Introduction
Discovered in the 1990s, protease-activated
receptors (PARs) are membrane-spanning proteins that belong to the
G protein-coupled receptor (GPCR) family and exist on the surface
of cells in a wide variety of tissues (1). The PARs contain 7 transmembrane
(TM1-7) helices, an extracellular amino terminal domain, 3
intracellular loops (ICL1-3), 3 extracellular loops (ECL1-3) and an
intracellular carboxyl terminal domain. As well as connecting TM4
and TM5, ECL2 also makes a disulfide bond with TM3 that is
conserved amongst GPCRs and contributes to receptor or structural
stability (2). The PAR family
consists of 4 members, PAR1-4. PAR1, PAR3 and PAR4 are thrombin
receptors, while PAR2 is a receptor for serine proteases (including
trypsin, mast cell tryptase, as well as factors Xa and VIIa, etc.)
(1,3,4).
Human PAR1 has been identified as a protein
containing 425 residues, including an amino-terminal signal
sequence of 20 residues and an extracellular amino-terminal domain
of 75 residues (5). PAR1 is
activated when cleaved by thrombin at a site between Arg41 and
Ser42, exposing a new N-terminal tethered ligand domain with the
sequence SFLLRN. The new N-terminus acts as a tethered ligand and
binds intramolecularly to the body of the receptor to effect TM
signaling (6,7).
Human PAR2 consists of 397 amino acids with a
molecular weight of 44 kDa. It has an extracellular N-terminal
domain of 75 amino acids tethered to a TM domain of 155 amino
acids, assembled in 7 pseudo-parallel helical sequences connected
by 3 extracellular and 3 intracellular loops of 117 amino acids,
with a small eighth helix within the intracellular C-terminus of 50
amino acids. The extracellular N-terminus of PAR2 is cleaved at a
site between Arg47 and Gly48 by protease (mainly serine protease),
and the newly exposed N-terminus is GYPGQV. The exposed N-terminus,
known as a tethered ligand, then folds back and self-activates PAR2
through binding to conserved regions of ECL2 and/or the PAR2 TM
region (1,8–10).
PAR3 is a typical GPCR with a thrombin cleavage site
between Lys38 and Thr39, which has 27% amino acid sequence
similarity to PAR1 and 28% similarity to PAR2 (11). After being cleaved by thrombin,
PAR3 exposes a new amino terminus (TFRGAP) which interacts with the
receptor as a tethered ligand (12).
Human PAR4 is a 385-amino acid protein with a
potential thrombin cleavage site in the extracellular
amino-terminal domain between Arg47 and Gly48, and has 33%
similarity to the other human PARs (13). PAR4 is one of the thrombin
receptors on human platelets and a potential target for the
management of thrombotic disorders. Briefly, when these receptors
are activated by proteinases at the specific cleavage site, the
extracellular N-terminus of the receptor, a new N-terminus, is
exposed. The new N-terminus has been found to act as a tethered
ligand and binds intramolecularly to initiate cellular signals
(13).
It has been demonstrated that following activation,
the PAR family is able to stimulate complex intracellular signaling
networks through classical G protein-mediated pathways and
β-arrestin signaling pathways (14). The signaling pathways induced by
the activation of PARs play a key role in a number of physiological
and pathological processes. For example, it has been demonstrated
that PARs expressed on cells are involved in immune responses and
inflammation, regulate endothelial-leukocyte interactions and
modulate the secretion of inflammatory mediators or neuropeptides
(4). PARs expressed on platelets
and the vascular endothelium play important roles in normal blood
vessel biology, which contribute to the pathogenesis of several
cardiovascular diseases, including atherosclerosis, restenosis and
thrombosis (15). Thus, it has
been suggested that the PARs are potential therapeutic targets for
the treatment of inflammation, hemostasis, thrombosis, vascular
dysfunction and cancer (16–19). Apart from studies on the structure
and function of PARs, few studies to date have focused on the
genetic evolution of the family of PARs. Kahn et al compared
the amino acid sequences, gene structures, locus organization and
chromosomal locations in human and mouse PARs (20). Xu et al found that the
function of PARs was conserved among vertebrates, after analyzing
the expression patterns in zebrafish and their mammalian human and
mouse counterparts (19). Thus
far, however, to the best of our knowleged, there are no studies
available on the global evolution of the PAR family in vertebrates.
In this study, we describe the evolutionary genetic association
between the members of the PAR family based on the analysis of
phylogenetic trees, chromosome location, selective pressure and
functional divergence.
Materials and methods
Sequence data collection
PAR gene sequences were obtained, based on their
orthologous and paralogous relationship, by querying the Ensembl
genome assemblies (http://www.ensembl.org/index.html) using the human PAR
gene sequences. The obtained PAR sequences were then used as
queries to obtain the best hit in BLAST at the NCBI database
(http://www.ncbi.nlm.nih.gov/pubmed).
Multiple alignment and phylogenetic
analysis
The protein coding sequences of the PAR gene family
were aligned using the ClustalW program in MEGA 5.1 (21). The aligned sequences were used for
the subsequent phylogenetic analysis. MEGA 5.1 was used to
construct a maximum likelihood (ML) tree of the PAR family with the
best fitting model of JTI+G+F that was selected by a model test in
the same package with a bootstrap value based on 1,000 repetitions.
MrBayes v3.1.2 was used to construct a Bayesian inference tree with
the nucleotide substitution model (22). The Markov chain Monte Carlo (MCMC)
technique was used for the PAR family. The repetitions run for
PAR1, PAR2, PAR3 and PAR4 were 2,000,000, 3,000,000, 1,000,000 and
1,000,000 generations, respectively. PAR1, PAR2, PAR3 and PAR4 were
sampled every 1,000 generations. The first 25% of the trees was
discarded as burn-in. Convergence was assessed by determining the
average standard deviation of split frequencies (<0.01). The
posterior probabilities for internal nodes were calculated from the
posterior density of trees.
Selective pressure analysis
In order to determine whether positive selection is
involved in the evolution of the PAR family, CodeML in PAML 4.7 was
used with the models M0 (one ratio), M1a (near neutral), M2a
(positive selection), M3 (discrete), M7 (β) and M8 (β and ω)
(23–25). M7 is a null model that does not
allow for any codons with ω>1, whereas the M8 model allows for
positive selective sites (ω>1). When the M8 model fit the data
significantly better (P-value <0.05) than the null model (M7),
the presence of positive selection sites was suggested. Conversely,
results with a P-value >0.05 were regarded as lacking any
positive selection sites. The non-synonymous/synonymous
substitutions rate ratio (ω=dN/dS) was also used to indicate
selective pressure;ω>1 indicates positive selection, ω<1
indicates negative selection and ω=1 indicates neutral evolution
(26). Twice the difference in
log likelihood between the M7 and M8 models (2Δl) was compared
against χ2 with critical values of 0.01 significance
levels. The number of non-synonymous substitutions (dN) per
non-synonymous site and the number of synonymous substitutions (dS)
per synonymous site were computed by MEGA 5.1 using the modified
Nei-Gojobori method. The transition/transversion ratio computed by
MEGA 5.1 with the ML method was 1.4, 1.58, 1.72 and 1.16
corresponding to PAR1, PAR2, PAR3 and PAR3, respectively.
Functional divergence analysis
The multiple alignment of amino acid sequences among
clusters of PAR1 and PAR2, PAR1 and PAR3, PAR1 and PAR4, PAR2 and
PAR3, PAR2 and PAR4, and PAR3 and PAR4, were input into a Clustal
format and type I functional divergence analysis was performed
using Diverge (v2.0) with the maximum-likelihood estimation (MLE)
and model-free method (MFE) (27,28). The co-efficient of functional
divergence θ (0<θ<1) was used to indicate the functional
divergence that occurred in different clusters of the PAR family. A
significant result, i.e., θ>0, suggests that it is desirable to
identify the amino acid residues in the protein which have
experienced a shift in their functional constraints. These sites
may be relevant to the functional-structural differences between
proteins.
Conservation of synteny analysis
To further study the evolution of the PAR gene
family, we investigated the chromosomal distribution of PAR1, PAR2,
PAR3 and PAR4 in eutheria, birds and teleosts. The location of
chromosomes and the number of exons in the PAR family were searched
in the Ensembl genome assemblies (http://www.ensembl.org/index.html).
Results
PAR gene repertoires in vertebrates
To examine the origin and genetic evolution of the
PAR family in vertebrates, we collected 57 vertebrate gene
sequences from Ensembl and tBLASTn (NCBI) using Homo sapiens
genes as queries. Following the elimination of uncompleted
sequences in Ensembl and NCBI, 169 functional gene sequences were
applied to this study (Table I).
The taxa comprised 18 non-mammals (9 teleost fish: coelacanth,
platyfish, tilapia, zebrafish, cod, fugu, medaka, stickleback and
tetraodon; 1 amphibian: xenopus; 3 reptiles: anole lizard,
armadillo and Chinese softshell turtle; 5 birds: duck, chicken,
flycatcher, turkey and zebra finch) and 38 mammals that include 10
primates (human, bushbaby, chimpanzee, gorilla, gibbon, orangutan,
macaque, marmoset, mouse lemur and tarsier), 26 other mammals
(alpaca, cow, cat, dog, dolphin, ferret, horse, megabat, microbat,
pig, panda, shrew, elephant, hyrax, wallaby, lesser hedgehog
tenrec, sloth, Tasmanian devil, guinea pig, mouse, rabbit, pika,
rat, kangaroo rat, squirrel and tree shrew), 1 metatheria (opossum)
and 1 prototheria (platypus). According to the completeness of
genes, we divided the collected genes into 2 groups as follows: i)
functional genes, which are sequences containing full-length open
reading frames (ORFs) and ii) uncompleted genes, meaning those that
lack start or stop codes or codes in the middle of sequences. Based
on these criteria, we identified 169 functional genes and 69
uncompleted genes from a total of 238 PAR family genes. The number
of PAR genes varied between the non-mammalian and mammalian
vertebrates; 58 functional genes were identified in the
non-mammalian species, ranging from 1 in the turkey to 11 in
zebrafish; 49 uncompleted genes were found in the non-mammalian
species, ranging from 1 in the duck, zebra finch, Chinese softshell
turtle, platyfish and zebrafish to 9 in the coelacanth. A total of
111 functional genes were identified in mammals and these ranged
from 1 in the mouse lemur, alpaca, hyrax, wallaby, and lesser
hedgehog tenrec to 5 in the opossum.
 | Table INumber of functional and uncompleted
genes in the vertebrate PAR family. |
Table I
Number of functional and uncompleted
genes in the vertebrate PAR family.
| Clade | Species | Name | Functional
genes | Uncompleted
genes | Total |
|---|
| Eutheria | Homo
sapiens | Human | 4 | | 4 |
| Otolemur
garnettii | Bushbaby | 4 | | 4 |
| Pan
troglodytes | Chimpanzee | 4 | | 4 |
| Nomascus
leucogenys | Gibbon | 3 | 1 | 4 |
| Gorilla
gorilla | Gorilla | 4 | | 4 |
| Pongo
abelii | Orangutan | 4 | | 4 |
| Macaca
mulatta | Macaque | 4 | | 4 |
| Callithrix
jacchus | Marmoset | 4 | | 4 |
| Microcebus
murinus | Mouse lemur | 1 | 2 | 3 |
| Tarsius
syrichta | Tarsier | | 1 | 1 |
| Vicugna
pacos | Alpaca | 1 | 2 | 3 |
| Bos
taurus | Cow | 4 | | 4 |
| Felis
catus | Cat | 4 | | 4 |
| Canis lupus
familiaris | Dog | 4 | | 4 |
| Tursiops
truncatus | Dolphin | 3 | 1 | 4 |
| Mustela putorius
furo | Ferret | 4 | | 4 |
| Equus
caballus | Horse | 3 | 1 | 4 |
| Pteropus
vampyrus | Megabat | 3 | 1 | 4 |
| Myotis
lucifugus | Microbat | 2 | 1 | 3 |
| Sus
scrofa | Pig | 4 | | 4 |
| Ailuropoda
melanoleuca | Panda | 3 | 1 | 4 |
| Sorex
araneus | Shrew | 2 | | 2 |
| Loxodonta
africana | Elephant | 3 | 1 | 4 |
| Procavia
capensis | Hyrax | 1 | 3 | 4 |
| Macropus
eugenii | Wallaby | 1 | 1 | 2 |
| Echinops
telfairi | Lesser hedgehog
tenrec | 1 | 1 | 2 |
| Choloepus
hoffmanni | Sloth | 2 | 1 | 3 |
| Sarcophilus
harrisii | Tasmanian
devil | 4 | | 4 |
| Cacia
porcellus | Guinea pig | 2 | 1 | 3 |
| Mus
musculus | Mouse | 4 | | 4 |
| Oryctolagus
cuniculus | Rabbit | 2 | 1 | 3 |
| Ochotona
princeps | Pika | 2 | | 2 |
| Dipodomys
ordii | Kangaroo rat | | 3 | 3 |
| Rattus
norvegicus | Rat | 4 | | 4 |
| Ictidomys
tridecemlineatus | Squirrel | 2 | | 2 |
| Tupaia
belangeri | Tree shrew | | 2 | 2 |
| Metatheria | Monodelphis
domestica | Opossum | 5 | | 5 |
| Prototheria | Ornithorhynchus
anatinus | Platypus | 3 | | 3 |
| Birds | Anas
platyrhynchos | Duck | 4 | 1 | 5 |
| Gallus
gallus | Chicken | 5 | | 5 |
| Ficedula
albicollis | Flycatcher | 5 | | 5 |
| Meleagris
gallopavo | Turkey | 1 | 4 | 5 |
| Taeniopygia
guttata | Zebra finch | 3 | 1 | 4 |
| Reptiles | Anolis
carolinensis | Anole lizard | 2 | | 2 |
| Dasypus
novemcinctus | Armadillo | 3 | | 3 |
| Pelodiscus
sinensis | Chinese softshell
turtle | 2 | 1 | 3 |
| Amphibia | Xenopus
tropicalis | Xenopus | 5 | | 5 |
| Teleost | Latimeria
chalumnae | Coelacanth | 7 | 9 | 16 |
| Xiphophorus
maculatus | Platyfish | 6 | 1 | 7 |
| Oreochromis
niloticus | Tilapia | 6 | 4 | 10 |
| Danio
rerio | Zebrafish | 11 | 1 | 12 |
| Gadus
morhua | Cod | | 6 | 6 |
| Takifugu
rubripes | Fugu | 2 | 3 | 5 |
| Oryzias
latipes | Medaka | 2 | 3 | 5 |
| Gasterosteus
aculeatus | Stickleback | | 6 | 6 |
| Tetraodon
nigroviridis | Tetraodon | | 4 | 4 |
| Total | | 57 | 169 | 69 | 238 |
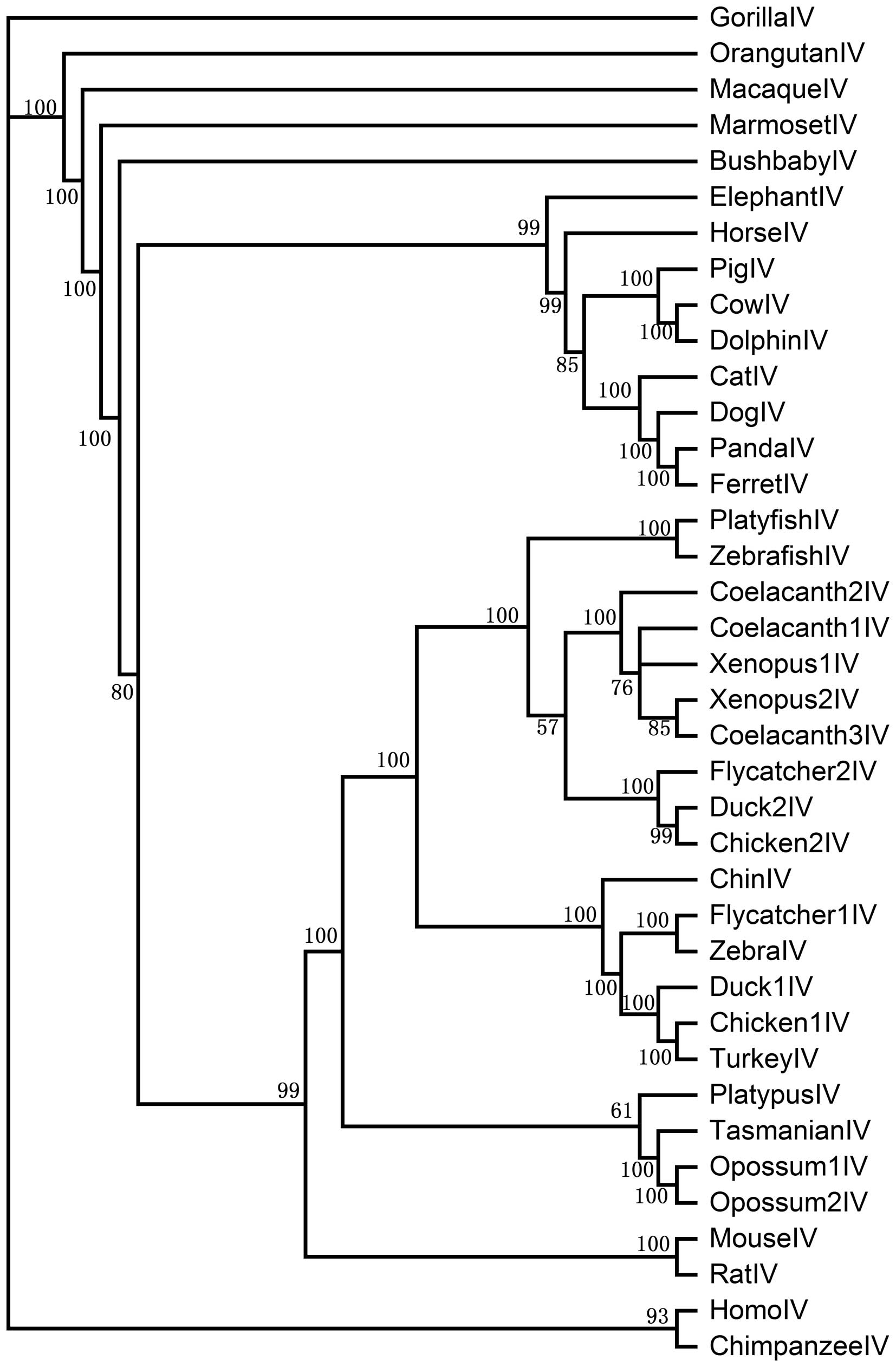
Phylogenetic analysis of the PAR
genes
To examine the evolutionary relationship of the PAR
gene family, the 169 functional genes that were identified from all
238 genes were analyzed by the ML method and Bayesian inference.
These yielded a similar result. The phylogenetic tree constructed
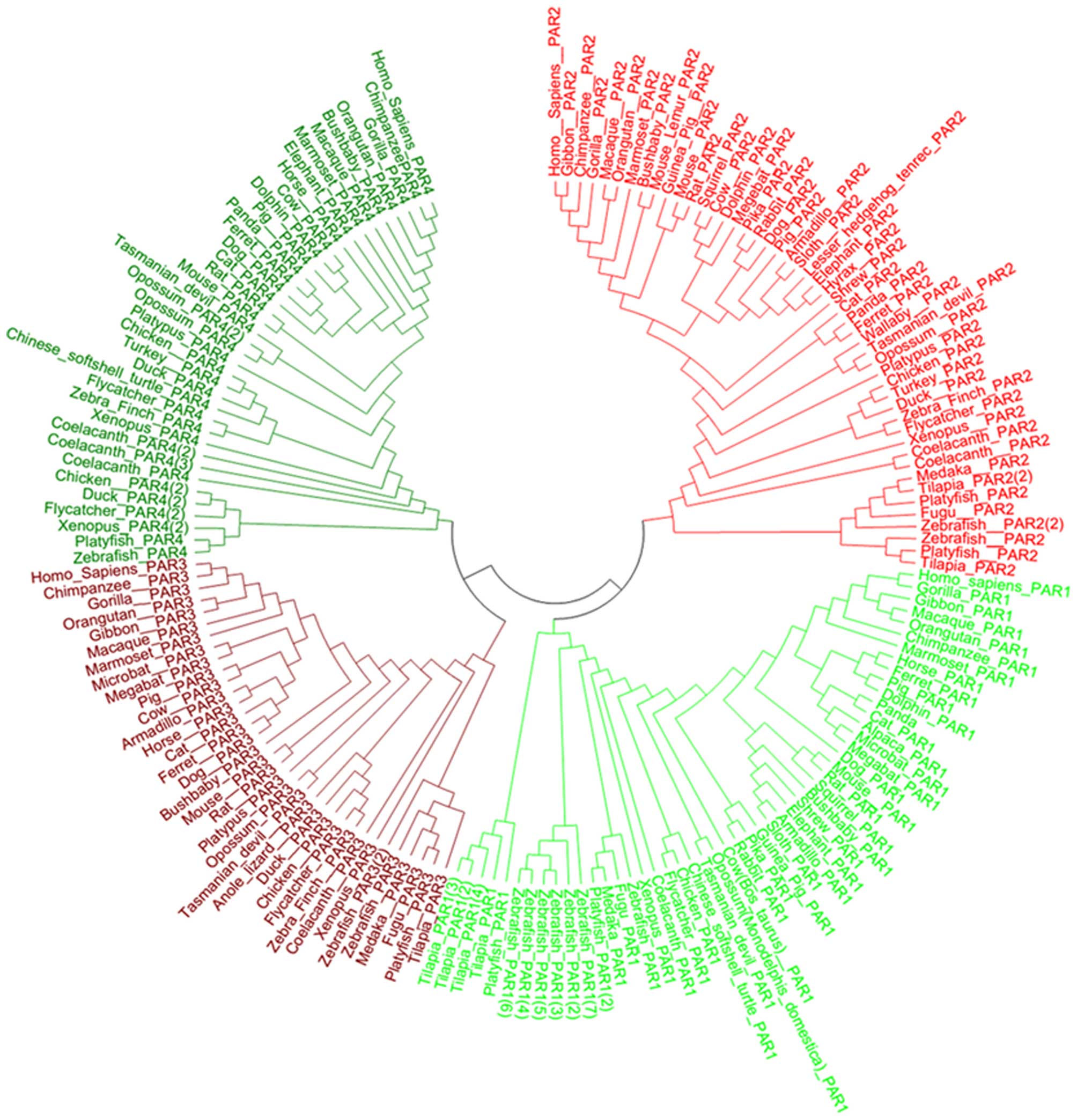
by ML is shown in Fig. 1. Based
on this model, the PAR gene family can be divided into 4 clades,
PAR1, PAR2, PAR3 and PAR4, suggesting that PAR gene sequences have
major differences. To further examine the phylogenetic relationship
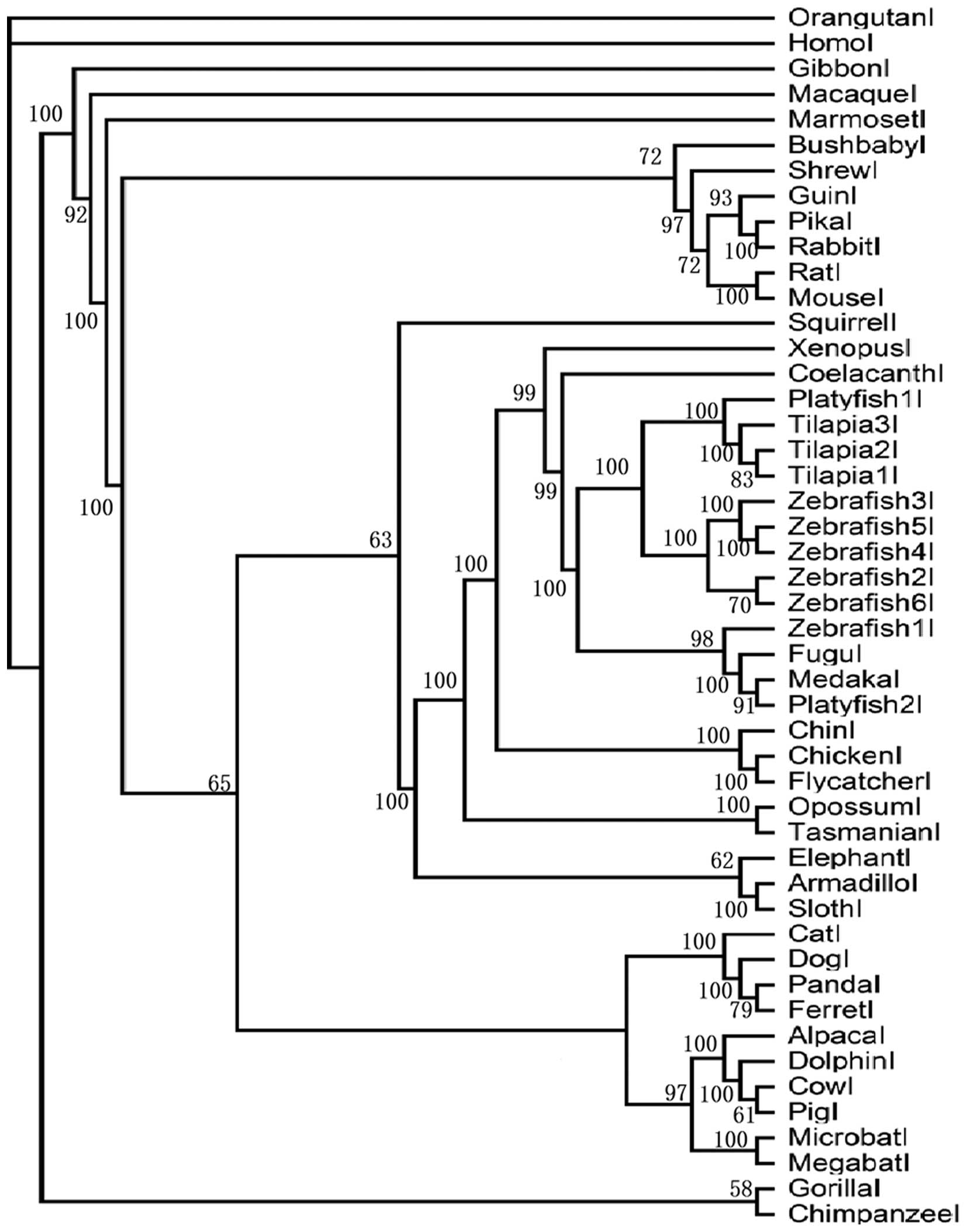
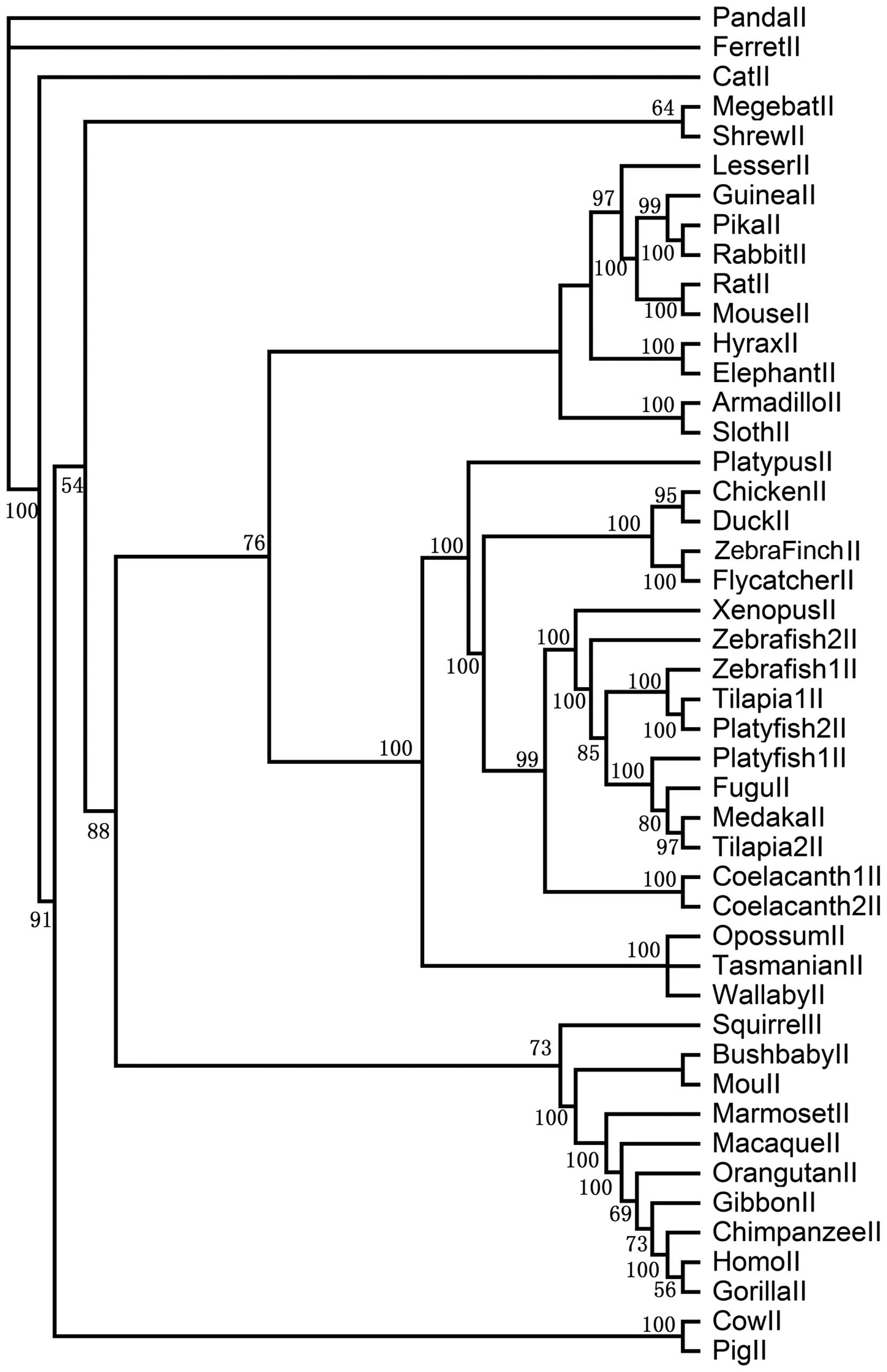
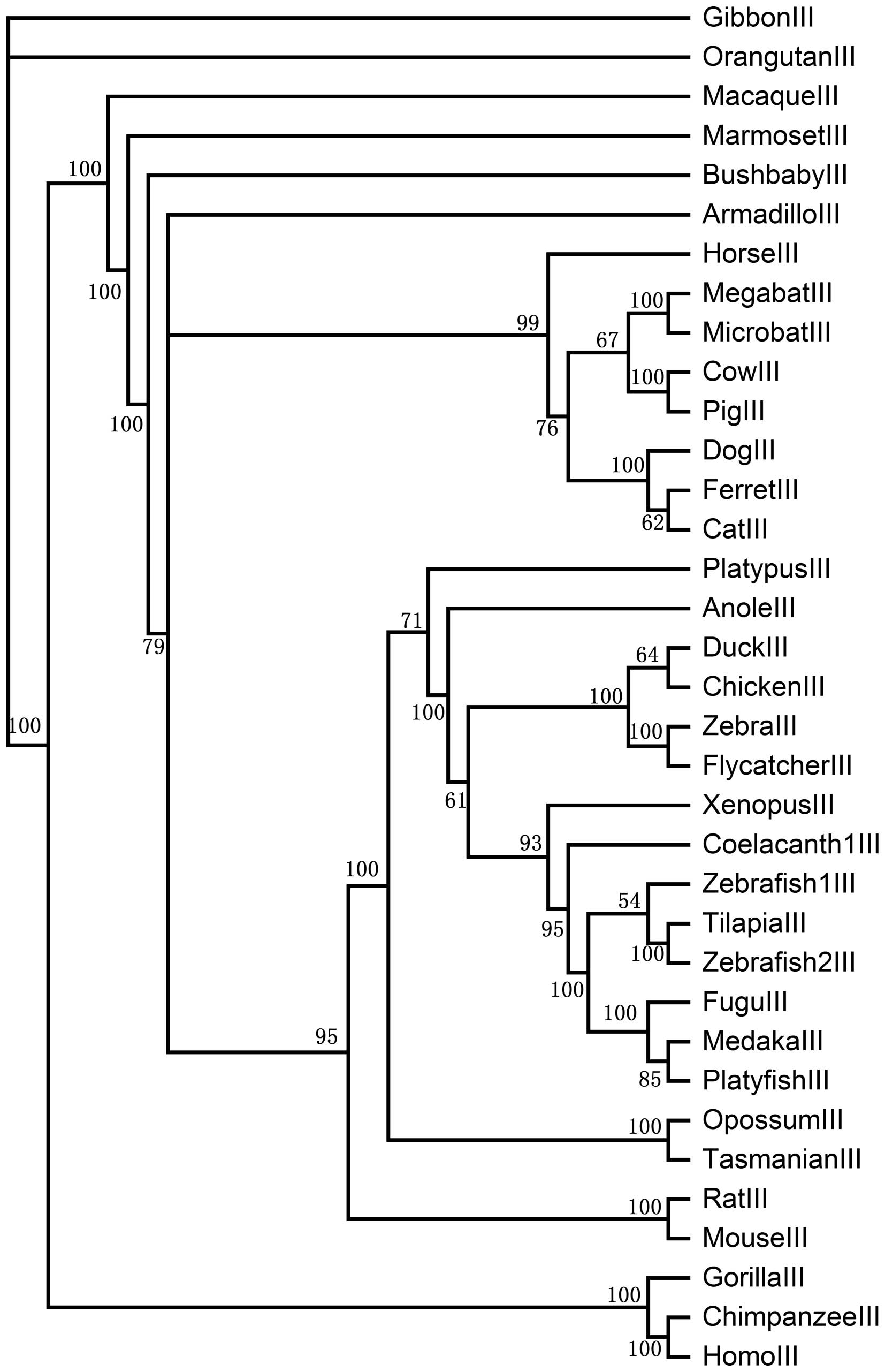
of each clade, we constructed 4 sub-trees for PAR1, PAR2, PAR3 and
PAR4 (Figs. 2Figure 3Figure 4–5) using Bayesian inference. In the PAR1
clade, the posterior probability is >58. Teleost PAR1 genes form
a group separate from eutherian and bird genes. We found that there
is only 1 AR1 copy in eutheria, metatheria, bird, amphibian and
reptile, whereas there are 6 copies in the zebrafish, 3 copies in
the tilapia and 2 copies in the platyfish. In addition, the
posterior probability of the teleost genes is higher than that of
eutherian and bird genes. In the PAR2 clade, the posterior
probability is >54. Primates, birds and teleost PAR2s form a
group separate from other vertebrate genes. There is only 1 PAR2
copy in most vertebrates apart from teleosts (2 copies in
zebrafish, 2 copies in tilapia, 2 copies in platyfish and 2 copies
in coelacanth). In the PAR3 clade, the posterior probability is
>54, with birds and teleosts each forming a group separate from
other species. There is only 1 PAR3 copy in most vertebrates apart
from zebrafish (2 copies). The posterior probability of primates is
100 and the posterior probabilities between megabat and microbat,
cow and pig, zebra finch and flycatcher, tilapia and zebrafish,
opossum and Tasmanian devil, rat and mouse are also 100. In the
PAR4 clade, the posterior probability is >57, with birds and
teleosts again forming a group separate from other species.
Teleosts, amphibians and birds have multiple copies in the PAR4
clade (3 copies in coelacanth, 2 copies in xenopus, 2 copies in
flycatcher and 2 copies in the chicken).
Functional divergence analysis
The 4 PAR clades were analyzed to determine whether
there is functional divergence between them (PAR1 vs. PAR2, PAR1
vs. PAR3, PAR1 vs. PAR4, PAR2 vs. PAR3, PAR2 vs. PAR4 and PAR3 vs.
PAR4). We estimated type I divergence using Diverge v2.0 (Table II). The results revealed that all
θ values were significant (>0), indicating that a site-specific
rate shift during gene duplication was a common phenomenon in the
evolution of the vertebrate PAR family. We also noticed that the 6
θ values of PAR1 vs. PAR2, PAR1 vs. PAR3, PAR1 vs. PAR4, PAR2 vs.
PAR3, PAR2 vs. PAR4 and PAR3 vs. PAR4 were all <0.5, suggesting
that they did not diverge markedly.
 | Table IIFunctional divergence of the PAR
family genes. |
Table II
Functional divergence of the PAR
family genes.
| θ value
| Standard error
|
|---|
| MFE method | MLE method | MFE method |
|---|
| PAR1 vs. PAR2 | 0.397 | 0.364 | 0.056 |
| PAR1 vs. PAR3 | 0.370 | 0.395 | 0.060 |
| PAR1 vs. PAR4 | 0.360 | 0.382 | 0.057 |
| PAR2 vs. PAR3 | 0.433 | 0.494 | 0.061 |
| PAR2 vs. PAR4 | 0.313 | 0.372 | 0.053 |
| PAR3 vs. PAR4 | 0.323 | 0.372 | 0.058 |
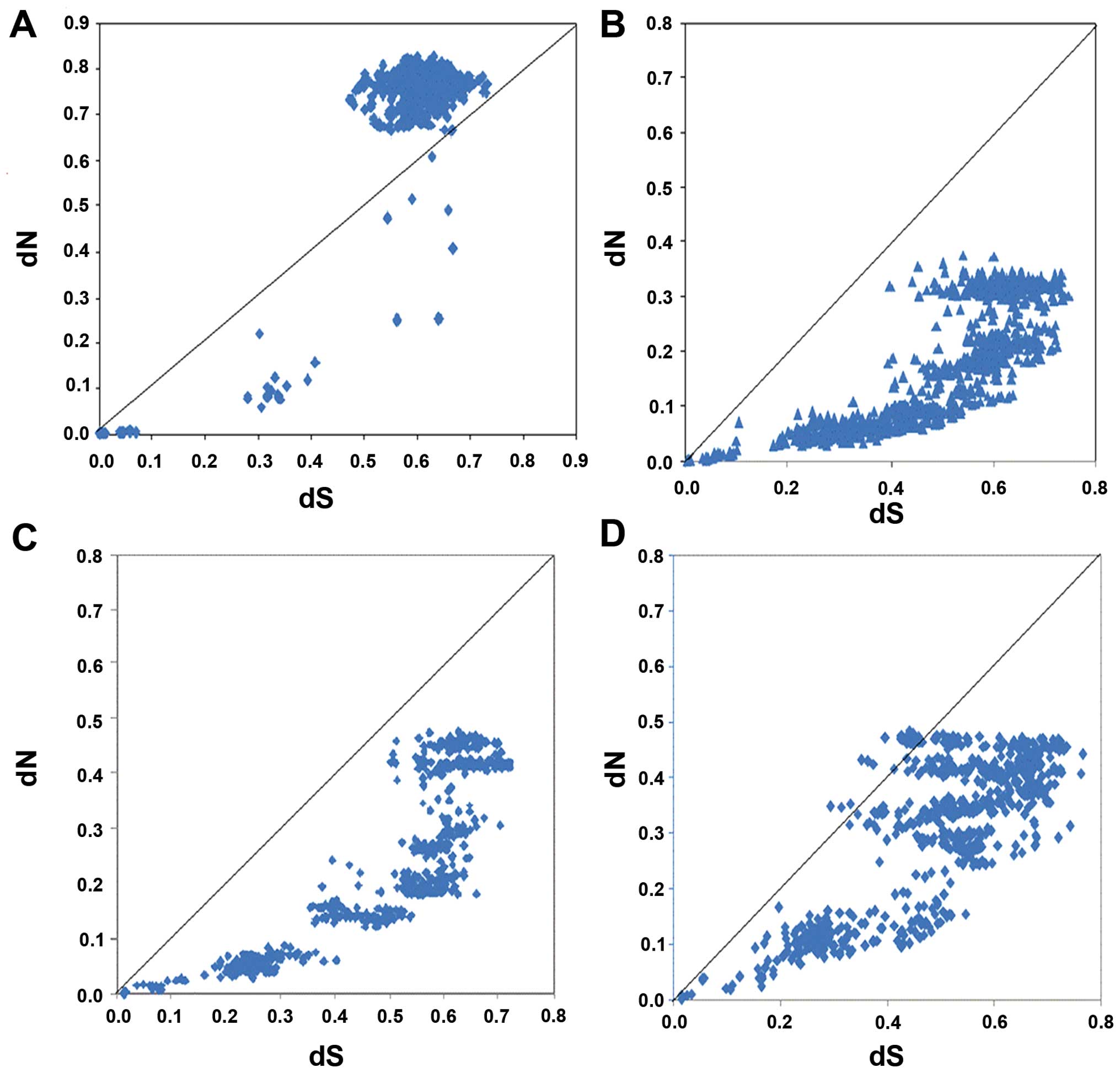
Selective pressure analysis
To determine whether positive selection drove the
evolution of the PAR gene family, we calculated the dN and dS
distances between each pair of the sequences from the 4 clades
(Fig. 6). The distance between dN
and dS is large in the pairwise comparison of the PAR1 sequences,
suggesting that positive selection is involved in PAR1 evolution.
The value of dN is not significantly higher than the value of dS in
the pairwise comparisons of the PAR2, PAR3 and PAR4 sequences. Most
values of dN/dS in these sequences were distributed below the
diagonal, suggesting that positive selection is not involved in the
evolution of these 3 clades. In addition, site-specific tests for
positive selection were performed on the vertebrate PAR gene family
using PAML4.7 (Table III) and
some positive selection sites for PAR1 were found. The amino acids,
65G** and 76K**, were identified as positive
selection sites (referring to Homo sapiens) in the PAR1
gene, whereas there were no positive selection sites found in the
PAR2, PAR3 and PAR4 genes. The positive selection sites identified
by PAML 4.7 were the same as those identified by dN/dS analysis
using MEGA 5.1.
 | Table IIISite-specific tests for positive
selection sites in the PAR gene family. |
Table III
Site-specific tests for positive
selection sites in the PAR gene family.
| Models | InL | Estimates of
parameters | 2ΔI | Positive selection
sites |
|---|
| PAR1 | M7 (β) | −18520.05 | p=0.42638,
q=1.45124 | 22.02
(P<0.01) | NA |
| M8 (β and ω) | −18498.03 | p0=0.95469,
p=0.54737, q=2.73176 (p1=0.04531) w=1.26332 | 22.02
(P<0.01) | 65G**
76K** |
| PAR2 | M7 (β) | −19806.32 | p=0.49569,
q=2.76102 | 0 (P=1.00) | NA |
| M8 (β and ω) | −19806.32 | p0=0.99999,
p=0.49569, q=2.76102 (p1=0.00001) w=2.89180 | 0 (P=1.00) | None |
| PAR3 | M7 (β) | −13970.25 | p=0.70619,
q=3.15181 | 0 (P=1.00) | NA |
| M8 (β and ω) | −13970.25 | p0=0.99999,
p=0.70622 q=3.15206 (p1=0.00001), w=1.00000 | 0 (P=1.00) | None |
| PAR4 | M7 (β) | −16496.19 | p=0.70253,
q=3.17231 | 0 (P=1.00) | NA |
| M8 (β and ω) | −16496.19 | p0=0.99999,
p=0.70253, q=3.17230, (p1=0.00001), w=5.42283 | 0 (P=1.00) | None |
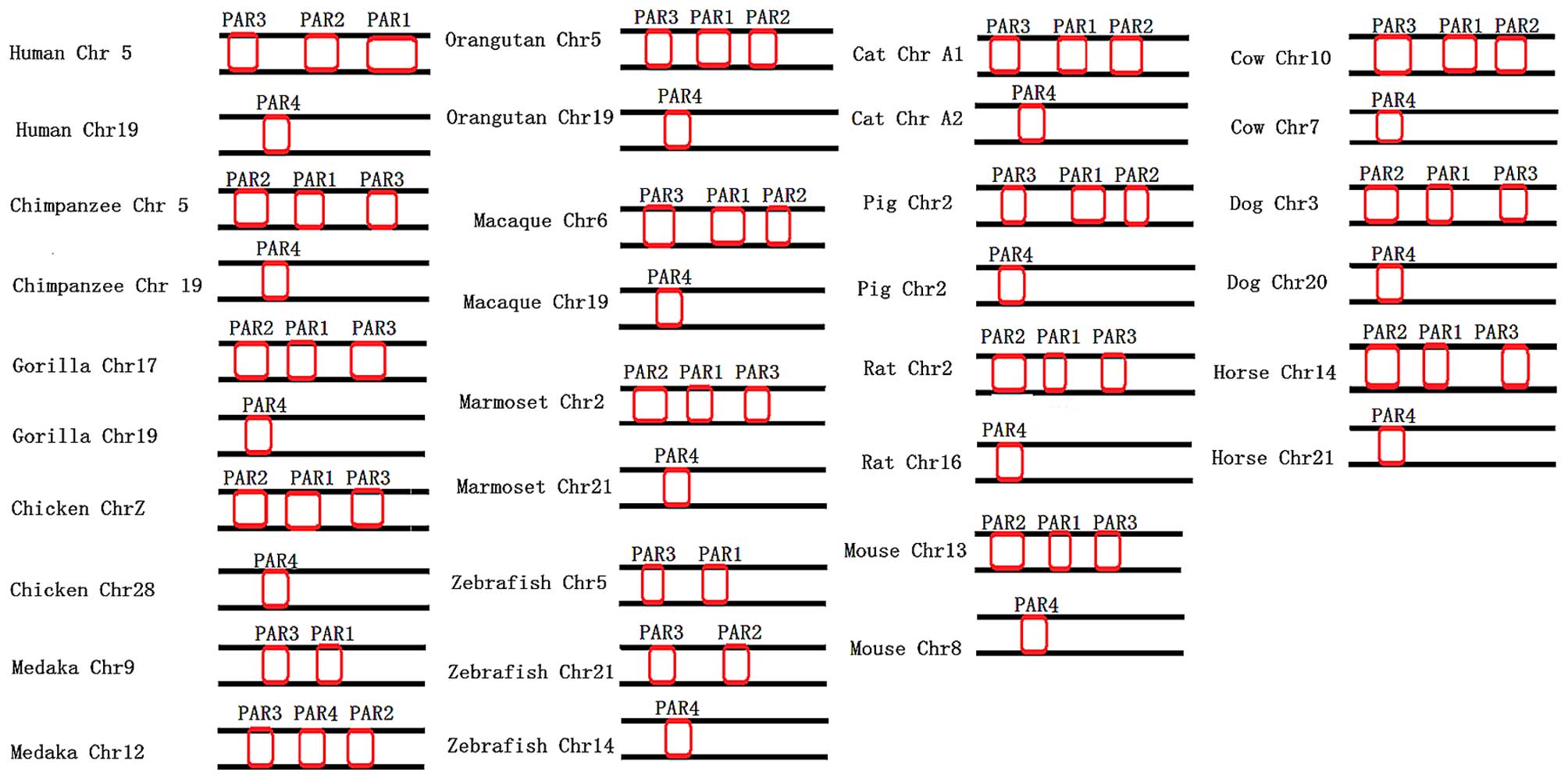
Conservation of synteny in the PAR gene
family
Based on the results shown in Fig. 7, we found that PAR1, PAR2, PAR3
are located on the same chromosome (F2R encoding PAR1, F2RL1
encoding PAR2 and F2RL2 encoding PAR3) and PAR4 is located on a
different chromosome (F2RL3 encoding PAR4). For example, PAR1, PAR2
and PAR3 are located on chromosome 5 and PAR4 on chromosome 19 in
humans, chimpanzees and orangutans. However, in teleosts, the
chromosomal distribution of PARs differs from that observed in
primates and birds. In this case, PAR1, PAR2 and PAR3 are not
located on just 1 chromosome, but are distributed among different
chromosomes. For example, in zebrafish, PAR1 is on chromosome 5,
PAR2 is on chromosome 21 and PAR4 is on chromosome 14, while PAR3
is found on chromosomes 5 and 21. In the medaka, PAR1 is found on
chromosome 9, PAR2 and PAR4 are found on chromosome 12 and PAR3 is
found on chromosomes 9 and 12. Most of the PAR genes have 2
orthologous exons. However, there are exceptions in the PAR genes
of many species (Table IV). The
dog, dolphin, horse and macaque have 3 exons, while the marmoset
and shrew have 4 exons in their PAR1 genes. The squirrel, ferret,
megabat, medaka and lesser hedgehog have 3 exons, while the cat,
wallaby and xenopus have 4 exons in their PAR2 genes. Megabat has
three exons in its PAR3 gene. The bushbaby and gibbon have 1 exon
in their PAR4 genes, while the orangutan has 4 exons and the ferret
has 3 exons in their PAR4 genes.
 | Table IVThe number of exons in the vertebrate
PAR gene family. |
Table IV
The number of exons in the vertebrate
PAR gene family.
| Species | No. of exons
|
|---|
| in PAR1 | in PAR2 | in PAR3 | in PAR4 |
|---|
| Human | 2 | 2 | 2 | 2 |
| Bushbaby | 2 | 2 | 2 | 1 |
| Chimpanzee | 2 | 2 | 2 | 2 |
| Gibbon | 2 | 2 | | 1 |
| Gorilla | 2 | 2 | 2 | 2 |
| Orangutan | | 2 | 2 | 4 |
| Macaque | 3 | 2 | 2 | 2 |
| Marmoset | 4 | 2 | 2 | 2 |
| Mouse lemur | | | 2 | |
| Alpaca | 2 | 2 | | |
| Cow | 2 | 2 | 2 | 2 |
| Cat | 2 | 4 | 2 | |
| Dog | 3 | 2 | | 2 |
| Dolphin | 3 | 2 | | 2 |
| Ferret | 2 | 3 | 2 | 3 |
| Horse | 3 | 1 | 2 | |
| Megabat | 2 | 3 | 3 | |
| Pig | 2 | 2 | 2 | 2 |
| Panda | | 3 | | 2 |
| Shrew | 4 | | | |
| Elephant | | 2 | | |
| Hyrax | | 2 | | |
| Wallaby | | 4 | | |
| Lesser
hedgehog | | 3 | | |
| tenrec | | | | |
| Sloth | 2 | 2 | | |
| Tasmanian
devil | 2 | 2 | 2 | 2 |
| Mouse | 2 | 2 | 2 | 2 |
| Rabbit | | 2 | | |
| Rat | | 2 | 2 | 2 |
| Squirrel | 2 | 3 | | |
| Opossum | 2 | 2 | 2 | 2 |
| Platypus | | 2 | 1 | |
| Chicken | 2 | 2 | 2 | |
| Flycatcher | 2 | | 2 | |
| Chinese
softshell | 2 | 1 | | 2 |
| turtle | | | | |
| Armadillo | 1 | | | |
| Xenopus | | 4 | 2 | |
| Coelacanth | 3 | 2 | | 2 |
| Platyfish | | 2 | | 2 |
| Tilapia | 2 | 2 | 2 | |
| Zebrafish 1 | 2 | 2 | 2 | 2 |
| Zebrafish 2 | 4 | | | |
| Zebrafish 3 | 3 | | | |
| Zebrafish 4 | 4 | | | |
| Zebrafish 5 | 3 | | | |
| Medaka | | 3 | | |
Discussion
The PAR family (PAR1-4) members belong to the GPCR
family and are found in vertebrates. Based on an analysis of a
number of different species, in the present study, we found that
specific-specific gene duplication did not occur in the vertebrate
PAR genes and that the lineage-specific gene expansion of PARs was
observed only in teleosts, indicating that gene duplication
occurred prior to the separation of these vertebrate species and
the teleost cluster evolved following a birth-and-death model
(4,14,29).
The PAR genes were divided into 4 clades (PAR1,
PAR2, PAR3 and PAR4) in the ML tree, suggesting that the PAR genes
originated from 4 ancestors. In the ML tree, PAR1 and PAR2 are
clustered into 1 subfamily and PAR3 and PAR4 are clustered into
another subfamily, which suggested that the genes encoding PAR1 and
PAR2 and the genes encoding PAR3 and PAR4 arose from a gene
duplication event that was relatively recent in the history of the
PAR family. In each of the 4 PAR clades, teleosts, primates and
other mammalian species are separated from the other vertebrates,
forming independent clusters. In the process of evolution, only
PAR1 was found to have arisen from positive selection, while no
evidence for positive selection was found with PAR2, PAR3 and PAR4.
The identified positive selection sites of PAR1 are 65G and 76K (a
signal peptide of approximately 20 amino acids in length is
cleaved). These two amino acids are located on the N-terminus of
PAR1. The thrombin cleavage site on the N-terminus of PAR1 is
located between Arg41 and Ser42, thus, the positive selection sites
are still present on the new N-terminus created following the
activation of PAR1. These positive selection sites may affect the
cleavage of PAR1 or the subsequent binding of the novel N-terminus
with the body of the receptor. The results of positive selection
analysis revealed that the PAR2, PAR3 and PAR4 lineages are highly
conserved in vertebrates, while PAR1 is associated with
environmental adaptation. The way in which PAR1 helps vertebrates
adapt to their environment warrants further investigation. Based on
the conservation of synteny analysis of the PAR gene family, we
found that the PAR4 genes are located on a chromosome distinct from
those of the genes encoding PAR1-3 in all vertebrates apart
fromteleosts, in which the PARs genes are distributed on at least
two chromosomes and PAR4 is not on a chromosome alone. This finding
suggested that chromosomal fusion may have occurred during the
evolution of teleosts. There are 2 exons in most of the vertebrate
PARs, implying that they may be derived from the same ancestor and
that exon rearrangement in PARs may not have occurred during the
evolution of the PAR gene family in vertebrates. Functional
divergences were found to have occurred between the PAR1, PAR2,
PAR3 and PAR4 genes following analysis by the software, Diverge
v2.0, implying that the PARs evolved individually in vertebrates.
Different PARs have various functions.
Studies have demonstrated that PAR1 is involved in
many functions, including the alteration of vascular tone and
permeability, angiogenesis, and smooth muscle cell proliferation
(30–32). As a receptor for serine proteases,
following cleavage at a special site on the N-terminus of PAR2,
activated PAR2 has been found to play a critical role in
inflammation, immunity and angiogenesis (33). It has been demonstrated that PAR3
reduces the platelet response to thrombin, thereby providing
protection from thrombosis, and mediates activated protein C
anti-apoptotic signaling (34).
PAR4 is involved in a number of functions, such as reducing
platelet response to thrombin, providing protection from thrombosis
in the cardiovascular system, regulating colonic nociception and
inhibiting hypersensitivity in the nervous system (35,36). Interaction also exists among the
members of the PAR family and may affect the individual functions
of the PARs. PAR3 can affect the signaling of PAR4 and strengthen
the activation of PAR4. In a study on mouse platelets, PAR3 was
activated at low thrombin concentrations, while PAR4 was activated
at high thrombin concentrations (37). It has been demonstrated that PAR3
serves as a co-factor for the activation of PAR4 at low thrombin
concentrations (38). In contrast
to PAR3 co-factoring with PAR4, PAR1 modulates the activity of PAR2
through a different mechanism. Following cleavage by thrombin, the
N-terminus of PAR1 was found to unmask a tethered ligand domain,
which binds in trans to activate PAR2 through an
intermolecular liganding mechanism that elicits a special signaling
response (39).
In conclusion, in this study, we examined the
evolution of the PAR family by constructing phylogenetic trees,
positive selection analysis, functional divergence analysis and
conservation of synteny analysis. We concluded that the 4 members
of the vertebrate PAR family originated from 4 ancestors and only
PAR1 evolved by positive selection. In addition, in their separate
evolution, there was no significant divergence of the functions of
individual PARs. The evolutionary rates of the 4 PAR members were
consistent. The findings of our study provide a theoretical
background for the evolution of the PAR gene family in
vertebrates.
Acknowledgments
This study was sponsored by the National Natural
Science Foundation of China (nos. 31340073, 81373128, 81273274 and
30972822), the Great Project (no. 2011ZX08011-005) from the Major
Program of National Science and Technology of China, the Special
Research Project (no. 201300000159) from the Science and
Information Technology of Guangzhou. The National Major Scientific
and Technological Special Project for 'Significant New Drugs
Development' (no. 2011ZX09302-003-02), the Jiangsu Province Major
Scientific and Technological Special Project (no. BM2011017) and a
project funded by the Priority Academic Program Development of
Jiangsu Higher Education Institutions.
References
|
1
|
Adams MN, Ramachandran R, Yau MK, Suen JY,
Fairlie DP, Hollenberg MD and Hooper JD: Structure, function and
pathophysiology of protease activated receptors. Pharmacol Ther.
130:248–282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ossovskaya VS and Bunnett NW:
Protease-activated receptors: contribution to physiology and
disease. Physiol Rev. 84:579–621. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vergnolle N, Ferazzini M, D'Andrea MR,
Buddenkotte J and Steinhoff M: Proteinase-activated receptors:
novel signals for peripheral nerves. Trends Neurosci. 26:496–500.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Steinhoff M, Buddenkotte J, Shpacovitch V,
Rattenholl A, Moormann C, Vergnolle N, Luger TA and Hollenberg MD:
Proteinase-activated receptors: transducers of proteinase-mediated
signaling in inflammation and immune response. Endocr Rev. 26:1–43.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hamm HE: How activated receptors couple to
G proteins. Proc Natl Acad Sci USA. 98:4819–4821. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Candia E: Mechanisms of platelet
activation by thrombin: a short history. Thromb Res. 129:250–256.
2012. View Article : Google Scholar
|
|
7
|
Kawabata A: Gastrointestinal functions of
proteinase-activated receptors. Life Sci. 74:247–254. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmidlin F and Bunnett NW:
Protease-activated receptors: how proteases signal to cells. Curr
Opin Pharmacol. 1:575–582. 2001. View Article : Google Scholar
|
|
9
|
Yau MK, Liu L and Fairlie DP: Toward drugs
for protease-activated receptor 2 (PAR2). J Med Chem. 56:7477–7497.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ramachandran R, Noorbakhsh F, Defea K and
Hollenberg MD: Targeting proteinase-activated receptors:
therapeutic potential and challenges. Nat Rev Drug Discov.
11:69–86. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Böhm SK, Grady EF and Bunnett NW:
Regulatory mechanisms that modulate signalling by G-protein-coupled
receptors. Biochem J. 322:1–18. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishihara H, Connolly AJ, Zeng D, Kahn ML,
Zheng YW, Timmons C, Tram T and Coughlin SR: Protease-activated
receptor 3 is a second thrombin receptor in humans. Nature.
386:502–506. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu WF, Andersen H, Whitmore TE, Presnell
SR, Yee DP, Ching A, Gilbert T, Davie EW and Foster DC: Cloning and
characterization of human protease-activated receptor 4. Proc Natl
Acad Sci USA. 95:6642–6646. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gieseler F, Ungefroren H, Settmacher U,
Hollenberg MD and Kaufmann R: Proteinase-activated receptors (PARs)
- focus on receptor-receptor-interactions and their physiological
and pathophysiological impact. Cell Commun Signal. 11:862013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leger AJ, Covic L and Kuliopulos A:
Protease-activated receptors in cardiovascular diseases.
Circulation. 114:1070–1077. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cirino G and Vergnolle N:
Proteinase-activated receptors (PARs): crossroads between innate
immunity and coagulation. Curr Opin Pharmacol. 6:428–434. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vergnolle N: Protease-activated receptors
as drug targets in inflammation and pain. Pharmacol Ther.
123:292–309. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee H and Hamilton JR: Physiology,
pharmacology, and therapeutic potential of protease-activated
receptors in vascular disease. Pharmacol Ther. 134:246–259. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu H, Echemendia N, Chen S and Lin F:
Identification and expression patterns of members of the
protease-activated receptor (PAR) gene family during zebrafish
development. Dev Dyn. 240:278–287. 2011. View Article : Google Scholar
|
|
20
|
Kahn ML, Hammes SR, Botka C and Coughlin
SR: Gene and locus structure and chromosomal localization of the
protease-activated receptor gene family. J Biol Chem.
273:23290–23296. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tamura K, Peterson D, Peterson N, Stecher
G, Nei M and Kumar S: MEGA5: molecular evolutionary genetics
analysis using maximum likelihood, evolutionary distance, and
maximum parsimony methods. Mol Biol Evol. 28:2731–2739. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ronquist F and Huelsenbeck JP: MrBayes 3:
Bayesian phylogenetic inference under mixed models. Bioinformatics.
19:1572–1574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Posada D: jModelTest: Phylogenetic model
averaging. Mol Biol Evol. 25:1253–1256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guindon S and Gascuel O: A simple, fast,
and accurate algorithm to estimate large phylogenies by maximum
likelihood. Syst Biol. 52:696–704. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Z: PAML 4: Phylogenetic analysis by
maximum likelihood. Mol Biol Evol. 24:1586–1591. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang Z, Nielsen R, Goldman N and Pedersen
AM: Codon-substitution models for heterogeneous selection pressure
at amino acid sites. Genetics. 155:431–449. 2000.PubMed/NCBI
|
|
27
|
Gu X and Vander Velden K: DIVERGE:
phylogeny-based analysis for functional-structural divergence of a
protein family. Bioinformatics. 18:500–501. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gu X: Statistical methods for testing
functional divergence after gene duplication. Mol Biol Evol.
16:1664–1674. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Piontkivska H and Nei M: Birth-and-death
evolution in primate MHC class I genes: divergence time estimates.
Mol Biol Evol. 20:601–609. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hirano K: The roles of
proteinase-activated receptors in the vascular physiology and
pathophysiology. Arterioscler Thromb Vasc Biol. 27:27–36. 2007.
View Article : Google Scholar
|
|
31
|
Weiss EJ, Hamilton JR, Lease KE and
Coughlin SR: Protection against thrombosis in mice lacking PAR3.
Blood. 100:3240–3244. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yue R, Li H, Liu H, Li Y, Wei B, Gao G,
Jin Y, Liu T, Wei L, Du J and Pei G: Thrombin receptor regulates
hematopoiesis and endothelial-to-hematopoietic transition. Dev
Cell. 22:1092–1100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo H, Liu D, Gelbard H, Cheng T, Insalaco
R, Fernández JA, Griffin JH and Zlokovic BV: Activated protein C
prevents neuronal apoptosis via protease activated receptors 1 and
3. Neuron. 41:563–572. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Busso N, Chobaz-Péclat V, Hamilton J, Spee
P, Wagtmann N and So A: Essential role of platelet activation via
protease activated receptor 4 in tissue factor-initiated
inflammation. Arthritis Res Ther. 10:R422008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Augé C, Balz-Hara D, Steinhoff M,
Vergnolle N and Cenac N: Protease-activated receptor-4 (PAR 4): a
role as inhibitor of visceral pain and hypersensitivity.
Neurogastroenterol Motil. 21:1189–e107. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Coughlin SR: Thrombin signalling and
protease-activated receptors. Nature. 407:258–264. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin H, Liu AP, Smith TH and Trejo J:
Cofactoring and dimerization of proteinase-activated receptors.
Pharmacol Rev. 65:1198–1213. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rothmeier AS and Ruf W: Protease-activated
receptor 2 signaling in inflammation. Semin Immunopathol.
34:133–149. 2012. View Article : Google Scholar
|
|
39
|
Hirano K and Kanaide H: Role of
protease-activated receptors in the vascular system. J Atheroscler
Thromb. 10:211–225. 2003. View Article : Google Scholar : PubMed/NCBI
|