Introduction
Spermatogonial stem cells (SSCs) are unipotent germ
cells which have been demonstrated to express many
pluripotency-associated genes as well as alkaline phosphatase (AP)
activity as they are pluripotent stem cells (PSCs) (1,2).
They also possess the potential ability to reacquire pluripotency
due to spontaneous epigenetic reprogramming (3). Epigenetic mechanisms are closely
associated with the induction and the maintenance of pluripotency
(4). Previous findings have
revealed the complex connection between epigenetic modification
factors and pluripotent transcription factors, both of which
control gene expression directly linked to pluripotency and
reprogramming (5). It has been
demonstrated that the generation of induced (i)PSCs relies on the
exogenous expression of transcription factors (such as Oct4,
Sox2, N-Myc and Klf4), which is an inefficient
and random reprogramming process (6). However, epigenetic factors have been
shown to provide a more powerful means of improving reprogramming
efficiency (7). In fact, the
molecular mechanism responsible for the in vitro
reprogramming of SSCs may provide insight into the epigenetic
reprogramming of iPSCs (5).
Although previous experiments have investigated the
differences in transcript and proteomic profiles between mouse
(m)SSCs and mouse embryonic stem cells (mESCs) (8,9),
differences in the expression of crucial transcription factors and
epigenetic factors remain unclear. A recent study has indicated
that the loss of Dmrt1, Dnmt1 and tumor protein
(Tp)53 expression, and the overexpression of Oct4
increased the rate of mSSC reprogramming (10). However, the mechanism of SSC
reprogramming to PSCs remains unknown, particularly due to the
difficulty of tracing orchestrated epigenetic changes during the
very low-efficiency reprogramming process (10). As a result, it becomes
increasingly important to determine the differential gene
expression of pluripotent factors and epigenetic factors in mSSCs
and mESCs in order to elucidate the mechanism of mSSC
reprogramming. Thus, we examined the relative mRNA expression of
ESC-associated transcription factors and epigenetic factors in
freshly isolated mSSCs [mSSCs (f)] and long-term propagated mSSCs
[mSSC (l)] versus mESCs.
Materials and methods
Isolation of mSSCs (f)
The mSSCs were isolated from 6-day-old imprinting
control region (ICR) male mouse testes at our laboratory by
two-step enzyme digestion and magnetic-activated cell sorting
(MACS) with CD90.2 microbeads (Miltenyi Biotec, Bergisch Gladbach,
Germany) as previously described (11). The experiment was repeated >3
times and 30 mice were used each time. The mice were sacrificed by
decapitation and the testes were removed for the isolation of
mSSCs. All procedures were performed in accordance with the animal
care guidelines of the Institutional Animal Care and Use Committee
of Guangzhou Medical University (Guangdong, China) and were
conducted in accordance with the National Research Council Guide
for the Care and Use of Laboratory Animals.
Culture of mSSCs and mESCs
The purified mSSCs (f) were cultured on mouse
embryonic fibroblast (MEF) feeder cells treated with mitomycin C
(Sigma, St. Louis, MO, USA). The cells were cultured in StemPro-34
SFM, a serum-free medium (Invitrogen, Carlsbad, CA, USA)
supplemented with 20 ng/ml recombinant rat glial cell line-derived
neurotrophic factor, 10 ng/ml recombinant human basic fibroblast
growth factor (both from PeproTech, Rocky Hill, NJ, USA), 10 ng/ml
mouse epidermal growth factor (Prospec-Tany TechnoGene, Ltd., East
Brunswick, NJ, USA), 1,000 U/ml recombinant mouse leukemia
inhibitory factor (LIF; Millipore, Billerica, MA, USA), 20 ng/ml
platelet-derived growth factor-BB (PeproTech), 1 mmol/l glutamine,
1X insulin-transferrin-selenium (ITS), and 1X B27 supplements (all
from Gibco, Grand Island, NY, USA). The mSSCs (f) (5×105/ml)
cultured in a 25 cm2 flask under these conditions were
passaged every 7 days and the culture medium was changed every 2
days. After culturing for 4 weeks, the mSSCs (f) were capable of
stably proliferating in vitro as mSSCs (l). Trypsin-EDTA
(0.25% Invitrogen) and Accutase (1 mg/ml, Sigma) were used to split
mSSCs clusters away from MEF feeder cells. To maintain the adherent
state of MEF feeder cells, the process of digestion was controlled
within no more than 1 min, observed under a light microscope and
stopped using the completed culture medium. The mSSC clusters were
transferred to a centrifuge tube and centrifuged under 69 x g at
4℃, 3 min after washing with phosphate-buffered saline (PBS).
The mESC (R1) cell line was kindly donated by Dr
Shaorong Gao at the School of Life Sciences and Technology at
Tongji University (Shanghai, China). The in vitro culture
and characterization of mESCs (R1) and the induced differentiation
of mSSCs into round spermatids (RSs) were performed as previously
described (11,12). Briefly, the mESC (R1) cell line
was cultured in DMEM (Gibco) supplemented with 1 mmol/l glutamine
(Gibco), 100X nucleotide (Millipore), 55 μM β-ME (Gibco), 15% fetal
bovine serum (FBS; Gibco) and 1,000 U/ml LIF (Millipore), on the
MEF feeder cells. For the induction of sperm differentiation, the
mSSCs were cultured in DMEM (Gibco) supplemented with 10% FBS
(Gibco), 500 ng/ml follicle-stimulating hormone (Sigma), 5 μM
vitamin A (Sigma), 0.1 mM testosterone (Sigma), 100X ITS (Gibco), 1
mmol/l glutamine (Gibco), 100X sodium pyruvate (Gibco), and 100X
nonessential amino acid (NEAA; Gibco) on mouse testicular
fibroblast feeder cells.
AP staining of mSSCs
The mSSC clusters were fixed in 4% paraformaldehyde
at room temperature for 20 min and then washed three times with PBS
for 15 min. The detector reagents from the AP detection kit
(Dingguo Changsheng Biotechnology Co., Ltd., Beijing, China) were
then added and the samples were incubated at room temperature (in
the dark) for 15 min. The reaction was terminated by performing
three PBS washes. Images were captured using a light microscope
(IX71 model with TH4-200 accessories; Olympus, Tokyo, Japan).
Immunohistochemical analysis
The mouse testes were fixed in 4% paraformaldehyde
for 24 h, embedded in paraffin, and processed for
immunohistochemical analysis. Briefly, 5-µm section slides
were dewaxed in xylene and rehydrated using a series of graded
alcohols. Immunostaining was performed by incubating the slides
with the mouse monoclonal anti-promyelocytic leukaemia zinc finger
(PLZF) antibody (sc-28319; 1:100) overnight at 4°C, followed by
incubation with goat anti-mouse IgG-HRP (sc-2005; 1:200) (both from
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at 37°C for 1
h. The sections of the mouse testes were counterstained with
hematoxylin after diaminobenzidine staining (both from Dingguo
Changsheng Biotechnology Co. Ltd., Beijing, China) and examined
under a light microscope (Olympus).
Immunofluorescence
The mSSC clusters were fixed with 4%
paraformaldehyde for 30 min, washed three times with PBS, and
blocked in 1% BSA (Sigma) for 30 min. The cells were incubated with
a mouse monoclonal anti-GFRα1 antibody (sc-271546; 1:200; Santa
Cruz Biotechnology, Inc.) and an anti-PLZF mouse IgG antibody
(sc-28319; 1:200; Santa Cruz Biotechnology, Inc.) at 4°C overnight
and washed three times in PBS. The secondary antibody, Alexa Fluor
568-labeled goat anti-mouse IgG (1:100; Invitrogen) was added and
incubated for 1 h at 37°C in the dark. The cell nuclei were stained
with 10 µg/ml Hoechst 33342 (Molecular Probes, Eugene, OR,
USA). The samples were observed under a fluorescent microscope
(IX71 with U-RFL-T accessories; Olympus).
Flow cytometric analysis
The mSSC clusters were digested with Accutase (Stem
Cell Technologies, Inc., Vancouver, BC, Canada) and the collected
cells were fixed in 4% paraformaldehyde for 20 min followed by
three washes with PBS. The cells were then stained with mouse
monoclonal anti-CD90.2-FITC (Miltenyi Biotec) for 30 min at 4°C in
the dark and detected by flow cytometry (FACSCalibur; BD
Biosciences, Franklin Lakes, NJ, USA).
RNA extraction, cDNA synthesis, and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted from mSSCs (f), mSSCs (l),
and mESCs using an RNeasy mini kit (Qiagen, Inc., Valencia, CA,
USA) according to the manufacturer's instructions. RNA was
transcribed to cDNA using a cDNA synthesis kit (Takara, Otsu,
Japan) with oligo-dT primers. The primer sequences used in this
study are listed in Tables I and
II. Relative mRNA expression
analyses were run in triplicate for each sample using a Power
SYBR-Green Realtime PCR kit (Toyobo Co., Ltd., Osaka, Japan) on a
qPCR machine (Illumina, Inc., San Diego, CA, USA). β-actin was used
as an internal control. The relative mRNA abundance of target genes
was expressed as 2−ΔΔCt.
 | Table IPrimer sequence, target product size
and accession number of target genes for regular PCR. |
Table I
Primer sequence, target product size
and accession number of target genes for regular PCR.
| Gene | Primer sequence
(5′→3′) | Product size
(bp) | Accession no. |
|---|
| β-actin | F:
TGCTGTCCCTGTATGCCTCTG | 222 | NM_007393.3 |
| R:
TGATGTCACGCACGATTTCC | | |
| Oct4 | F:
GGGATGGCATACTGTGGACC | 837 | NM_013633.3 |
| R:
CAGAGCAGTGACGGGAACAGA | | |
| Sox2 | F:
AAACCACCAATCCCATCCAA | 459 | U31967.1 |
| R:
TTGCCTTAAACAAGACCACGAA | | |
| Nanog | F:
CTGATTCTTCTACCAGTCCCAAAC | 380 | XM_006506651.1 |
| R:
AGATGCGTTCACCAGATAGCC | | |
| Lin28 | F:
CCAAAGGAGACAGGTGCTACAA | 167 | XM_006539317.1 |
| R:
GGCAGGCTTTCCCTGAGAA | | |
| N-Myc | F:
GGTGGGTCGTCGAGTGCTAG | 393 | M36277.1 |
| R:
AGTGGTTACCGCCTTGTTGTTA | | |
| Klf4 | F:
ACTAACCGTTGGCGTGAGGA | 625 | BC010301.1 |
| R:
TGCTAACACTGATGACCGAAGG | | |
| Tert | F:
AGCATTTCACCCAGCGTCTC | 436 | XM_006517210.1 |
| R:
TGCTCGATGACAACGGAGTTC | | |
| Plzf | F:
ACCCATACTGGCACGGACAT | 346 | XM_006510258.1 |
| R:
TGTGAACCCTGTAGTGCGTCTC | | |
| Vasa | F:
AGCATTCCCATTGTATTAGCAGG | 573 | NM_001145885.1 |
| R:
CACTTGCCCAACAGCGACA | | |
| Dazl | F:
GTTAGGATGGATGAAACCGAAAT | 739 | NM_010021.5 |
| R:
CAGATTTAAGCACTGCCCGAC | | |
| Nanos3 | F:
CGAGTCCCGTGCCATCTATC | 302 | NM_194059.2 |
| R:
GGGGCTTCCTGCCACTTT | | |
| Stra8 | F:
AGGCAACCAACCCAGTGATG | 156 | XM_006505829.1 |
| R:
TCCTGTTCCTGAATATGAATCTTTGT | | |
| Cldn6 | F:
GGCAACAGCATCGTCGTGG | 333 | NM_018777.4 |
| R:
GAAGTCCTGGATGATAGAGTGGGC | | |
| Pdgfrα | F:
GTTCAAGACCAGCGAGTTTAATGT | 376 | NM_011058.2 |
| R:
GCCAAAGGTGGGCTCAATC | | |
| Gsg2 | F:
CTTTAGTGATTGCCTTTCCACG | 612 | D87326.1 |
| R:
GTGGGAATGGTGCTCGTTTT | | |
| Acrosin | F:
TCTTGGCAGTGTCCGTGGTT | 309 | D00754.1 |
| R:
TGTTTCTTCCATATTCGATTTCTTGT | | |
 | Table IIPrimer sequence, target product size
and accession number of target genes for RT-qPCR. |
Table II
Primer sequence, target product size
and accession number of target genes for RT-qPCR.
| Gene | Primer sequence
(5′→3′) | Product size
(bp) | Accession no. |
|---|
| β-actin | F:
TGCTGTCCCTGTATGCCTCTG | 222 | NM_007393.3 |
| R:
TGATGTCACGCACGATTTCC | | |
| Oct4 | F:
GTGTTCAGCCAGACCACCATC | 112 | NM_013633.3 |
| R:
CATTGTTGTCGGCTTCCTCC | | |
| Sox2 | F:
CAAGGAAGGAGTTTATTCGGATTT | 178 | U31967.1 |
| R:
ATCAACCTGCATGGGCATTT | | |
| Nanog | F:
CTGATTCTTCTACCAGTCCCAAAC | 156 | XM_006506651.1 |
| R:
GCTTCTGAAACCTGTCCTTGAGT | | |
| Lin28 | F:
CCAAAGGAGACAGGTGCTACAA | 167 | XM_006539317.1 |
| R:
GGCAGGCTTTCCCTGAGAA | | |
| N-Myc | F:
TCCTCTAACAACAAGGCGGTAA | 130 | M36277.1 |
| R:
TGTGCTGCTGATGGATGGG | | |
| Klf4 | F:
ACTAACCGTTGGCGTGAGGA | 175 | BC010301.1 |
| R:
CGTTGAACTCCTCGGTCTCC | | |
| Esrrb | F:
CATGAAATGCCTCAAAGTGGG | 186 | NM_011934.4 |
| R:
TCCTGCTCAACCCCTAGTAGATT | | |
| Utf1 | F:
TCCTCTTACGAGCACCGACAC | 146 | NM_009482.2 |
| R:
GAGCAACCTGCGGGGAA | | |
| Dppa2 | F:
GAGGAGCCAAACACAGACTACG | 138 | AF490346 |
| R:
CGGAGGACAGGTGCTTGGT | | |
| Tbx3 | F:
GGAACCCGAAGAAGACGTAGAA | 160 | NM_011535.3 |
| R:
CTTTTTATCCAGTCCAGAGCACC | | |
| Nr5a2 | F:
TCCCACACCTGATACTGGAACTT | 114 | NM_030676.3 |
| R:
GCTTTTCTTGCCTGTTTCGG | | |
| Prdm14 | F:
GAGTGAGATTTGGACCCTTTCG | 165 | NM_001081209 |
| R:
ACCGAGCACAGTTGACATAGGAC | | |
| Klf2 | F:
CCCAGGAAAGAAGACAGGAGTCT | 122 | NM_008452.2 |
| R:
ACTCAAAGGCATTTCTCACAAGG | | |
| Prmt5 | F:
CCTTTGCCGACAACGAGC | 179 | NM_013768.3 |
| R:
AAACTGTGCCTCAGGATCGC | | |
| Dmrt1 | F:
GGAGCGACAGCGGGTGA | 142 | AF202778.1 |
| R:
CGGGTTGCTGGCATTATTCT | | |
| Tet1 | F:
CCTATCTTCCTTCCTAAGCCTCC | 164 | NM_001253857.1 |
| R:
TCAGGGTTTGGTGGGAGTTG | | |
| Tet2 | F:
AATGGAAGCCCGTTAGCAGA | 150 | XM_006501281.1 |
| R:
GCACCTGGAATACCCTCTGTCT | | |
| Tet3 | F:
GCTCGTCTGGAAGATGCCC | 120 | XM_006505773.1 |
| R:
CTCACGACTCATCTCACGGTTG | | |
| Parp1 | F:
CGTCAACTACGAGAAACTCAAAACT | 120 | NM_007415.2 |
| R:
AGGTCATAGGCGTTGTGCG | | |
| Dnmt1 | F:
AGTCGGACAGTGACACCCTTTC | 118 | NM_001199431.1 |
| R:
GGTTTCCGTTTAGTGGGGC | | |
| Kdm2b | F:
ACTCACCTTACCGAATTTGAACTG | 149 | NM_001003953.1 |
| R:
ACGTGCTCTTTCAGTACATTCTTTAC | | |
| Dot1l | F:
CTGGCAAGCCTGTCTCCTACTAT | 149 | NM_199322.1 |
| R:
CGTGGTCGCATTGCTCTTG | | |
| Max | F:
CTCTACACCAACGCCAAGGG | 178 | NM_001146176.1 |
| R:
CAGAAGGAGGATGCGACGAG | | |
| Tert | F:
TGCTGGACACTCAGACTTTGGA | 102 | XM_006517210.1 |
| R:
TTCAACCGCAAGACCGACA | | |
| Trf1 | F:
AAGAACGCCTTATCGCAGTTAA | 120 | NM_009352.3 |
| R:
TCCACTGGTTCTTCGGTTCC | | |
| Zscan4c | F:
GCAAATGTTGGTGAAAGCTGTAGT | 175 | NM_001013765.2 |
| R:
TAGTCGGAGCACTCGGGAAG | | |
Western blot analysis
Proteins were extracted from mSSCs (l) and mESCs
using RIPA lysis buffer (Beyotime, Shanghai, China) containing 1%
protease inhibitor cocktail (Roche, Mannheim, Germany). The lysed
samples were centrifuged at 4°C, 10,000 × g for 15 min to obtain
the supernatants. Protein concentrations in the supernatants were
determined using the BCA protein assay kit (Bio-Rad, Hercules, CA,
USA). The supernatant proteins were denatured, separated by
SDS-PAGE, and transferred to nitrocellulose membranes (Bio-Rad).
The membranes were blocked with 5% non-fat dry milk powder in 1X
PBS containing 0.1% Tween-20 (TBST) for 1 h at room temperature.
The blots were incubated with primary antibodies [rabbit anti-mouse
PRMT5 (ab2538; 1:200; MultiSciences Biotech Co., Ltd., Hangzhou,
China); rabbit anti-mouse LIN28 homolog A (LIN28) (sc-67266;
1:200); rabbit anti-mouse β-actin (sc-130656; 1:1,000) in TBST with
5% non-fat milk overnight at 4°C with gentle shaking, followed by
incubation with peroxidase-conjugated secondary antibody (goat
anti-rabbit IgG-HRP; sc-2030; 1:1000) (all from Santa Cruz
Biotechnology, Inc.) in TBST with 5% non-fat milk for 2 h at room
temperature. Chemiluminescence signals were detected using
SuperSignal West Dura HRP detection kits (Pierce, Rockford, IL,
USA). The images were captured using a ChemiDoc XRS system equipped
with Quantity One software (Bio-Rad).
DNA methylation analysis
Genomic DNA was extracted from mSSCs (l) and mESCs
using a Genomic DNA kit (Tiangen Biotechnology, Beijing, China) and
treated with an EZ DNA Methylation-Gold kit (Zymo Research, Irvine,
CA, USA) to deaminate unmethylated cytosines to uracils. The DNA
templates were used to amplify differentially methylated regions
(DMRs) by specific primers (forward,
5′-TGGTTGTTTTGTAGGATTTGTTAGA-3′ and reverse,
5′-AAAACTTCCCTCTTCCCTCTTAATAT-3′). The amplified products were then
purified using a Gel Extraction kit (Omega Bio-Tek, Inc., Norcross,
GA, USA), subcloned into pMD™18-T vectors (Takara) and sequenced by
M13R primers.
Statistical analysis
The differences between groups were assessed using
ANOVA and Student's t-tests with SPSS v.11 software. The results
are presented as the means ± standard error. A p<0.05 was
considered to indicate a statistically significant difference.
Results
Isolation of mSSCs (f)
Immunohistochemical staining of sections of
6-day-old male ICR mouse testes showed that the PLZF-positive mSSCs
were localized to the basal membrane of the testicular seminiferous
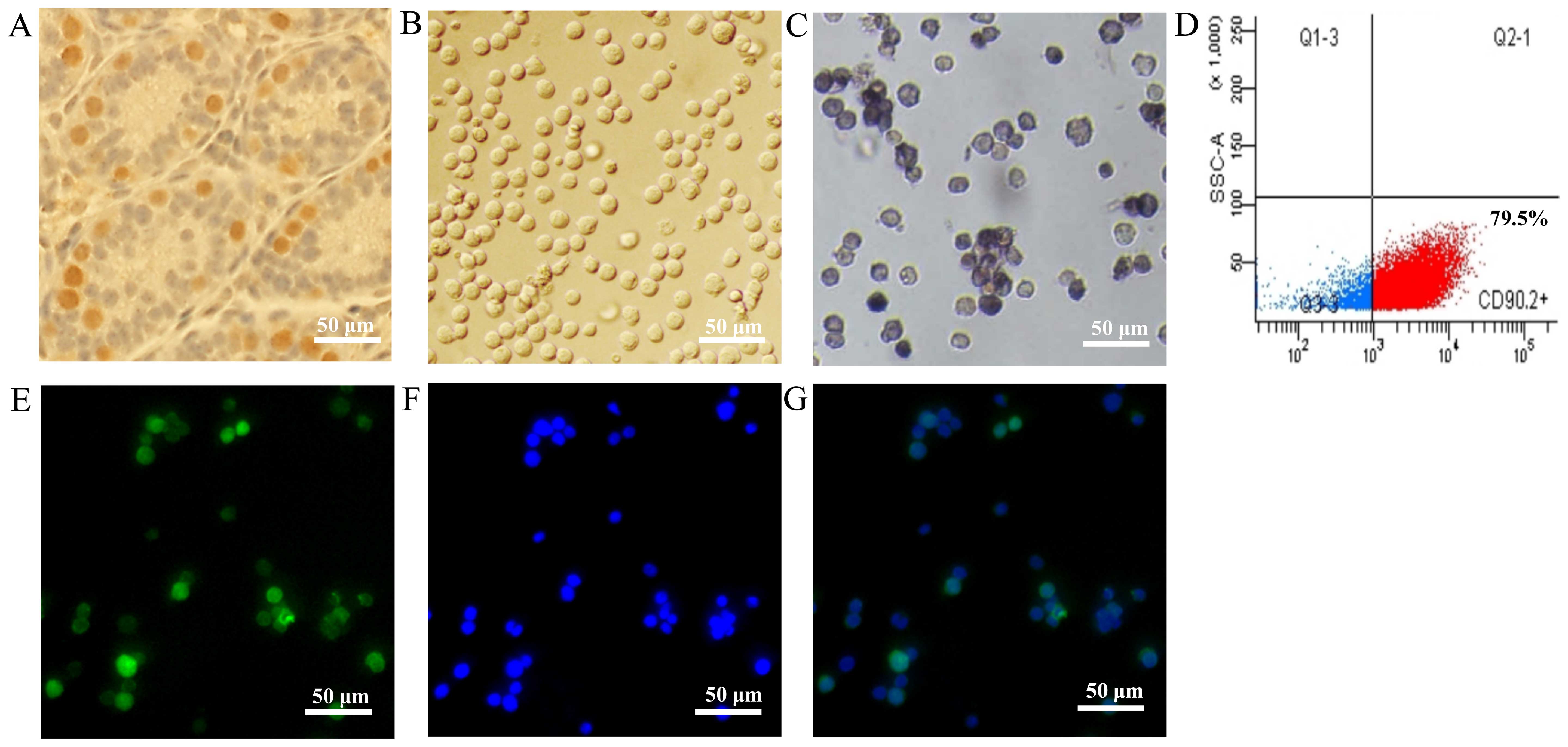
tubules (Fig. 1A). The mSSCs (f),
enriched by CD90.2 microbeads, displayed a unified morphological
appearance (Fig. 1B) and AP
staining activity (Fig. 1C).
These mSSCs (f) had a purity of 79.5%, as detected by flow
cytometry (Fig. 1D), and
immunofluorescence staining confirmed that they expressed the SSC
marker, PLZF protein (Fig.
1E–G).
Propagation and characterization of mSSCs
(l)
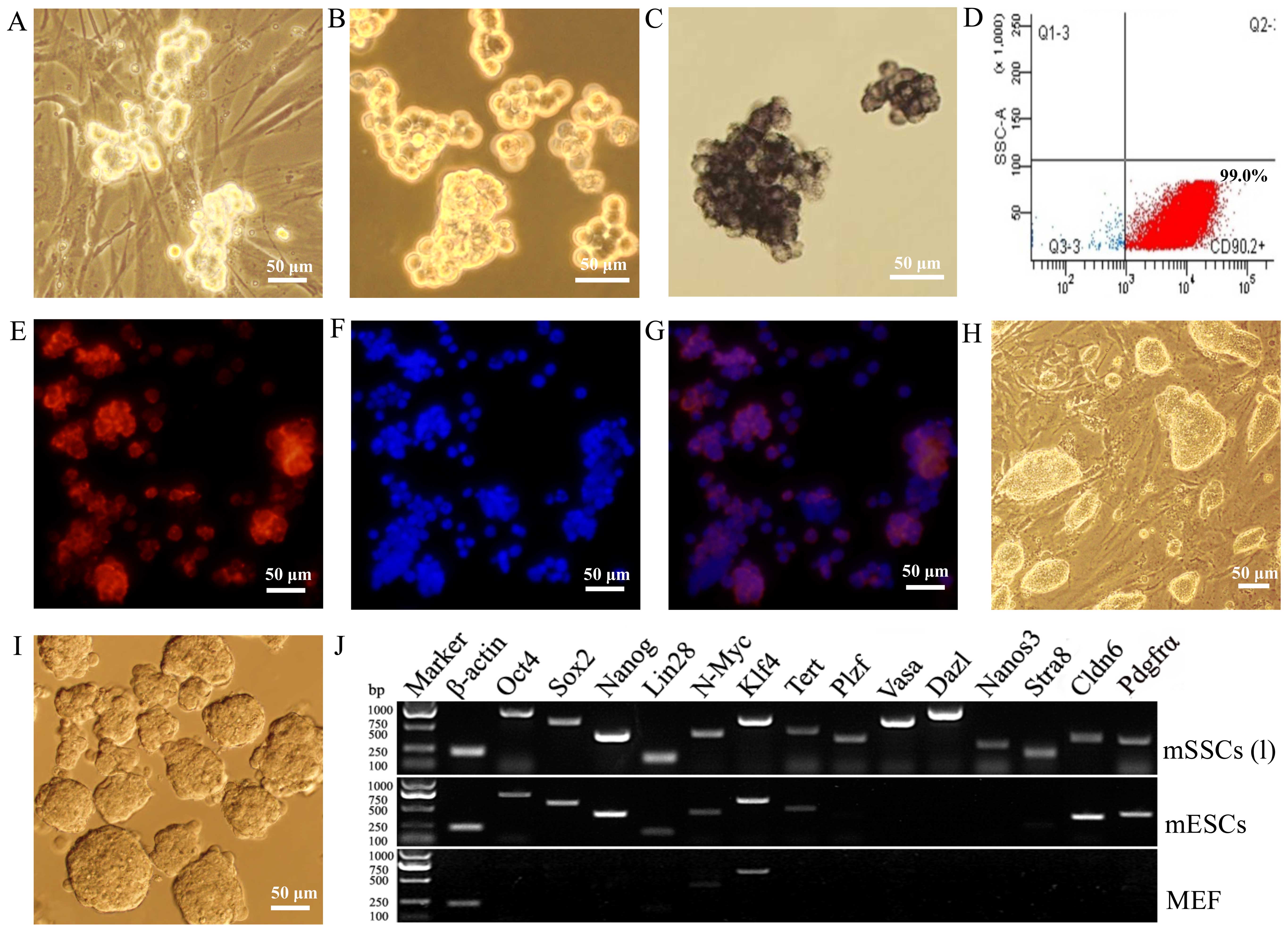
The self-renewal capacity of mSSCs (f) was
maintained in vitro for >5 months [to produce mSSCs (l)]
on MEF feeder cells (Fig. 2A and
B). The mSSCs (l) displayed AP activity (Fig. 2C) and expressed CD90.2 (Fig. 2D) and GFRα1 (Fig. 2E–G). These colonies of mSSCs (l)
were quite different from the colonies of mESCs (Fig. 2H and I). Furthermore, RT-PCR
revealed that the mSSCs (l) expressed germline factors
(Plzf, Vasa, Dazl, Nanos3 and
Stra8), ESC pluripotency factors (Oct4, Sox2,
Nanog, Lin28, N-Myc, Klf4 and
Tert) and Cldn6 and Pdgfrα surface markers,
whereas MEFs only expressed N-Myc and Klf4 (Fig. 2J).
Differentiation of mSSCs (l)
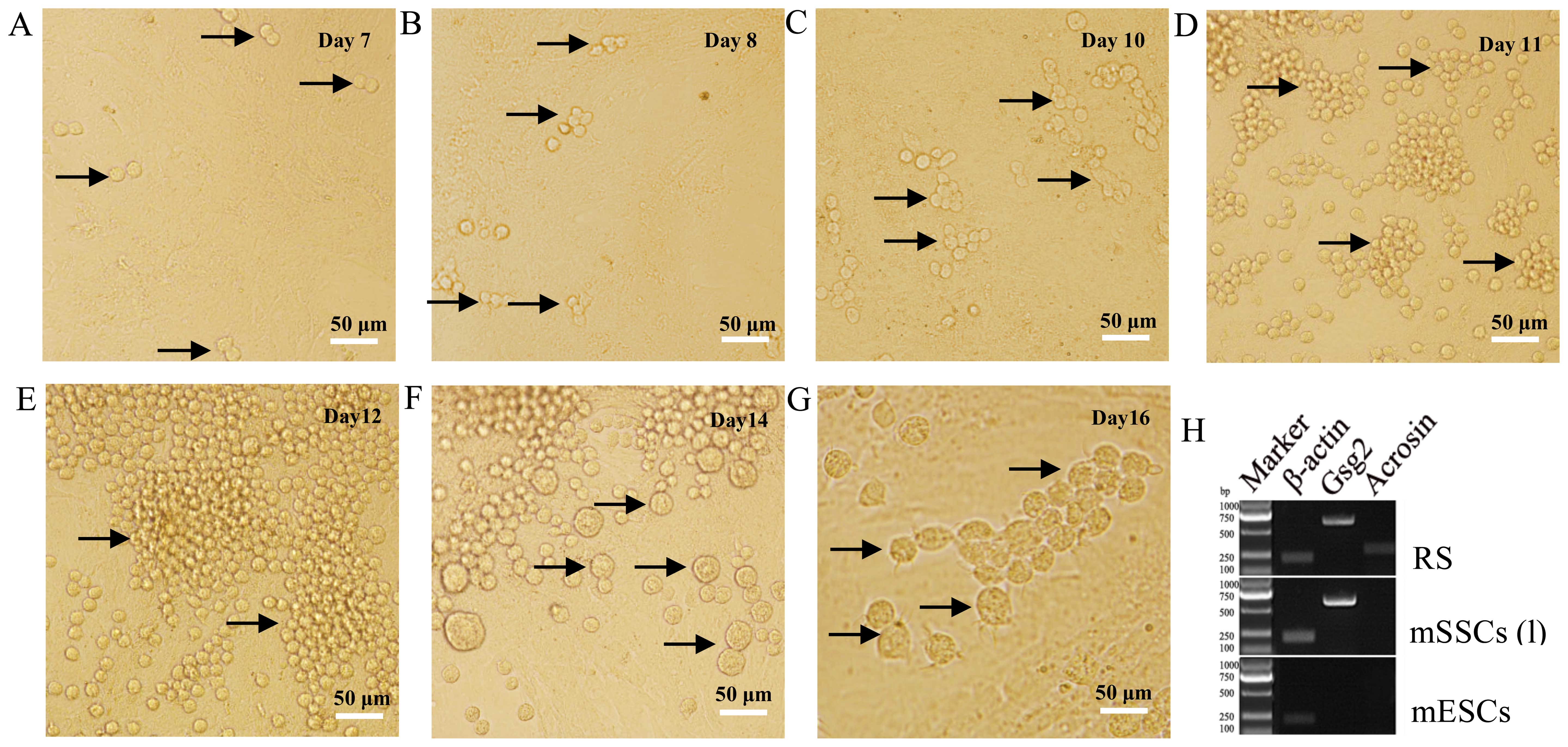
Our results indicated that mSSCs (l) were capable of
differentiating into sperm in vitro. After 7 days of
differentiation culture, A-paired (Apr) spermatogonia were observed
(Fig. 3A). Subsequently,
A-aligned (Aal) spermatogonia of 4- (Aal-4) (Fig. 3B), 8- (Aal-8) (Fig. 3C) and 16-cells (Aal-16) (Fig. 3D) emerged on days 8, 10 and 11,
respectively. Next, A1, A2, A3, A4, intermediate (In), and B
spermatogonia began to appear from days 12 to 14 (Fig. 3E and F). During this pivotal
developmental time frame, differentiated spermatogonia (A2 to B)
derived from A1 cells were synthesized in bulk in preparation for
meiosis. Round spermatids (RSs) were formed on day 16 (Fig. 3G) after meiosis. These RSs
expressed sperm markers (Gsg2 and Acrosin), whereas
mESCs did not express either gene (Fig. 3H).
Relative mRNA expression of transcription
factors in mSSCs
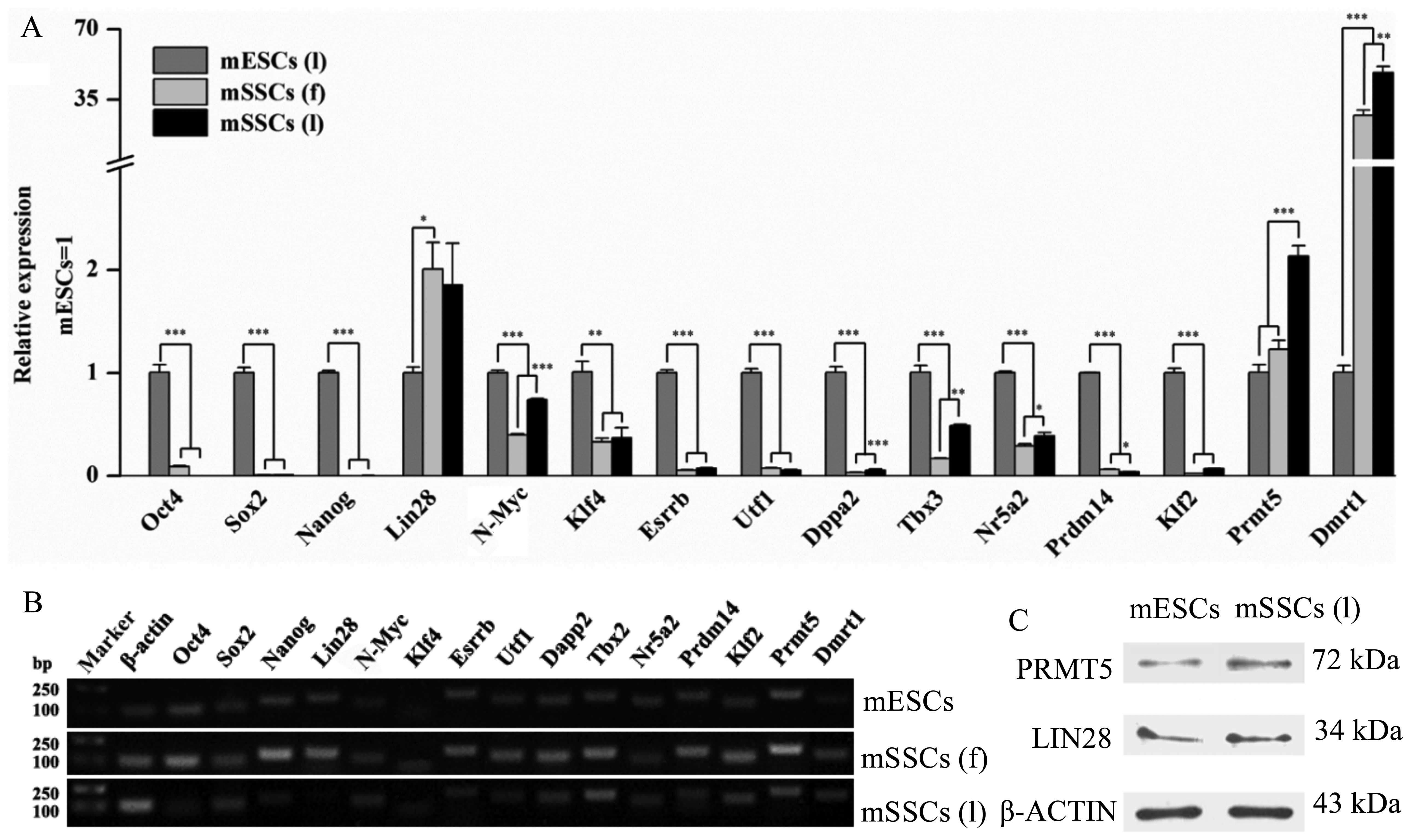
The relative mRNA expression of transcription
factors (Oct4, Sox2, Nanog, N-Myc,
Klf4, Esrrb, Utf1, Dppa2, Tbx3,
Nr5a2, Prdm14 and Klf2) in both types of mSSC
was significantly lower than those in the mESCs (Fig. 4A). For example, the expression of
Oct4, Sox2 and Nanog in the mESCs was
significantly higher than in the mSSCs (l). Notably, the expression
level of Prmt5 and Lin28 was significantly higher in
the mSSCs (l) versus the mESCs. Western blot analysis also
confirmed that the mSSCs (l) and the mESCs expressed LIN28 and
PRMT5 proteins (Fig. 4C). The
mRNA expression of Dmrt1 in both the mSSC types was higher
compared with that in the mESCs (Fig.
4A). Additionally, our results indicated that the expression of
N-Myc, Dppa2, Tbx3, Nr5a2 and
Prmt5 in the mSSCs (l) was markedly upregulated in
comparison with the mSSCs (f) (Fig.
4A). Confirmation of the qPCR products of the transcription
factors was also demonstrated (Fig.
4B).
Relative mRNA expression of epigenetic
factors in mSSCs
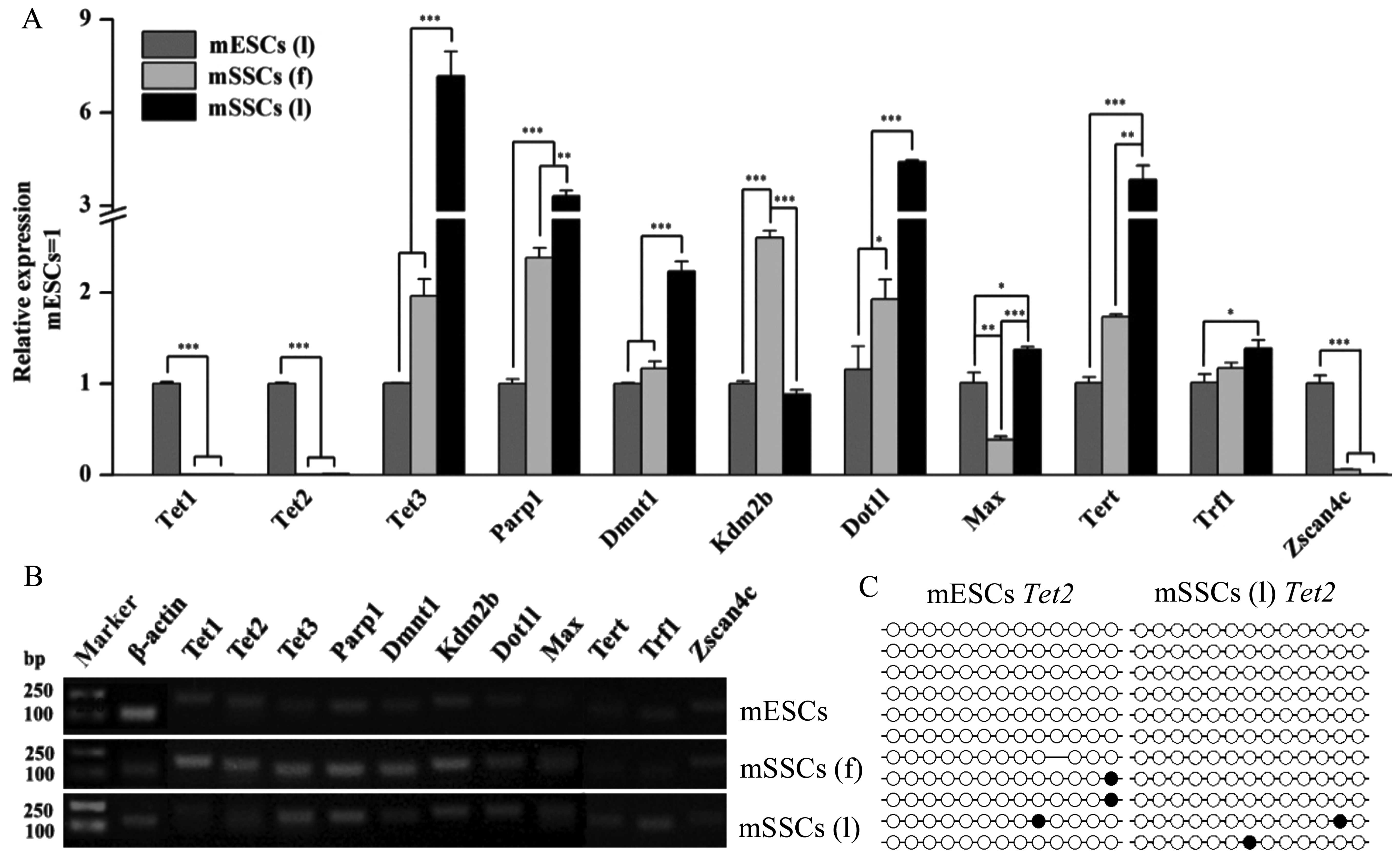
Epigenetic factors critical for promoting
pluripotency and reprogramming were investigated (Fig. 5), including the genes responsible
for genomic methylation regulation (Tet1, Tet2,
Tet3, Parp1 and Dnmt1, histone modification
(Kdm2b, Dot1l and Max), and telomere
maintenance (Tert, Trf1 and Zscan4c). The
results of RT-qPCR revealed that the mSSCs and the mESCs exhibited
different expression levels of these factors (Fig. 5A). Tet1, Tet2 and
Zscan4c were abundantly expressed in the mESCs but not in
the mSSCs (l), whereas the levels of Tet3, Parp1,
Dnmt1, Dot1l and Tert were significantly
higher in the mSSCs than in the mESCs (Fig. 5A). To further examine the possible
association between the low expression of Tet2 and DNA
methylation, we determined the DNA methylation state of the
Tet2 promoter. However, the Tet2 promoter in the
mSSCs (l) did not show a high DNA methylation level by bisulfite
sequencing PCR analysis (Fig.
5C). Furthermore, Kdm2b expression was significantly
higher in the mSSCs (f) than in the mESCs and the mSSCs (l)
(Fig. 5A). All three cell types
exhibited different expression levels of Max (Fig. 5A). Lower levels of Trf1
were expressed in the mESCs than in the mSSCs (l) (Fig. 5A). Confirmation of the qPCR
products of the epigenetic factors was also demonstrated (Fig. 5B).
Discussion
It has been previously demonstrated that the
membrane protein CD90.2 was extensively expressed on the surface of
mSSCs (13). In addition, the
enrichment of mSSCs using CD90.2 microbeads was more efficient than
the conventional isolation methods (13). Herein, we observed that the mSSCs
(l) exhibited AP activity and expressed the SSC markers, GFRα1 and
CD90.2, which is in agreement with previous findings (14). Further experiments demonstrated
that the mSSCs (l) expressed germ genes (Plzf, Vasa,
Dazl, Nanos3 and Stra8) and pluripotency genes
(Oct4, Sox2, Nanog, Lin28,
N-Myc, Klf4 and Tert). Cldn6 has been
identified as a novel surface marker for mouse PSCs (15), and Pdgfrα was found to be
involved in the regulation of cell division and migration (16). Our results showed that
Cldn6 and Pdgfrα were expressed on the mSSCs (l). The
successful establishment of mSSCs is characterized by their
self-renewal potential and ability to differentiate into sperm
(17). Herein, we showed that the
mSSCs (l) were capable of differentiating into sperm, by observing
the morphological characteristics of mSSCs (l) as well as by
determining the expression of the sperm markers, Gsg2 and
Acrosin. Collectively, our results suggested that the mSSCs
(f) isolated from 6-day-old ICR mouse testes using CD90.2
microbeads may be cultured long-term and maintain the ability to
differentiate into sperm.
On the one hand, pluripotency transcriptional
networks have been found to be crucial for controlling ESC
pluripotency and for somatic cell reprogramming (5,18).
Well-known transcription factors, Oct4, Sox2,
Nanog, Lin28, N-Myc and Klf4, have been
used to induce pluripotency (6,19).
However, recent evidence has suggested that the downstream factors,
Esrrb, Utf1, Lin28 and Dppa2, may also
promote iPSC production (20). It
has been demonstrated that Tbx3 is essential for
pluripotency regulation by regulating the expression of
Tet2, Dnmt3b and Zscan4 (21). Furthermore, high expression of
Nr5a2 [also known as liver receptor homolog-1 (Lrh1)]
had the capacity to replace Oct4 to facilitate reprogramming
(22,23). In addition, the germline factors
(Prdm14, Klf2 and Prmt5) were necessary for
primordial germ cell (PGC) specialization and they simultaneously
shared the ability to reprogramme PGCs and somatic cells into PSCs
(24,25). Our results indicated that the
mSSCs (f) and the mSSCs (l) exhibited low expression of most
transcription factors (Oct4, Sox2, Nanog,
N-Myc, Klf4, Esrrb, Utf1, Dppa2,
Tbx3, Nr5a2, Prdm14 and Klf2) in
contrast with the mESCs. However, using RT-qPCR and western blot
analysis, we found a very high expression of Prmt5 and
Lin28 in the mSSCs (l) indicating that they may be critical
for supporting mSSC reprogramming in vitro. A previous study
has shown that Lin28, an abundant protein in ESCs, may
repress let-7 microRNA processing, thereby controlling ESC
self-renewal and differentiation (26). Prmt5 may mediate histone
methylation and interacted with Stat3 to stimulate the
conversion of the inner cell mass, primordial germ cells, epiblast
stem cells, and somatic cells into PSCs (25,27,28). Moreover, it has been demonstrated
that the knockdown of Dmrt1 facilitated mSSC reprogramming
(10). Our results also revealed
that Drmt1 was expressed at a high level in both types of
mSSCs.
On the other hand, epigenetic mechanisms are
important for mammalian development and cellular reprogramming
(5). The maintenance of
particular gene expression patterns has been attributed to DNA
methylation and certain histone modifications (5). Epigenetic factors (Tet1,
Tet2, Tet3, Parp1, Dnmt1, Kdm2b,
Dot1l, Max, Tert, Trf1 and
Zscan4c) may alter genomic methylation and chromatin
structure, which is directly associated with pluripotency and
reprogramming (5).
The genomic methylation enzymes, Tet1,
Tet2, Tet3, Parp1 and Dnmt1, are
essential regulators of gene expression and reprogramming.
Specifically, Tet2 and Parp1 were found to be
required for early-stage epigenetic modifications during somatic
cell reprogramming (29). In
addition, a recent study found that Tet3 played a possible
role in germ cell modification of the zygotic paternal genome
(30). We have shown that
Tet3 and Parp1, genes involved in genomic
methylation, were expressed at a higher level in mSSCs (l) compared
with the mSSCs (f) and the mESCs; this may be key to mSSC
epigenetic reprogramming. Furthermore, it has been demonstrated
that Parp1 was engaged in the modulation of DNA damage
repair and gene transcription, and it promoted epigenetic
reprogramming during the early stages of iPSC formation (31). Dnmt1, which was found to be
involved in sustaining genomic DNA methylation and regarded as a
barrier to iPSC reprogramming (10), exhibited higher expression in the
mSSCs than in the mESCs in this study. Notably, we found a
significantly lower level of Tet2 in the mSSCs (l) versus
the mESCs, which may play a key role in SSC reprogramming. However,
this low expression was not due to DNA methylation of the
Tet2 promoter according to our bisulfite sequencing PCR
analysis.
Histone-associated modified enzymes (Kdm2b,
Dot1l and Max) may change the structure of chromatin
to influence gene expression. It has been demonstrated that
Kdm2b plays a role in anti-senescence and pluripotency and
may improve iPSC generation (32,33). A recent study found that histone
H3 lysine 79 (H3K79) methytransferase, a crucial epigenetic enzyme
for transcriptional regulation, served as a barrier to
reprogramming and restrained the expression of Nanog and
Lin28 (34). Evidence
suggests that Max interacts with histone H3K9 methyltransferases
and negatively controls germ cell-specific genes in mESCs (35). We found that there were similar
expression levels of Kdm2b and Max in the mSSCs (l)
and the mESCs, indicating their potential roles in facilitating SSC
reprogramming. However, Dot1l was more highly expressed in
the mSSCs (l) implying its possible inhibitory effect in SSC
reprogramming. In addition, the lower expression of Max in
the mSSCs (f) versus the mESCs and the mSSCs (l) may contribute to
sustained high levels of germline factor expression for
gametogenesis.
Telomere maintenance is essential for chromosome
stability, cell replicative capacity, and the induction and
establishment of pluripotency (36,37). It has been demonstrated that
Tert (38), Trf1
(36) and Zscan4c
(37) were involved in the
modulation of telomere length, thus, markedly improving
reprogramming efficiency and iPSC quality (39). We observed the high expression of
Tert and Trf1 in the mSSCs (l) and Zscan4c in
the mESCs; this may provide new insights into mSSC
reprogramming.
Taken together, our results suggested that the
mSSCs exhibited high expression of pluripotency-associated factors
(Lin28 and Prmt5), as well as the expression of
crucial epigenetic factors (Tet3, Parp1, Max,
Tert and Trf1) that may promote reprogramming.
However, the high expression of Dnmt1, Dmrt1 and
Dot1l, and the low expression of Tet1 and Tet2
in mSSCs (l) may be an obstacle for mSSC reprogramming.
Abbreviations:
|
Dnmt1
|
DNA methyltransferase 1
|
|
Dmrt1
|
doublesex and mab-3 related
transcription factor 1
|
|
Dot1l
|
disruptor of telomeric silencing
1-like
|
|
Dppa2
|
developmental pluripotency associated
2
|
|
Esrrb
|
estrogen-related receptor b
|
|
iPSCs
|
induced pluripotent stem cells
|
|
Kdm2b
|
lysine (K)-specific demethylase
2b
|
|
Klf2
|
Krüppel-like factor 2
|
|
Klf4
|
Krüppel-like factor 4
|
|
Lin28
|
Lin-28 homolog A
|
|
MACS
|
magnetic-activated cell sorting
|
|
Max
|
Myc associated factor x
|
|
MEF
|
mouse embryonic fibroblast
|
|
mESCs
|
mouse embryonic stem cells
|
|
mSSCs (f)
|
freshly isolated mouse spermatogonial
stem cells
|
|
mSSCs (l)
|
long-term propagated mouse
spermatogonial stem cells
|
|
Nr5a2
|
nuclear receptor subfamily 5, group
A, member 2
|
|
Oct4
|
octamer-binding transcription factor
4
|
|
Parp1
|
poly[ADP-ribose] polymerase 1
|
|
Prdm14
|
PR domain containing 14
|
|
Prmt5
|
protein Arg N-methyltransferase 5
|
|
RS
|
round spermatid
|
|
Sox2
|
Sry (sex determining region Y)-box
2
|
|
SSCs
|
spermatogonial stem cells
|
|
Tbx3
|
T-box 3
|
|
Tert
|
telomerase reverse transcriptase
|
|
Tet1
|
ten-eleven translocation
methylcytosine dioxygenase 1
|
|
Tet2
|
ten-eleven translocation
methylcytosine dioxygenase 2
|
|
Tet3
|
ten-eleven translocation
methylcytosine dioxygenase 3
|
|
Trf1
|
telomeric repeat binding factor 1
|
|
Utf1
|
undifferentiated embryonic cell
transcription factor 1
|
|
Zscan4c
|
zinc finger and SCAN domain
containing 4c
|
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (nos. 81170623 and 31402072) and by the
China Postdoctoral Science Foundation.
References
|
1
|
Pirouz M, Klimke A and Kessel M: The
reciprocal relationship between primordial germ cells and
pluripotent stem cells. J Mol Med Berl. 90:753–761. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim HJ, Lee HJ, Lim JJ, Kwak KH, Kim JS,
Kim JH, Han YM, Kim KS and Lee DR: Identification of an
intermediate state as spermatogonial stem cells reprogram to
multipotent cells. Mol Cells. 29:519–526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saitou M, Kagiwada S and Kurimoto K:
Epigenetic reprogramming in mouse pre-implantation development and
primordial germ cells. Development. 139:15–31. 2012. View Article : Google Scholar
|
|
4
|
Gifford CA, Ziller MJ, Gu H, Trapnell C,
Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R,
et al: Transcriptional and epigenetic dynamics during specification
of human embryonic stem cells. Cell. 153:1149–1163. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Orkin SH and Hochedlinger K: Chromatin
connections to pluripotency and cellular reprogramming. Cell.
145:835–850. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Onder TT, Kara N, Cherry A, Sinha AU, Zhu
N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al:
Chromatin-modifying enzymes as modulators of reprogramming. Nature.
483:598–602. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kurosaki H, Kazuki Y, Hiratsuka M, Inoue
T, Matsui Y, Wang CC, Kanatsu-Shinohara M, Shinohara T, Toda T and
Oshimura M: A comparison study in the proteomic signatures of
multipotent germline stem cells, embryonic stem cells, and germline
stem cells. Biochem Biophys Res Commun. 353:259–267. 2007.
View Article : Google Scholar
|
|
9
|
Fujino RS, Ishikawa Y, Tanaka K,
Kanatsu-Shinohara M, Tamura K, Kogo H, Shinohara T and Hara T:
Capillary morphsx-ogenesis gene (CMG)-1 is among the genes
differentially expressed in mouse male germ line stem cells and
embryonic stem cells. Mol Reprod Dev. 73:955–966. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takashima S, Hirose M, Ogonuki N, Ebisuya
M, Inoue K, Kanatsu-Shinohara M, Tanaka T, Nishida E, Ogura A and
Shinohara T: Regulation of pluripotency in male germline stem cells
by Dmrt1. Genes Dev. 27:1949–1958. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kubota H, Avarbock MR and Brinster RL:
Culture conditions and single growth factors affect fate
determination of mouse spermatogonial stem cells. Biol Reprod.
71:722–731. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu M, Wei H, Zhang J, Bai Y, Gao F, Li L
and Zhang S: Efficient production of chimeric mice from embryonic
stem cells injected into 4- to 8-cell and blastocyst embryos. J
Anim Sci Biotechnol. 4:122013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Li L, Bai Y, Shi R, Wei H and
Zhang S: Mouse undifferentiated spermatogonial stem cells cultured
as aggregates under simulated microgravity. Andrologia.
46:1013–1021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Godmann M, May E and Kimmins S: Epigenetic
mechanisms regulate stem cell expressed genes Pou5f1 and Gfra1 in a
male germ cell line. PLoS One. 5:e127272010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Xue Y, Shen Y, Li W, Cheng Y, Yan
X, Shi W, Wang J, Gong Z, Yang G, et al: Claudin 6: A novel surface
marker for characterizing mouse pluripotent stem cells. Cell Res.
22:1082–1085. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Eberhart JK, He X, Swartz ME, Yan YL, Song
H, Boling TC, Kunerth AK, Walker MB, Kimmel CB and Postlethwait JH:
MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat
Genet. 40:290–298. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ventelä S, Mäkelä JA, Kulmala J,
Westermarck J and Toppari J: Identification and regulation of a
stage-specific stem cell niche enriched by Nanog-positive
spermatogonial stem cells in the mouse testis. Stem Cells.
30:1008–1020. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim J, Chu J, Shen X, Wang J and Orkin SH:
An extended transcriptional network for pluripotency of embryonic
stem cells. Cell. 132:1049–1061. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu J, Vodyanik MA, Smuga-Otto K,
Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA,
Ruotti V, Stewart R, et al: Induced pluripotent stem cell lines
derived from human somatic cells. Science. 318:1917–1920. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Buganim Y, Faddah DA, Cheng AW, Itskovich
E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A and Jaenisch
R: Single-cell expression analyses during cellular reprogramming
reveal an early stochastic and a late hierarchic phase. Cell.
150:1209–1222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Han J, Yuan P, Yang H, Zhang J, Soh BS, Li
P, Lim SL, Cao S, Tay J, Orlov YL, et al: Tbx3 improves the
germ-line competency of induced pluripotent stem cells. Nature.
463:1096–1100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang
H, Wang W, Liu R, George J, Ng HH, Perera RJ, et al: MicroRNA-134
modulates the differentiation of mouse embryonic stem cells, where
it causes post-transcriptional attenuation of Nanog and LRH1. Stem
Cells. 26:17–29. 2008. View Article : Google Scholar
|
|
23
|
Heng JC1, Feng B, Han J, Jiang J, Kraus P,
Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al: The nuclear
receptor Nr5a2 can replace Oct4 in the reprogramming of murine
somatic cells to pluripotent cells. Cell Stem Cell. 6:167–174.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gillich A, Bao S, Grabole N, Hayashi K,
Trotter MW, Pasque V, Magnúsdóttir E and Surani MA: Epiblast stem
cell-based system reveals reprogramming synergy of germline
factors. Cell Stem Cell. 10:425–439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nagamatsu G, Kosaka T, Kawasumi M,
Kinoshita T, Takubo K, Akiyama H, Sudo T, Kobayashi T, Oya M and
Suda T: A germ cell-specific gene, Prmt5, works in somatic cell
reprogramming. J Biol Chem. 286:10641–10648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong X, Li N, Liang S, Huang Q, Coukos G
and Zhang L: Identification of microRNAs regulating reprogramming
factor LIN28 in embryonic stem cells and cancer cells. J Biol Chem.
285:41961–41971. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang J, van Oosten AL, Theunissen TW, Guo
G, Silva JC and Smith A: Stat3 activation is limiting for
reprogramming to ground state pluripotency. Cell Stem Cell.
7:319–328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tee WW, Pardo M, Theunissen TW, Yu L,
Choudhary JS, Hajkova P and Surani MA: Prmt5 is essential for early
mouse development and acts in the cytoplasm to maintain ES cell
pluripotency. Genes Dev. 24:2772–2777. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doege CA, Inoue K, Yamashita T, Rhee DB,
Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al:
Early-stage epigenetic modification during somatic cell
reprogramming by Parp1 and Tet2. Nature. 488:652–655. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deplus R, Delatte B, Schwinn MK, Defrance
M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E,
et al: TET2 and TET3 regulate GlcNAcylation and H3K4 methylation
through OGT and SET1/COMPASS. EMBO J. 32:645–655. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Villani P, Fresegna AM, Ranaldi R,
Eleuteri P, Paris L, Pacchierotti F and Cordelli E: X-ray induced
DNA damage and repair in germ cells of PARP1(−/−) male mice. Int J
Mol Sci. 14:18078–18092. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liang G, He J and Zhang Y: Kdm2b promotes
induced pluripotent stem cell generation by facilitating gene
activation early in reprogramming. Nat Cell Biol. 14:457–466. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He J, Shen L, Wan M, Taranova O, Wu H and
Zhang Y: Kdm2b maintains murine embryonic stem cell status by
recruiting PRC1 complex to CpG islands of developmental genes. Nat
Cell Biol. 15:373–384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen J, Liu H, Liu J, Qi J, Wei B, Yang J,
Liang H, Chen Y, Chen J, Wu Y, et al: H3K9 methylation is a barrier
during somatic cell reprogramming into iPSCs. Nat Genet. 45:34–42.
2013. View Article : Google Scholar
|
|
35
|
Maeda I, Okamura D, Tokitake Y, Ikeda M,
Kawaguchi H, Mise N, Abe K, Noce T, Okuda A and Matsui Y: Max is a
repressor of germ cell-related gene expression in mouse embryonic
stem cells. Nat Commun. 4:17542013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schneider RP, Garrobo I, Foronda M,
Palacios JA, Marión RM, Flores I, Ortega S and Blasco MA: TRF1 is a
stem cell marker and is essential for the generation of induced
pluripotent stem cells. Nat Commun. 4:19462013.PubMed/NCBI
|
|
37
|
Zalzman M, Falco G, Sharova LV, Nishiyama
A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al:
Zscan4 regulates telomere elongation and genomic stability in ES
cells. Nature. 464:858–863. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Winkler T, Hong SG, Decker JE, Morgan MJ,
Wu C, Hughes WM, Yang Y, Wangsa D, Padilla-Nash HM, Ried T, et al:
Defective telomere elongation and hematopoiesis from
telomerase-mutant aplastic anemia iPSCs. J Clin Invest.
123:1952–1963. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang
H, Okuka M, Zhou C, Zhang X, Liu L and Li J: Zscan4 promotes
genomic stability during reprogramming and dramatically improves
the quality of iPS cells as demonstrated by tetraploid
complementation. Cell Res. 23:92–106. 2013. View Article : Google Scholar :
|