Introduction
Pediatric asthma is one of the most common
respiratory diseases in children. A survey of asthma morbidity and
mortality conducted by the Centers for Disease Control and
Prevention reported that the incidence of this disease was 8.0%
during 2006–2010 in the United States, with a prevalence of 9.4%
among children (1). A recent
study of childhood asthma in urban areas of China described an
increasing incidence of asthma, by approximately 10% in one decade,
and the overall prevalence of asthma in children <14 years of
age in urban areas was 3.02% in 2009–2010 (2). Etiologically, pediatric asthma may
result from gene-environment interactions (3). The common environmental factors
include air pollution, pollen, fungi and dust mites (4–6).
Up to 80% of asthmatics are allergic to house dust mites, although
the exact rate depends on the geographic region (7). Currently, the pathogenic mechanism
of childhood asthma remains unknown. As a heterogeneous disease
with strong genetic factors, the pathogenesis of pediatric asthma
is complex (8–10). It is well known that immune
regulation is closely associated with the inflammatory response in
asthma. Previous studies showed that the abnormal regulation of
immune and inflammatory responses occurred in asthma patients, as
well as an imbalance in T helper type 1 (Th1)/T helper type 2 (Th2)
cells, which increased the inflammatory factors interleukin (IL)-4,
IL-6 and IL-13 (mediated by Th2) and decreased IL-12 and
γ-interferon (γ-IFN) (mediated by Th1) to cause chronic airway
inflammation and hyperresponsiveness (11–13). These inflammatory responses are
closely associated with several signaling pathways, such as the
phosphoinositide 3-kinase (PI3K)-AKT, Janus kinase (JAK)-signal
transducers and activators of transcription (STAT),
mitogen-activated protein kinase (MAPK) and nuclear factor-ĸB
(NF-ĸB) pathways (14–16), which resulted in alterations in
cytokine secretion.
Previous findings have shown that PI3K-AKT and NF-ĸB
are associated with Th1/Th2 differentiation and cytokine secretion
(e.g., IL and IFN) (17). It was
found that several genes, including Cbl proto-oncogene, E3
ubiquitin protein ligase (CBL) and estrogen receptor 1
(ESR1), are involved in the PI3K-AKT and/or NF-ĸB signal
pathway(s) (18,19). These genes may influence
downstream genes such as spleen tyrosine kinase (SYK) and
epidermal growth factor receptor (EGFR) in order to regulate
the PI3K-AKT and NF-κB pathway(s) (20,21). However, the underlying mechanism
of regulation in asthma has not yet been clearly illustrated.
MicroRNAs (miRNAs or miRs), a group of small,
non-coding RNAs 21–25 nucleotides in length, have been reported to
be transcriptional regulators involved in many complex human
disorders, and in biological processes including cell proliferation
and apoptosis (22,23). miR-223 was reported to regulate
the maturation, function and differentiation of neutrophils
(24). Major histocompatibility
complex, class I, G (HLA-G), an asthma susceptibility gene,
has been found to be a common target of miR-148a, miR-148b, and
miR-152 (25). miR-146b, miR-223,
miR-29b, miR-29c, miR-483, miR-5745p, miR-672 and miR-690 are
differentially expressed in asthma patients (26–28). The miRNA regulation of target
genes appears to play an important role in immune and inflammatory
responses as well as in the development of asthma (29). As miRNAs are regulators of the
inflammatory response in asthma, it is likely that miRNAs may
regulate their targeted messenger RNA(s) [mRNA(s)] that are
involved in the asthma-associated inflammatory pathway, thereby
leading to an asthma attack. In the present study, we conducted a
genome-wide investigation of novel miRNA(s) that may have
regulatory functions in dust mite-induced asthma.
Subjects and methods
Subjects
Dust mites have been determined to be the
predominant allergen in asthma (7). In the present study, we focused on
dust mite-induced asthma. The study was approved by the Bioethics
Committee of the Shanghai Children's Hospital (Shanghai, China),
and written informed consent was obtained from the participants or
their guardians.
A total of 62 pediatric patients with asthma were
recruited, as the case group, from our Asthma Clinic at Shanghai
Children's Hospital (Shanghai, China). A diagnosis of asthma was
made according to the Global Initiative for Asthma (GINA)
guidelines (10). We focused on
patients with intermittent-mild asthma in this study, as the
pathogenesis of severe asthma is more complex (30). Detailed information regarding
patient demographics and enrollment inclusion and/or exclusion
criteria is provided in Table I.
Forty-one boys and 21 girls (average age, 6.05±2.15 years) were
recruited for the asthma case group. During the time that they were
enrolled in this study and their blood specimens were being
collected, these patients did not have current or recent
infections, were not taking any medication, such as inhaled
corticosteroid (ICS) (31), and
did not experience any acute asthma exacerbations or attacks, in
order to avoid some disturbing factors. An independent, unrelated
group of 37 boys and 25 girls (average age, 7.83±3.30 years)
showing no clincial phenotypes was enrolled as normal controls. No
significant difference was observed between the groups of cases and
controls, in terms of gender and age (χ2=0.667,
p=0.414).
 | Table IDemographics of asthma and control
groups. |
Table I
Demographics of asthma and control
groups.
| Group | Gendera
| Agea (years) | IgE (kU/l) | EOS (%) |
|---|
| Male | Female |
|---|
| Asthma | 41 | 21 | 6.05±2.15 | 453.29±284.28 | 4.60±2.80 |
| Control | 37 | 25 | 7.83±3.30 | 38.16±34.51 | 3.72±3.40 |
Immunoglobulin E (IgE) serology
Total IgE and specific serum IgE, which were
thresholds of enrollment inclusion (Table I), were assessed using the
ImmunoCAP System (Thermo Fisher Scientific/Phadia AB, Uppsala,
Sweden). The cut-off of total IgE was 60 kU/l. In the asthma group,
total IgE was >60 kU/l, and IgE serology atopic sensitization
was indicated if the child had only dust mite allergen-specific
serum IgE (≥0.35 kU/l) (data not shown) without food allergens of
Fx5 (egg white, milk, fish, wheat, peanut and soy) (32). In the control group, no
allergen-specific IgE was >0.35 kU/l, and total IgE was <60
kU/l. The percentage of eosinophils in peripheral blood (EOS) was
also counted in order to confirm the patient's atopy. The cut-off
value was 4%.
Array hybridization
Twelve pairs of gender- and age-matched children
with asthma and normal control individuals were subjected to
initial microarray-based discovery analysis. Peripheral blood (5
ml) was collected from each participant using
ethylenediaminetetraacetic acid (EDTA) as the anticoagulant. Total
RNA was isolated using TRIzol (Invitrogen, Grand Island, NY, USA)
and an miRNeasy mini kit (Qiagen, Valencia, CA, USA) kits according
to the manufacturer's instructions, which efficiently recovered all
RNA species, including miRNAs. The quality and quantity of RNAs
collected from all participants were measured using a NanoDrop
spectrophotometer ND-1000 (NanoDrop Technologies, Wilmington, DE,
USA), and RNA integrity was determined by gel electrophoresis.
After RNA was isolated from the samples, the miRCURY LNA™ microRNA
Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark) was used
according to the manufacturer's instructions for miRNA labeling.
Briefly, 1 μg of each sample was 3′-end-labeled with Hy3
fluorescent dye, using T4 RNA ligase according to the following
procedure: RNA in 2.0 μl of water was combined with 1.0
μl CIP buffer and CIP (Exiqon). The mixture was incubated
for 30 min at 37°C and was terminated at 95°C for 5 min. Then, 3.0
μl labeling buffer, 1.5 μl fluorescent label (Hy3),
2.0 μl dimethyl sulfoxide (DMSO), and 2.0 μl labeling
enzyme were added into the mixture. The labeling reaction was
incubated for 1 h at 16°C and was terminated by incubation at 65°C
for 15 min. After termination, the Hy3-labeled samples were
hybridized on the miRCURY LNA array (v.18.0; Exiqon) according to
the manufacturer's instructions. The total 25 μl mixture
from Hy3-labeled samples together with 25 μl hybridization
buffer were denatured at 95°C for 2 min, incubated in ice for 2
min, and then hybridized to the microarray at 56°C for 16–20 h with
a 12-Bay Hybridization system (Nimblegen Systems, Inc., Madison,
WI, USA). After hybridization, the slides were prepared, washed
using a wash buffer kit (Exiqon), and dried by centrifugation at
200 × g for 5 min. The slides were then scanned using the Axon
GenePix 4000B microarray scanner (Axon Instruments, Union City, CA,
USA).
miRNA profiling and differential
expression analysis
The signal quantification of scanned microarray
images was captured using GenePix Pro 6.0 software (Molecular
Devices, Sunnyvale, CA, USA). Replicated miRNAs were averaged, and
miRNAs whose intensities were ≥30 in all samples were selected for
calculation of the normalization factor. Expressed data were
normalized using the median normalization. Significant
differentially expressed miRNAs were identified through volcano
plot filtering. Hierarchical clustering was performed to show
distinguishable miRNA expression profiling among samples using MEV
software (v4.6, TIGR; www.tm4.org/).
A threshold (fold-change ≥2.0 and p-value ≤0.05) was used to
determine the significance of differences of up- or downregulated
miRNAs. The heat map diagram shows the result of the two-way
hierarchical clustering of miRNAs and samples.
Bioinformatics analysis
Web-based programs, namely miRBase (http://mirbase.org/), PicTar (http://pictar.mdc-berlin.de/), miRDB (http://mirdb.org/miRDB/), miRanda (http://www.microrna.org/microrna/) and TargetScan
(http://www.targetscan.org/), were
employed to predict miRNA target(s). The miRNA-targeted
transcripts, predicted by a minimum of three programs as the
miRNA-targeted candidates, were selected for further analysis. The
functional enrichment was analyzed using the DAVID program
(http://david.abcc.ncifcrf.gov/), in
which gene ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways were provided for analysis. Venny software
(http://bioinfogp.cnb.csic.es/tools/venny/) was used to
show the predicted target genes that are co-regulated by
multi-miRNA.
Validation of miRNAs and mRNA using
reverse transciption-quantitative polymerase chain reaction
To validate the microarray results, miRNA expression
was quantified in all 124 samples using RT-qPCR. The primers used
for reverse transcription are presented in Table II, and the PCR conditions are
shown in Table III. miRNA-U6
(33) was applied as an internal
control. Briefly, reverse transcription was performed using MMLV
Reverse Transcriptase and RNase inhibitor (both from Epicentre,
Madison, WI, USA), 10X buffer (250 mM Tris-HCl, pH 8.3; 200 mM KCl;
40 mM MgCl2; 5 mM DTT), 2.5 mM dNTP with RT primers at
16°C for 30 min, followed by 42°C for 40 min and 85°C for 5 min.
Twenty microliters of RNA (final concentration 50 ng/μl) was
used as a template. The cDNA products from reverse transcription
reactions were quantified using a real-time PCR System 7500
(Applied Biosystems, Foster City, CA, USA). The resulting amplicon
was quantified by RT-qPCR with end-point SYBR-Green fluorescence.
Each assay was performed in triplicate. RT-qPCR was also performed
to determine the expression of the target genes regulated by the
miRNAs that were discovered in our microarray analysis. The reverse
transcription reaction was performed as descibed above except using
SuperScript™ III reverse transcriptase (Invitrogen). RT primers
were added into the reaction mixture at 37°C for 1 min, followed by
50°C for 60 min and 75°C for 15 min. Twenty microliters of RNA
(final concentration 75 ng/μl) was used as a template. The
cDNA products from the reverse transcription reactions were
quantified using a real time PCR system 7500 (Applied Biosystems).
The resulting amplicon was quantified by RT detection of end-point
SYBR-Green fluorescence. Each assay was performed in triplicate.
The primers and reaction conditions are presented in Table IV. The relevant quantification
was obtained for each sample with normalization to the internal
control β-actin.
 | TablePrimers for miRNA sequences. |
Table
Primers for miRNA sequences.
| Name of miRNA | RT primer |
|---|
| U6 |
5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| hsa-miR-675-3p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTGAGCG-3′ |
| hsa-let-7d-3p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGAAAG-3′ |
|
hsa-miR-550b-3p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCAGTGC-3′ |
| hsa-miR-501-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCTCAC-3′ |
| hsa-miR-4312 |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTGGGGA-3′ |
|
hsa-miR-151a-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACTAGA-3′ |
| hsa-miR-625-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGGACTA-3′ |
| hsa-miR-22-3p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACAGTT-3′ |
| hsa-miR-126-3p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCGCATT-3′ |
|
hsa-miR-513a-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACATGACA-3′ |
 | Table IIIPrimers used for RT-qPCR of miRNAs
and PCR conditions. |
Table III
Primers used for RT-qPCR of miRNAs
and PCR conditions.
| Name of miRNA | Bidirectional
primer sequence | Annealing
temperature (°C) | Amplicom length
(bp) |
|---|
| U6 | F:
5′-GCTTCGGCAGCACATATACTAAAAT-3′ | | |
| R:
5′-CGCTTCACGAATTTGCGTGTCAT-3′ | 60 | 89 |
| hsa-miR-675-3p | GSP:
5′-GAAGGCTGTATGCCCTCACC-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
| hsa-let-7d-3p | GSP:
5′-GGGCTATACGACCTGCTGC-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
|
hsa-miR-550b-3p | GSP:
5′-GGGGGGTCTTACTCCCTCAG-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 64 |
| hsa-miR-501-5p | GSP:
5′-GGAGAATCCTTTGTCCCTGG-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 64 |
| hsa-miR-4312 | GSP:
5′-GGGGTAAGCCTTGTTCCTGT-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
|
hsa-miR-151a-5p | GSP:
5′-GGGGTCGAGGAGCTCACAG-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
| hsa-miR-625-5p | GSP:
5′-GGGGAGGGGGAAAGTTCTA-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
| hsa-miR-22-3p | GSP:
5′-GGGAAGCTGCCAGTTGAAG-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
| hsa-miR-126-3p | GSP:
5′-GGGGTCGTACCGTGAGTAAT-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 64 |
|
hsa-miR-513a-5p | GSP:
5′-GGAGGGTTCACAGGGAGGT-3′ | | |
| R:
5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 62 |
 | Table IVPrimers used for RT-qPCR of target
genes regulated by miRNAs. |
Table IV
Primers used for RT-qPCR of target
genes regulated by miRNAs.
| Gene | Primer
sequence | Annealing
temperature (°C) | Amplicom length
(bp) |
|---|
| β-actin | F:
5′-CCTGTACGCCAACACAGTGC-3′ | | |
| R:
5′-ATACTCCTGCTTGCTGATCC-3′ | 60 | 211 |
|
PPARGC1B | F:
5′-GGCGGTAGACGAATGACAGAC-3′ | | |
| R:
5′-CTTAGTGGAGGCAGCAGAAAGA-3′ | 60 | 199 |
| ESR1 | F:
5′-CCTTGCTATGTTACTAAGCGTGAG-3′ | | |
| R:
5′-GGGTGCTATAATAAACCCTTGAC-3′ | 60 | 247 |
| CBL | F:
5′-GGACAAGAAGATGGTGGAGAAGT-3′ | | |
| R:
5′-CTTGACAAGATAGTACGGAGATGC-3′ | 60 | 150 |
Measurement of inflammatory
cytokines
The plasma concentrations of IL-4, -6, -10, -12 and
-13; γ-IFN; and tumor necrosis factor-α (TNF-α) were determined
using an enzyme-linked immunosorbent assay (ELISA) kit
(Multiscience Biotech Co. Ltd., Shanghai, China).
Statistical analysis
The differences between the groups were analyzed
using the Student's t-test when two groups were compared or by
one-way ANOVA when > two groups were compared. Analyses were
performed using SPSS 19.0 software (IBM, Armonk, NY, USA). A
p-value <0.05 was considered to indicate a statistically
significant difference.
Results
Differential expression of miRNAs in
children with asthma
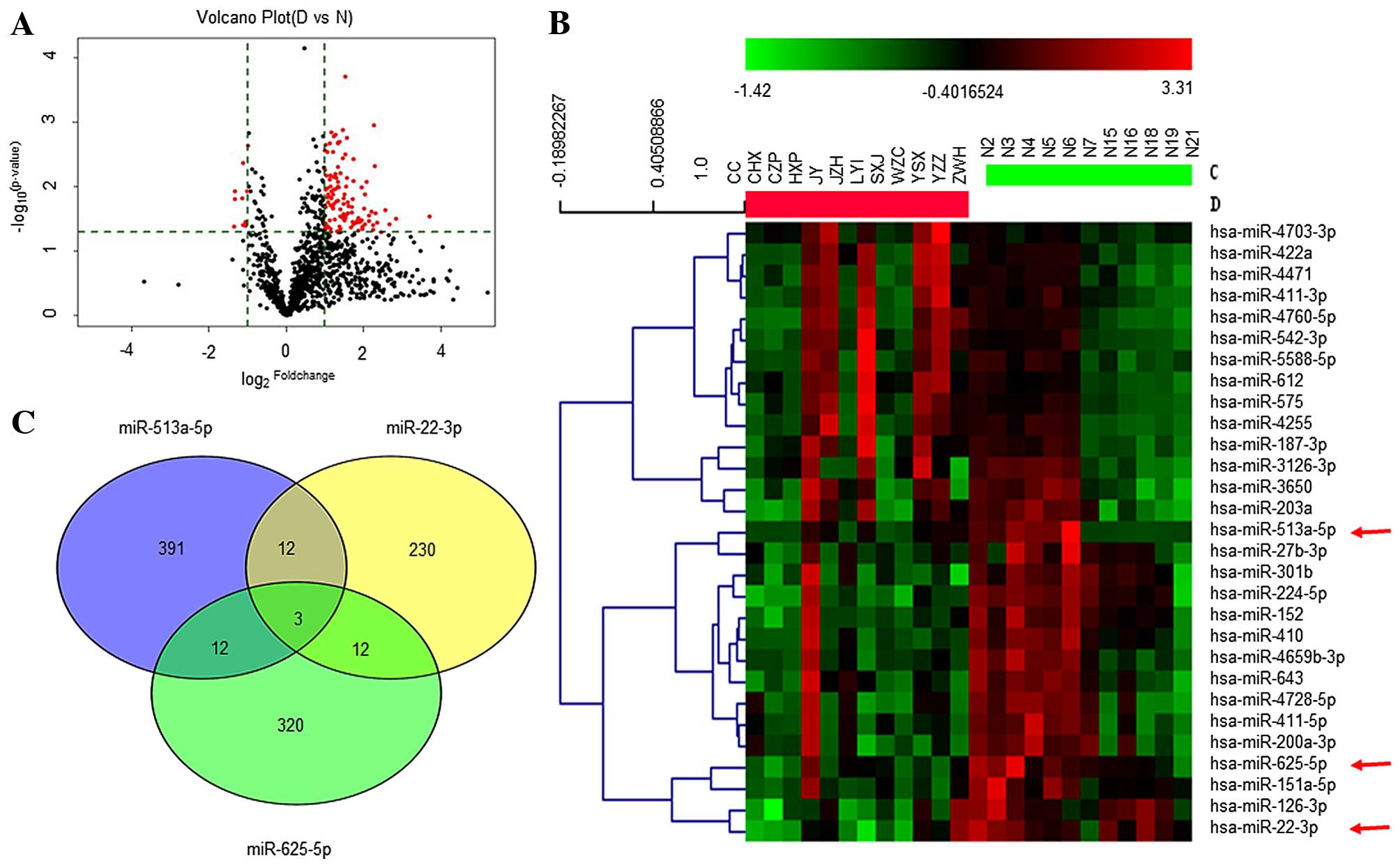
A total of 122 differentially expressed miRNAs were
identified from 12 children with asthma in a discovery study with a
micro-array that detects 3,100 genome miRNAs. Among differentially
expressed miRNAs, 112 were identified as upregulated, and 10 were
downregulated (Fig. 1A and B). To
confirm the micro-array results, five upregulated (miR-let-7d-3p,
miR-550b-3p, miR-501-5p, miR-4312 and miR-675-3p) and five
downregulated (miR-126-3p, miR-22-3p, miR-513a-5p, miR-151a-5p and
miR-625-5p) miRNAs were randomly subjected to RT-qPCR with the 12
pairs of RNA samples that were subjected to the original
array-based discovery study. The differential expression of four
upregulated (miR-let-7d-3p, miR-550b-3p, miR-501-5p and miR-675-3p)
and all five downregulated miRNAs were validated to be significant
(p<0.05) (Table V). Three
downregulated miRNAs (miR-625-5p with 2.17-fold decrease,
miR-513a-5p with 2.07-fold-change, and miR-22-3p with
2.21-fold-change) (Table VI),
which were statistically significant differences (p<0.05) and
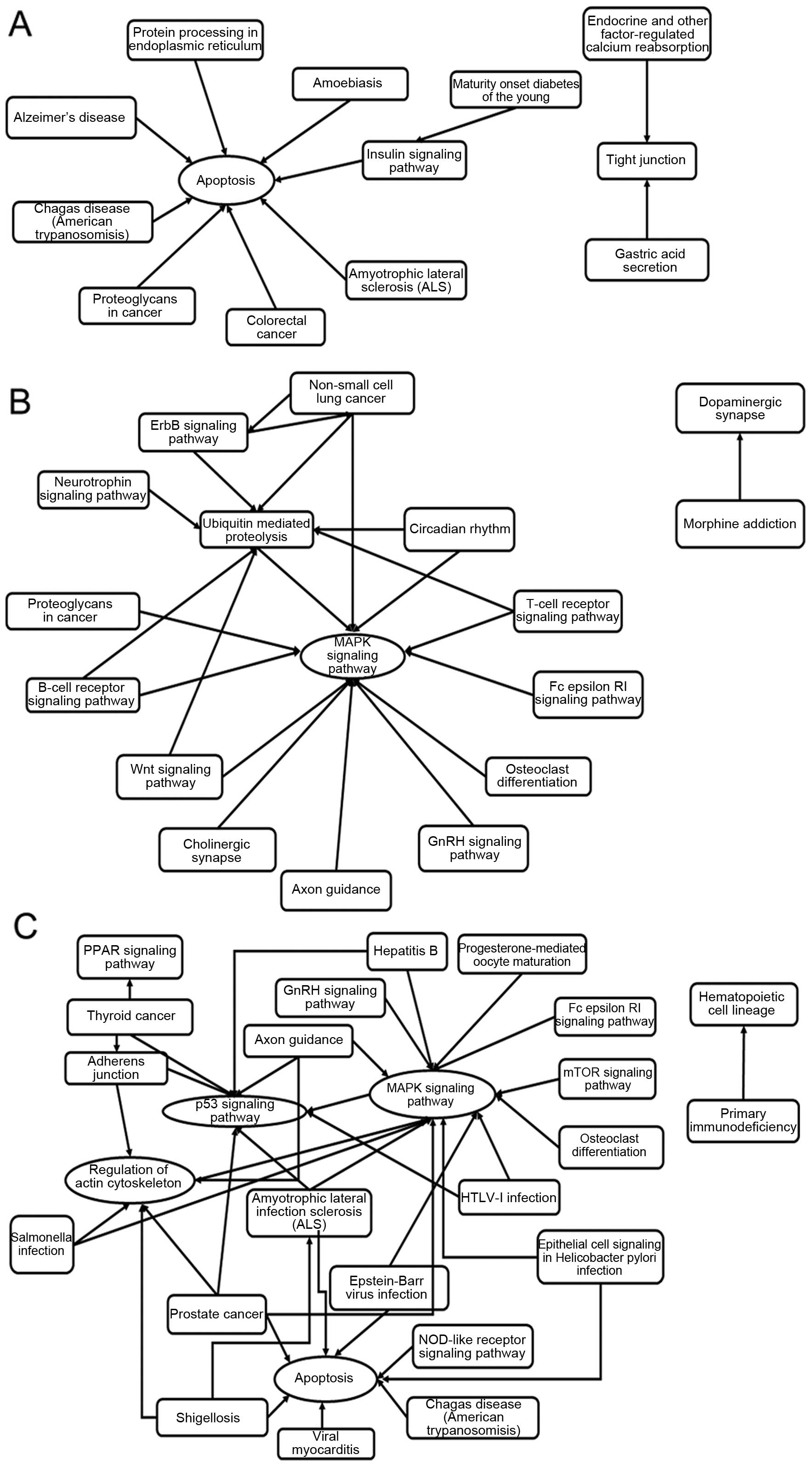
functionally linked to inflammation and apoptosis by bioinformatics
analysis (Fig. 2), were applied
for prediction of targeting transcripts.
 | Table VUp- and downregulated miRNAs
determined using RT-qPCR (2−ΔΔCT). |
Table V
Up- and downregulated miRNAs
determined using RT-qPCR (2−ΔΔCT).
| miRNA | Asthma (n=12) | Control (n=12) | t-value | p-value | 95% confidence
interval of difference |
|---|
| Upregulated |
| let-7d-3p | 0.943±0.120 | 0.434±0.089 | 8.650 | 0.000 | 0.385 to 0.632 |
| miR-550b-3p | 0.937±0.130 | 0.472±0.056 | 7.632 | 0.000 | 0.337 to 0.593 |
| miR-501-5p | 1.252±0.244 | 0.786±0.183 | 3.893 | 0.001 | 0.215 to 0.717 |
| miR-675-3p | 1.063±0.172 | 0.660±0.044 | 5.092 | 0.000 | 0.237 to 0.569 |
| miR-4312 | 1.268±0.282 | 1.130±0.207 | 0.999 | 0.331 | −0.152 to
0.428 |
| Downregulated |
| miR-151a-5p | 1.006±0.193 | 2.024±0.374 | −8.028 | 0.000 | −1.284 to
−0.751 |
| miR-625-5p | 1.130±0.222 | 2.206±0.168 | −9.858 | 0.000 | −1.305 to
−0.847 |
| miR-126-3p | 0.992±0.142 | 1.790±0.161 | −24.619 | 0.000 | −0.957 to
−0.639 |
| miR-513a-5p | 1.031±0.140 | 2.288 ±.02565 | −14.089 | 0.000 | −1.445 to
−1.070 |
| miR-22-3p | 0.836±0.175 | 1.580±0.269 | −7.215 | 0.000 | −0.961 to
−0.527 |
 | Table VIDifferential expression of miR-22-3p,
miR-513a-5p and miR-625-5p. |
Table VI
Differential expression of miR-22-3p,
miR-513a-5p and miR-625-5p.
| miRNA | Fold-change (A vs.
C) | p-value | CV-value
|
|---|
| A | C |
|---|
| miR-22-3p | 0.452 | 0.015 | 0.422 | 0.693 |
| miR-513a-5p | 0.484 | 0.040 | 0.426 | 0.792 |
| miR-625-5p | 0.460 | 0.004 | 0.646 | 0.506 |
To further confirm the microarray results, an
additional independent 100 RNA samples, including 50 from
individuals with asthma and 50 from control subjects, were
subjected to RT-qPCR in order to validate the expression of
miR-625-5p, miR-22-3p and miR-513a-5p. These three miRNAs were
selected on the basis of predicting their targeted mRNAs:
CBL, peroxisome proliferator-activated receptor gamma,
coactivator 1 beta (PPARGC1B) and ESR1, which is
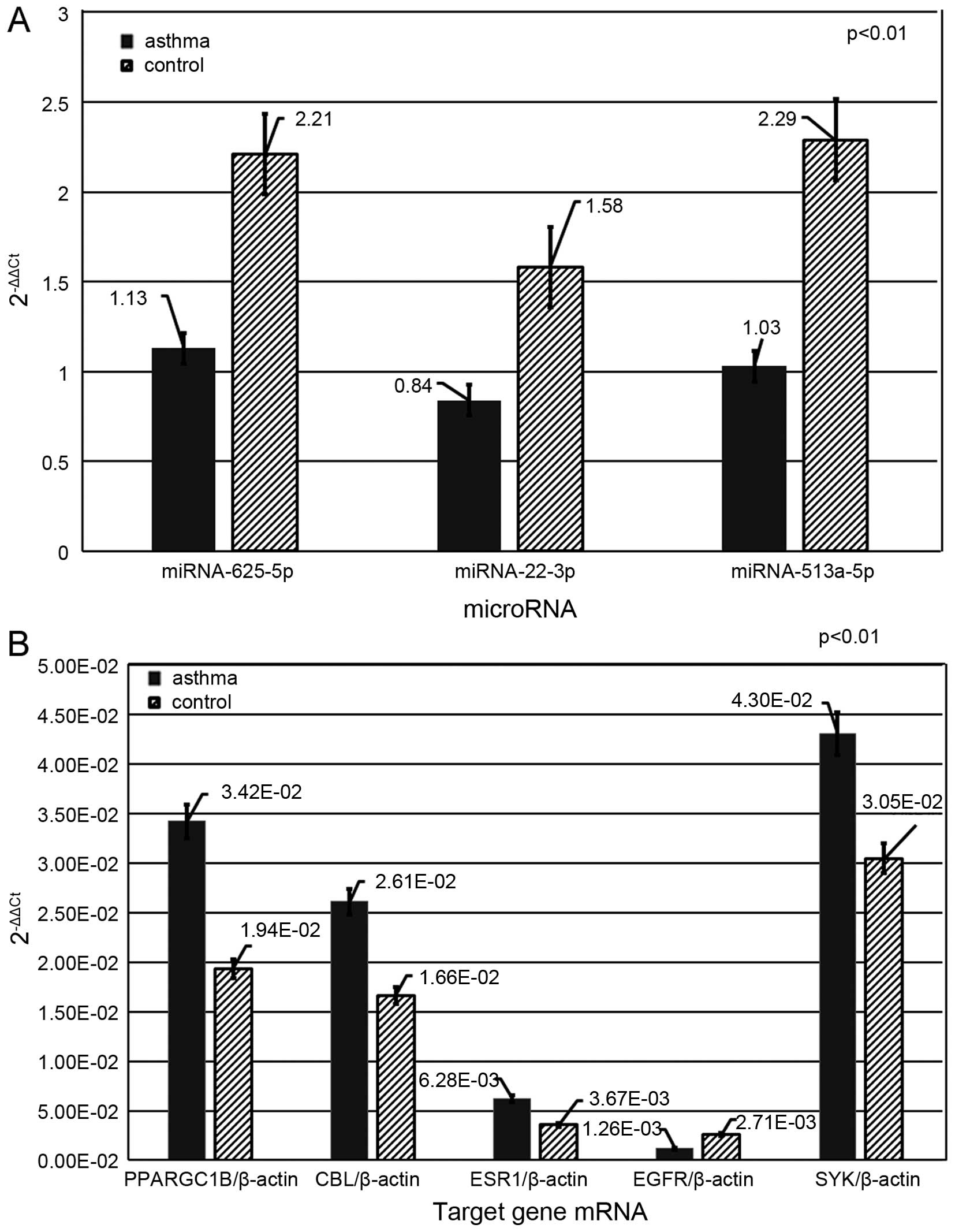
involved in the PI3K-AKT and NF-ĸB pathways (18,19). Our results showed that the average
levels of miR-625-5p, miR-22-3p and miR-513a-5p in the asthma group
were significantly lower than those in the control group (1.13±0.22
vs. 2.21±0.17, 0.84±0.17 vs. 1.58±0.27 and 1.03±0.14 vs. 2.29±0.26,
respectively; p<0.01) (Fig.
3A). No statistical significance was observed, regarding the
expression levels of these three miRNAs, among individual
participants within either the asthma group or the control group
(p>0.05).
Predicted target gene mRNAs
A total of 418 genes were predicted as the targets
of miRNA-513a-5p, 257 targeted by miRNA-22-3p, and 347 targeted by
miRNA-625-5p, respectively, in three web-based programs (miRDB,
miRBase and miRanda). These listed target genes were subjected to
further analysis with Venny software, through which miRNA targets
were found to be overlapped (Fig.
1C). Three mRNAs (PLCXD3, MSL2 and BRWD3) are the overlapped
targets of miRNA-513a-5p, miRNA-22-3p and miRNA-625-5p. Three pairs
of any two miRNAs combined may have 12 target genes co-regulated.
However, with bioinformatics functional analysis using the GO and
KEGG pathways, only three transcripts of mRNAs, CBL,
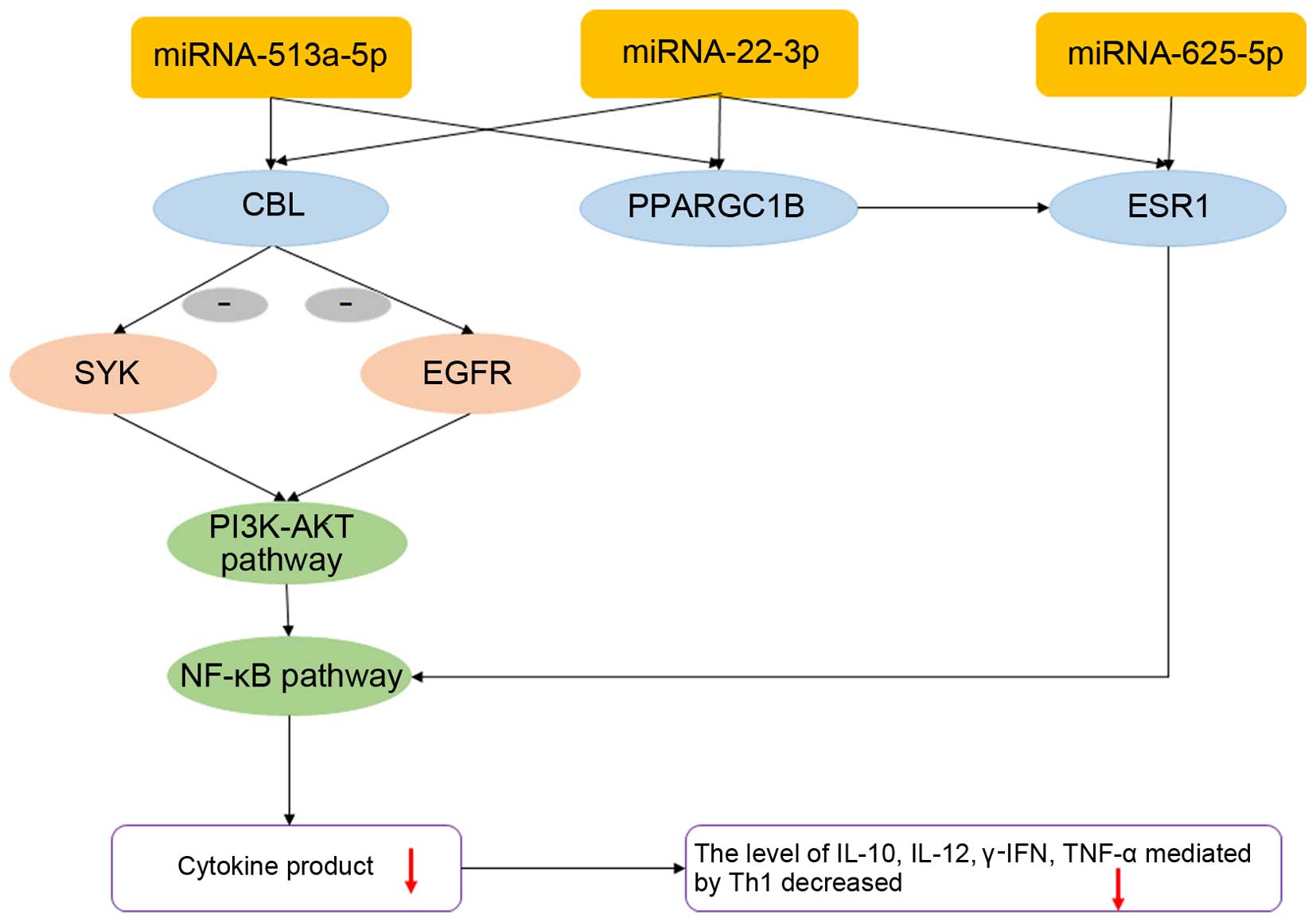
PPARGC1B and ESR1, were predicted as the targets of
miR-513a-5p, miR-22-3p and miR-625-5p, respectively (34). Transcripts of CBL and
PPARGC1B were dually targeted by miR-513a-5p and miR-22-3p,
and that of ESR1 was targeted by miR-625-5p and miR-22-3p.
mRNAs of CBL, PPARGC1B and ESR1 were
differentially elevated in the asthma group (CBL, 2.61E-02
vs. 1.66E-02; PPARGC1B, 3.42E-02 vs. 1.94E-02; ESR1,
6.28E-03 vs. 3.67E-03; p<0.01), as compared with the control
group (Fig. 3B) and have been
found to be associated with the PI3K-AKT and NF-ĸB signaling
pathways (Fig. 4) (35,36). It was also confirmed by GO and
KEGG pathway analysis in DAVID software that the CBL was
significantly associated with T- and B-cell differentiation and
inflammatory factor signaling pathways (GO enrichment value
>1.9, p<0.05).
SYK and EGFR are two molecules downstream
from CBL and may be mediated by the activation of CBL through the
PI3K-AKT and NF-ĸB pathways
Our assessment showed that in the asthma group,
levels of EGFR declined (1.26E-03 vs. 2.71E-03; p<0.01),
whereas levels of the mRNA of SYK involved in this pathway
were elevated (4.30E-02 vs. 3.05E-02; p<0.01) (Fig. 3B).
ELISA analysis of the plasma
concentrations of associated inflammatory cytokines
We further examined the concentrations of
inflammatory cytokine factors (γ-IFN, TNF-α, IL-13, IL-12, IL-4,
IL-6 and IL-10) which may be associated with the Th1, Th2 and
inflammation pathways PI3K-1KT and NF-κB that are mediated by the
miRNA-targeted genes CBL, PPARGC1B and ESR1
(18,19,37) (Table VII). Decreased concentrations of
γ-IFN, TNF-α, IL-12 and IL-10 in the plasma were found in the
asthma group, compared with those in the control group (3.44±2.87
vs. 7.08±4.30 pg/ml, 93.80±83.41 vs. 221.06±150.33 pg/ml, 1.50±0.66
vs. 3.43±1.37 pg/ml, and 2.39±2.47 vs. 3.74±1.98 pg/ml,
respectively, p<0.05). These decreased concentrations may be
associated with the different expression of upstream genes with a
regulatory function. The cytokine concentrations of IL-4, IL-6 and
IL-13 in the asthma group were neither changed nor showing
statistically significant differences compared with those in the
control group (p>0.05).
 | Table VIIPlasma concentrations of inflammatory
factors in dust mite–induced asthma. |
Table VII
Plasma concentrations of inflammatory
factors in dust mite–induced asthma.
| Inflammatory
factor | Asthma (pg/ml)
(n=62) | Control (pg/ml)
(n=62) | t-value | p-value |
|---|
| γ-IFN | 3.44±2.87 | 7.08±4.30 | 8.178 | 0.000 |
| TNF-α | 93.80±83.41 | 221.06±150.33 | 6.767 | 0.045 |
| IL-13 | 6.24±3.58 | 7.59±5.04 | 1.528 | 0.109 |
| IL-12 | 1.50±0.66 | 3.43±1.37 | 2.898 | 0.007 |
| IL-4 | 0.83±0.64 | 0.91±0.78 | 6.920 | 0.623 |
| IL-6 | 19.83±18.79 | 18.79±12.64 | 10.717 | 0.918 |
| IL-10 | 2.39±2.47 | 3.74±1.98 | 2.598 | 0.006 |
Discussion
Previous studies demonstrated that the abnormal
regulation of immune and inflammatory responses occur in asthma
patients (11–13). House dust mite allergen (Der f 2)
may activate the PI3K-AKT pathway (14), which subsequently activates the
NF-ĸB pathway and induces IL-13 expression in human airway
epithelial cells according to in vitro research. PI3K-AKT
was also involved in the induction of IL-6 in Der-p1-stimulated
nasal epithelial cells (NECs) from patients with allergic rhinitis
(AR), and may have potential implications for the prevention and
treatment of AR and asthma (38).
There was an interaction between the PI3K-AKT and NF-ĸB pathways
(38). NF-κB activation is the
crucial step between contact with an allergen and the downstream
manifestations of asthma (16).
In the present study, we found that these pathways may be regulated
by CBL, PPARGC1B and ESR1, which may be
mediated by miRNA, and alter the concentrations of downstream
cytokines.
As an immune regulator, miRNA appears to play an
important role in the regulation of immune responses and the
inflammatory signaling pathway in pediatric asthma (39). However, the mechanism responsible
for the involvement of miRNAs in the development of asthma has not
yet been fully elucidated. In the present study, children with
asthma were enrolled in a genome-wide investigation of novel
miRNA(s) that may have a regulatory function in pediatric asthma.
The results showed that the expression of three miRNAs
(miR-513a-5p, miR-22-3p and miR-625-5p) was downregulated in
samples from the asthma group and they were closely associated with
the immune response and cytokine secretion. GO and KEGG pathway
analysis using DAVID software demonstrated that the target gene
CBL, regulated by miR-513a-5p and miR-22-3p, was
significantly associated with T- and B-cell differentiation and
inflammatory factor signaling pathways (GO enrichment value above
1.9, p<0.05) (data not shown). The signaling pathways that were
mainly involved included the PI3K-AKT and NF-ĸB as well as the
T-cell receptor pathway (14–16,18). The PI3K-AKT pathway, in
particular, may affect NF-ĸB in the T- and B-cell receptor signal
pathways in the KEGG pathway maps [map04660 (http://www.genome.jp/kegg-bin/show_pathway?map04660),
map04062 (http://www.genome.jp/kegg-bin/show_pathway?map04062)
and map04668 (http://www.genome.jp/kegg-bin/show_pathway?map04668)].
However, the mechanism through which miRNAs regulate their
targeting genes to affect the inflammatory pathway as well as
identifying which pathway is principally involved needs to be
further elucidated.
CBL is an adapter protein that functions as a
negative regulator of many signaling pathways that are triggered by
the activation of cell surface receptors (34). CBL alters the inflammatory
signaling pathways in pediatric asthma by inhibiting the expression
of SYK and EGFR, which mediate the PI3K-AKT, MAPK and
NF-ĸB signaling pathways (21,40), resulting in the onset of asthma
(35,36). Data from our present study provide
evidence that miR-513a-5p and miR-22-3p are involved in dust
mite-induced pediatric asthma through regulating CBL, which
possibly alters the gene expression of EGFR and SYK
and further decreases levels of the cytokines γ-IFN, TNF-α, IL-12
and IL-10.
Additionally, PPARGC1B, predicted to be
regulated by miR-513a-5p and miR-22-3p, and ESR1, regulated
by miR-625-5p and miR-22-3p, were found by GO and KEGG analysis to
be significantly associated with inflammatory factor signaling
pathways. PPARGC1B is a protein that stimulates the activity
of transcription factors and nuclear receptors, and ESR1 is a
nuclear hormone receptor. There is an interaction between PPARGC1B
and ESR1 (34). ESR1, a negative
regulator of several inflammatory signaling pathways, directly
affects the transcription factor signaling pathway by inhibiting
NF-κB (18), thereby mediating
changes in the secretion of several inflammatory cytokines such as
IL-10, TNF, IL-6, γ-IFN and others in the NF-κB signaling pathway.
Some studies reported that ESR1 gene expression affected the
airway resistance of asthmatic patients (18,41). In this study, RT-qPCR determined
that the expression of the target genes PPARGC1B and
ESR1 regulated by miRNAs was significantly upregulated in
the samples from asthma patients.
The present study showed that levels of the
inflammatory cytokines TNF-α, IL-12, γ-IFN and IL-10 were lower in
the asthma group than in the control group. We assumed that the
decreased secretion of these cytokines was a result of the
upregulated expression of the associated target genes, which may be
the negative regulators of several inflammatory signaling pathways
such as PI3K-ATK, MAPK and NF-κB, and subsequently impacted on the
pathways mediating inflammatory cytokine secretion that lead to
pediatric asthma attacks, although this assumption requires further
investigation. In this study, no statistically significant
differences were observed in the concentrations of IL-4, IL-6 and
IL-13 between the asthma group and the control group, which does
not concur with the findings of previously mentioned in
vitro research (14). This
study demonstrated that miRNAs and their target genes principally
affected the secretion of the inflammatory cytokines TNF-α, IL-12,
γ-IFN, IL-10, and not the secretion of IL-4, IL-6 and IL-13. On the
other hand, we wondered whether our results may be associated with
the fact that the enrolled patients were not receiving any
medication or experiencing any acute asthma attacks. This is due to
the fact that different cytokine levels have been associated with
different phenotypes of asthma and medication therapy (30,31). Therefore, in future studies, the
enrollment of additional patients, including those receiving
medication or experiencing acute asthma attacks, would further
validate these findings.
In conclusion, we have identified three significant
miRNAs - miR-513a-5p, miR-22-3p and miR-625-5p - that were
differentially expressed in pediatric asthma. These miRNAs
regulated their target genes (CBL, PPARGC1B and
ESR1) and are likely have an impact on the regulation of
immune reactions and inflammatory cytokine pathways. We believe
that these three miRNAs, particularly miR-513a-5p and miR-22-3p,
and their target gene CBL, as reported in this study, may
play important roles in the epigenetic mechanisms underlying
pediatric asthma.
Acknowledgments
The present study was supported by the Shanghai
Municipal Health Bureau's Research Grant (no. 20124132). We thank
Kangchen Biotech (www.kangchen.com) for their technical assistance in
performing the miRNA microarray study.
References
|
1
|
Moorman JE, Person CJ and Zahran HS;
Centers for Disease Control and Prevention (CDC): Asthma attacks
among persons with current asthma - United States, 2001–2010. MMWR
Surveill Summ. 62(Suppl 3): 93–98. 2010.
|
|
2
|
National Cooperative Group on Childhood
Asthma; Institute of Environmental Health and Related Product
Safety; Chinese Center for Disease Control Prevention; Chinese
Center for Disease Control Prevention: Third nationwide survey of
childhood asthma in urban areas of China. Zhonghua Er Ke Za Zhi.
51:729–735. 2013.In Chinese.
|
|
3
|
Kabesch M: Gene by environment
interactions and the development of asthma and allergy. Toxicol
Lett. 162:43–48. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carlsten C and Melén E: Air pollution,
genetics, and allergy: an update. Curr Opin Allergy Clin Immunol.
12:455–460. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tham R, Dharmage SC, Taylor PE, Katelaris
CH, Vicendese D, Abramson MJ and Erbas B: Outdoor fungi and child
asthma health service attendances. Pediatr Allergy Immunol.
25:439–449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cecchi L: From pollen count to pollen
potency: the molecular era of aerobiology. Eur Respir J.
42:898–900. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gregory LG and Lloyd CM: Orchestrating
house dust mite-associated allergy in the lung. Trends Immunol.
32:402–411. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anderson GP: Endotyping asthma: new
insights into key pathogenic mechanisms in a complex, heterogeneous
disease. Lancet. 372:1107–1119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barnes PJ: Immunology of asthma and
chronic obstructive pulmonary disease. Nat Rev Immunol. 8:183–192.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
lobal Initiative for Asthma (GINA): Global
Strategy for Asthma Management and Prevention. 2014:2014.
|
|
11
|
Schuijs MJ, Willart MA, Hammad H and
Lambrecht BN: Cytokine targets in airway inflammation. Curr Opin
Pharmacol. 13:351–361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van Panhuys N, Le Gros G and McConnell MJ:
Epigenetic regulation of Th2 cytokine expression in atopic
diseases. Tissue Antigens. 72:91–97. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang X, Tang L, Wang F and Song G:
Astragaloside IV attenuates allergic inflammation by regulation
Th1/Th2 cytokine and enhancement CD4(+)CD25(+)Foxp3 T cells in
ovalbumin-induced asthma. Immunobiology. 219:565–571. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ro EJ, Cha PH, Kim HY, Cho YH, Park JW,
Han JS and Choi KY: House dust mite allergen Der f 2 induces
interleukin-13 expression by activating the PI3K/Akt pathway.
Immunol Res. 56:181–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou X, Hu H, Balzar S, Trudeau JB and
Wenzel SE: MAPK regulation of IL-4/IL-13 receptors contributes to
the synergistic increase in CCL11/eotaxin-1 in response to TGF-β1
and IL-13 in human airway fibroblasts. J Immunol. 188:6046–6054.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tully JE, Hoffman SM, Lahue KG, Nolin JD,
Anathy V, Lundblad LK, Daphtary N, Aliyeva M, Black KE, Dixon AE,
et al: Epithelial NF-κB orchestrates house dust mite-induced airway
inflammation, hyperresponsiveness, and fibrotic remodeling. J
Immunol. 191:5811–5821. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mora E, Guglielmotti A, Biondi G and
Sassone-Corsi P: Bindarit: an anti-inflammatory small molecule that
modulates the NFκB pathway. Cell Cycle. 11:159–169. 2012.
View Article : Google Scholar :
|
|
18
|
Wang X, Belguise K, O'Neill CF,
Sánchez-Morgan N, Romagnoli M, Eddy SF, Mineva ND, Yu Z, Min C,
Trinkaus-Randall V, et al: RelB NF-kappaB represses estrogen
receptor alpha expression via induction of the zinc finger protein
Blimp1. Mol Cell Biol. 29:3832–3844. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Li Y, Zhang L, Li M, Li C, Xue C,
Huang X and Zhou P: NF-κB downregulates Cbl-b through binding and
suppressing Cbl-b promoter in T cell activation. J Immunol.
194:3778–3783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Truscott M, Islam AB, Lightfoot J,
López-Bigas N and Frolov MV: An intronic microRNA links Rb/E2F and
EGFR signaling. PLoS Genet. 10:e10044932014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han C, Jin J, Xu S, Liu H, Li N and Cao X:
Integrin CD11b negatively regulates TLR-triggered inflammatory
responses by activating Syk and promoting degradation of MyD88 and
TRIF via Cbl-b. Nat Immunol. 11:734–742. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang B and Farwell MA: microRNAs: a new
emerging class of players for disease diagnostics and gene therapy.
J Cell Mol Med. 12:3–21. 2008. View Article : Google Scholar
|
|
23
|
Pauley KM, Cha S and Chan EK: MicroRNA in
autoimmunity and autoimmune diseases. J Autoimmun. 32:189–194.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johnnidis JB, Harris MH, Wheeler RT,
Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD and
Camargo FD: Regulation of progenitor cell proliferation and
granulocyte function by microRNA-223. Nature. 451:1125–1129. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tan Z, Randall G, Fan J, Camoretti-Mercado
B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF,
Nicolae D and Ober C: Allele-specific targeting of microRNAs to
HLA-G and risk of asthma. Am J Hum Genet. 81:829–834. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu TX, Munitz A and Rothenberg ME:
MicroRNA-21 is upregulated in allergic airway inflammation and
regulates IL-12p35 expression. J Immunol. 182:4994–5002. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Williams AE, Larner-Svensson H, Perry MM,
Campbell GA, Herrick SE, Adcock IM, Erjefalt JS, Chung KF and
Lindsay MA: MicroRNA expression profiling in mild asthmatic human
airways and effect of corticosteroid therapy. PLoS One.
4:e58892009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garbacki N, Di Valentin E, Huynh-Thu VA,
Geurts P, Irrthum A, Crahay C, Arnould T, Deroanne C, Piette J,
Cataldo D and Colige A: MicroRNAs profiling in murine models of
acute and chronic asthma: A relationship with mRNAs targets. PLoS
One. 6:e165092011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu TX and Rothenberg ME: Diagnostic,
functional, and therapeutic roles of microRNA in allergic diseases.
J Allergy Clin Immunol. 132:3–13. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bell MC and Busse WW: Severe asthma: an
expanding and mounting clinical challenge. J Allergy Clin Immunol
Pract. 1:110–121. 2013. View Article : Google Scholar
|
|
31
|
Cheng D, Xue Z, Yi L, Shi H, Zhang K, Huo
X, Bonser LR, Zhao J, Xu Y, Erle DJ and Zhen G: Epithelial
interleukin-25 is a key mediator in Th2-high,
corticosteroid-responsive asthma. Am J Respir Crit Care Med.
190:639–648. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rø AD, Simpson MR, Storrø O, Johnsen R,
Videm V and Øien T: The predictive value of allergen skin prick
tests and IgE tests at pre-school age: the PACT study. Pediatr
Allergy Immunol. 25:691–698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McDermott AM, Kerin MJ and Miller N:
Identification and validation of miRNAs as endogenous controls for
RQ-PCR in blood specimens for breast cancer studies. PLoS One.
8:e837182013. View Article : Google Scholar
|
|
34
|
Gene Cards The Human gene compendium.
http://www.genecards.org/.
|
|
35
|
Ishmael S and MacGlashan D Jr: Early
signal protein expression profiles in basophils: A population
study. J Leukoc Biol. 86:313–325. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Till SJ, Raynsford EJ, Reynolds CJ,
Quigley KJ, Grzybowska-Kowalczyk A, Saggar LR, Goldstone A,
Maillere B, Kwok WW, Altmann DM, et al: Peptide-induced immune
regulation by a promiscuous and immunodominant CD4T-cell epitope of
Timothy grass pollen: a role of Cbl-b and Itch in regulation.
Thorax. 69:335–345. 2014. View Article : Google Scholar
|
|
37
|
Qu D, Xu XM, Zhang M, Jiang TS, Zhang Y
and Li SQ: Cbl participates in shikonin-induced apoptosis by
negatively regulating phosphoinositide 3-kinase/protein kinase B
signaling. Mol Med Rep. 12:1305–1313. 2015.PubMed/NCBI
|
|
38
|
Shi J, Luo Q, Chen F, Chen D, Xu G and Li
H: Induction of IL-6 and IL-8 by house dust mite allergen Der p1 in
cultured human nasal epithelial cells is associated with
PAR/PI3K/NFkappaB signaling. ORL J Otorhinolaryngol Relat Spec.
72:256–265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Di Valentin E, Crahay C, Garbacki N,
Hennuy B, Guéders M, Noël A, Foidart JM, Grooten J, Colige A,
Piette J and Cataldo D: New asthma biomarkers: lessons from murine
models of acute and chronic asthma. Am J Physiol Lung Cell Mol
Physiol. 296:L185–L197. 2009. View Article : Google Scholar
|
|
40
|
Kadera BE, Toste PA, Wu N, Li L, Nguyen
AH, Dawson DW and Donahue TR: Low expression of the E3 ubiquitin
ligase CBL confers chemoresistance in human pancreatic cancer and
is targeted by epidermal growth factor receptor inhibition. Clin
Cancer Res. 21:157–165. 2015. View Article : Google Scholar :
|
|
41
|
Koppelman GH and Sayers I: Evidence of a
genetic contribution to lung function decline in asthma. J Allergy
Clin Immunol. 128:479–484. 2011. View Article : Google Scholar : PubMed/NCBI
|