Introduction
Extensive bone defects continue to pose a major
challenge in orthopedic and trauma patients. Autologous and
allogenous bone grafts obtained from the iliac crest and fibula are
considered to be the gold standard for treating these defects.
However, in cases of multi-fragmentary fractures, pseudarthrosis,
peri-prosthetic fractures, infection and tumor resection, the
resulting defects are large and the volumes of bone available for
grafting are insufficient. Furthermore, bone harvesting is often
accompanied by complications, such as donor site pain, limited bone
availability and extended operative times (1). The combination of mesenchymal stem
cells (MSCs) with an appropriate biomaterial scaffold has led to
the extensive regeneration of bone tissue (2–4).
These findings suggest the possibility of engineering large volumes
of bone tissue.
MSCs are easily isolated from the iliac crest by
fine needle puncture, whereas MSCs located in the marrow of the
femur are only accessible during a surgical intervention, such as
intramedullary nailing that is the treatment of choice for the
stabilization of fractures of long tubular bones. The application
of the novel technique, Reamer/Irrigator/Aspirator (RIA), allows
the harvest of vital bone marrow from the femur by continuous
irrigation and simultaneous aspiration of the irrigation fluid. The
irrigation fluid, as well as the osseous particles within the
irrigation fluid are harvested using a filter. It has been
demonstrated that the reaming debris obtained using the RIA
technique contains elevated levels of fibroblast growth factor
(FGF)-1, platelet-derived growth factor (PDGF), insulin-like growth
factor (IGF)-1, transforming- growth factor (TGF)-β1 and bone
morphogenetic protein (BMP)-1 in comparison with samples obtained
from the iliac crest using the needle puncture/aspiration technique
(5). Moreover, it was also
demonstrated that human reaming debris is a rich source of
multipotent stem cells. The yielded cells exhibit a phenotype and a
plasticity commonly attributed to MSCs in culture (6). However, little is known about the
quantitative and qualitative properties of isolated stem cells. In
our previous study, we found that the MSCs harvested using the RIA
technique demonstrated sustained and higher osteogenic properties
than the MSCs harvested from the iliac crest using the traditional
aspiration technique (7).
Thus, it can be hypothesized that the mechanical
stress of the RIA procedure may cause cellular activation, leading
to an altered differentiation response. Moreover, the high daily
mechanical environment load of the femur may contribute to the
alteration of the methylation pattern of genes that induces the
osteogenic differentiation of the MSCs compared with the iliac
crest. The DNA methylation pattern is inherited by daughter cells
(8); this may explain why
femur-derived MSCs are capable of sustaining osteogenic
capabilities in comparison with MSCs derived from other tissue
sites.
In order to determine the effects of the harvest
procedure, we compared MSCs isolated from the femur harvested using
hte RIA technique with MSCs isolated from the femur harvested by
gentle scraping with a spoon. To determine the influence of the
tissue site, we compared femur-derived MSCs with iliac crest
bone-derived MSCs, both harvested by gentle scraping using a spoon.
We analyzed osteogenic differentiation by means of von Kossa
staining and by determination of the expression of genes involved
in the osteogenic differentiation of MSCs (BMP-2 signaling, WNT-3
pathway and osteogenic marker genes). Moreover, we examined whether
the tissue site-specific gene methylation pattern of MSC is
associated with a specific harvest site as regards osteogenic
differentiation potential and whether putative differences in
osteogenic differentiation between the MSC samples were maintained
over multiple passaging.
Subjects and methods
Ethics
The sampling of bone marrow from the various tissue
sites using either a spoon or the RIA technique was approved and
performed in accordance with the regulations set forth by the
Ethics Committee Goethe University Hospital (Frankfurt am Main,
Germany; project no. 94/10, samples from patients; project no.
329/10, anonymized samples from healthy donors), and in accordance
with German law. Written informed consent was obtained from all
patients and healthy donors prior to obtaining the samples.
Group sizes and demographic
characteristics of the study participants
A total of 13 bone marrow samples were obtained from
the femur (by RIA, from 7 males and 6 females), 7 samples were
obtained from the femur (using a spoon, from 5 males and 2
females), 14 samples were obtained from the iliac crest (using a
spoon, from 7 males and 7 females) and 8 samples were obtained from
the iliac crest (by fine-needle aspiration) were analyzed. The mean
age of the patients did not differ significantly between the groups
[age is presented as median and interquartiles (25% quartile / 75%
quartile)]: RIA [51 (42/54)], femur [48 (31/69)], iliac crest [51
(36/63)]. The gender distribution was also comparable between the
groups and did not differ significantly. The age and gender of the
iliac crest (aspirate) donors were unavailable.
Isolation of MSCs obtained from different
sources
MSCs from the iliac crest were isolated as
previously described (9).
Briefly, bone marrow aspirates were washed once using
phosphate-buffered saline (PBS). The pellet was resuspended in PBS
and layered on a Ficoll density gradient (d=1,077 g/ml, Biochrom,
Berlin, Germany). Following centrifugation (30 min, 1,100 × g), the
mononuclear cells in the interphase were collected and washed twice
using PBS (10 min, 900 × g) containing 2% fetal bovine serum (FBS).
The cells were resuspended in 3 ml MesenCult + Supplements (Cell
Systems, St. Katharinen, Germany) and were counted.
The cell suspension was then divided. One million
mononuclear cells were used for the colony forming unit-fibroblast
(CFU-F) assay and the remaining cells were cultured in order to
obtain MSCs for further analysis (osteogenic differentiation, gene
expression of osteogenic marker genes and methylation analysis).
The mononuclear cells were cultured in MesenCult + Supplements at
37°C in 5% CO2. The medium was changed 3 times each
week.
Femoral reaming debris, particularly the spongy bone
from the iliac crest or the femur, was placed in a sterile Petri
dish. Spongy bone was cut into small sections (2–3 mm) using a
sterile scalpel. The bone marrow was flushed out from the bony
fragments several times with PBS and the collected cells were
pelleted once.
The pellet was resuspended in PBS and filtered
through 100-, 70- and 40-µm mesh (BD Biosciences,
Heidelberg, Germany). Subsequently, the cell suspension was layered
on a Ficoll density gradient. Further processing was performed as
described for the isolation of pelvic MSCs obtained by fine-needle
aspiration.
Spongious bone from the iliac crest was carefully
obtained using a spoon during the bone harvest procedure for
autologous bone transplantation. Spongious bone from the femur was
carefully obtained using a spoon during the surgical treatment of
femur fractures from the defect site. The isolation of MSCs from
the spongious bone for was carried out analogously as described for
MSCs harvested from femoral reaming debris.
When the cells reached 80% confluency, the MSCs were
subdivided in a 1:1 ratio. The cells were rinsed with PBS and
subsequently incubated with Accutase (PAA-Laboratories, Linz,
Austria) for 10 min. The detached MSCs were washed twice in
MesenCult medium and further subcultivated. For the experiments,
the cells from the 1st, 3rd and 6th culture passages were used. A
period of approximately 2–3 weeks was required to obtain a
sufficient number of MSCs to begin the experiments (passage no. 1)
and at least 3 further weeks for cells of passage no. 3, and a
further 6 weeks, respectively for cells of passage no. 6.
CFU-F assay and osteogenic
differentiation
The concentration of the MSCs was assessed by CFU-F
assay. Mononuclear cells were seeded at a density of
1×106 cells per 25 cm2 culture flask and
cultivated for 14 days in MesenCult medium + Supplements at 37°C in
5% CO2 and 100% humidity. The medium was changed thrice
a week. The colonies were stained using Diff-Quick (Medion
Diagnostics, Düdingen, Switzerland) and digitized using an image
analysis system (BioRad, Munich, Germany). The colonies were
subsequently counted.
Osteogenic differentiation was induced as previously
described (9). Briefly, the cells
were seeded at a density of 3×103/cm2 in
MesenCult +10% fetal calf serum (FCS) containing dexamethasone
(1×10−7 M), ascorbic acid-2-phosphate (5×10−5
M) and β-glycerophosphate (1×10−2 M). All substances
were obtained from Sigma-Aldrich (Munich, Germany). The medium was
changed thrice a week. After 14 days, the extent of calcium
deposition was assessed by von Kossa staining. At least 5 random
microscopic fields were selected at a magnification of ×50 for
analysis. Subsequently, the area of stained calcium deposits was
determined using Cell Explorer software (BioSciTec, Frankfurt,
Germany).
Gene expression analysis by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the MSCs using the
RNeasy system (Qiagen, Hilden, Germany) according to the
manufacturer's instructions. The quality and the amount of RNA were
determined using a NanoDrop ND-1000 device (NanoDrop Technologies,
Wilmington, DE, USA). Contaminating genomic DNA was removed by
digestion with the RNase-free DNase-Kit according to the
manufacturer's instructions (Qiagen).
Each 250 ng of RNA was reverse transcribed using the
Affinity script QPCR-cDNA synthesis kit (Stratagene, La Jolla, CA,
USA) according to the manufacturer's instructions. Subsequently,
SYBR-Green-based quantitative (real-time) (qPCR) was performed on a
Stratagene Mx3005P qPCR system (Stratagene). PCR was performed
using target-specific primers for alkaline phosphatase,
liver/bone/kidney (ALPL, NM_000478; catalog number, PAHS-026),
catenin beta 1 (CTNNB1, NM_001904; catalog number, PPH00643F),
BMP-2 (NM_00120; catalog number, PPH00549C), collagen, type I,
alpha 1 (COL1A, NM_000088; catalog number, PPH01299F), dickkopf-1
(DKK1, NM_012242; catalog number, PPH01752C), low-density
lipoprotein receptor-related protein 5 (LRP5, NM_002335; catalog
number, PPH02315F), osteocalcin [also known as bone
gamma-carboxyglutamate protein (BGLAP), NM 199173; catalog number,
PPH01898A], runt-related transcription factor 2 (RUNX2, NM_004348;
catalog number, PPH01897C), SMAD-5 (NM_005903; catalog number,
PPH01940C) and Wnt-3a (WNT3A, NM_033131; catalog number, PPH02772B)
(all purchased from Qiagen). The expression of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH, NM_002046.3,
catalog number, PPH00150E) was measured as a reference gene. The
PCR cycling conditions were as follows: initial denaturation at
95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and
finally a melting curve at 60°C for 1 min. The relative
quantification of the mRNA levels of the target genes was
determined using the comparative (CT; threshold cycle values)
method (2−ΔCT method). The results are presented as the
fold change relative to GAPDH gene expression.
Gene methylation analysis
Total DNA was extracted from 5×105 MSCs
using the QIAamp DNA Mini kit (Qiagen) according to the
manufacturer's instructions. The DNA concentration and purity were
measured using a NanoDrop device (NanoDrop Instruments/Thermo
Scientific, Schwerte, Germany). EpiTect Methyl qPCR assays (Qiagen)
were perfomed to determine the methylation status of ALP (ALPL),
catenin beta 1 (CTNNB1), BMP-2 (BMP2), collagen-1a (COL1A),
dickkopf-1 (DKK1), LRP5 (LRP5), osteocalcin (BGLAP), runx-2
(RUNX2), smad-5 (SMAD5) and Wnt3a (WNT3A). The assay procedures
were performed according to the manufacturer's instructions.
Briefly, DNA (250 ng/µl) was distributed to 4 reaction tubes
containing either methylation-sensitive restriction enzymes,
methylation-resistant enzymes, or a mixture of both
methylation-sensitive and methylation-resistant enzymes and
incubated at 37°C overnight in a heating block. Following the heat
inactivation of the enzymes (65°C, 20 min), the remaining DNA was
amplified using SYBR-Green-based primer assays for the detection of
the above-mentioned genes. The Ct values of each digest were
compared with mock digest and the relative amounts of
hypermethylated, intermediate methylated and unmethylated DNA were
calculated using an Excel sheet provided by the manufacturer
(Qiagen).
Statistical analysis
The Kruskal-Wallis test followed by Bonferroni-Holm
correction for multiplicity were used for group comparisons and
comparisons between the different culture passages. A
Bonferroni-adjusted Fisher's Exact test was used to calculate
significance in gender distribution between the groups. The
Spearman-Rank correlation was applied for correlational analysis. A
p-value <0.05 was considered to indicate a statistically
significant difference.
Results
Cell numbers and MSC concentrations
The median number of mononuclear cells per ml sample
differed significantly between the cells obtained from the femoral
reaming debris (6.8×106 cells), iliac crest spongiosa
(3.3×106 cells) and iliac crest aspirates
(1.4×106 cells) compared to the cells obtained from the
femur spongiosa using a spoon (5.0×105 cells).
The concentration of MSCs per 1.0×106
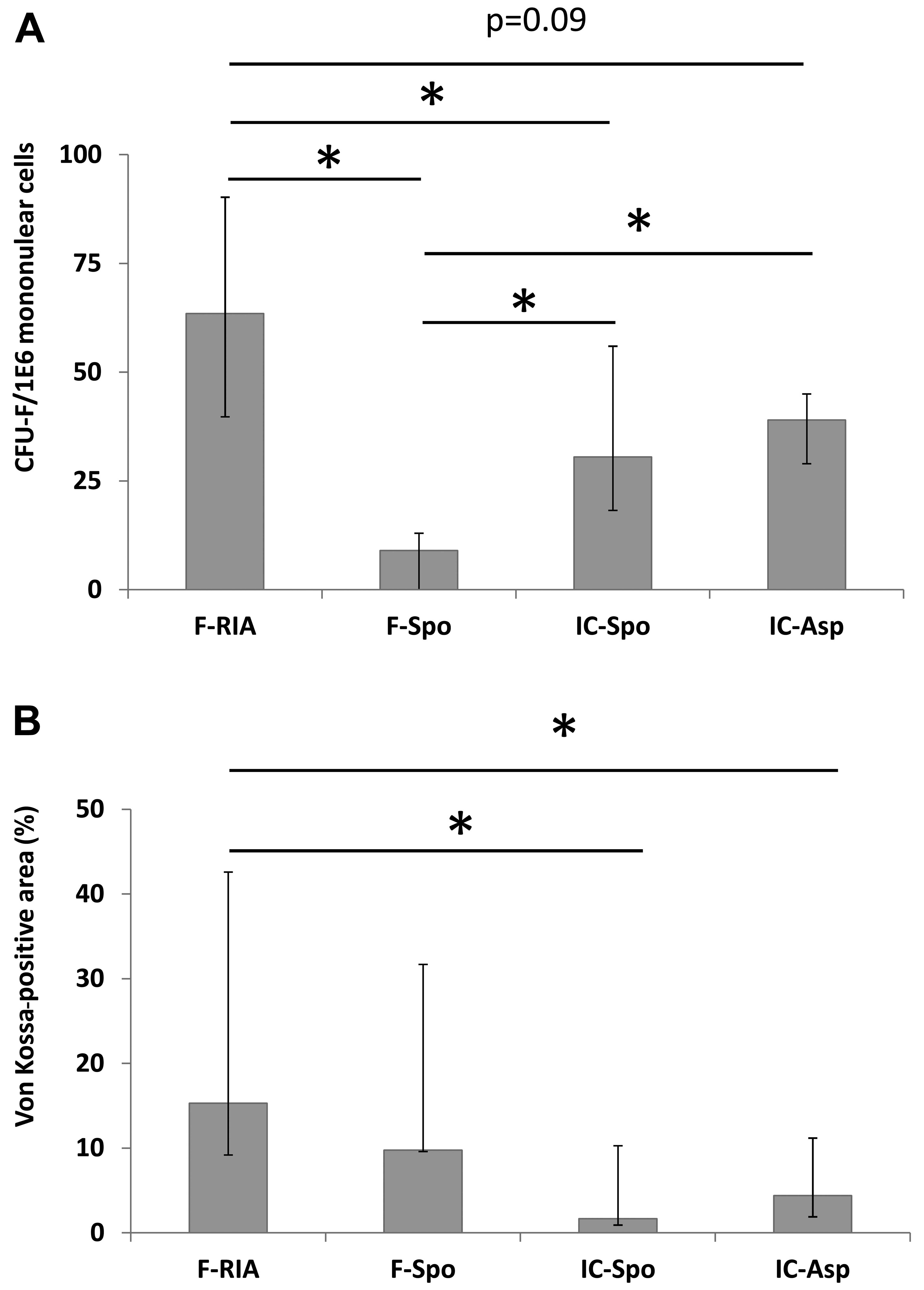
mononuclear cells was measured by CFU-F assay. We observed a higher
CFU-F number in the cells obtained from the femoral reaming debris
(63.5), iliac crest aspirate (39.0) and spongy iliac crest bone
(30.0) in comparison with those from the spongy bone from the femur
(9.0, p<0.05). Furthermore, the CFU-F-number in the cells
obtained from the femoral reaming debris was higher than that in
the cells from the spongy bone from the iliac crest and the iliac
crest aspirates (trend, p<0.09) (Fig. 1A).
Osteogenic differentiation is dependent
on the tissue site and isolation procedure
The MSCs of the first culture passage were analyzed.
The MSCs obtained using the RIA technique exhibited a higher
calcium deposition in comparison with the MSCs isolated from the
iliac crest (by fine-needle aspiration) and the MSCs obtained from
the iliac crest (using a spoon) (p<0.05). No significant
differences were observed between the MSCs derived from the femur
(using a spoon), the iliac crest (using a spoon) and the iliac
crest aspirates (Fig. 1B).
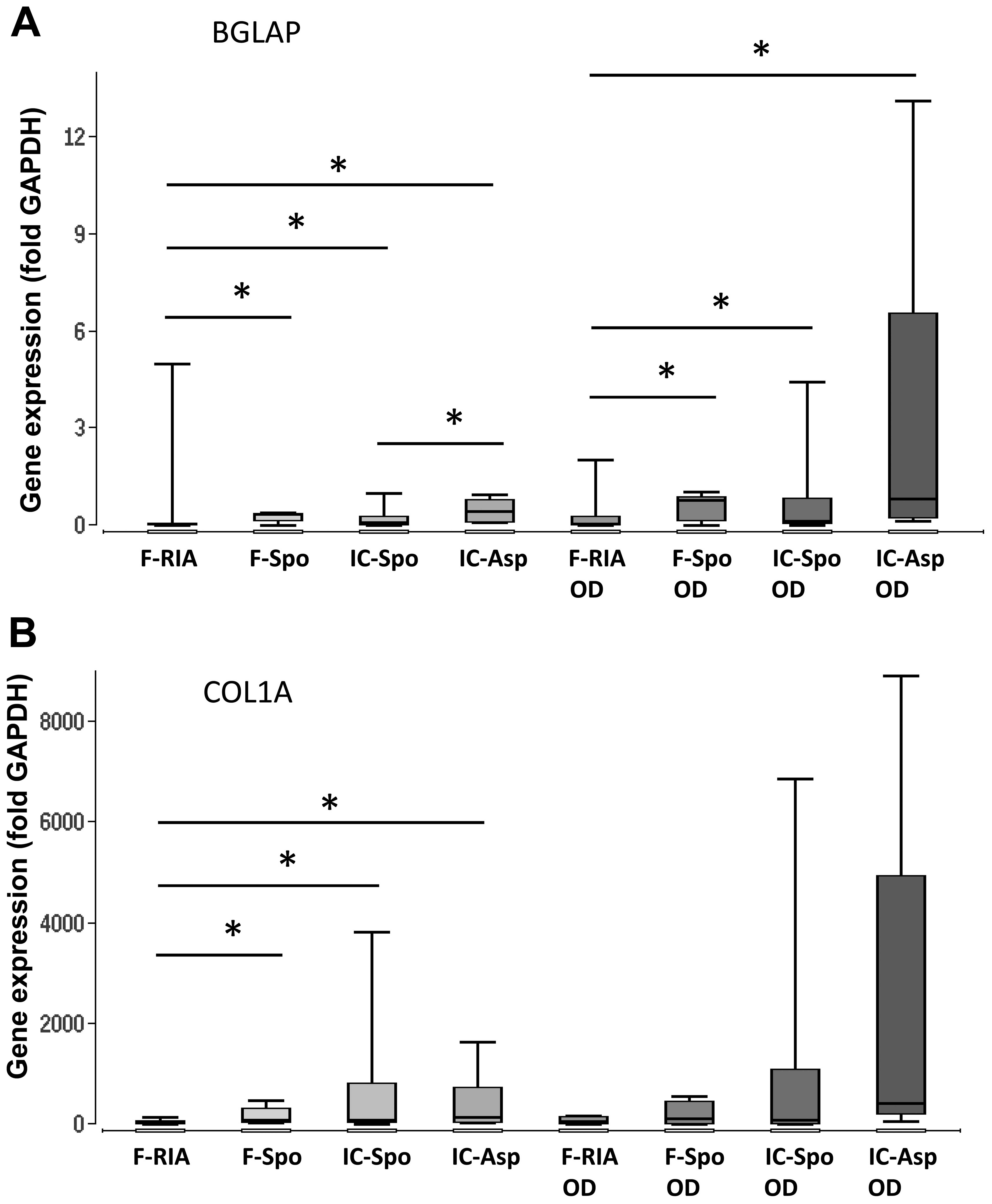
Additionally, the expression of the osteogenic marker genes, BGLAP,
COL1A and ALPL, was analyzed. The overall expression of BGLAP and
COL1A was highest in the MSCs obtained from the iliac crest
aspirates both in the controls and following incubation with
osteogenic differentiation medium. The gene expression in the MSCs
isolated using a spoon from the iliac crest and the femur did not
differ significantly. However, the gene expression in the MSCs
obtained from the femur (using a spoon) was consistently higher in
comparison with that in the MSCs obtained using the RIA technique.
The gene expression of ALPL did not differ significantly between
the groups. Osteogenic differentiation over a period of 14 days did
not alter the gene expression levels significantly in comparison
with those in the MSCs under control conditions (Fig. 2 and Table I).
 | Table IExpression of genes involved in the
osteogenic differentiation of mesenchymal stem cells (MSCs)
cultured under (A) control (A) and (B) osteogenic-inducing
conditions, as well as (C) intergroup comparisons according to the
harvest site and harvest technique.a |
Table I
Expression of genes involved in the
osteogenic differentiation of mesenchymal stem cells (MSCs)
cultured under (A) control (A) and (B) osteogenic-inducing
conditions, as well as (C) intergroup comparisons according to the
harvest site and harvest technique.a
| A, Control
conditions |
|---|
|
|---|
| Gene | Femur (RIA) | Femur (spoon) | Iliac crest
(spoon) | Iliac crest
(aspirates) |
|---|
| BMP2 | | | p=0.05 vs. RIA | p=0.009 vs.
RIA |
| SMAD5 | | | | p=0.02 vs. RIA |
| RUNX2 | | | p=0.01 vs. RIA | p=0.004 vs.
RIA |
| WNT3A | | | | p=0.04 vs. RIA |
| LRP5 | | | | p=0.03 vs. RIA |
| | | | p=0.05 vs. iliac
crest (spoon) |
| DKK1 | | | | p=0.04 vs. RIA |
| CTNNB1 | | | | p=0.04 vs. RIA |
| BGLAP | | p=0.05 vs. RIA | p=0.04 vs. RIA | p=0.006 vs.
RIA |
| | | | p=0.02 vs. iliac
crest (spoon) |
| COL1A | | p=0.02 vs. RIA | p=0.02 vs. RIA | p=0.002 vs.
RIA |
| ALPL | | | | |
| B,
Osteogenic-inducing conditions |
|---|
|
|---|
| Gene | Femur (RIA) | Femur (spoon) | Iliac crest
(spoon) | Iliac crest
(aspirates) |
|---|
| BMP2 | | p=0.03 vs. RIA | p=0.004 vs.
RIA | p=0.001 vs.
RIA |
| SMAD5 | | p=0.05 vs. RIA | | p=0.02 vs. RIA |
| RUNX2 | | | | p=0.02 vs. RIA |
| WNT3A | | | | |
| LRP5 | | | | p=0.03 vs. RIA |
| DKK1 | | | | |
| CTNNB1 | | | | |
| BGLAP | | p=0.02 vs. RIA | p=0.04 vs. RIA | p=0.002 vs.
RIA |
| COL1A | | | | |
| ALPL | | | | |
| C, Comparison
between control conditions (cont) and osteogenic-inducing
conditions (ost. diff) |
|---|
|
|---|
| Gene | Femur (RIA) cont.
vs. ost. diff. | Femur (spoon) cont.
vs. ost. diff. | Iliac crest (spoon)
cont. vs. ost. diff. | Iliac crest (asp.)
cont. vs. ost. diff. |
|---|
| BMP2 | 0.21, n.s. | 0.46, n.s. | 0.71, n.s. | 0.66, n.s. |
| SMAD5 | 0.48, n.s. | 0.81, n.s. | 0.83, n.s. | 0.46, n.s. |
| RUNX2 | 0.91, n.s. | 0.91, n.s. | 0.36, n.s. | 0.83, n.s. |
| WNT3A | 0.73, n.s. | 0.51, n.s. | 0.20, n.s. | 0.02,
significant |
| LRP5 | 0.43, n.s. | 0.77, n.s. | 0.58, n.s. | 0.43, n.s. |
| DKK1 | 0.53, n.s. | 0.75, n.s. | 0.11, n.s. | 0.19, n.s. |
| CTNNB1 | 0.68, n.s. | 0.24, n.s. | 0.81, n.s. | 0.13, n.s. |
| BGLAP | 0.47, n.s. | 0.34, n.s. | 0.22, n.s. | 0.24, n.s. |
| COL1A | 0.26, n.s. | 0.91, n.s. | 0.85, n.s. | 0.12, n.s. |
| ALPL | 0.51, n.s. | 0.65, n.s. | 0.16, n.s. | 0.41, n.s. |
Gene expression of BMP-2 and Wnt
signaling components
One of the main objectives of the present
observational study was to determine the expression of genes
involved in osteogenic differentiation pathways, depending on the
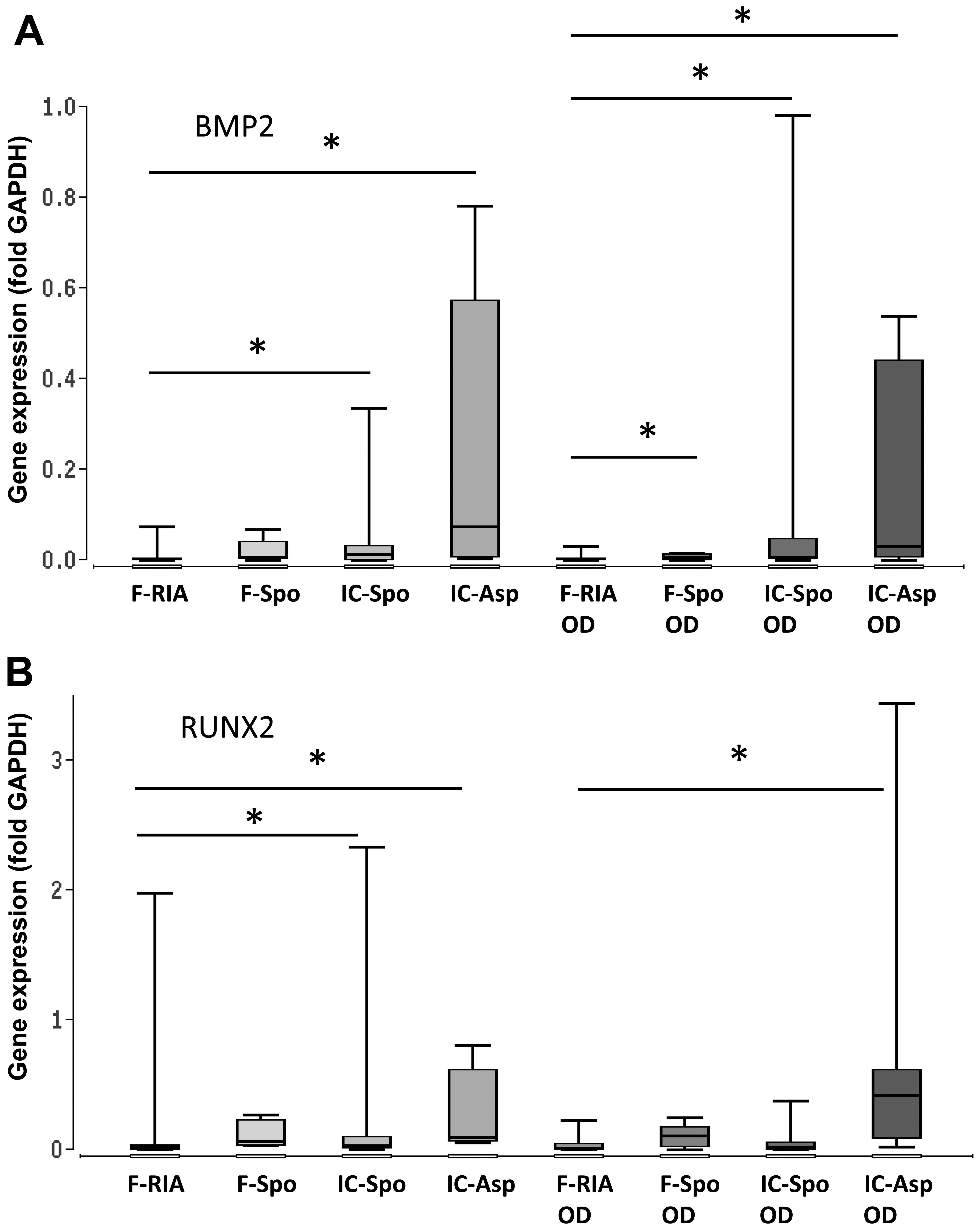
tissue site and the isolation procedure being applied. The highest
expression of BMP2, SMAD5 and RUNX2 was observed in the iliac
crest-derived MSCs (by fine-needle aspiration) and the lowest
expression in the femur-derived MSCs (by RIA). The differences in
gene expression between both groups were statistically significant.
Gene expression in the femur- derived MSCs (using a spoon) was also
significantly increased in comparison with that in the
femur-derived MSCs (by RIA) (Fig.
3 and Table I).
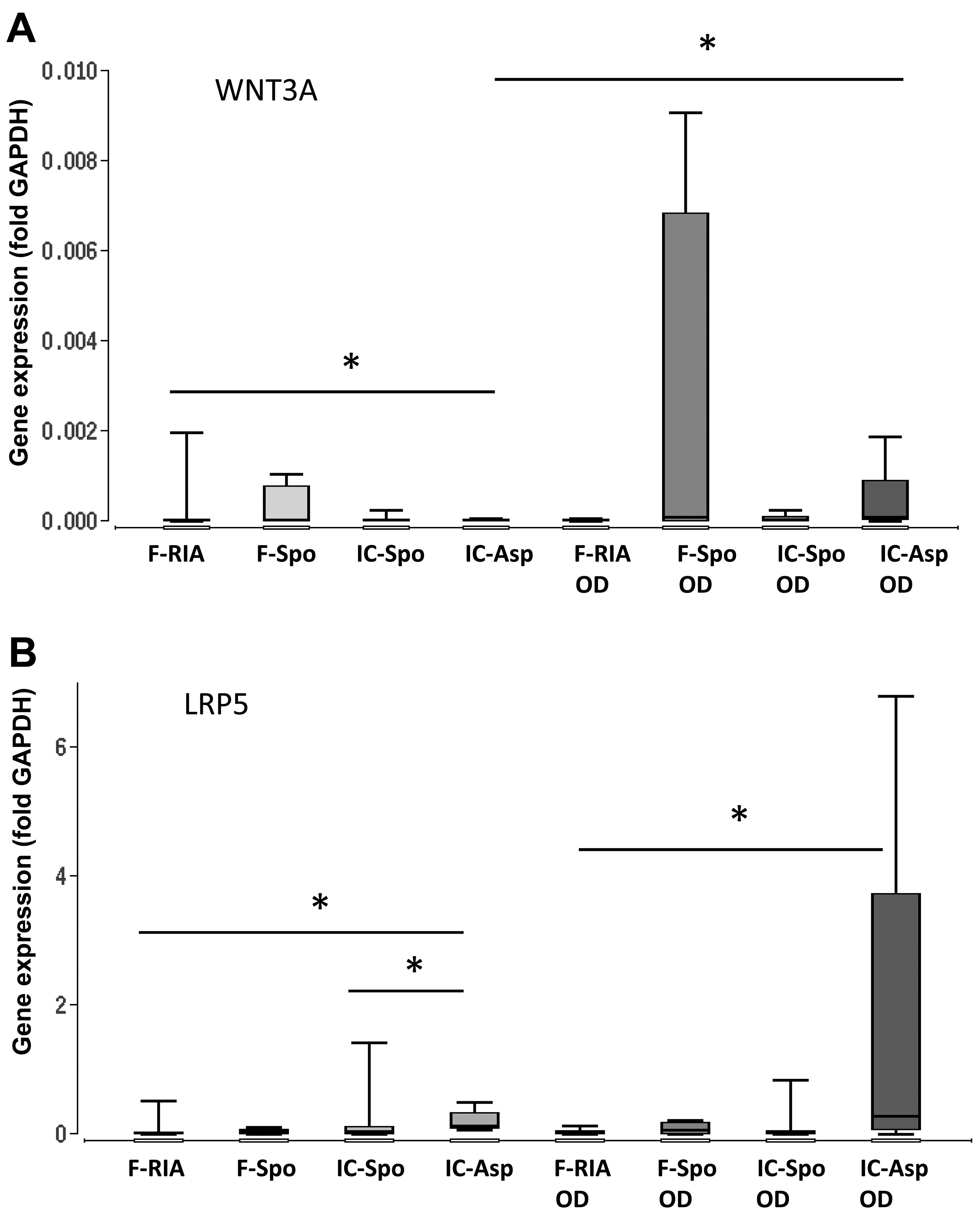
Furthermore, we analyzed the gene expression of
WNT3A, LRP5, DKK1 and CTNNB1, the key components of the canonical
Wnt signaling pathway. Our results demonstrated a consistent and
significantly higher expression of the iliac crest-derived MSCs (by
fine-needle aspiration) in comparison with the femur-derived MSCs
(by RIA). No significant differences were observed between the MSCs
obtained from the iliac crest (using a spoon) and those obtained
from the femur (using a spoon) (Fig.
4 and Table I).
Heterogeneous gene methylation
We analyzed the percentage of intermediate
methylated and hypermethylated DNA sequences from the broad
selection of our target genes in the MSCs at the 1st culture
passage. The analysis revealed a heterogeneous methylation of
osteogenic genes between the groups. The iliac crest-derived MSCs
(by fine-needle aspiration) exhibited a decreased methylation of
the RUNX2 gene in comparison with the femur-derived MSCs (using the
RIA technique and using a spoon) and a decreased methylation of the
SMAD5 gene in comparison with the iliac crest-derived MSCs (using a
spoon). The WNT3A gene in the iliac crest-derived MSCs (by
fine-neelde aspiration) exhibited an increased methylation in
comparison with the femur-derived MSCs (by RIA) (Table II). No correlation was observed
between the gene methylation and gene expression of the respective
genes in either the control MSCs or in the osteogenically
differentiated MSCs (data not shown).
 | Table IIDegree of intermediate DNA
methylation (%) in mesenchymal stem cells (MSCs) obtained from
either the iliac crest or the femur using different harvest
techniques.a |
Table II
Degree of intermediate DNA
methylation (%) in mesenchymal stem cells (MSCs) obtained from
either the iliac crest or the femur using different harvest
techniques.a
| A, Gene
methylation |
|---|
|
|---|
| Gene | Femur (RIA) | Femur (spoon) | Iliac crest
(spoon) | Iliac crest
(aspirates) |
|---|
| COL1A | M: 0.0 | M: 0.0 | M: 25.0 | M: 0.0 |
| LQ: 0.0 | LQ: 0.0 | LQ: 0.0 | LQ: 0.0 |
| UQ: 30.1 | UQ: 17.6 | UQ: 34.1 | UQ: 01 |
| CTNNB1 | M: 0.0 | M: 5.5 | M: 9.6 | M: 5.3 |
| LQ: 0.0 | LQ: 0.0 | LQ: 0.0 | LQ: 0.0 |
| UQ: 80.5 | UQ: 77.7 | UQ: 38.2 | UQ: 14.9 |
| RUNX2 | M: 11.4 | M: 40.6 | M: 0.0 | M: 0.0 |
| LQ: 0.0 | LQ: 0.0 | LQ: 0.0 | LQ: 0.0 |
| UQ: 40.3 | UQ: 99.8 | UQ: 6.6 | UQ: 2.6 |
| SMAD5 | M: 6.5 | M: 48.6 | M: 82.6 | M: 1.6 |
| LQ: 0.0 | LQ: 0.0 | LQ: 31.3 | LQ: 0.0 |
| UQ: 100.0 | UQ: 100.0 | UQ: 100.0 | UQ: 43.5 |
| WNT3A | M: 0.0 | M: 0.0 | M: 0.0 | M: 8.6 |
| LQ: 0.0 | LQ: 0.0 | LQ: 0.0 | LQ: 0.0 |
| UQ: 0.0 | UQ: 54.4 | UQ: 0.0 | UQ: 11.7 |
| B, Statistical
evaluation (intergroup comparison between different MSC
sources) |
|---|
|
|---|
| Gene | Femur (RIA) | Femur (spoon) | Iliac crest
(spoon) | Iliac crest
(aspirates) |
|---|
| COL1A | | | | |
| CTNNB1 | | | | |
| RUNX2 | p(nc)=0.08 vs.
aspirates | p(nc)=0.05 vs.
aspirates | | |
| | p(nc)=0.08 vs.
iliac | | |
| | crest (spoon) | | |
| SMAD5 | | | p=0.05 (nc) vs.
aspirates | |
| WNT3A | | | | p(nc)=0.05 vs.
RIA |
The percentage of hypermethylated DNA sequences was
generally lower compared with some intermediate methylation. A
significantly increased hypermethylation of the COL1A gene was
observed in the MSCs obtained from the femur (using the RIA
technique and using a spoon), as well as in the MSCs from the iliac
crest (using a spoon) in comparison with the MSCs obtained from the
iliac crest by fine-needle aspiration. The increased
hypermethylation of the WNT3A gene was observed in the MSCs
obtained from the femur by RIA in comparison with the MSCs obtained
from the iliac crest by fine-needle aspiration (Table III).
 | Table IIIPercentage of hypermethylated DNA (%)
in mesenchymal stem cells (MSCs) obtained either from the iliac
crest or from the femur using different harvest
techniques.a |
Table III
Percentage of hypermethylated DNA (%)
in mesenchymal stem cells (MSCs) obtained either from the iliac
crest or from the femur using different harvest
techniques.a
| A, Gene
methylation |
|---|
|
|---|
| Gene | Femur (RIA) | Femur (spoon) | Iliac crest
(spoon) | Iliac crest
(aspirate) |
|---|
| COL1A | M: 0.6 | M: 1.0 | M: 2.0 | M: 0.1 |
| LQ: 0.5 | LQ: 0.7 | LQ: 0.6 | LQ: 0.1 |
| UQ: 2.5 | UQ: 8.5 | UQ: 2.9 | UQ: 0.7 |
| CTNNB1 | M: 0.2 | M: 0.1 | M: 2.6 | M: 0.2 |
| LQ: 0.0 | LQ: 0.0 | LQ: 0.9 | LQ: 0.1 |
| UQ: 1.2 | UQ: 0.3 | UQ: 10.9 | UQ: 0.4 |
| RUNX2 | M: 2.1 | M: 1.8 | M: 4.3 | M: 0.6 |
| LQ: 0.4 | LQ: 0.4 | LQ: 2.5 | LQ: 0.3 |
| UQ: 30.4 | UQ: 14.0 | UQ: 19.5 | UQ: 2.2 |
| SMAD5 | M: 7.9 | M: 0.4 | M: 2.2 | M: 0.3 |
| LQ: 0.0 | LQ: 0.0 | LQ: 0.0 | LQ: 0.2 |
| UQ: 26.7 | UQ: 40.8 | UQ: 17.0 | UQ: 3.6 |
| WNT3A | M: 4.9 | M: 1.7 | M: 2.4 | M: 0.4 |
| LQ: 1.1 | LQ: 0.1 | LQ: 0.2 | LQ: 0.2 |
| UQ: 82.7 | UQ: 25.3 | UQ: 5.4 | UQ: 0.5 |
| B, Statistical
evaluation (intergroup comparison between MSCs obtained from
different sources) |
|---|
|
|---|
| Gene | Femur (RIA) | Femur (spoon) | Iliac crest
(spoon) | Iliac crest
(aspirates) |
|---|
| COL1A | p=0.1 vs.
iliac | p=0.05 vs.
iliac | p=0.05 vs.
iliac | |
| crest
aspirates | crest
aspirates | crest
aspirates | |
| CTNNB1 | | | | |
| RUNX2 | | | p=0.08 vs.
iliac | |
| | | crest
aspirates | |
| SMAD5 | | | | |
| WNT3A | p=0.04 vs.
iliac | | | |
| crest
aspirates | | | |
Negative correlations between the degree of RUNX2
hypermethylation and the gene expression of the RUNX2-regulated
genes, LRP5 (rho = −0.42, p=0.03), collagen-1α (rho = −0.37,
p=0.04) and smad5 (rho = −0.36, p=0.05) were recorded. The
expression of the RUNX2 gene correlated in trend with the
hypermethylation of the RUNX2 gene (rho = −0.29, p=0.13) whereas no
correlations were observed between RUNX2-independent gene
expression (WNT3A, CTNNB1, DKK) and RUNX2 gene
hypermethylation.
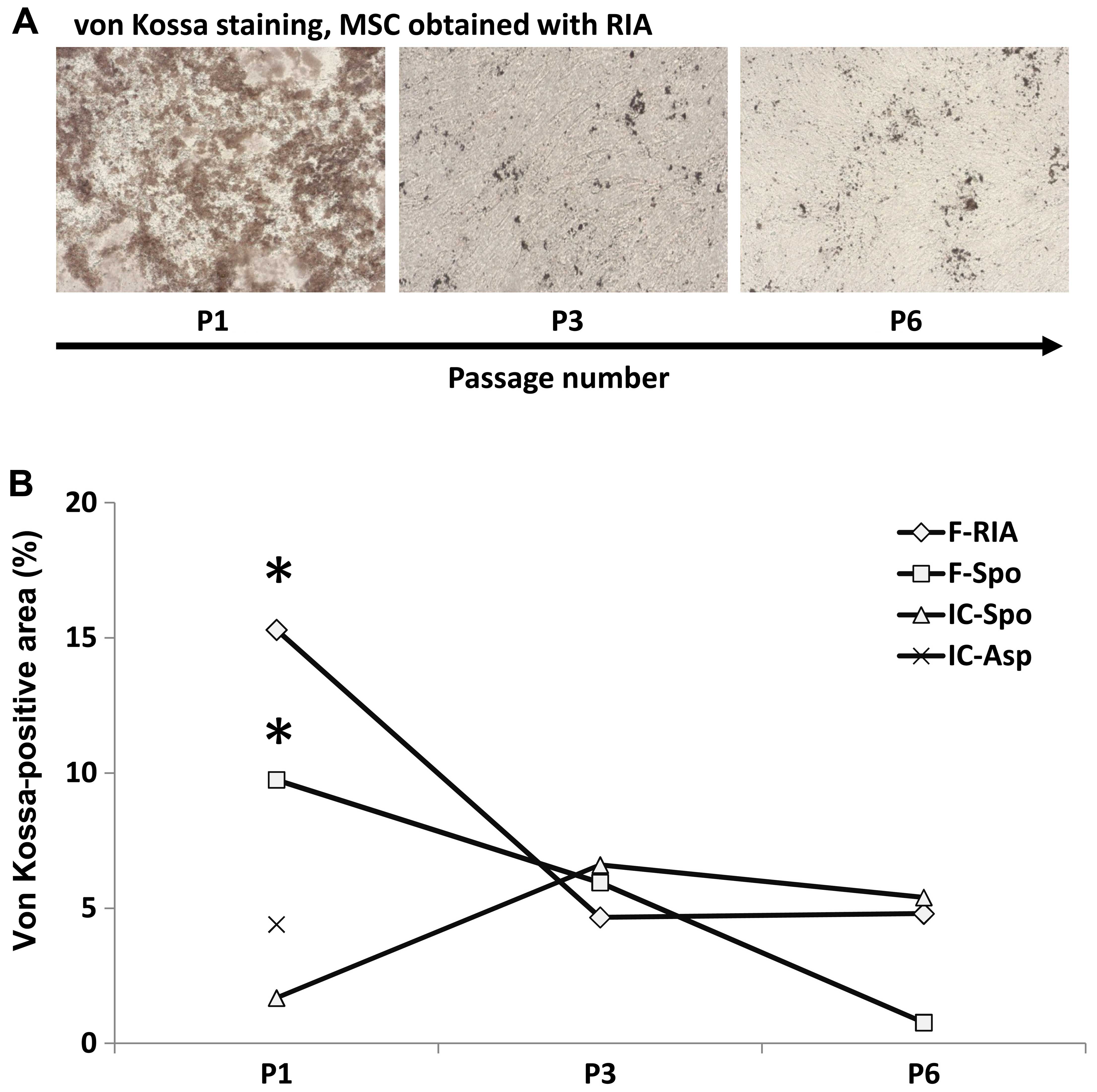
Significant decrease in the osteogenic
differentiation capacity with increasing passage number
The calcium deposition capacity and gene expression
were also assessed at the 3rd and 6th MSC culture passage,
respectively except in the MSCs obtained from the iliac crest (by
fine-needle aspiration), which were only assessed at the 1st
passage (calcium deposition) and 3rd passage (gene expression).
Higher passage numbers led to a marked decrease in the capacity of
MSCs for osteogenic differentiation. Calcium deposition decreased
significantly from the 1st to the 3rd passage. MSCs at the 6th
passage demonstrated the lowest values. A decline in calcium
deposition was observed in all the MSC cultures regardless of the
source and isolation procedure (Fig.
5).
The expression of all analyzed genes decreased
significantly in all the groups from the 1st to the 6th culture
passage, except for the iliac crest aspirate-derived MSCs which
were analyzed only up to the 3rd passage and no significant
differences were observed between the 3rd and 6th passage.
Osteogenic stimulation over 14 days did not enhance the gene
expression of the analyzed genes in comparison with that in the
untreated controls; however, the decrease in gene expression with
the increasing passage number was partially less pronounced for
BMP2 (femur spoon, iliac crest spoon), DKK1 (femur spoon, iliac
crest spoon), BGLAP (RIA, femur spoon, iliac crest spoon), COL1A
(RIA) and ALPL (femur spoon, iliac crest spoon) when compared with
their respective controls (Table
IV).
 | Table IVImpairment of gene expression with
the increasing passage number.a |
Table IV
Impairment of gene expression with
the increasing passage number.a
| A, Control
conditions |
|---|
|
|---|
| Harvest sites | BGLAP | COL1A | ALP | BMP2 | RUNX2 | SMAD5 | WNT3A | LRP5 | DKK1 | CTNNB1 |
|---|
| Femur, RIA | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 | 53±31 | 106±35 | 8±4 | 52±38 | 289±134 | 15±7 | 0±0 | 4±2 | 17±13 | 16±3 |
| Pas. 6 |
16±7b |
10±3b |
10±6b |
2±1b |
9±4b |
3±2b |
0±0b |
0±0b |
3±2b |
1±0b |
| Femur, spoon | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 | 13±5 | 16±8 | 9±5 | 16±5 | 37±15 | 5±2 | 0±0 | 0±0 | 0±0 | 11±3 |
| Pas. 6 |
18±17b |
5±1b |
5±5b |
53±51b |
5±4b |
3±3b |
0±0b |
12±12b |
1±1b |
5±5b |
| Il. cre. spoon | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 | 19±5 | 4±1 | 11±6 | 14±5 | 39±11 | 0±0 | 0±0 | 0±0 | 15±14 | 7±3 |
| Pas. 6 |
12±5b |
6±3b |
31±16b |
5±2b |
38±16b |
1±1b |
13±11b |
1±0b |
10±7b |
2±1b |
| Il. cre. asp. | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 |
0±0b |
1±1b |
0±0b |
0±0b |
0±0b |
0±0b |
0±0b |
0±0b |
0±0b |
1±1b |
| B,
Osteogenic-inducing conditions |
|---|
|
|---|
| Harvest sites | BGLAP | COL1A | ALP | BMP2 | RUNX2 | SMAD5 | WNT3A | LRP5 | DKK1 | CTNNB1 |
|---|
| Femur, RIA | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 | 2±3 | 112±21 | 17±12 | 24±22 | 11±3 | 0±0 | 0±0 | 1±1 | 1±1 | 2±1 |
| Pas. 6 | 28±23 | 107±96 | 6±5b | 1±1b | 5±3b | 0±0b | 0±0b | 0±0b | 0±0b | 0±0b |
| Femur, spoon | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 | 62±49 | 24±17 | 160±113 | 13±9 | 31±21 | 0±0 | 0±0 | 61±12 | 56±56 | 61±32 |
| Pas. 6 | 32±30 | 10±2b | 47±35 | 49±48 | 23±6 | 7±7b | 8±8b | 0±0b | 33±33 | 1±1b |
| Il. cre. spoon | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 | 992±910 | 28±21 | 14±8 | 613±552 | 13±4 | 1±0 | 0±0 | 1±1 | 1±1 | 9±3 |
| Pas. 6 | 117±49 | 7±4b | 22±15 | 32±16 | 16±10 | 282±274 | 8±3b | 0±0b | 36±15 | 14±14b |
| Il. cre. asp. | | | | | | | | | | |
| Pas. 1 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Pas. 3 | 3±4b | 4±3b | 3±3b | 0±0b | 5±5b | 0±0b | 0±0b | 0±0b | 1±1b | 0±0b |
Discussion
Previously, we observed that the MSCs obtained from
the femur had a greater potential for osteogenic differentiation in
comparison with the MSCs obtained from the iliac crest using the
fine-needle aspiration technique (7). In order to conduct the experiments,
the MSCs had to be expanded from a few hundred to at least
1×105 cells. The inheritance of the improved osteogenic
capacity over several cell divisions suggests that osteogenic
pathways undergo long-lasting alterations in the femur-derived
MSCs. Thus, we hypothesized that these differences were due to the
activation processes mediated by the different harvest procedures
or the different environmental mechanical load of the MSCs at their
respective body site. The latter may lead to a prevalence of
osteogenic signaling pathways in the MSCs obtained from the femur
due to the distinct methylation pattern of osteogenic genes.
Hence, the present study was conducted to examine
the effects of the tissue site and the harvest procedure on the
osteogenic functions and the gene regulation of osteogenic pathways
in human MSCs. We were able to demonstrate that functional aspects
(calcium deposition) and gene expression in MSCs were more affected
by the harvest technique than the tissue site. However, the
diminishing differences were observed after 6 culture passages and
no significant evidence was found that differential methylation
patterns of osteogenic genes play an important role in the
functional alterations of the MSCs.
Influence of the harvest technique and
mechanical strain on osteogenic differentiation
The MSCs obtained from the femor demonstrated an
increased mean calcium deposition activity in comparison with the
MSCs obtained from the iliac crest. This difference was significant
for the MSCs obtained using the RIA technique. This finding is in
line with previously published results from our group comparing the
osteogenic differentiation capabilities of the MSCs obtained from
the iliac crest by fine-needle aspiration with those isolated from
the human femur (7), and also
MSCs obtained from porcine femurs by RIA, as shown by another group
(10). Contrary results were
reported by Sanchez-Guijo et al (11). They reported the slightly, but not
significantly increased osteogenic differentiation of MSCs obtained
from the femoral head through a milling procedure compared with
those obtained from the iliac crest aspirates. The milling
procedure may not be comparable with the RIA process and thus, may
explain the comparatively low difference between the femor-derived
MSCs and the iliac crest-derived MSCs in terms of osteogenesis
(11).
RIA technology was developed to avoid the
disadvantages of conventional techniques used for intramedullary
reaming, such as marked increases in intramedullary pressure with
pressure peaks of up to 2.7×104 Pa to 2.0×105
Pa and temperatures at the reaming head up to 67°C (12). Using a porcine and a sheep model,
respectively, Husebye et al and Pape et al observed
that the application of RIA indeed led to a significant reduction
of intramedullary pressure in the femur to almost baseline levels
and to a significant decrease in systemic inflammation in
comparison with conventional medullary nailing (13,14). The reaming process itself is a
harsh procedure, exerting high shear forces on the cells due to the
continuous drilling and aspiration process. Many different cell
types have been demonstrated to be highly mechanosensitive in
vitro (15). As regards MSCs,
it has been further described that tensile strain, shear stress and
compressive loading support the osteogenic differentiation of MSCs
and inhibit adipogenesis (16–18).
Another factor which contributes to the improved
osteogenic differentiation of femor-derived MSCs compared to iliac
crest-derived MSCs may be the differential daily mechanical load of
the bones from which the cells are isolated. The association
between mechanical load and tissue formation has roots going back
to to the work of Wolff (1892), who suggested that the daily
mechanical environment influences the morphology of the skeletal
tissue architecture. Previous research has confirmed this and
suggests that this phenomenon plays a crucial role in creating,
maintaining or repairing the skeleton (19,20). The 'mechanosensitivity' of the
skeleton is likely to exert a direct effect on the gene expression
pattern, molecular architecture and tissue type during the process
of development and healing (21).
It is also feasible to assume that the high daily mechanical
loading of the femur will lead to distinct methylation patterns of
osteogenic genes in femor-derived MSCs in order to prime the cells
for their putative development in regards to the osteoblastic
lineage. Tissue site-specific differences in gene methylation were
described for MSCs derived from the iliac crest in comparison with
MSCs derived from adipose tissue. Non-adipogenic lineage-specific
promoters are hypermethylated, raising the hypothesis that
adipose-derived MSCs are epigenetically pre-programmed for
adipogenesis (22).
Our gene expression analysis frequently revealed the
reduced expression of BMP2 and the WNT family in the MSCs obtained
using the RIA technique from the femur in comparison with MSCs
obtained from the iliac crest by fine-needle aspiration, although
it has been reported that the mechanical load induces the rapid
expression of BMP-2, cbfa-1 and smad5, which further enhances the
gene expression of ALP, collagen type 1, osteocalcin and
osteopontin (23). Furthermore,
more recent research suggests that pathways such as the
Wnt/β-catenin signaling pathway are also triggered by mechanical
stimuli. The prevention of the adipogenic differentiation and the
improvement of the osteogenic differentiation of MSCs through
mechanical loading is due to the activation of β-catenin (24).
Of note, we observed the reduced expression of
osteogenic genes in the MSCs isolated using the RIA technique, the
cells that putatively received the highest mechanical stimuli in
the present study. However, this finding does not necessarily imply
the denial of the hypothesis that the mechanical stimulation of
MSCs supports osteogenic differentiation for the following reasons.
An important factor is the time that passes between the isolation
of the cells and the measurements. Whereas other researchers
stimulated cultured MSCs in vitro and directly measured gene
expression or cell functions (15,25,26), the MSCs used in our study received
mechanical stimuli a long time prior to the measurement, while
residing in their respective bone and during their subsequent
harvest procedure. Furthermore, a review of the literature revealed
that the duration of the stimulus and the type of stimulus are
critical factors for the induction of osteogenesis. For example, it
has been demonstrated that a 10% global strain of human bone
marrow-derived MSCs led to tenogenesis, whereas a '3% strain
favoured osteogenic differentiation' (15).
Compression loading, on the other hand, seems to
support the formation of mineralized bone-like matrix (25). One might argue that the
compression load is a major force affecting MSCs residing in the
femur. This may explain the improved calcium deposition that was
noted for the femor-derived MSCs in comparison with the MSCs
obtained from the iliac crest. The higher calcium deposition in the
femor-derived MSCs isolated using the RIA technique may be due to
further stimulation mediated by additional shear stress, which is
supposed to enhance osteogenesis, as summarized by McCoy and
O'Brien (26). Moreover, it is
also conceivable that molecular danger signals released from
necrotic cells during the reaming procedure stimulate the
osteogenic activity of MSCs. Evidence has indicated that
mechanical, as well as alarmin-induced stimulation leads to the
activation of mitogen-activated protein (MAP) kinases and that the
activation of MAP kinases supports the process of osteogenesis
(reviewed in ref. 27;28).
Furthermore, it has been described that alarmins, such as high
mobility group box 1 (HMGB1), support the osteogenic
differentiation of MSCs (29).
Hence, it may be assumed that the further increase in calcium
deposition in MSCs isolated using the RIA technique reflects the
combined stimulation with mechanical and molecular stimuli (by
different types of alarmins).
The long-lasting changes in MSC functions in
relation to the tissue site and the isolation procedure also
suggest a role of the epigenetic pre-programming of major
osteogenic pathways. DNA methylation is the most well-characterized
epigenetic modification. CpG dinucleotides in mammalian genomic DNA
may be methylated at cytosine moieties. Hypermethylation results in
the repression of gene expression, whereas hypomethylation enables
the transcription of certain genes. Upon replication, the same
methylation pattern is established on the newly synthesized DNA
strand by DNA methyltransferase-1, thus the methylation pattern is
inherited by both daughter cells. Thus, the effects mediated by the
alteration of gene methylation patterns may be effective over
several cell generations (8).
Methylation may affect gene transcription by directly interfering
with transcription factors or methyl-CpG-binding proteins that
modify histones and thereby inactivate the respective promoter
region (30). Studies have
demonstrated that epigenetic alterations are associated with the
level of differentiation potential in human stem cells (22,31). With regard to MSCs, it was
recently demonstrated that 'DNA methylation restricts the
spontaneous multi-lineage differentiation of mesenchymal progenitor
cells, but is stable during growth factor-induced terminal
differentiation' (31).
We observed trends towards the increased methylation
of the RUNX2 gene, and the decreased methylation of the SMAD5 and
WNT3A genes in the MSCs obtained using the RIA technique in
comparison with the MSCs obtained from the iliac crest either by
using a spoon or by fine-needle aspiration. The findings suggest
that gene methylation may be altered, depending on the mechanical
stress associated with the respective isolation method. This
suggestion is supported by results published in the study by
Arnsdorf et al (32); the
methylation of the osteopontin gene decreased by 35% with exposure
to oscillatory flow, whereas the methylation of the collagen I and
osteocalcin genes remained unaltered (32). Based on our results, we cannot
rule out that the tissue site may influence DNA methylation as
well. For example, evidence suggests that MSCs derived from adipose
tissue display an altered epigenetic pattern in comparison with the
MSCs derived from the iliac crest. Non-adipogenic lineage-specific
promoters are hypermethylated, raising the hypothesis that
adipose-derived MSCs are epigenetically pre-programmed for
adipogenesis (22).
Certainly, we could not calculate any correlation
between the methylation/hypermethylation and the expression of the
respective genes with the exception of the RUNX2 gene. The lack of
correlations are not in conflict with current research. Evidence
suggests that gene expression does not necessarily correlate with
gene methylation. For example, van Eijk et al analyzed the
DNA methylation and gene expression levels in the whole blood of
148 healthy human volunteers and reported '522 negative
associations and 276 positive associations between methylation and
expression levels' (33). As
already mentioned, we observed that particularly the degree of
hypermethylation of the RUNX-2 gene inversely correlated with the
gene expression of the RUNX2-regulated genes, COL1A, LRP5, SMAD5,
and in trend with RUNX2. This indicates that even slight
percentages of hypermethylation of the RUNX2 gene may affect RUNX2
expression and thus, the expression of the RUNX-2 regulated genes.
Conversly, Uehara et al reported that the hypermethylation
of RUNX2 may be involved in the the lipopolysaccharide
(LPS)-induced inhibition of osteoblastic cell differentiation
(34). These results underline
the importance of RUNX2 for osteogenesis (35).
Surprisingly, we did not observe a relevant increase
in the levels of typical osteogenic genes after 14 days of
incubation in osteogenic differentiation medium. However, the gain
of calcium deposition following osteogenic stimulation indicated
that osteogenic processes had occurred. The kinetics of osteogenic
gene expression have been extensively analyzed by Kulterer et
al. The authors observed that the gene expression of ALPL and
BMP2 peaked around day 4 and the gene expression of COL1A on day 7.
BGLAP as a marker of matrix mineralization was 'maximally expressed
on day 21′. Their 'results show that MSCs on day 7 of their
differentiation were most similar to osteoblasts' (36). Hence, with regard to the present
study, it appears most likely that the maximum expression levels of
the analyzed genes had already passed by day 14, and thus they did
not reach peak levels on day 14.
Effect of cell age on osteogenic
differentiation
We observed a marked decrease in osteogenic
differentiation with the increasing passage number, independently
of the source of MSCs, as measured by means of calcium deposition
and the expression of osteogenic genes. The cell age-dependent
decline in the osteogenic capabilities of MSCs was first reported
by Banfi et al (37). They
observed that 'MSCs had a markedly diminished proliferation rate
after the first passage and gradually lost their multiple
differentiation potential' which was accompanied by a functional
loss with regard to their bone formation capacity in vivo
(37). Similar results were
reported by Kretlow et al (38). They observed a significant decline
in the calcium deposition capacity of MSCs from the first to the
sixth culture passage (38).
Reasons for the deprivation of osteogenic potential with the
increasing passage number may be found in the loss of initial
heterogenity of the adherent MSCs during ongoing cultivation due to
the preferential proliferation of one over the other cell types as
suggested by Kretlow et al (38). It is feasible to assume that the
same effect is responsible for the decline in osteogenic
differentiation independently of the harvest site and harvest
procedure observed in the present study.
In conclusion, this observational study demonstrated
that femor-derived MSCs isolated using the RIA technique exhibited
a significantly improved calcium deposition. Furthermore, it was
noted that calcium deposition, the expression of osteogenic genes
and the gene methylation of the MSCs obtained from different tissue
sites were similar to a great extent, when comparable tissue types
were analyzed (cancellous bone) and comparable isolation procedures
were applied (using a spoon). The latter results further suggest
that the daily environmental mechanical load of the bone does not
necessarily lead to significant alterations in the methylation and
the expression of genes involved in osteogenesis in MSCs. Hence,
the significantly altered gene expression and gene methylation of
the MSCs harvested from the femur using the RIA technique may be
attributed to the harsh isolation procedure. Our correlation
analysis underlined the importance of the methylation state of the
central osteogenic transcription factor, RUNX2, and the expression
of RUNX2-regulated genes. However, the lack of correlation between
the methylation and the expression of any other of the analyzed
genes suggests that other cellular processes override the effects
of gene methylation to a certain extent. In conclusion, we provide
evidence herein that primarily, the harvest procedure and not the
type of bone tissue determines the osteogenic capabilities of MSCs
in vitro. Albeit, any differences in osteogenic capabilities
were diminished with the increasing passage number. The variable
differentiation ability of the MSCs, which depends on the harvest
site and harvest technique, as well as the rapid loss of osteogenic
differentiation capacity with the increasing culture time should be
taken into consideration when using MSCs as a potential therapeutic
application for bone tissue engineering.
Acknowledgments
This study was founded by the AORF: S-10-47-H.
References
|
1
|
Hannouche D, Petite H and Sedel L: Current
trends in the enhancement of fracture healing. J Bone Joint Surg
Br. 83:157–164. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feinberg SE, Hollister SJ, Halloran JW,
Gabe Chu TM and Krebsbach PH: A tissue engineering approach to
site-specific reconstruction of skeletal structures of the
maxillofacial region:part I. Shanghai Kou Qiang Yi Xue. 9:34–38.
2000.In Chinese.
|
|
3
|
Feinberg SE, Hollister SJ, Halloran JW,
Gabe Chu TM and Krebsbach PH: A tissue engineering approach to site
specific reconstruction of skeletal structures of the maxillofacial
region: part II. Shanghai Kou Qiang Yi Xue. 9:88–93. 2000.In
Chinese.
|
|
4
|
Petite H, Viateau V, Bensaïd W, Meunier A,
de Pollak C, Bourguignon M, Oudina K, Sedel L and Guillemin G:
Tissue- engineered bone regeneration. Nat Biotechnol. 18:959–963.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmidmaier G, Herrmann S, Green J, Weber
T, Scharfenberger A, Haas NP and Wildemann B: Quantitative
assessment of growth factors in reaming aspirate, iliac crest, and
platelet preparation. Bone. 39:1156–1163. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wenisch S, Trinkaus K, Hild A, Hose D,
Herde K, Heiss C, Kilian O, Alt V and Schnettler R: Human reaming
debris: a source of multipotent stem cells. Bone. 36:74–83. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Henrich D, Seebach C, Sterlepper E,
Tauchmann C, Marzi I and Frank J: RIA reamings and hip aspirate: a
comparative evaluation of osteoprogenitor and endothelial
progenitor cells. Injury. 41(Suppl 2): S62–S68. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Djupedal I and Ekwall K: Epigenetics:
heterochromatin meets RNAi. Cell Res. 19:282–295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seebach C, Henrich D, Tewksbury R, Wilhelm
K and Marzi I: Number and proliferative capacity of human
mesenchymal stem cells are modulated positively in multiple trauma
patients and negatively in atrophic nonunions. Calcif Tissue Int.
80:294–300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van der Bel R and Blokhuis TJ: Increased
osteogenic capacity of Reamer/Irrigator/Aspirator-derived
mesenchymal stem cells. Injury. 45:2060–2064. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanchez-Guijo FM, Blanco JF, Cruz G,
Muntion S, Gomez M, Carrancio S, Lopez-Villar O, Barbado MV,
Sanchez-Abarca LI, Blanco B, et al: Multiparametric comparison of
mesenchymal stromal cells obtained from trabecular bone by using a
novel isolation method with those obtained by iliac crest
aspiration from the same subjects. Cell Tissue Res. 336:501–507.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Henry SL, Adcock RA, Von Fraunhofer JA and
Seligson D: Heat of intramedullary reaming. South Med J.
80:173–176. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Husebye EE, Lyberg T, Madsen JE, Eriksen M
and Roise O: The influence of a one-step reamer-irrigator-aspirator
technique on the intramedullary pressure in the pig femur. Injury.
37:935–940. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pape HC, Zelle BA, Hildebrand F,
Giannoudis PV, Krettek C and van GM: Reamed femoral nailing in
sheep: does irrigation and aspiration of intramedullary contents
alter the systemic response? J Bone Joint Surg Am. 87:2515–2522.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Delaine-Smith RM and Reilly GC:
Mesenchymal stem cell responses to mechanical stimuli. Muscles
Ligaments Tendons J. 2:169–180. 2012.
|
|
16
|
Delaine-Smith RM and Reilly GC: The
effects of mechanical loading on mesenchymal stem cell
differentiation and matrix production. Vitam Horm. 87:417–480.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sen B, Xie Z, Case N, Ma M, Rubin C and
Rubin J: Mechanical strain inhibits adipogenesis in mesenchymal
stem cells by stimulating a durable beta-catenin signal.
Endocrinology. 149:6065–6075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Simmons CA, Matlis S, Thornton AJ, Chen S,
Wang CY and Mooney DJ: Cyclic strain enhances matrix mineralization
by adult human mesenchymal stem cells via the extracellular
signal-regulated kinase (ERK1/2) signaling pathway. J Biomech.
36:1087–1096. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Beaupré GS, Stevens SS and Carter DR:
Mechanobiology in the development, maintenance, and degeneration of
articular cartilage. J Rehabil Res Dev. 37:145–151. 2000.PubMed/NCBI
|
|
20
|
Carter DR, Beaupré GS, Giori NJ and Helms
JA: Mechanobiology of skeletal regeneration. Clin Orthop Relat Res
(Suppl). S41–S55. 1998. View Article : Google Scholar
|
|
21
|
Cullinane DM, Salisbury KT, Alkhiary Y,
Eisenberg S, Gerstenfeld L and Einhorn TA: Effects of the local
mechanical environment on vertebrate tissue differentiation during
repair: does repair recapitulate development? J Exp Biol.
206:2459–2471. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boquest AC, Noer A and Collas P:
Epigenetic programming of mesenchymal stem cells from human adipose
tissue. Stem Cell Rev. 2:319–329. 2006. View Article : Google Scholar
|
|
23
|
Rath B, Nam J, Knobloch TJ, Lannutti JJ
and Agarwal S: Compressive forces induce osteogenic gene expression
in calvarial osteoblasts. J Biomech. 41:1095–1103. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sen B, Styner M, Xie Z, Case N, Rubin CT
and Rubin J: Mechanical loading regulates NFATc1 and beta-catenin
signaling through a GSK3beta control node. J Biol Chem.
284:34607–34617. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sittichokechaiwut A, Edwards JH, Scutt AM
and Reilly GC: Short bouts of mechanical loading are as effective
as dexamethasone at inducing matrix production by human bone marrow
mesenchymal stem cell. Eur Cell Mater. 20:45–57. 2010.PubMed/NCBI
|
|
26
|
McCoy RJ and O'Brien FJ: Influence of
shear stress in perfusion bioreactor cultures for the development
of three-dimensional bone tissue constructs: a review. Tissue Eng
Part B Rev. 16:587–601. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thompson WR, Rubin CT and Rubin J:
Mechanical regulation of signaling pathways in bone. Gene.
503:179–193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bianchi ME: DAMPs, PAMPs and alarmins: all
we need to know about danger. J Leukoc Biol. 81:1–5. 2007.
View Article : Google Scholar
|
|
29
|
Pistoia V and Raffaghello L:
Damage-associated molecular patterns (DAMPs) and mesenchymal stem
cells: a matter of attraction and excitement. Eur J Immunol.
41:1828–1831. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: how the genome integrates intrinsic
and environmental signals. Nat Genet. 33(Suppl): 245–254. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hupkes M, van Someren EP, Middelkamp SH,
Piek E, van Zoelen EJ and Dechering KJ: DNA methylation restricts
spontaneous multi-lineage differentiation of mesenchymal progenitor
cells, but is stable during growth factor-induced terminal
differentiation. Biochim Biophys Acta. 1813:839–849. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Arnsdorf EJ, Tummala P, Castillo AB, Zhang
F and Jacobs CR: The epigenetic mechanism of mechanically induced
osteogenic differentiation. J Biomech. 43:2881–2886. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
van Eijk KR, de JS, Boks MP, Langeveld T,
Colas F, Veldink JH, de Kovel CG, Janson E, Strengman E, Langfelder
P, et al: Genetic analysis of DNA methylation and gene expression
levels in whole blood of healthy human subjects. BMC Genomics.
13(636)2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Uehara O, Abiko Y, Saitoh M, Miyakawa H
and Nakazawa F: Lipopolysaccharide extracted from Porphyromonas
gingivalis induces DNA hypermethylation of runt-related
transcription factor 2 in human periodontal fibroblasts. J
Microbiol Immunol Infect. 47:176–181. 2014. View Article : Google Scholar
|
|
35
|
Schroeder TM, Jensen ED and Westendorf JJ:
Runx2: A master organizer of gene transcription in developing and
maturing osteoblasts. Birth Defects Res C Embryo Today. 75:213–225.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kulterer B, Friedl G, Jandrositz A,
Sanchez-Cabo F, Prokesch A, Paar C, Scheideler M, Windhager R,
Preisegger KH and Trajanoski Z: Gene expression profiling of human
mesenchymal stem cells derived from bone marrow during expansion
and osteoblast differentiation. BMC Genomics. 8(70)2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Banfi A, Muraglia A, Dozin B,
Mastrogiacomo M, Cancedda R and Quarto R: Proliferation kinetics
and differentiation potential of ex vivo expanded human bone marrow
stromal cells: implications for their use in cell therapy. Exp
Hematol. 28:707–715. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kretlow JD, Jin YQ, Liu W, Zhang WJ, Hong
TH, Zhou G, Baggett LS, Mikos AG and Cao Y: Donor age and cell
passage affects differentiation potential of murine bone
marrow-derived stem cells. BMC Cell Biol. 9(60)2008. View Article : Google Scholar : PubMed/NCBI
|