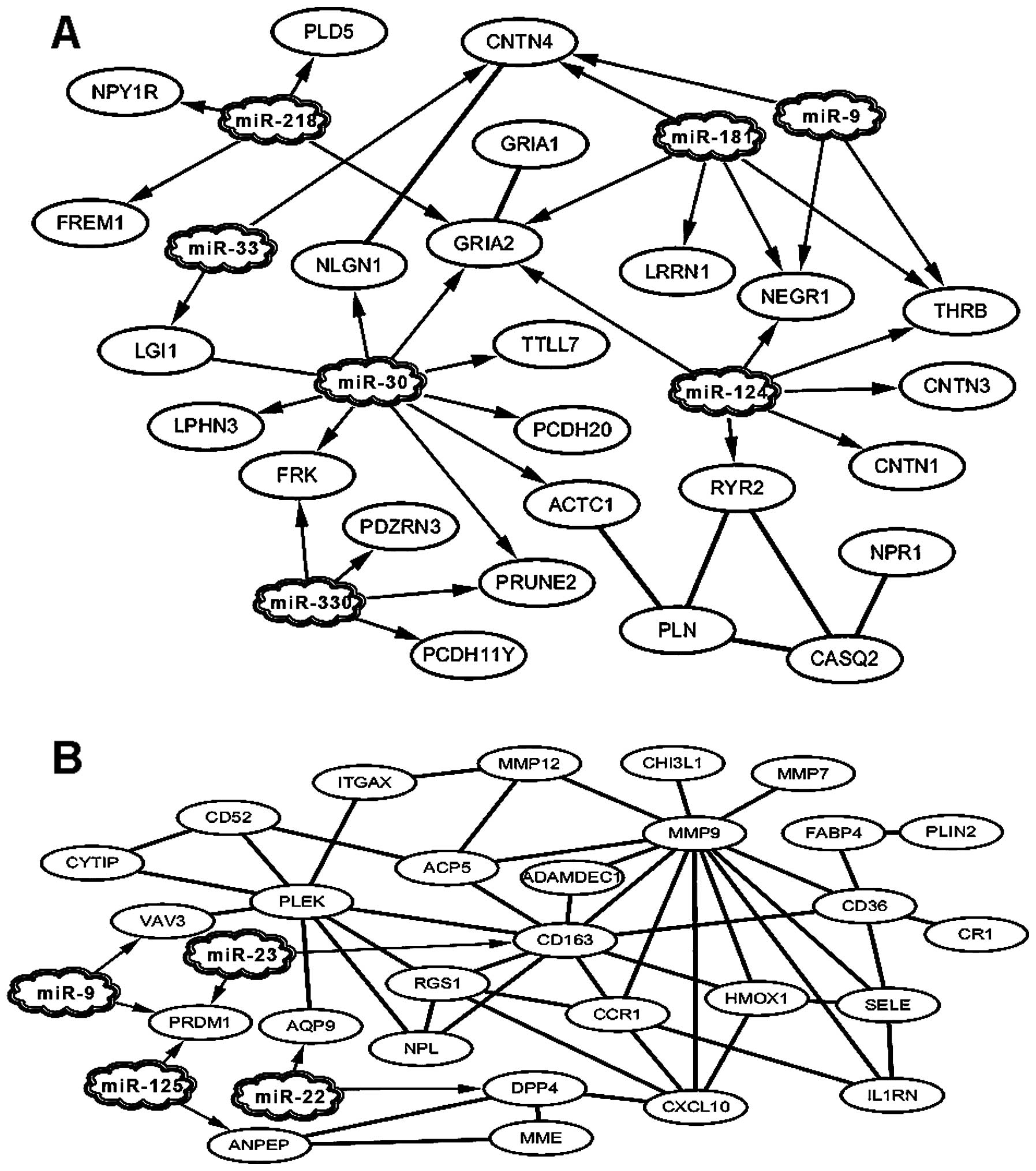
Confirmed miRNA-targeted DEGs interaction
networks and important DEG nodes
PPIs of DGEs were constructed from the STRING
databases. Interactions associated with confirmed miRNAs were
retained, and confirmed miRNA-targeted DEGs interaction networks
were subsequently visualized using Cytoscape software (Fig. 2). Four miRNAs were linked to 25
downregulated DEGs (Fig. 2A) and
7 miRNAs were linked to 28 upregulated DEGs (Fig. 2B) forming miRNAs-DEGs interaction
networks. Table II shows the
DEGs in the interaction networks. The most important nodes in the
miRNA interaction networks were downregulated DEGs including THRB,
CNTN4, NEGR1 and glutamate receptor 2 (GRIA2) (Fig. 1A), and upregulated DEGs including
VAV3, PRDM1, CD163, matrix metallopeptidase 9 (MMP9), pleckstrin
(PLEK), CD36, chemokine (C-X-C motif) ligand 10 (CXCL10), chemokine
(C-C motif) receptor 1 (CCR1), regulator of G-protein signaling 1
(RGS1), HMOX1, tartrate resistant acid phosphatase 5 (ACP5),
dipeptidyl peptidase 4 (DPP4) and selectin E (SELE) (Fig. 2B).
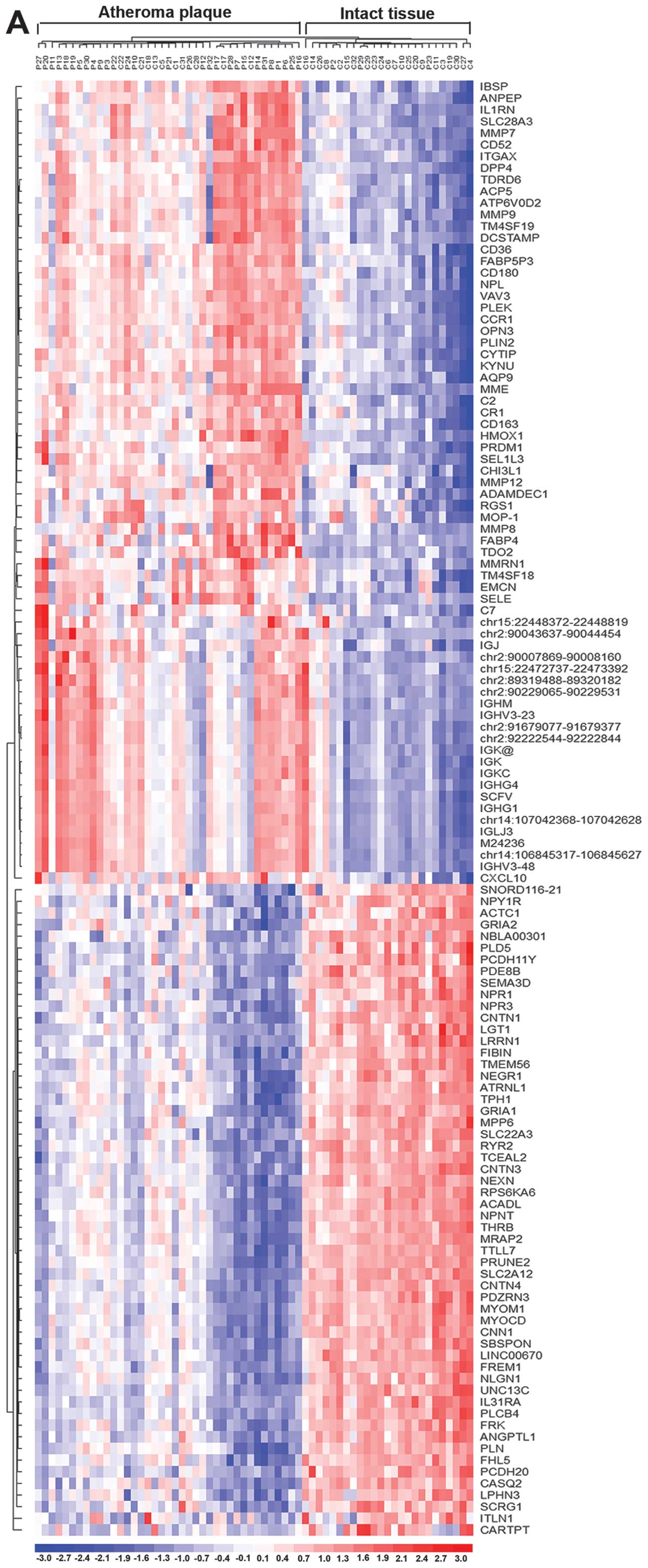
Hierarchical clustering of expressional
values of DEGs
Using the algorithm based on Euclidean distance
metric and centroid linkage rule, the hierarchical clustering of
expressional values of DEGs and the confirmed miRNA-targeted DEGs
were divided into the atheromatous plaque and intact tissue groups
(Fig. 3). Fig. 3 shows atheromatous plaque and
intact tissue samples separated with the exception of 13 paired
samples that were not well classified. There were 10 intact tissues
clustered in the atheromatous plaque group, and three atheromatous
plaques in the intact tissue group, indicating that the differences
in gene expression between intact tissues and atheromatous plaques
from those patients are extremely close and difficult to
distinguish, and that there was a high risk of atheromatous in the
10 patients. Nevertheless, two clustering maps showed identical
resampling, suggesting that the confirmed miRNA-targeted DEGs are
important representative genes in the interaction networks, and
that screening is extremely significant.
GO function and pathway analysis of
confirmed miRNA- targeted DEG interaction
GO function suggested that the confirmed miRNAs
targeting downregulated DEGs were predominately associated with
signal transduction, the circulatory system, biological adhesion
and striated muscle contraction, and that the targeted upregulated
DEGs were most significantly associated with signal transduction,
wound healing and the immune system (Table III).
 | Table IIIGO enrichment analysis of confirmed
miRNAs and targeted DEGs interaction networks. |
Table III
GO enrichment analysis of confirmed
miRNAs and targeted DEGs interaction networks.
Downregulated DEGs
|
|---|
| GO-ID | Description | Genes in test
set |
|---|
| 44057 | Regulation of
system process |
PLN|NLGN1|RYR2|NPR1|CNTN4|NPY1R|LGI1 |
| 8015 | Blood
circulation |
ACTC1|PLN|RYR2|NPR1|NPY1R |
| 3013 | Circulatory system
process |
ACTC1|PLN|RYR2|NPR1|NPY1R |
| 7155 | Cell adhesion |
PCDH11Y|FREM1|PCDH20|NLGN1|CNTN1|CNTN4|CNTN3|NEGR1 |
| 22610 | Biological
adhesion |
PCDH11Y|FREM1|PCDH20|NLGN1|CNTN1|CNTN4|CNTN3|NEGR1 |
| 6941 | Striated muscle
contraction |
ACTC1|RYR2|CASQ2 |
|
Upregulated DEGs
|
| GO-ID | Description | Genes in test
set |
|
| 9611 | Response to
wounding |
CR1|CD36|PLEK|HMOX1|CCR1|IL1RN|MME|SELE|MMP12|CD163|CXCL10 |
| 2376 | Immune system
process | CR1 RGS1 PLEK AQP9
CCR1 MMP9 IL1RN ACP5 SELE DPP4 CXCL10 |
| 6950 | Response to
stress | CR1 CD36 PLEK AQP9
HMOX1 CCR1 IL1RN MME ACP5 SELE MMP12 DPP4 CD163 CXCL10 |
| 6954 | Inflammatory
response | CR1 HMOX1 CCR1
IL1RN SELE CD163 CXCL10 |
| 7229 | Integrin-mediated
signaling pathway | VAV3 PLEK ITGAX
ADAMDEC1 |
| 48583 | Regulation of
response to stimulus | CR1 PLEK HMOX1
FABP4 ACP5 SELE DPP4 CXCL10 |
| 42221 | Response to
chemical stimulus | PLIN2 AQP9 HMOX1
CCR1 IL1RN FABP4 MME ACP5 SELE MMP12 DPP4 CXCL10 |
| 30155 | Regulation of cell
adhesion | CD36 VAV3 CYTIP
ADAMDEC1 DPP4 |
| 6952 | Defense
response | CR1 HMOX1 CCR1
IL1RN ACP5 SELE CD163 CXCL10 |
| 32101 | Regulation of
response to external stimulus | PLEK FABP4 ACP5
SELE CXCL10 |
| 42060 | Wound healing | CD36 PLEK HMOX1 MME
MMP12 |
| 60054 | Positive regulation
of epithelial cell proliferation involved in wound healing | MME MMP12 |
| 2685 | Regulation of
leukocyte migration | HMOX1 SELE
CXCL10 |
| 34383 | Low-density
lipoprotein particle clearance | CD36 HMOX1 |
| 50896 | Response to
stimulus | CR1 AQP9 PLEK CCR1
IL1RN ACP5 MME MMP12 CD163 CXCL10 CD36 RGS1 PLIN2 HMOX1 FABP4 SELE
DPP4 |
| 15718 | Monocarboxylic acid
transport | CD36 PLIN2
AQP9 |
| 48518 | Positive regulation
of biological process | CR1 CD36 VAV3 PLEK
HMOX1 MMP9 FABP4 MME PRDM1 SELE MMP12 DPP4 CXCL10 |
| 42493 | Response to
drug | PLIN2 AQP9 FABP4
MME MMP12 |
| 6955 | Immune
response | CR1 RGS1 AQP9 CCR1
IL1RN ACP5 CXCL10 |
| 10033 | Response to organic
substance | PLIN2 AQP9 HMOX1
IL1RN FABP4 ACP5 SELE CXCL10 |
| 8284 | Positive regulation
of cell proliferation | HMOX1 FABP4 MME
MMP12 DPP4 CXCL10 |
Discussion
The predominant etiology of stroke is
atherosclerosis (atheroma) (39).
The present study provides a list of potential clinical diagnoses
and prognostic biomarkers and therapeutic targets for acute
ischemic stroke via the identification of a list of DEGs associated
with miRNA targets, the validation of expression of predicted
miRNAs in acute ischemic stroke, and the GO function analysis of
the confirmed miRNAs targeted DEGs in the advanced plaque using the
microarray data from Ayari and Bricca (23). Furthermore, a new molecular
mechanism causing atherosclerotic plaque rupture and intraplaque
hemorrhage, and subsequently ischemic stroke remains to be
determined.
Whole-transcript expression profile of human
atherosclerotic arteries (endarterectomy specimens) was performed
by Ayari and Bricca (23). As it
is impossible to obtain normal artery tissue, the intra-patient
comparison on the expression profile of atheromatous plaque and
intact tissues from the individual was carried out to examine the
atherogenic process per se (23).
Ayari and Bricca analyzed the expression of two genes (CD163 and
HMOX1) involved in the homeostasis of iron and heme, and their role
in atheromatous plaques (23). In
the present study, we identified 56 downregulated DEGs and 69
upregulated DEGs (including CD163 and HMOX1). A 2-fold change with
FDR<0.05 was set as the threshold criterion. Using these DEGs,
we screened the associated Top 10 miRNA. Six miRNAs were linked to
the upregulated DEGs including miR-9, -22, -23, -27, -125 and -524,
and 10 miRNAs were linked to the downreg ulated DEGs including
miR-9, -30, -33, -124, -135, -181, -197, -218, -330 and -452
(Table I). Of these miRNAs,
miR-33 and miR-125 were found to associate with hyperlipidemia
(19). Notably, miR-9 is an
important miRNA associated with down- and upregulated DEGs. The
significant change in expression of predicted serum miRNAs (miR-9,
-30, -33, -124, -181, -218, -330, -22, -23 and -125) in acute
ischemic stroke was validated by RT-qPCR. The results confirmed
that miRNAs may be important in acute ischemic stroke.
PPIs of DGEs were constructed from the STRING
databases, and any interactions that were not associated with any
miRNA targets were discarded, such as the interaction between
calponin 1 (CNN1) and myocardin (MYOCD), which played crucial roles
in cardiogenesis and differentiation of the smooth muscle cell
lineage (40), as well as the
interaction between tryptophan 2,3-dioxygenase (TDO2) and
kynureninase (KYNU), which were involved in tryptophan metabolism
(41). Interactions associated
with confirmed miRNAs were retained, and the confirmed
miRNA-targeted DEGs interaction networks were constructed. In
total, 25 downregulated DEGs and 28 upregulated DEGs were involved
in the miRNAs-DEGs interaction networks.
In the present study, the intact tissues were type I
and II lesions, and the atheromatous plaque was type IV lesion and
later lesion according to the definition and classification
suggested by the Nomenclature Committee of the American Heart
Association (24). Of note, type
I and II lesions may be combined under the term early lesions, and
type IV–VI lesions combined under the term advanced lesions. The
early lesions generally are those that occur in infants and
children, although they also occur in adults (24). Following the early lesions onset,
advanced lesions may subsequently evolve following thickening of
the arterial wall and therefore narrowing of the lumen or
obstruction or modification of the blood flow. Thus, common gene
expression in early and advanced lesions from some individuals may
be identical. This hypothesis was supported by the hierarchical
clustering maps which did not separate all the endarterectomy
specimens into intact tissue and atheromatous plaque groups.
Nevertheless, two clustering maps showed identical resampling,
indicating that the miRNA-target DEGs were important representative
genes in the interaction networks, and that screening is
essential.
In the confirmed miRNAs-targeted DEGs interaction
networks, the most important nodes identified were THRB, CNTN4,
NEGR1 and GRIA2 in the downregulated network, and VAV3, PRDM1,
CD163, MMP9, PLEK, CD36, CXCL10, CCR1, RGS1, HMOX1, ACP5, DPP4 and
SELE in the upregulated network. Of note, the miR-9 target DEGs
including THRB, CNTN4, NEGR1, VAV3 and PRDM1 were important nodes
in the miRNAs-target DEGs interaction networks, suggesting that
miR-9 plays a central role in the atheroma. The important nodes
were enriched in the GO term.
Ischemic stroke occurs when an artery is obstructed
(19), which is most often caused
by atheroma. Atheroma (atherosclerosis), which is also known as the
type VI lesion, is characterized by larger, confluent and more
disruptive core of extracellular lipid (24). Almost 70% of plaque specimens
(carotid endarterectomy specimens) demonstrated fissures, hematoma
or hemorrhage, and/or thrombus, 64% demonstrated neovascularization
(14,24). Arteries with thinned or ruptured
fibrous caps, intraplaque hemorrhage, larger lipid-rich necrotic
cores and larger wall thickness were associated with the occurrence
of subsequent clinical events, especially the stroke (19,42). In addition, recurrent stroke is
associated with a higher incidence of large-artery atherosclerosis
than the first stroke (19). The
exact mechanisms causing plaque are not yet completely known
(15). The plaques that are rich
in soft extracellular lipid (43), neoformed vessels and inflammatory
infiltration (44) are
vulnerable. The fibrous cap at the site of the rupture/erosion had
an eroded surface characterized by loss of the endothelial lining
(45). As demonstrated in GO
annotations: CD36 was associated with lipid storage (GO: 19915);
and HOMOX1, CCR1, SELE, CD163 and CXCL10 were associated with the
inflammatory response (GO: 6950). miR-9 targeted downregulated DEGs
including CNTN4, NEGR1 and THRB were predominately associated with
biological adhesion and regulation of the neurological system, and
targeted upregulated DEGs including VAV3 and PRDM1 were most
significantly associated with signal transduction and vasculature
development, showing a crucial role of miR-9 in the vulnerability
of plaque. By contrast, the miRNA expression profile in young
stroke patients (18–49 years) have not shown a difference in miR-9
(22). Our results supported that
miR-9 is a potential biomarker for acute ischemic stroke. We also
observed significant changes in miR-30, -33, -124, -181, -218,
-330, -22, -23 and -125 expression. However, further investigation
focusing on their role in acute ischemic stroke should be
conducted. Furthermore, investigation into the molecular mechanism
underlying the formation and rupture of atherosclerotic plaque may
provide a new therapeutic strategy for the prevention, diagnosis,
treatment and prognosis of ischemic strokes.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant no. 11402153), and the Talent
Introduction Scientific Research Projects Funded Start-Up Funds of
Sichuan University of China (no. 2082204174089).
References
|
1
|
Lopez AD, Mathers CD, Ezzati M, Jamison DT
and Murray CJ: Global and regional burden of disease and risk
factors, 2001: Systematic analysis of population health data.
Lancet. 367:1747–1757. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization: The Global
Burden of Disease. 2004.Update, 2008.
|
|
3
|
Ding Z, Tong WC, Lu XX and Peng HP:
Hyperbaric oxygen therapy in acute ischemic stroke: a review.
Interv Neurol. 2:201–211. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krafft PR, Bailey EL, Lekic T, Rolland WB,
Altay O, Tang J, Wardlaw JM, Zhang JH and Sudlow CL: Etiology of
stroke and choice of models. Int J Stroke. 7:398–406. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Warlow C, Sudlow C, Dennis M, Wardlaw J
and Sandercock P: Stroke. Lancet. 362:1211–1224. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herson PS, Palmateer J and Hurn PD:
Biological sex and mechanisms of ischemic brain injury. Transl
Stroke Res. 4:413–419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Go AS, Mozaffarian D, Roger VL, Benjamin
EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, et al:
American Heart Association Statistics Committee and Stroke
Statistics Subcommittee: Heart disease and stroke statistics - 2014
update: a report from the American Heart Association. Circulation.
129:e28–e292. 2014. View Article : Google Scholar
|
|
8
|
Yong H, Foody J, Linong J, Dong Z, Wang Y,
Ma L, Meng HJ, Shiff S and Dayi H: A systematic literature review
of risk factors for stroke in China. Cardiology Rev. 21:77–93.
2013. View Article : Google Scholar
|
|
9
|
He J, Klag MJ, Wu Z and Whelton PK: Stroke
in the People's Republic of China. II. Meta-analysis of
hypertension and risk of stroke. Stroke. 26:2228–2232.
1995.PubMed/NCBI
|
|
10
|
Janardhan V and Qureshi AI: Mechanisms of
ischemic brain injury. Curr Cardiol Rep. 6:117–123. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kikuchi K, Miura N, Kawahara KI, Murai Y,
Morioka M, Lapchak PA and Tanaka E: Edaravone (Radicut), a free
radical scavenger, is a potentially useful addition to thrombolytic
therapy in patients with acute ischemic stroke. Biomed Rep. 1:7–12.
2013.PubMed/NCBI
|
|
12
|
Golledge J, Greenhalgh RM and Davies AH:
The symptomatic carotid plaque. Stroke. 31:774–781. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Milei J, Parodi JC, Alonso GF, Barone A,
Grana D and Matturri L: Carotid rupture and intraplaque hemorrhage:
immunophenotype and role of cells involved. Am Heart J.
136:1096–1105. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Milei J, Parodi JC, Ferreira M, Barrone A,
Grana DR and Matturri L: Atherosclerotic plaque rupture and
intraplaque hemorrhage do not correlate with symptoms in carotid
artery stenosis. J Vasc Surg. 38:1241–1247. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van der Wal AC, Becker AE, van der Loos CM
and Das PK: Site of intimal rupture or erosion of thrombosed
coronary atherosclerotic plaques is characterized by an
inflammatory process irrespective of the dominant plaque
morphology. Circulation. 89:36–44. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Russo G and Giordano A: miRNAs: from
biogenesis to networks. Methods. Mol Biol. 563:303–352. 2009.
|
|
17
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: a novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lorenzen JM, Volkmann I, Fiedler J,
Schmidt M, Scheffner I, Haller H, Gwinner W and Thum T: Urinary
miR-210 as a mediator of acute T-cell mediated rejection in renal
allograft recipients. Am J Tranplant. 11:2221–2227. 2011.
View Article : Google Scholar
|
|
19
|
Rink C and Khanna S: MicroRNA in ischemic
stroke etiology and pathology. Physiol Genomics. 43:521–528. 2011.
View Article : Google Scholar :
|
|
20
|
Tan JR, Koo YX, Kaur P, Liu F, Armugam A,
Wong PT and Jeyaseelan K: microRNAs in stroke pathogenesis. Curr
Mol Med. 11:76–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bates DJ, Li N, Liang R, Sarojini H, An J,
Masternak MM, Bartke A and Wang E: MicroRNA regulation in Ames
dwarf mouse liver may contribute to delayed aging. Aging Cell.
9:1–18. 2010. View Article : Google Scholar :
|
|
22
|
Tan KS, Armugam A, Sepramaniam S, Lim KY,
Setyowati KD, Wang CW and Jeyaseelan K: Expression profile of
MicroRNAs in young stroke patients. PLoS One. 4:e76892009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ayari H and Bricca G: Identification of
two genes potentially associated in iron-heme homeostasis in human
carotid plaque using microarray analysis. J Biosci. 38:311–315.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stary HC, Chandler AB, Dinsmore RE, Fuster
V, Glagov S, Insull W Jr, Rosenfeld ME, Schwartz CJ, Wagner WD and
Wissler RW: A definition of advanced types of atherosclerotic
lesions and a histological classification of atherosclerosis A
report from the Committee on Vascular Lesions of the Council on
Arteriosclerosis, American Heart Association. Circulation.
92:1355–1374. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fujita A, Sato JR, Rodrigues LO, Ferreira
CE and Sogayar MC: Evaluating different methods of microarray data
normalization. BMC Bioinformatics. 7:4692006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Butte AJ, Ye J, Häring HU, Stumvoll M,
White MF and Kohane IS: Determining significant fold differences in
gene expression analysis. Pac Symp Biocomput. 2001:6–17. 2001.
|
|
27
|
Gentleman R, Carey V, Huber W, Irizarry RA
and Dudoit S: Bioinformatics and computational biology solutions
using R and Bioconductor. 746718470. Springer-Verlag; New York, NY:
2005, View Article : Google Scholar
|
|
28
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc. 57:289–300. 1995.
|
|
29
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
An integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33(Suppl 2): W741–W748. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duncan D, Prodduturi N and Zhang B:
WebGestalt2: An updated and expanded version of the Web-based Gene
Set Analysis Toolkit. BMC Bioinformatics. 11(Suppl 4): 102010.
View Article : Google Scholar
|
|
31
|
Lodes MJ, Caraballo M, Suciu D, Munro S,
Kumar A and Anderson B: Detection of cancer with serum miRNAs on an
oligonucleotide microarray. PLoS One. 4:e62292009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar :
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Suppl 1): D561–D568. 2011. View Article : Google Scholar :
|
|
35
|
Smoot ME, Ono K, Ruscheinski J, Wang P-L
and Ideker T: Cytoscape 2.8: new features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar :
|
|
36
|
Li C and Hung Wong W: Model-based analysis
of oligonucleotide arrays: model validation, design issues and
standard error application. Genome Biol. 2:RESEARCH0032.
2001.PubMed/NCBI
|
|
37
|
Deza MM and Deza E: Encyclopedia of
distances. Springer; Berlin: 2009, View Article : Google Scholar
|
|
38
|
Maere S, Heymans K and Kuiper M: BiNGO: a
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim JS and Bonovich D: Research on
intracranial atherosclerosis from the East and west: why are the
results different? J Stroke. 16:105–113. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lv B, Zhao J, Yang F, Huang X, Chen G,
Yang K, Liu S, Fan C, Fu H and Chen Z: Phenotypic transition of
corpus cavernosum smooth muscle cells subjected to hypoxia. Cell
Tissue Res. 357:823–833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miller CL, Murakami P, Ruczinski I, Ross
RG, Sinkus M, Sullivan B and Leonard S: Two complex genotypes
relevant to the kynurenine pathway and melanotropin function show
association with schizophrenia and bipolar disorder. Schizophr Res.
113:259–267. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Takaya N, Yuan C, Chu B, Saam T, Underhill
H, Cai J, Tran N, Polissar NL, Isaac C, Ferguson MS, et al:
Association between carotid plaque characteristics and subsequent
ischemic cerebrovascular events: a prospective assessment with MRI
- initial results. Stroke. 37:818–823. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Falk E: Why do plaques rupture?
Circulation. 86(Suppl): III30–III42. 1992.PubMed/NCBI
|
|
44
|
Ribatti D, Levi-Schaffer F and Kovanen PT:
Inflammatory angiogenesis in atherogenesis - a double-edged sword.
Ann Med. 40:606–621. 2008. View Article : Google Scholar
|
|
45
|
Milei J, Parodi JC, Fernandez Alonso G,
Barone A, Beigelman R, Ferreira LM, Arrigoni G and Matturri L:
Carotid atherosclerosis. Immunocytochemical analysis of the
vascular and cellular composition in endarterectomies. Cardiologia.
41:535–542. 1996.PubMed/NCBI
|