Introduction
Skeletal muscle differentiation is a highly complex
and coordinated biological process which involves a broad spectrum
of signaling molecules. It firstly begins with the commitment of
satellite cells (muscle stem cells) to myogenic precursor cells
known as myoblasts. Subsequently, myoblasts gradually become
terminally differentiated myocytes coordinated by a series of
regulatory factors. Finally, mononucleated myocytes specifically
fuse to form multinucleated myotubes (1,2).
To date, many efforts have been devoted to exploring and
elaborating the precise regulation of myogenic differentiation. A
number of transcription factors and muscle-specific genes, such as
paired box (Pax)3/Pax7 (3–5),
myogenic differentiation (MyoD) (6), Myogenic factor 5 (MYF5) (7), myogenin (8,9)
and myosin heavy chain (MyHC) (10–12) have been confirmed as muscle
determination factors. Myogenin is a member of the MyoD family,
which is suggested to function in myogenesis. Previous studies have
found that myogenin is expressed during myoblast differentiation,
and its expression directly affects the progression of myoblasts
into skeletal muscle (13,14).
Recent studies have demonstrated that several regulators, such as
miR-186 (9), multiple EGF like
domains 10 (MEGF10) (15) and p53
(16) are involved in myoblast
differentiation through the regulation of myogenin. These results
provide evidence for a key role of myogenin as a critical regulator
of myoblast differentiation. During myogenesis, myogenic regulatory
factors (MRFs) are actived and regulate the transcription of genes,
such as MyHC (17). In adult
skeletal muscle, MyHC mRNA isoforms are expressed in a distinct
patterns, including MyHC-I, MyHC-IIa, MyHC-IIx, MyHC-IIb, embryonic
(emb) and neonatal (neo) (10,18). It has been confirmed that MyHC is
expressed in late and terminal differentiation, and that it is the
most suitable marker of muscle fibre (1).
A series of signaling molecules, including p38
(19), Wnt (20,21), extracellular signal-regulated
kinase 1/2 (Erk1/2) (22,23), c-Jun N-terminal kinase (JNK)
(24) and mitogen-activated
protein kinase kinase kinase kinase 4 (MAP4K4) (25), have been shown to be involved in
myogenesis. However, the precise molecular mechanisms of myogenic
differentiation remain largely unknown, and a number of novel genes
involved in this process remain to be identified.
Microarray technology provides us with a unique
opportunity to examine gene expression patterns in a whole genome.
However, the heterogeneity of gene expression data could exist
across different laboratories, different ChIP platforms or
different experimental operations, which can be partly circumvented
by meta-analysis so as to yield a more robust result.
In this study, we found that the leucine-rich
repeat-containing 75B (Lrrc75b), also known as AI646023, was
downregulated during myogenesis by performing a meta-analysis of
C2C12 myogenic differentiation microarray data in the GEO database.
It has been demonstrated that many proteins containing leucine-rich
repeat (LRR) domains participate in important biological processes,
such as signal transduction, cell adhesion, cell development and
DNA repair (26). Importantly,
studies have revealed the involvement of LRR proteins in cell
differentiation. LRRC8 (also known as FAD158) is expressed in
differentiating 3T3-L1 cells, and the knockdown of LRRC8 has been
shown to significantly inhibit 3T3-L1 adipocyte differentiation
(27). Another LRR protein,
LRRC17, functions as an inhibitor of RANKL-induced osteoclast
differentiation (28).
The aim of this study was to elucidate the potential
function of Lrrc75b in myogenesis. Using knockdown and
overexpression techniques, we found that Lrrc75b significantly
regulated the activity of muscle marker genes and the
phosphorylation of Erk. Our results demonstrated that Lrrc75b is a
novel negative regulator of myogenesis.
Materials and methods
Meta-analysis of C2C12 myogenic
differentiation microarray data
To obtain the differentially expressed genes in
C2C12 myogenic differentiation, the GEO database was used (29). Three datasets (listed in Table I) were used and we also used the
Affymetrix mouse expression array (including 430 2.0 array, 430A
and B array). To the best of our knowledge, these arrays contain
more abundant gene probesets. The raw data from each experiment
were normalized using ChIP analysis tools and the following
thresholds were then used to obtain sets of differentially
expressed genes: i) E/B >1.5 or B/E >1.5, use lower 90%
confidence bound of fold; ii) E-B >50 or B-E >50 and iii)
P-value of 0.05. The upregulated or downregulated gene probesets
were converted to official gene symbols using DAVID [National
Institute of Allergy and Infectious Diseases (NIAID), National
Institutes of Health (NIH), USA], a functional annotation tool
(30). The differentially
expressed genes in at least 2 experiments in all the above 3
experiments were designated as potential myogenesis upregulated or
downregulated genes.
 | Table IList of the datasets used in this
research. |
Table I
List of the datasets used in this
research.
| GEO accession
no. | ChIP samples used
to annlyze | Gene ChIP type | Submitted by |
|---|
| GSE4694 | GSM106142,
GSM106143, GSM106144, GSM106145, GSM106146, GSM106147 | | |
| Mouse 430 2.0
array | Ihsiung Brandon
Chen |
| PMID: 17062158 |
| GSE5447 | GSM124854,
GSM124866 | Mouse 430 2.0
array | Christof Burek |
| | | PMID: 17045206 |
| GSE5305 | GSM119558,
GSM119559, GSM119560, GSM119563, GSM119564, GSM119565, GSM119568,
GSM119571, GSM119576, GSM119578, GSM119579, GSM119580 | Mouse 430A and B
array | Eric Hoffman |
Cell culture
C2C12 myoblasts (CRL-1772; ATCC, Manassas, VA, USA)
were maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS; both from HyClone
Laboratories, Inc., Logan, UT, USA) at 37°C with 5% CO2.
To induce myogenic differentiation, the cells were plated on tissue
culture plates and grown to 95% confluence before switching to
differentiation medium (DM) (DMEM and 2% horse serum; HyClone
Laboratories, Inc.). The cells were replenished with fresh DM every
other day until day 5.
Transfection with Lrrc75b siRNA
Stealth RNAi™ pre-designed siRNAs specific for mouse
Lrrc75b (MSS208271; Gibco Life Technologies, Rockville, MD, USA)
were synthesized by Gibco Life Technologies. Stealth RNAi medium GC
control was used as a negative control. siRNA duplexes were
transfected into the cells using 2.5 µl/ml Lipofectamine
RNAiMAX (Gibco Life Technologies) according to the manufacturer's
instructions. Briefly, the C2C12 cells at 40–60% confluence were
transfected with 0.2 µM siRNA.
Adenoviral infection
The C2C12 cells were plated in culture dishes at a
density of 1.3×104 cells/cm2. Adenoviral
shuttle vector expressing either pDC316-mCMV-EGFP-CMV-Lrrc75b
(C-terminal Myc-tagged) or the empty pDC316-mCMV-EGFP and
adenovirus packaging were completed by a professional company
(Biowit Technologies, Shenzhen, China). Briefly, mouse Myc-tagged
Lrrc75b was synthetized and inserted into vector pDC316-mCMV-EGFP
using the restriction enzymes NheI and HindIII. The
adenoviral shuttle vector and virus backbone plasmid
pBHGloxdeltaE13Cre were then co-transfected into 293 cells by
polyfectamine. Adenoviruses were generated following the
instructions of AdMax™ Adenoviral Vector Creation System and the
recombinant adenoviruses were collected and amplified in 293 cells.
When the cells grew to 50–70% confluency, they were infected with
Ad-Lrrc75b or Ad-GFP at an MOI of 200 for 12 h in growth medium and
this was then changed to fresh growth medium. At 48 h
post-infection, the cells were harvested for western blot analysis
or, the medium was changed to to DM and the cells were cultured for
the indicated periods of time (0, 1, 3, 5 days) before
harvesting.
Reverse transcription-quantitative
(real-time) PCR (RT-qPCR)
Total RNA was extracted from the C2C12 cells with
TRIzol Reagent (Gibco Life Technologies) following the
manufacturer's instructions. Each sample was reverse transcribed
into cDNA using the PrimeScript™ RT reagent kit with gDNA Eraser
(Perfect Real Time; Takara Bio Inc., Otsu, Japan). This was
followed by quantitative PCR (qPCR) using an Applied Biosystems
7500 Real-time PCR system (Applied Biosystems Life Technologies,
Foster City, CA, USA) with the One-Step SYBR PrimeScript RT-PCR kit
II (Takara Bio Inc.) according to the manufacturer's instructions.
The housekeeping gene, GAPDH, was used as an internal normalization
control to obtain the relative fold changes using the comparative
CT method. The sequences of the primers used were as follows: mouse
Lrrc75b forward, 5′-ggaccatgagctctggaagt-3′ and reverse,
5′-atccacagtctcccctacca-3′; and mouse GAPDH forward,
5′-cgtgttcctacccccaatgt-3′ and reverse, 5′-gcttcaccacc
ttcttgatgtc-3′.
Western blot analysis
The whole-cell lysates were harvested for 30 min on
ice in RIPA lysis buffer containing 100 mM PMSF (Beyotime Institute
of Biotechnology, Haimen, China) and then centrifuged at 12,000 rpm
for 15 min at 4°C. Total protein concentrations were measured by
BCA protein assay and equal amounts of proteins were then separated
by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a polyvinylidene fluoride (PVDF) membranes. The
membranes were blocked with 5% non-fat milk for 1.5 h at room
temperature, and then incubated overnight at 4°C with the following
primary antibodies: as anti-myogenin (SC-12732, 1:200; F5D; Santa
Cruz Biotechnology Inc., Santa Cruz, CA, USA), anti-MyHC (m4276,
1:1,000; Sigma-Aldrich, St. Louis, MO, USA), anti-phospho-p44/42
MAPK (4377,1:2,000; p-Erk1/2) (Thr202/Tyr204), anti-p44/42 MAPK
(4695, 1:2,000; Erk1/2) and anti-Myc-tagged (2278, 1:300) (all from
Cell Signaling Technology, Inc., Beverly, MA, USA) and anti-tubulin
(AT819, 1:2,000; Beyotime Institute of Biotechnology). After being
washed with TBST 3 times, the membranes were incubated with
HRP-labeled goat anti-mouse IgG (A0216, 1:1,000; Beyotime Institute
of Biotechonogy,) or HRP-labeled goat anti-rabbit IgG (AB6721,
1:5,000; Abcam, Cambridge, MA, USA) for 1 h at 37°C. Bands were
visualized using the Luminata Crescendo Western HRP substrate
(Millipore, Billerica, MA, USA). The quantification of the band
intensities was performed using ImageJ software (National
Institutes of Health, Bethesda, MD, USA).
Immunofluorescence microscopy and myotube
analysis
The cells were fixed with 4% formaldehyde for 20 min
and washed 3 times in PBS, and then incubated with 0.3% Triton
X-100 in PBS for 20 min and blocked in 10% donkey serum at 37°C for
30 min. Subsequently, the cells were incubated overnight at 4°C
with anti-myosin primary antibody against MyHC (M4276, 1:150;
Sigma-Aldrich). The cells were then incubated with Alexa Fluor
594-conjugated secondary antibody (A-21203, 1:200, Gibco Life
Technologies) for 1 h at room temperature. The nuclei of the cells
were visualized using 4′,6-diamidino-2-phenylindole dihydrochloride
(DAPI) staining for 10 min. Images of samples were also captured
using a fluorescence microscope (DP73; Olympus Corp., Tokyo,
Japan). Finally, 5 random fields with representative images per
sample were used to calculate the myotube area (area occupied by
myotubes relative to the total area) and the fusion index (the
ratio of nuclei in MyHC-positive myotubes with ≥2 nuclei to the
total number of nuclei in the field).
Statistical analysis
The experiments were repeated 3 times, and
statistical analyses were performed using the Student's t-test or
ANOVA. Statistical comparisons were considered significant at
P<0.05.
Results
Meta-analysis of myogenic differentiation
microarray data indicating the downregulation of Lrrc75b
To overcome the weakness of conventional
microarray-based data analysis, we designed a meta-analysis
strategy for microarray datasets used in this research to build up
lists of meta-genes in C2C12 myogenesis. The differentially
expressed genes in at least 2 datasets in all the above-mentioned 3
experiments were designated as potential myogenesis upregulated or
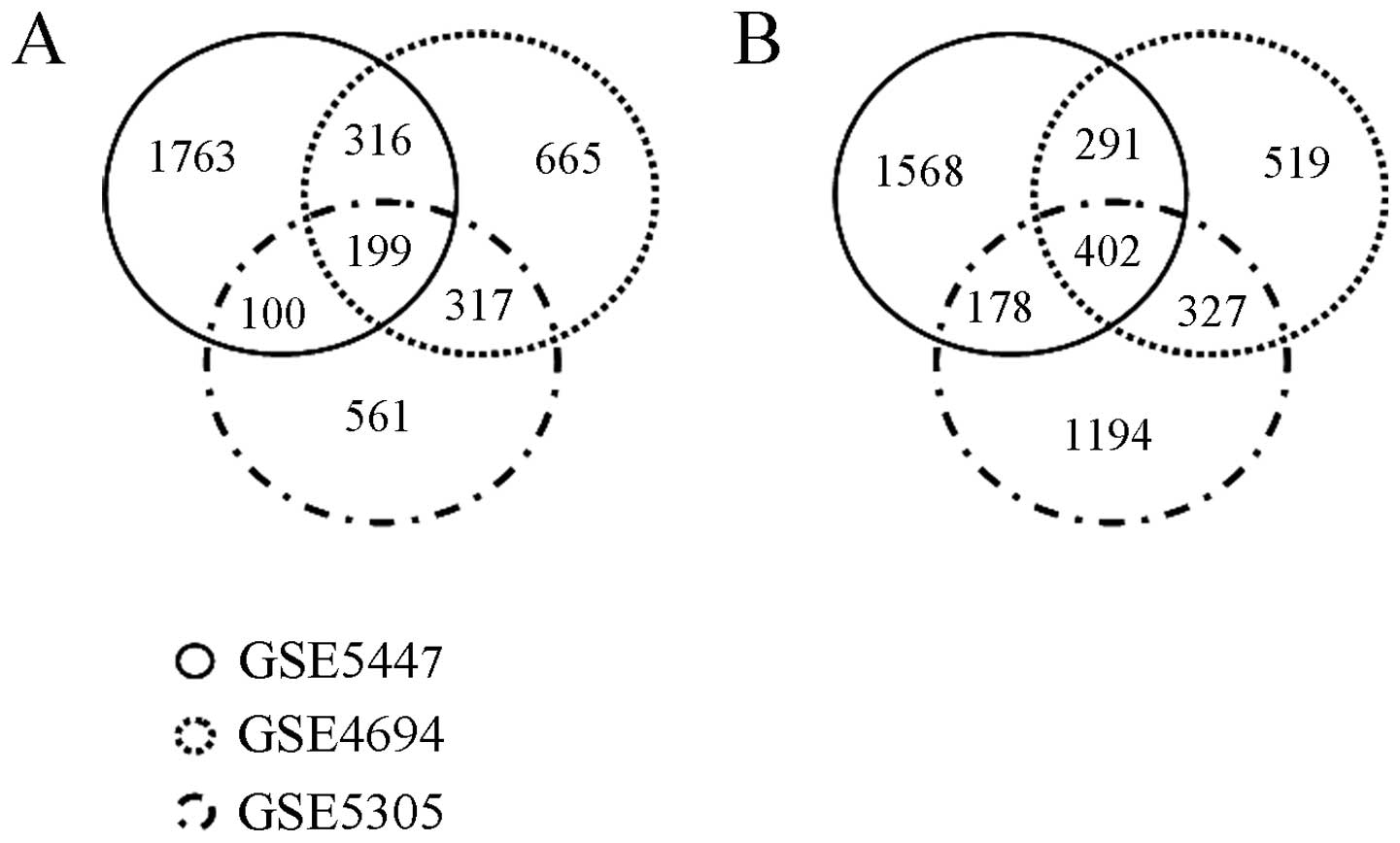
downregulated genes. The Venn diagram in Fig. 1 shows the distribution of
differentially expressed (upregulated or downregulated) genes among
the GSE5447, GSE4694 and GSE5305 datasets. In brief, among a total
of 3,921 upregulated genes, only 932 genes were upregulated in more
than 2 different experiments, of which 199 were upregulated in all
3 datasets. Of the 1,198 myogenesis downregulated genes in more
than 2 different experiments, 402 genes were downregulated in all 3
datasets. Lrrc75b was one of the myogenesis downregulated genes in
our meta-analysis of myogenic differentiation microarray data.
Lrrc75b expression is inhibited during
myogenic differentiation
To investigate the roles of Lrrc75b in myogenic
differentiation, we first established a C2C12 myoblast
differentiation model. C2C12 cells are derived from satellite cells
of C3H mice, which is the classical muscle differentiation model
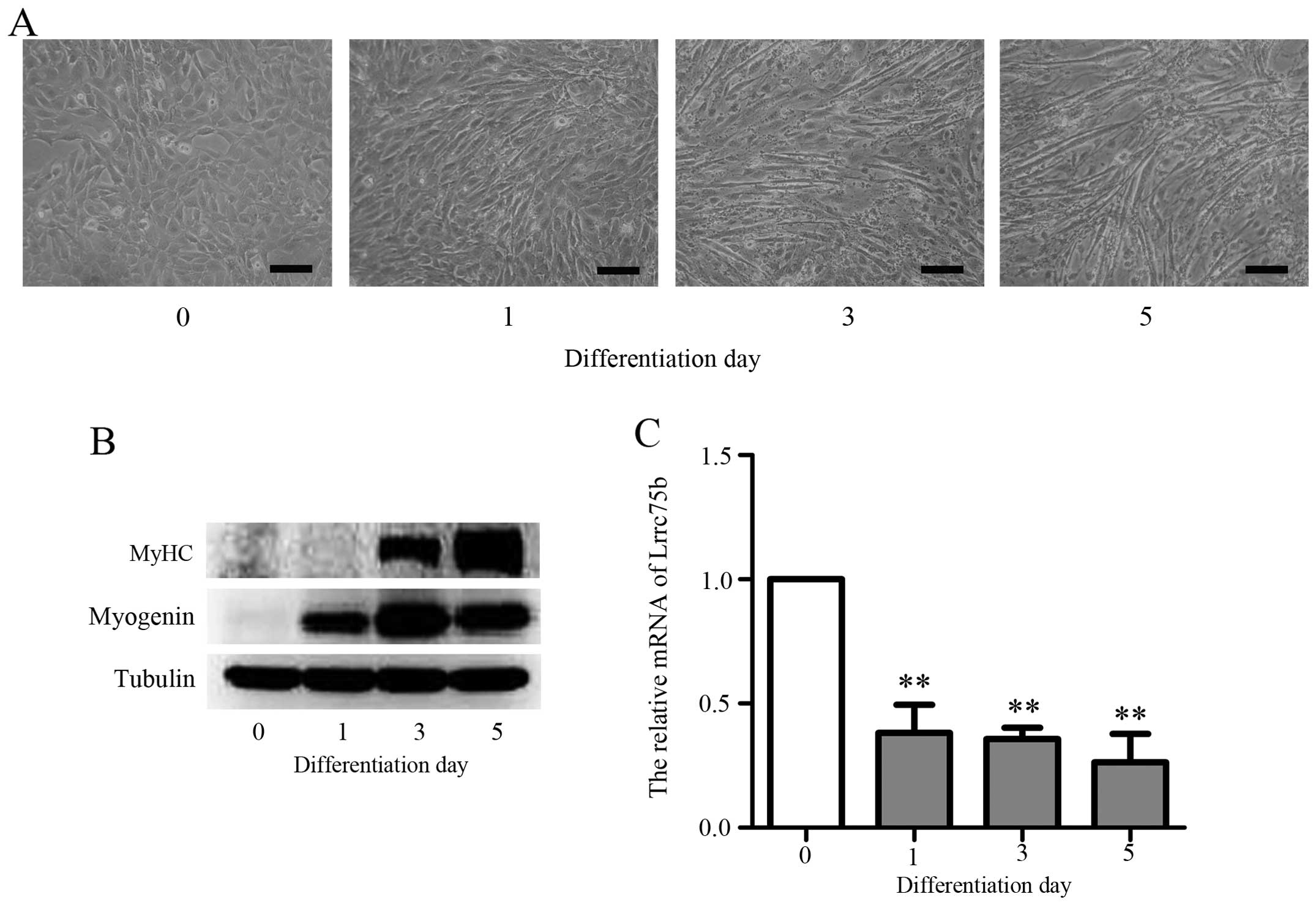
(8,31). The results presented in Fig. 2A show that the exposure of C2C12
cells to DM resulted in the formation of myotubes within 3 days and
further fusion, forming multinucleased myotubes within 5 days.
Moreover, as shown in Fig. 2B,
the expression of muscle-specific genes, such as myogenin and MyHC
increased upon the induction of differentiation. These results are
consistent with those of a previous study (1). To confirm that Lrrc75b is indeed
required for myoblast differentiation, RT-qPCR was performed. As
shown in Fig. 2C, the mRNA
expression of Lrrc75b was markedly decreased during
differentiation; similar results were obtained by the analysis of
microarray data. This result suggests that Lrrc75b plays a role in
the differentiation of C2C12 myoblasts into myotubes.
Knockdown of Lrrc75b promotes the
myoblast differentiation of C2C12
To clarify the possible role of Lrrc75b in
myogenesis, we established C2C12 cells in which Lrrc75b expression
was depleted by using stealth siRNA that target the specific region
of the mouse Lrrc75b gene and then examined the cells for their
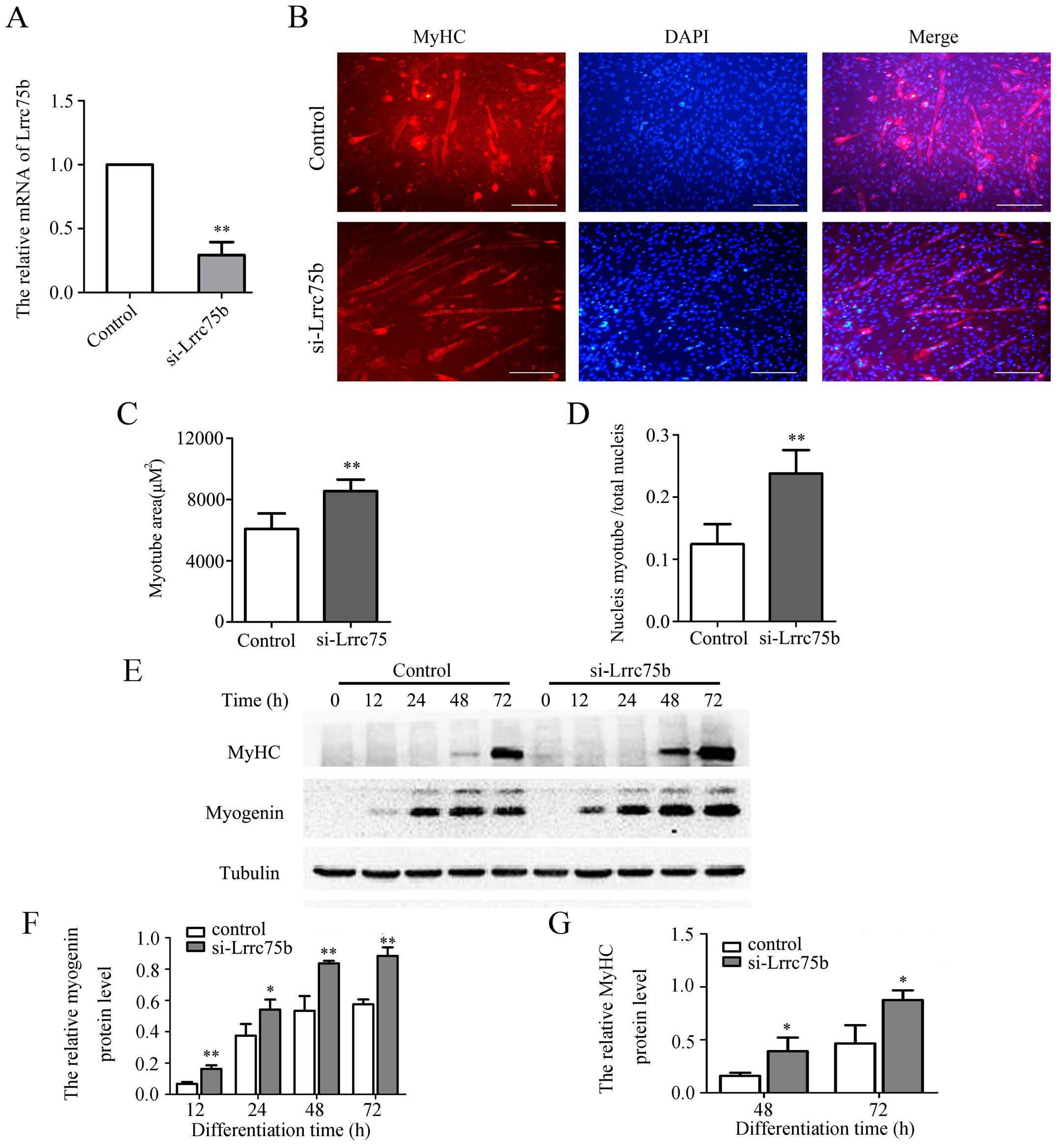
myoblast differentiation ability. As shown in Fig. 3A, the mRNA expression of Lrrc75b
significantly decreased by ~70% in the cells transfected with siRNA
targeting Lrrc75b (si-Lrrc75b) compared with the cells transfected
with the control siRNA. Subsequently, immunofluorescence staining
using anti-MyHC antibody was used to detect the formation of
myotubes. As shown in Fig. 3B,
the knockdown of Lrrc75b significantly promoted myogenic
differentiation and resulted in a higher myogenic index, such as a
greater myotube area and fusion index (Fig. 3C and D). In addition, as shown in
Fig. 3E–G, the results of western
blot analysis confirmed that the knockdown of Lrrc75b markedly
increased the expression of myogenin and MyHC.
Overexpression of Lrrc75b inhibits
myogenic differentiation
Having clarified that the knockdown of Lrrc75b
promotes myogenic differentiation, we then wished to determine
whether the upregulation of Lrrc75b in C2C12 cells would affect
myogenesis. To examine this hypothesis, we induced the
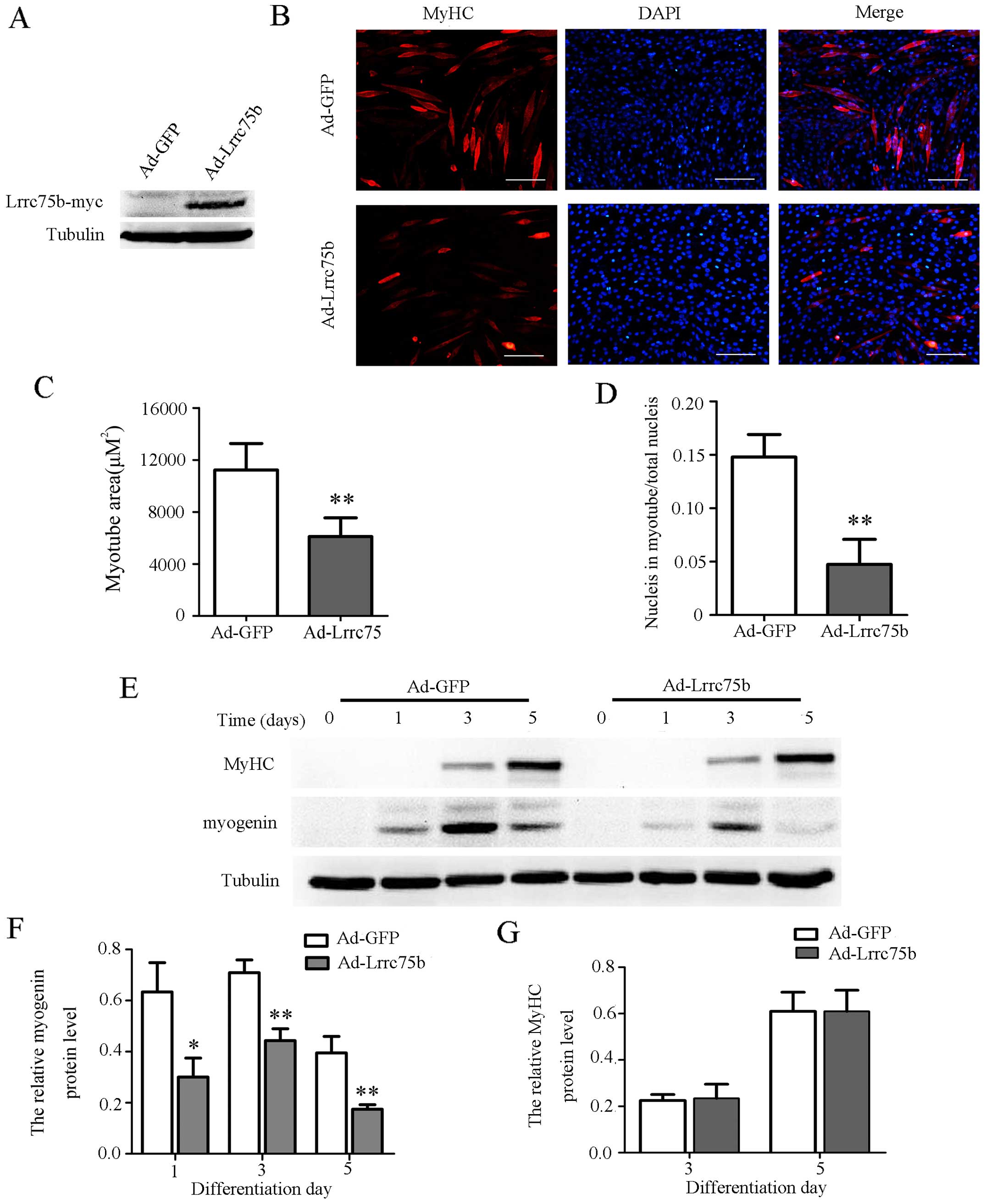
overexpression of Lrrc75b using Myc-tagged Lrrc75b adenovirus. As
compared to the C2C12 cells infected with GFP adenovirus, the
expression of Myc-tagged Lrrc75b emerged in the cells infected with
the Lrrc75b adenovirus (Fig. 4A).
Moreover, as shown in Fig. 4B–D,
in the cells infected with Lrrc75b adenovirus, the number of
myotubes and myogenic index were markedly lower than those observed
in the cells infected with the GFP adenovirus. Furthermore, the
results of western blot analysis revealed that the overexpression
of Lrrc75b significantly decreased the expression of myogenin
(Fig. 4E and F). As regards the
expression of MyHC, infection of the cells with Lrrc75b adenovirus
did not exert a significant effect on its expression, neither on
day 3 nor day 5. We hypothesized that the reason for this may be
that the expression of muscle-specific genes is essential but not
sufficient for terminally differentiated myoblast fuse into
multinuclear myotubes (32).
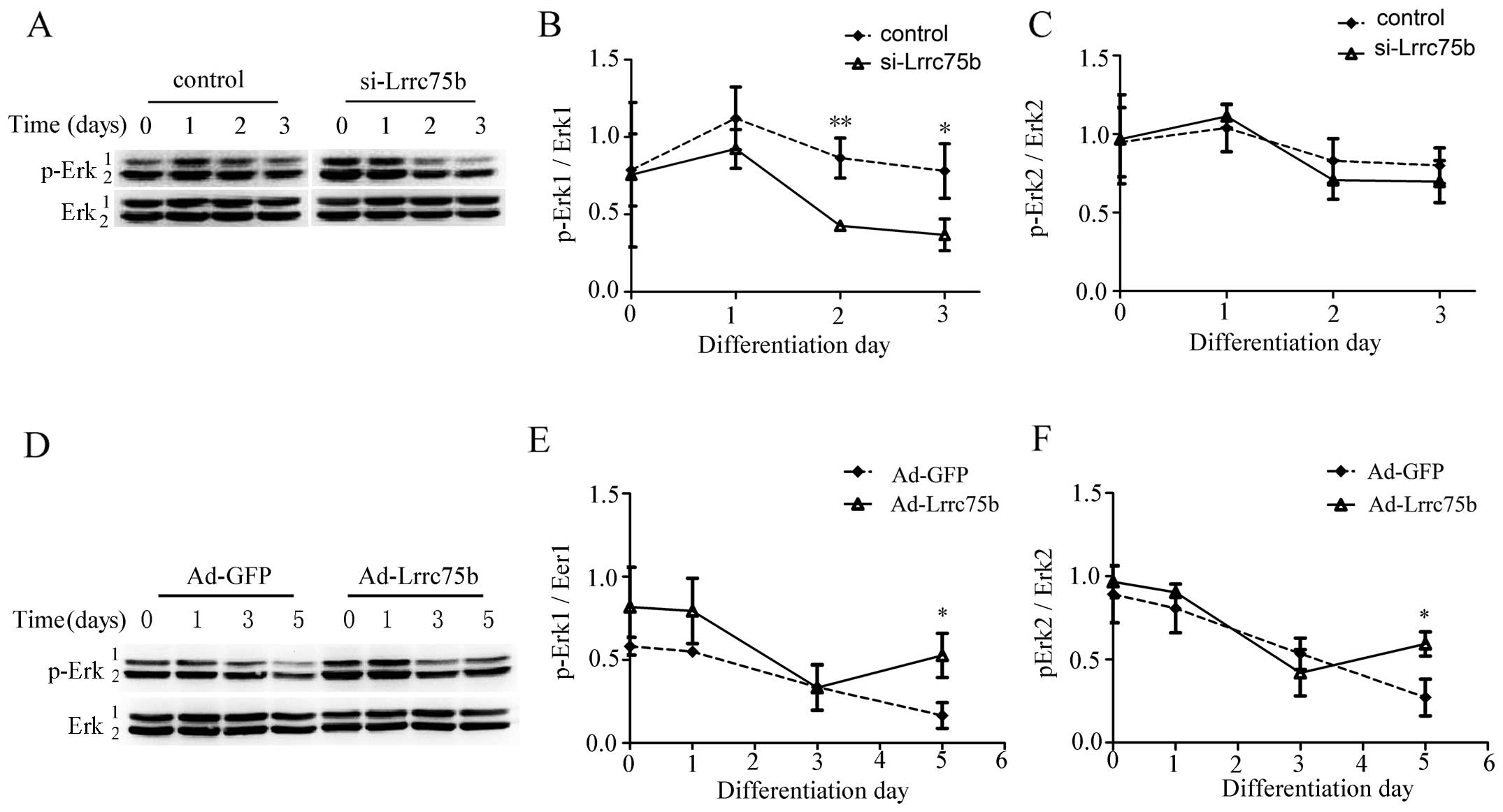
Lrrc75b affects the phosphorylation of
Erk1/2
To further elucidate the mechanisms involved in the
regulation of myogenic differentiation by Lrrc75b, we investigated
the target effector of the MAPK pathway, p-Erk1/2. At 24 h
post-transfection with control or si-Lrrc75b, the C2C12 cells
underwent myogenic differentiation. The phosphorylation of Erk1/2
was analyzed in the 2 groups of cells at 0, 1, 2 and 3 days. As
shown in Fig. 5A–C, the
phosphorylation level of Erk1/2 was gradually decreased in both
groups of cells at the later stage of differentiation. However, the
phosphorylation of Erk1/2 was much weaker in the
si-Lrrc75b-transfected cells on days 2 and 3. On the contrary, the
C2C12 cells infected with GFP or Lrrc75b adenovirus exhibited an
inhibition of myogenic differentiation. As shown in Fig. 5D–F, the phosphorylation of Erk1/2
was increased in the Lrrc75b adenovirus-infected cells on day
5.
Discussion
Whole genome microarray analyses, viewed as a
non-biased, genome wide molecular taxonomy, is helpful for the
understanding of the molecular mechanisms of myogenesis. As
previously reported (33,34), probably due to
platform-to-platform or laboratory-to-laboratory variability, there
was a great heterogeneity in the diffrerentially expressed genes of
C2C12 myogenesis in the 3 datasets. For example, among a total of
3,921 upregulated genes, only 932 genes were upregulated in more
than 2 different experiments. Thus, meta-analysis provides a list
of more pertinent genes for further study.
To date, many efforts have been devoted to exploring
and elaborating the precise regulation of myogenic differentiation.
However, the precise molecular mechanisms of myogenic
differentiation remain largely unknown and many novel genes
involved in this process remain to be identified. In this study, we
found that Lrrc75b, a completely unknown function gene, was
downregulated during myoblast differentiation. The knockdown of
Lrrc75b using siRNA markedly enhanced C2C12 myoblast
differentiation, as evidenced by an increase in the myotube area
and fusion index. By contrast, the overexpression of Lrrc75b
attenuated the differentiation of C2C12 myoblasts into myotubes,
decreasing the myotube area and fusion index. Therefore, these
findings indicate that Lrrc75b plays an essential role in C2C12
myoblast differentiation.
Skeletal muscle development is a complex and orderly
progress wherein mesodermal cells commit to myoblasts and
subsequently fuse into multinuclear myotubes (35). This progress is mainly controlled
by a family of MRFs, such as myogenin (25). It has been reported that activated
myogenin in early differentiation can drive terminal
differentiation, the formation of myotubes and muscle fiber
maturation (25,36). Our observations that the knockdown
of Lrrc75b using siRNA resulted in an increase in myogenin
expression, whereas the opposite was observed with the
overexpression of Lrrc75b by infection with adenovirus, indicate
that Lrrc75b is involved in signaling upstream of myogenin during
C2C12 myoblast differentiation into myotubes.
Given that the Erk1/2 signaling pathway plays an
important role during myoblast differentiation (37–42), we examined the changes in Erk1/2
phosphorylatoin in response to myogenic differentiation and found
that the phosphorylation of Erk1/2 was decreased. This result is
consistent with the findings of a previous study (23). Our results also demonstrated that
the phosphorylation of Erk1/2 was decreased following the knockdown
of Lrrc75b using siRNA, whereas it was increased in late-stage
differentiation with the overexpression of Lrrc75b by infection
with adenovirus. Taken together, these data suggest that Lrrc75b
plays a negative role in myogenic differentiation and this role is
mediated at least partly via Erk1/2.
In conclusion, in this study, we obtained a more
pertinent gene set for C2C12 myogenic differentiation through
meta-analysis of three microarray datasets from different
laboratories. Our results also demonstrate that Lrrc75b is a novel
suppressor of C2C12 myogenic differentiation by modulating myogenin
and Erk1/2 signaling.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (no. 81101357), the Science and
Technological Program for Dongguan's Higher Education, Science and
Research and Health Care Institutions (no. 2011 108102029).
References
|
1
|
Ge Y, Waldemer RJ, Nalluri R, Nuzzi PD and
Chen J: Flt3L is a novel regulator of skeletal myogenesis. J Cell
Sci. 126:3370–3379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miyake M, Hayashi S, Iwasaki S, Uchida T,
Watanabe K, Ohwada S, Aso H and Yamaguchi T: TIEG1 negatively
controls the myoblast pool indispensable for fusion during myogenic
differentiation of C2C12 cells. J Cell Physiol. 226:1128–1136.
2011. View Article : Google Scholar
|
|
3
|
Calhabeu F, Hayashi S, Morgan JE, Relaix F
and Zammit PS: Alveolar rhabdomyosarcoma-associated proteins
PAX3/FOXO1A and PAX7/FOXO1A suppress the transcriptional activity
of MyoD-target genes in muscle stem cells. Oncogene. 32:651–662.
2013. View Article : Google Scholar
|
|
4
|
Zhang RP, Liu HH, Wang HH, Wang Y, Han CC,
Li L, He H, Xu HY, Xu F and Wang JW: Silencing Pax3 by shRNA
inhibits the proliferation and differentiation of duck (Anas
platyrhynchos) myoblasts. Mol Cell Biochem. 386:211–222. 2014.
View Article : Google Scholar
|
|
5
|
Dey BK, Gagan J and Dutta A: miR-206 and
-486 induce myoblast differentiation by downregulating Pax7. Mol
Cell Biol. 31:203–214. 2011. View Article : Google Scholar :
|
|
6
|
Wu SL, Li GZ, Chou CY, Tsai MS, Chen YP,
Li CJ, Liou GG, Chang WW, Chen SL and Wang SH: Double homeobox
gene, Duxbl, promotes myoblast proliferation and abolishes myoblast
differentiation by blocking MyoD transactivation. Cell Tissue Res.
358:551–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Averous J, Gabillard JC, Seiliez I and
Dardevet D: Leucine limitation regulates myf5 and MyoD expression
and inhibits myoblast differentiation. Exp Cell Res. 318:217–227.
2012. View Article : Google Scholar
|
|
8
|
Naka A, Iida KT, Nakagawa Y, Iwasaki H,
Takeuchi Y, Satoh A, Matsuzaka T, Ishii KA, Kobayashi K, Yatoh S,
et al: TFE3 inhibits myoblast differentiation in C2C12 cells via
down-regulating gene expression of myogenin. Biochem Biophys Res
Commun. 430:664–669. 2013. View Article : Google Scholar
|
|
9
|
Antoniou A, Mastroyiannopoulos NP, Uney JB
and Phylactou LA: miR-186 inhibits muscle cell differentiation
through myogenin regulation. J Biol Chem. 289:3923–3935. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brown DM, Parr T and Brameld JM: Myosin
heavy chain mRNA isoforms are expressed in two distinct cohorts
during C2C12 myogenesis. J Muscle Res Cell Motil. 32:383–390. 2012.
View Article : Google Scholar
|
|
11
|
Wang L, Chen X, Zheng Y, Li F, Lu Z, Chen
C, Liu J, Wang Y, Peng Y, Shen Z, et al: MiR-23a inhibits myogenic
differentiation through down regulation of fast myosin heavy chain
isoforms. Exp Cell Res. 318:2324–2334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang M, Yu H, Kim YS, Bidwell CA and Kuang
S: Myostatin facilitates slow and inhibits fast myosin heavy chain
expression during myogenic differentiation. Biochem Biophys Res
Commun. 426:83–88. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brunetti A and Goldfine ID: Role of
myogenin in myoblast differentiation and its regulation by
fibroblast growth factor. J Biol Chem. 265:5960–5963.
1990.PubMed/NCBI
|
|
14
|
Hasty P, Bradley A, Morris JH, Edmondson
DG, Venuti JM, Olson EN and Klein WH: Muscle deficiency and
neonatal death in mice with a targeted mutation in the myogenin
gene. Nature. 364:501–506. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park SY, Yun Y, Kim MJ and Kim IS:
Myogenin is a positive regulator of MEGF10 expression in skeletal
muscle. Biochem Biophys Res Commun. 450:1631–1637. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang ZJ, Broz DK, Noderer WL, Ferreira JP,
Overton KW, Spencer SL, Meyer T, Tapscott SJ, Attardi LD and Wang
CL: p53 suppresses muscle differentiation at the myogenin step in
response to genotoxic stress. Cell Death Differ. 22:560–573. 2015.
View Article : Google Scholar :
|
|
17
|
Braun T and Gautel M: Transcriptional
mechanisms regulating skeletal muscle differentiation, growth and
homeostasis. Nat Rev Mol Cell Biol. 12:349–361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harrison BC, Allen DL and Leinwand LA: IIB
or not IIB? Regulation of myosin heavy chain gene expression in
mice and men. Skelet Muscle. 1:52011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu H, Wang X, Liu S, Wu Y, Zhao T, Chen X,
Zhu L, Wu Y, Ding X, Peng X, et al: Sema4C participates in myogenic
differentiation in vivo and in vitro through the p38 MAPK pathway.
Eur J Cell Biol. 86:331–344. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Terada K, Misao S, Katase N, Nishimatsu S
and Nohno T: Interaction of Wnt signaling with BMP/Smad signaling
during the transition from cell proliferation to myogenic
differentiation in mouse myoblast-derived cells. Int J Cell Biol.
2013:6162942013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
von Maltzahn J, Chang NC, Bentzinger CF
and Rudnicki MA: Wnt signaling in myogenesis. Trends Cell Biol.
22:602–609. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Harada T, Horinouchi T, Higa T, Hoshi A,
Higashi T, Terada K, Mai Y, Nepal P, Horiguchi M, Hatate C, et al:
Endothelin-1 activates extracellular signal-regulated kinases 1/2
via trans-activation of platelet-derived growth factor receptor in
rat L6 myoblasts. Life Sci. 104:24–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feng Y, Niu LL, Wei W, Zhang WY, Li XY,
Cao JH and Zhao SH: A feedback circuit between miR-133 and the
ERK1/2 pathway involving an exquisite mechanism for regulating
myoblast proliferation and differentiation. Cell Death Dis.
4:e9342013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hindi SM, Tajrishi MM and Kumar A:
Signaling mechanisms in mammalian myoblast fusion. Sci Signal.
6:re22013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang M, Amano SU, Flach RJR, Chawla A,
Aouadi M and Czech MP: Identification of Map4k4 as a novel
suppressor of skeletal muscle differentiation. Mol Cell Biol.
33:678–687. 2013. View Article : Google Scholar :
|
|
26
|
Kajava AV: Structural diversity of
leucine-rich repeat proteins. J Mol Biol. 277:519–527. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tominaga K, Kondo C, Kagata T, Hishida T,
Nishizuka M and Imagawa M: The novel gene fad158, having a
transmembrane domain and leucine-rich repeat, stimulates adipocyte
differentiation. J Biol Chem. 279:34840–34848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim T, Kim K, Lee SH, So HS, Lee J, Kim N
and Choi Y: Identification of LRRc17 as a negative regulator of
receptor activator of NF-κB ligand (RANKL)-induced osteoclast
differentiation. J Biol Chem. 284:15308–15316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wilhite SE and Barrett T: Strategies to
explore functional genomics data sets in NCBI's GEO database.
Methods Mol Biol. 802:41–53. 2012. View Article : Google Scholar
|
|
30
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: database for annotation,
visualization, and integrated discovery. Genome Biol. 4:32003.
View Article : Google Scholar
|
|
31
|
Kocić J1, Santibañez JF, Krstić A,
Mojsilović S, Dorđević IO, Trivanović D, Ilić V and Bugarski D:
Interleukin 17 inhibits myogenic and promotes osteogenic
differentiation of C2C12 myoblasts by activating ERK1, 2. Biochim
Biophys Acta. 1823:838–849. 2012. View Article : Google Scholar
|
|
32
|
Bennett AM and Tonks NK: Regulation of
distinct stages of skeletal muscle differentiation by
mitogen-activated protein kinases. Science. 278:1288–1291. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Assou S, Le Carrour T, Tondeur S, Ström S,
Gabelle A, Marty S, Nadal L, Pantesco V, Réme T, Hugnot JP, et al:
A meta-analysis of human embryonic stem cells transcriptome
integrated into a web-based expression atlas. Stem Cells.
25:961–973. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen X, Liang S, Zheng W, Liao Z, Shang T
and Ma W: Meta-analysis of nasopharyngeal carcinoma microarray data
explores mechanism of EBV-regulated neoplastic transformation. BMC
Genomics. 9:3222008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Katase N, Terada K, Suzuki T, Nishimatsu S
and Nohno T: miR-487b, miR-3963 and miR-6412 delay myogenic
differentiation in mouse myoblast-derived C2C12 cells. BMC Cell
Biol. 16:132015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ge X, Zhang Y, Park S, Cong X, Gerrard DE
and Jiang H: Stac3 inhibits myoblast differentiation into myotubes.
PLoS One. 9:e959262014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gredinger E, Gerber AN, Tamir Y, Tapscott
SJ and Bengal E: Mitogen-activated protein kinase pathway is
involved in the differentiation of muscle cells. J Biol Chem.
273:10436–10444. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jones NC, Fedorov YV, Rosenthal RS and
Olwin BB: ERK1/2 is required for myoblast proliferation but is
dispensable for muscle gene expression and cell fusion. J Cell
Physiol. 186:104–115. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li J and Johnson SE: ERK2 is required for
efficient terminal differentiation of skeletal myoblasts. Biochem
Biophys Res Commun. 345:1425–1433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Wang X, Zhang P, Zhu L, Zhao T, Liu
S, Wu Y, Chen X and Fan M: Extracellular signal-regulated kinase
1/2 mitogen-activated protein kinase pathway is involved in
inhibition of myogenic differentiation of myoblasts by hypoxia. Exp
Physiol. 97:257–264. 2012. View Article : Google Scholar
|
|
41
|
Riuzzi F, Sorci G and Donato R: S100B
stimulates myoblast proliferation and inhibits myoblast
differentiation by independently stimulating ERK1/2 and inhibiting
p38 MAPK. J Cell Physiol. 207:461–470. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang W, Chen Y, Zhang Y, Wang X, Yang N
and Zhu D: Extracellular signal-regulated kinase 1/2
mitogen-activated protein kinase pathway is involved in
myostatin-regulated differentiation repression. Cancer Res.
66:1320–1326. 2006. View Article : Google Scholar : PubMed/NCBI
|