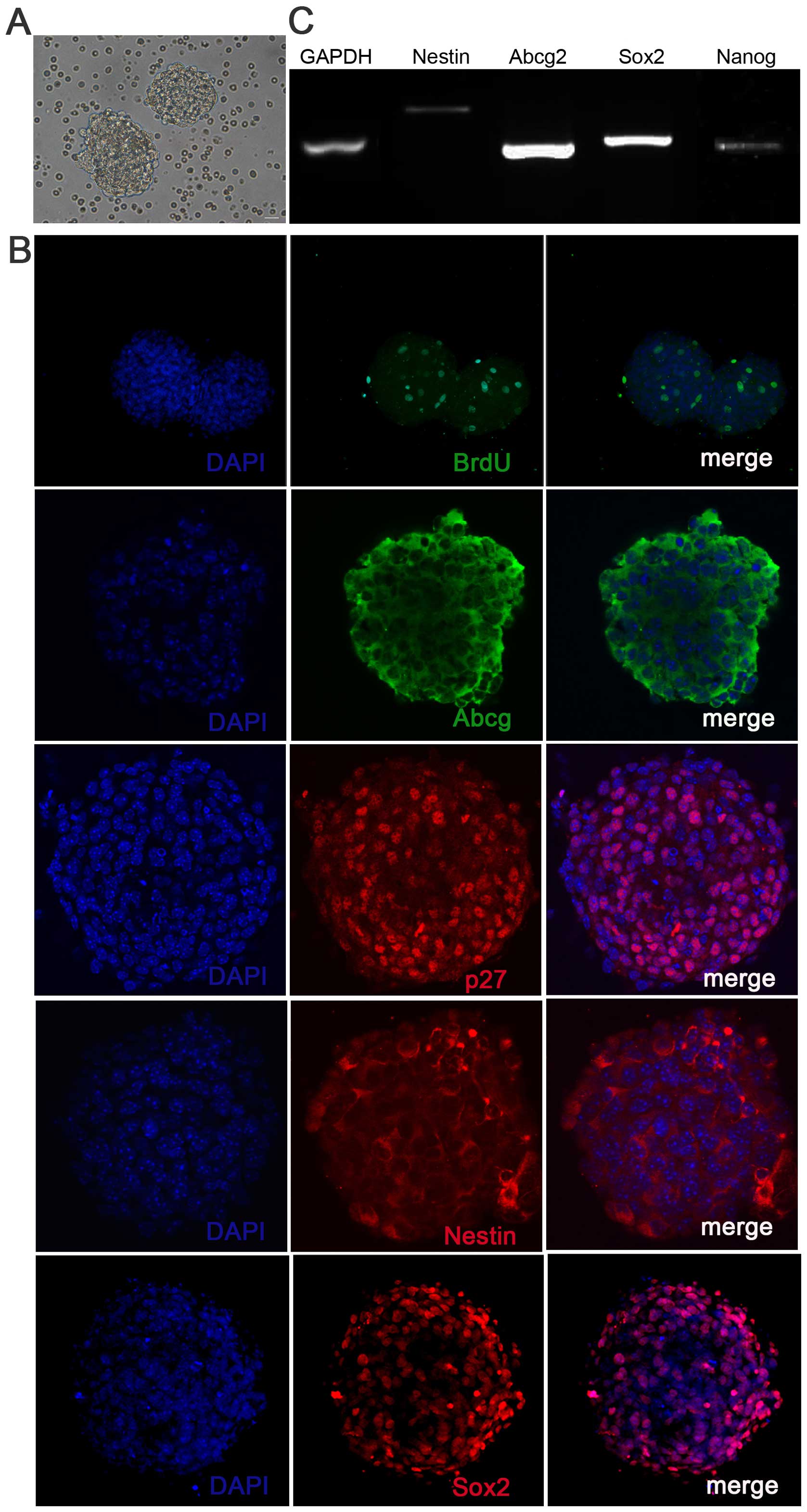
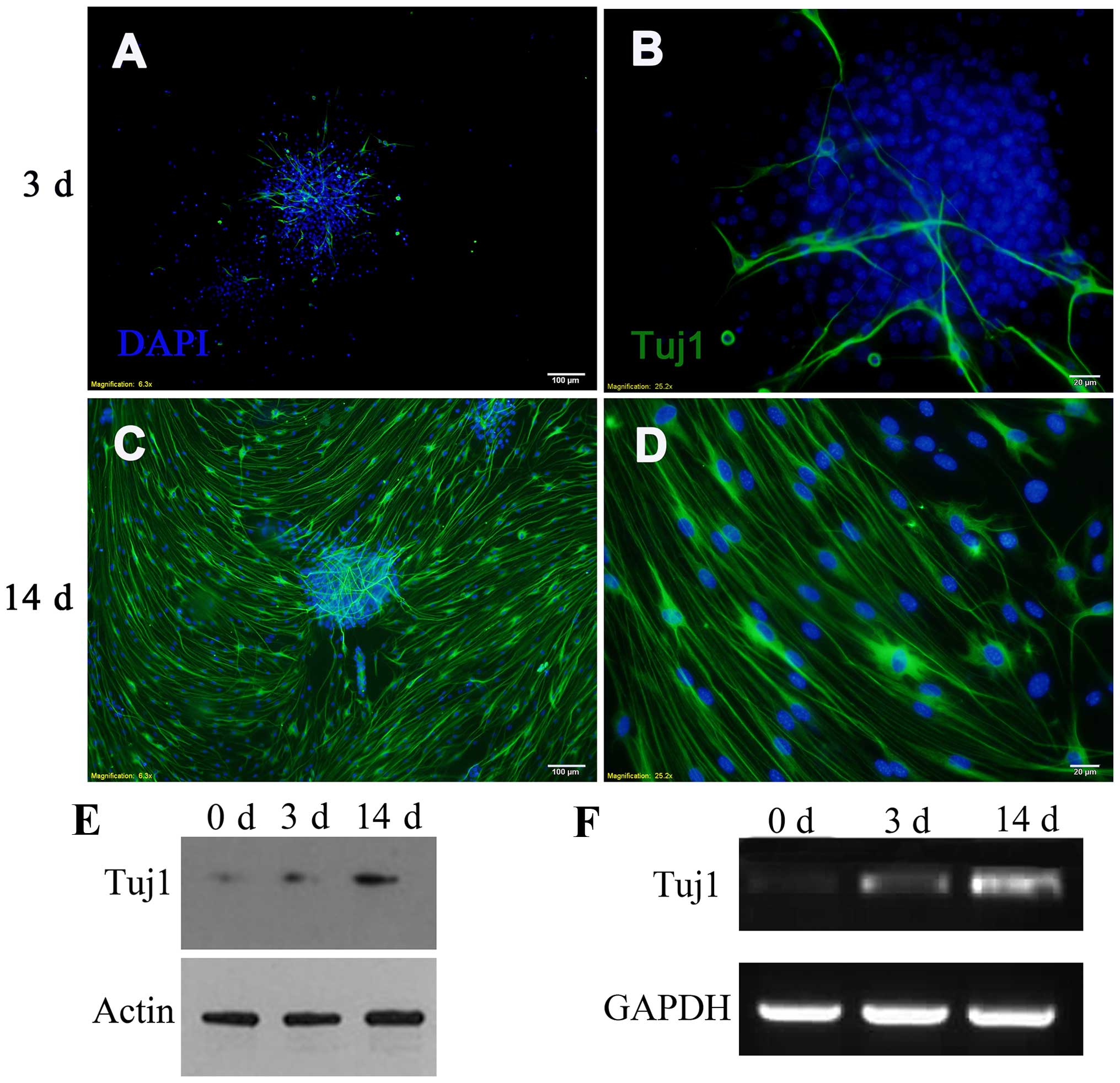
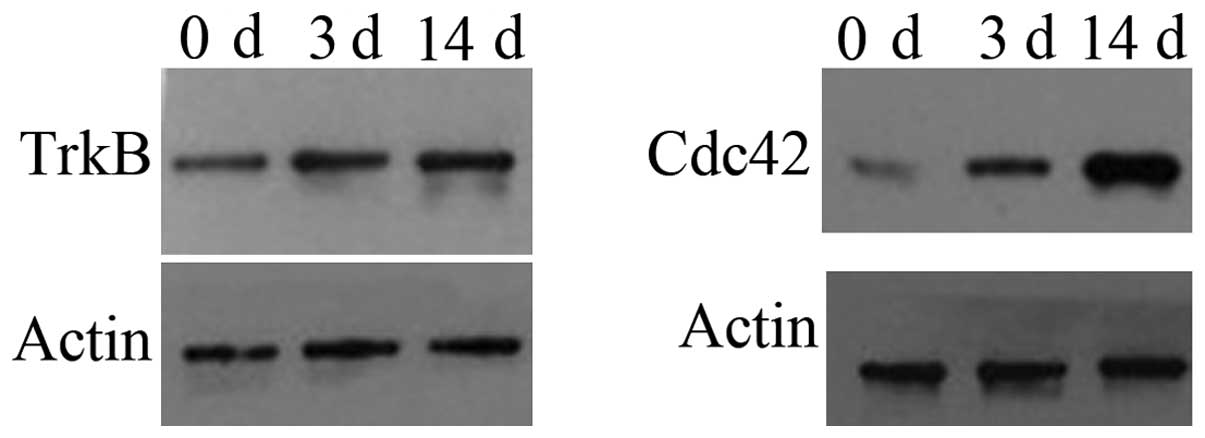
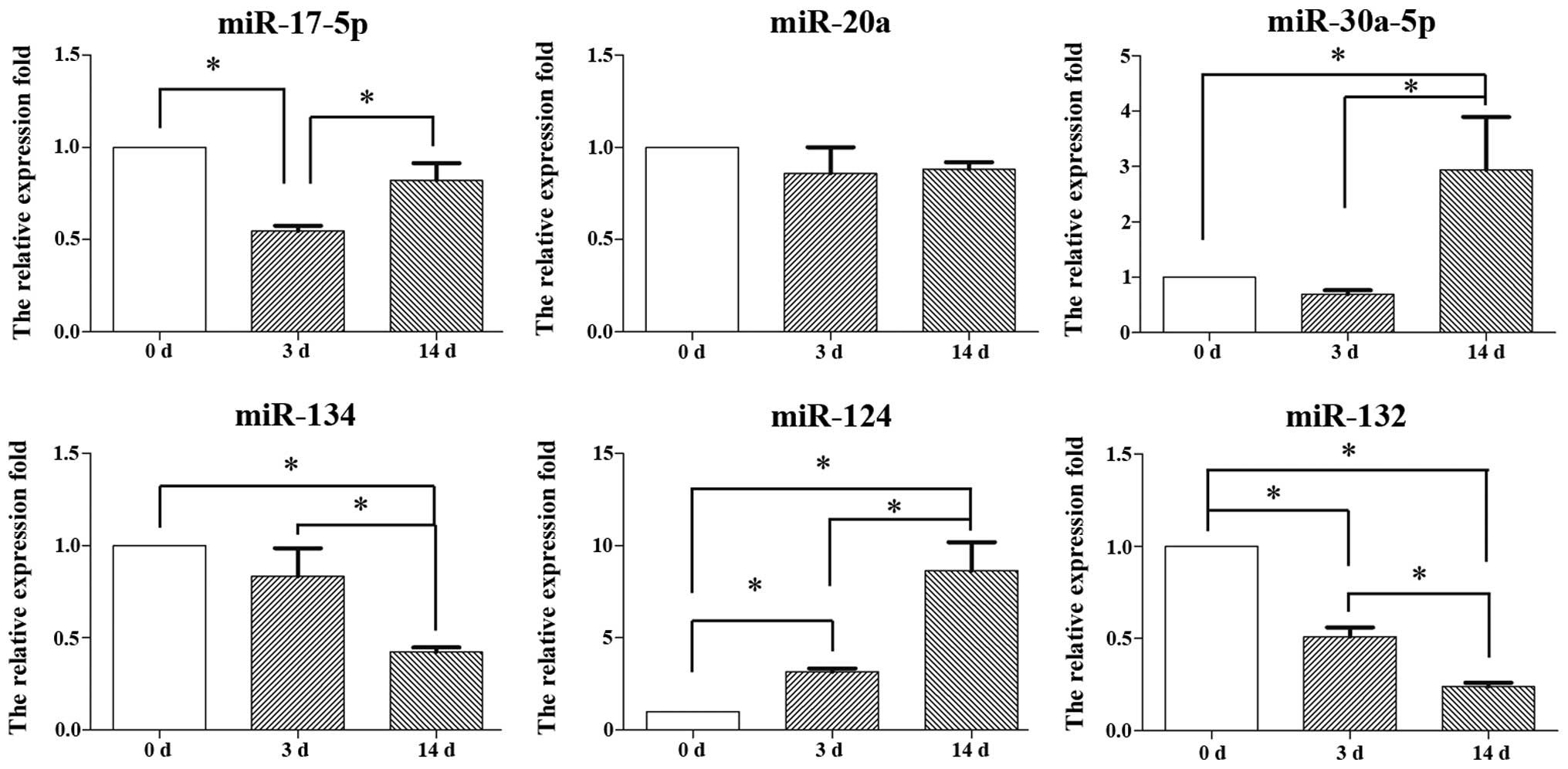
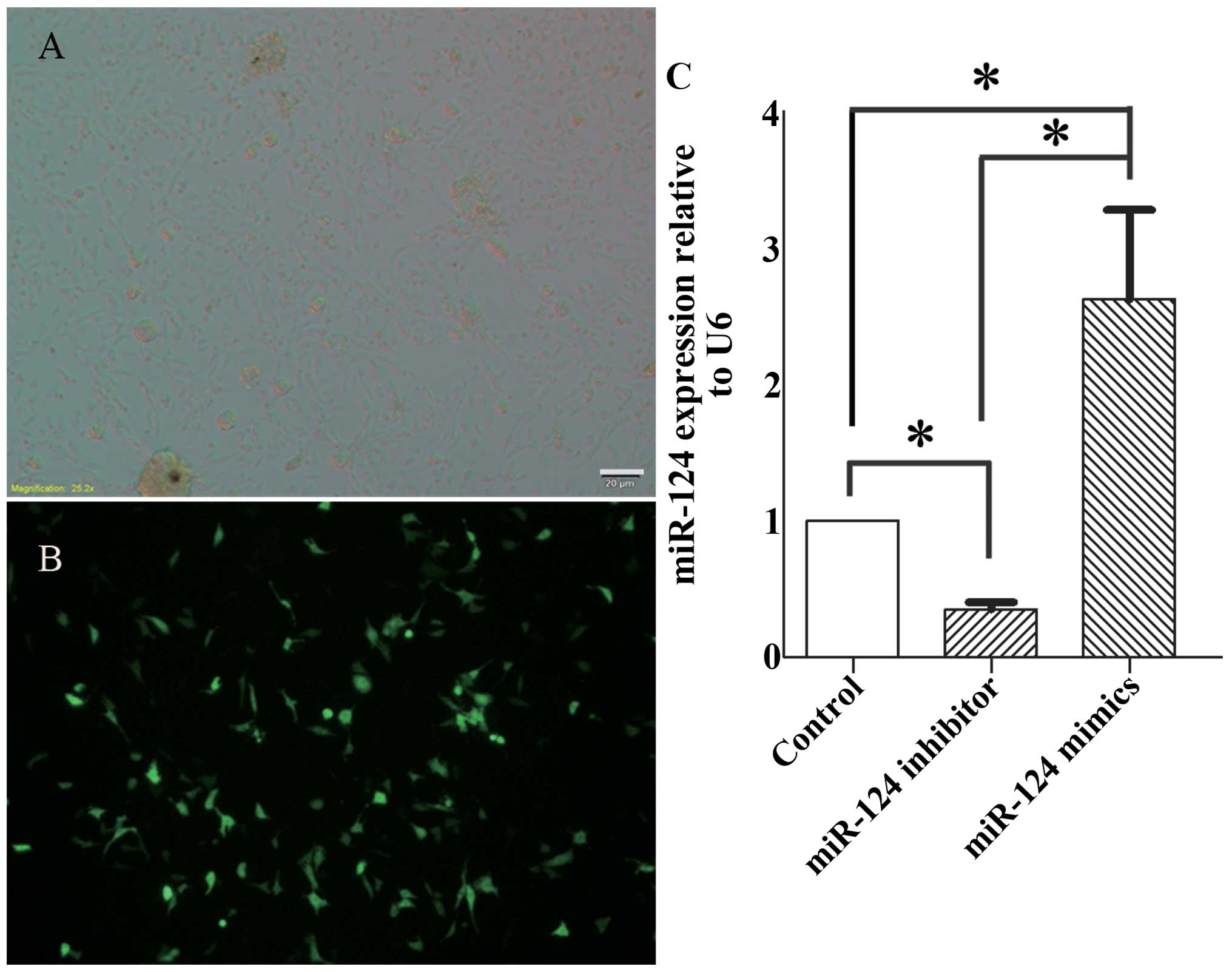
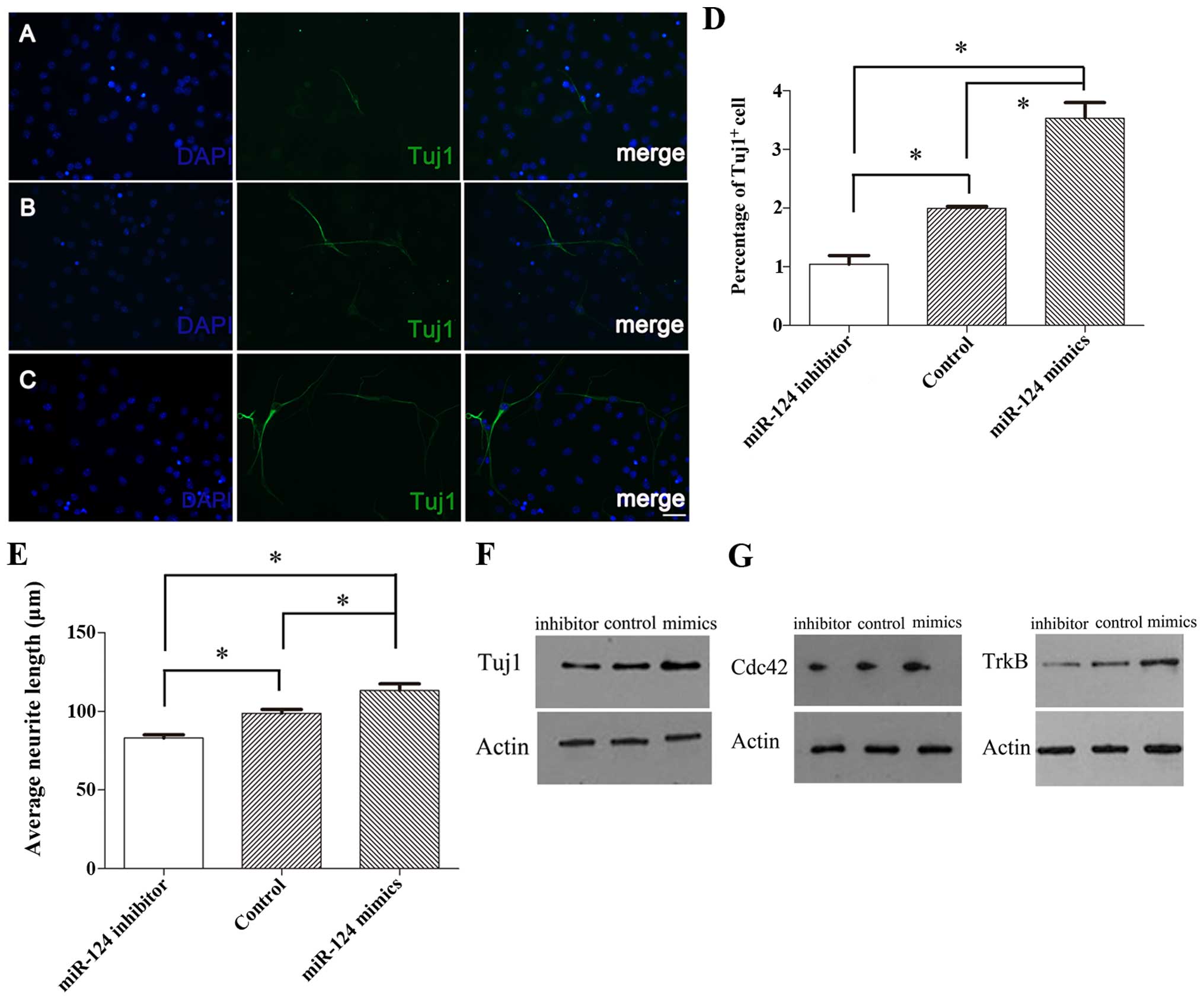
|
1
|
Spoendlin H: Retrograde degeneration of
the cochlear nerve. Acta Otolaryngol. 79:266–275. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hawkins JE Jr: Comparative otopathology:
aging, noise, and ototoxic drugs. Adv Otorhinolaryngol. 20:125–141.
1973.PubMed/NCBI
|
|
3
|
Zhao C, Deng W and Gage FH: Mechanisms and
functional implications of adult neurogenesis. Cell. 132:645–660.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oshima K, Grimm CM, Corrales CE, Senn P,
Martinez Monedero R, Géléoc GS, Edge A, Holt JR and Heller S:
Differential distribution of stem cells in the auditory and
vestibular organs of the inner ear. J Assoc Res Otolaryngol.
8:18–31. 2007. View Article : Google Scholar
|
|
5
|
Oshima K, Senn P and Heller S: Isolation
of sphere-forming stem cells from the mouse inner ear. Methods Mol
Biol. 493:141–162. 2009. View Article : Google Scholar
|
|
6
|
Rask-Andersen H, Boström M, Gerdin B,
Kinnefors A, Nyberg G, Engstrand T, Miller JM and Lindholm D:
Regeneration of human auditory nerve. In vitro/in video
demonstration of neural progenitor cells in adult human and guinea
pig spiral ganglion. Hear Res. 203:180–191. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gu S, Jin L, Zhang F, Sarnow P and Kay MA:
Biological basis for restriction of microRNA targets to the 3′
untranslated region in mammalian mRNAs. Nat Struct Mol Biol.
16:144–150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lai EC: MicroRNAs are complementary to 3′
UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vaucheret H: Post-transcriptional small
RNA pathways in plants: Mechanisms and regulations. Genes Dev.
20:759–771. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Darnell DK, Kaur S, Stanislaw S, Konieczka
JH, Yatskievych TA and Antin PB: MicroRNA expression during chick
embryo development. Dev Dyn. 235:3156–3165. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Deo M, Yu JY, Chung KH, Tippens M and
Turner DL: Detection of mammalian microRNA expression by in situ
hybridization with RNA oligonucleotides. Dev Dyn. 235:2538–2548.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kapsimali M, Kloosterman WP, de Bruijn E,
Rosa F, Plasterk RH and Wilson SW: MicroRNAs show a wide diversity
of expression profiles in the developing and mature central nervous
system. Genome Biol. 8:R1732007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krichevsky AM, Sonntag KC, Isacson O and
Kosik KS: Specific microRNAs modulate embryonic stem cell-derived
neurogenesis. Stem Cells. 24:857–864. 2006. View Article : Google Scholar
|
|
17
|
Mansfield JH, Harfe BD, Nissen R, Obenauer
J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli
AE, Ruvkun G, et al: MicroRNA-responsive 'sensor' transgenes
uncover Hox-like and other developmentally regulated patterns of
vertebrate microRNA expression. Nat Genet. 36:1079–1083. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nelson PT, Baldwin DA, Kloosterman WP,
Kauppinen S, Plasterk RH and Mourelatos Z: RAKE and LNA-ISH reveal
microRNA expression and localization in archival human brain. RNA.
12:187–191. 2006. View Article : Google Scholar :
|
|
19
|
Fineberg SK, Kosik KS and Davidson BL:
MicroRNAs potentiate neural development. Neuron. 64:303–309. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao FB: Context-dependent functions of
specific microRNAs in neuronal development. Neural Dev. 5:252010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Beveridge NJ, Tooney PA, Carroll AP, Tran
N and Cairns MJ: Down-regulation of miR-17 family expression in
response to retinoic acid induced neuronal differentiation. Cell
Signal. 21:1837–1845. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hohjoh H and Fukushima T: Marked change in
microRNA expression during neuronal differentiation of human
teratocarcinoma NTera2D1 and mouse embryonal carcinoma P19 cells.
Biochem Biophys Res Commun. 362:360–367. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um
M, Udolph G, Yang H, Lim B and Lodish HF: MicroRNA-125b promotes
neuronal differentiation in human cells by repressing multiple
targets. Mol Cell Biol. 29:5290–5305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain-specific alternative pre-mRNA splicing. Mol
Cell. 27:435–448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Visvanathan J, Lee S, Lee B, Lee JW and
Lee SK: The microRNA miR-124 antagonizes the anti-neural REST/SCP1
pathway during embryonic CNS development. Genes Dev. 21:744–749.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sacheli R, Nguyen L, Borgs L, Vandenbosch
R, Bodson M, Lefebvre P and Malgrange B: Expression patterns of
miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr
Patterns. 9:364–370. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang XR, Zhang XM, Zhen J, Zhang PX, Xu G
and Jiang H: MicroRNA expression in the embryonic mouse inner ear.
Neuroreport. 21:611–617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weston MD, Pierce ML, Rocha-Sanchez S,
Beisel KW and Soukup GA: MicroRNA gene expression in the mouse
inner ear. Brain Res. 1111:95–104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Friedman LM, Dror AA, Mor E, Tenne T,
Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E
and Avraham KB: MicroRNAs are essential for development and
function of inner ear hair cells in vertebrates. Proc Natl Acad Sci
USA. 106:7915–7920. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lewis MA, Quint E, Glazier AM, Fuchs H, De
Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M,
Redshaw N, et al: An ENU-induced mutation of miR-96 associated with
progressive hearing loss in mice. Nat Genet. 41:614–618. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li H, Kloosterman W and Fekete DM:
MicroRNA-183 family members regulate sensorineural fates in the
inner ear. J Neurosci. 30:3254–3263. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mencía A, Modamio-Høybjør S, Redshaw N,
Morín M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I,
Steel KP, Dalmay T, et al: Mutations in the seed region of human
miR-96 are responsible for nonsyndromic progressive hearing loss.
Nat Genet. 41:609–613. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Soukup GA, Fritzsch B, Pierce ML, Weston
MD, Jahan I, McManus MT and Harfe BD: Residual microRNA expression
dictates the extent of inner ear development in conditional Dicer
knockout mice. Dev Biol. 328:328–341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Martinez-Monedero R, Yi E, Oshima K,
Glowatzki E and Edge AS: Differentiation of inner ear stem cells to
functional sensory neurons. Dev Neurobiol. 68:669–684. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Roehm PC, Xu N, Woodson EA, Green SH and
Hansen MR: Membrane depolarization inhibits spiral ganglion neurite
growth via activation of multiple types of voltage sensitive
calcium channels and calpain. Mol Cell Neurosci. 37:376–387. 2008.
View Article : Google Scholar
|
|
36
|
Cheung ZH, Chin WH, Chen Y, Ng YP and Ip
NY: Cdk5 is involved in BDNF-stimulated dendritic growth in
hippocampal neurons. PLoS Biol. 5:e632007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Endo M, Antonyak MA and Cerione RA:
Cdc42-mTOR signaling pathway controls Hes5 and Pax6 expression in
retinoic acid-dependent neural differentiation. J Biol Chem.
284:5107–5118. 2009. View Article : Google Scholar
|
|
38
|
Sciarretta C, Fritzsch B, Beisel K,
Rocha-Sanchez SM, Buniello A, Horn JM and Minichiello L:
PLCγ-activated signalling is essential for TrkB mediated sensory
neuron structural plasticity. BMC Dev Biol. 10:1032010. View Article : Google Scholar
|
|
39
|
Sosa L, Dupraz S, Laurino L, Bollati F,
Bisbal M, Cáceres A, Pfenninger KH and Quiroga S: IGF-1 receptor is
essential for the establishment of hippocampal neuronal polarity.
Nat Neurosci. 9:993–995. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lai KO, Wong AS, Cheung MC, Xu P, Liang Z,
Lok KC, Xie H, Palko ME, Yung WH, Tessarollo L, et al: TrkB
phosphorylation by Cdk5 is required for activity-dependent
structural plasticity and spatial memory. Nat Neurosci.
15:1506–1515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gage FH: Mammalian neural stem cells.
Science. 287:1433–1438. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Elkan-Miller T, Ulitsky I, Hertzano R,
Rudnicki A, Dror AA, Lenz DR, Elkon R, Irmler M, Beckers J, Shamir
R and Avraham KB: Integration of transcriptomics, proteomics, and
microRNA analyses reveals novel microRNA regulation of targets in
the mammalian inner ear. PLoS One. 6:e181952011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Patel M, Cai Q, Ding D, Salvi R, Hu Z and
Hu BH: The miR-183/Taok1 target pair is implicated in cochlear
responses to acoustic trauma. PLoS One. 8:e584712013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Frucht CS, Santos-Sacchi J and Navaratnam
DS: MicroRNA181a plays a key role in hair cell regeneration in the
avian auditory epithelium. Neurosci Lett. 493:44–48. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hertzano R, Elkon R, Kurima K, Morrisson
A, Chan SL, Sallin M, Biedlingmaier A, Darling DS, Griffith AJ,
Eisenman DJ and Strome SE: Cell type-specific transcriptome
analysis reveals a major role for Zeb1 and miR-200b in mouse inner
ear morphogenesis. PLoS Genet. 7:e10023092011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gu C, Li X, Tan Q, Wang Z, Chen L and Liu
Y: MiR-183 family regulates chloride intracellular channel 5
expression in inner ear hair cells. Toxicol In Vitro. 27:486–491.
2013. View Article : Google Scholar
|
|
47
|
Wang XR, Zhang XM, Du J and Jiang H:
MicroRNA-182 regulates otocyst-derived cell differentiation and
targets T-box1 gene. Hear Res. 286:55–63. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mellios N, Huang HS, Grigorenko A, Rogaev
E and Akbarian S: A set of differentially expressed miRNAs,
including miR-30a-5p, act as post-transcriptional inhibitors of
BDNF in prefrontal cortex. Hum Mol Genet. 17:3030–3042. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gaughwin P, Ciesla M, Yang H, Lim B and
Brundin P: Stage-specific modulation of cortical neuronal
development by Mmu-miR-134. Cereb Cortex. 21:1857–1869. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mellios N, Sugihara H, Castro J, Banerjee
A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, et
al: miR-132, an experience-dependent microRNA, is essential for
visual cortex plasticity. Nat Neurosci. 14:1240–1242. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Clovis YM, Enard W, Marinaro F, Huttner WB
and De Pietri Tonelli D: Convergent repression of Foxp2 3′UTR by
miR-9 and miR-132 in embryonic mouse neocortex: Implications for
radial migration of neurons. Development. 139:3332–3342. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lin LF, Chiu SP, Wu MJ, Chen PY and Yen
JH: Luteolin induces microRNA-132 expression and modulates neurite
outgrowth in PC12 cells. PLoS One. 7:e433042012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pathania M, Torres-Reveron J, Yan L,
Kimura T, Lin TV, Gordon V, Teng ZQ, Zhao X, Fulga TA, Van Vactor D
and Bordey A: miR-132 enhances dendritic morphogenesis, spine
density, synaptic integration, and survival of newborn olfactory
bulb neurons. PLoS One. 7:e381742012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Aranha MM, Santos DM, Xavier JM, Low WC,
Steer CJ, Solá S and Rodrigues CM: Apoptosis-associated microRNAs
are modulated in mouse, rat and human neural differentiation. BMC
Genomics. 11:5142010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Trompeter HI, Abbad H, Iwaniuk KM, Hafner
M, Renwick N, Tuschl T, Schira J, Müller HW and Wernet P: MicroRNAs
MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F
activity on cell cycle arrest during neuronal lineage
differentiation of USSC. PLoS One. 6:e161382011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Miska EA, Alvarez-Saavedra E, Townsend M,
Yoshii A, Sestan N, Rakic P, Constantine-Paton M and Horvitz HR:
Microarray analysis of microRNA expression in the developing
mammalian brain. Genome Biol. 5:R682004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Smirnova L, Gräfe A, Seiler A, Schumacher
S, Nitsch R and Wulczyn FG: Regulation of miRNA expression during
neural cell specification. Eur J Neurosci. 21:1469–1477. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yu JY, Chung KH, Deo M, Thompson RC and
Turner DL: MicroRNA miR-124 regulates neurite outgrowth during
neuronal differentiation. Exp Cell Res. 314:2618–2633. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lee MR, Kim JS and Kim KS: miR-124a is
important for migratory cell fate transition during gastrulation of
human embryonic stem cells. Stem Cells. 28:1550–1559. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Maisel M, Habisch HJ, Royer L, Herr A,
Milosevic J, Hermann A, Liebau S, Brenner R, Schwarz J, Schroeder M
and Storch A: Genome-wide expression profiling and functional
network analysis upon neuroectodermal conversion of human
mesenchymal stem cells suggest HIF-1 and miR-124a as important
regulators. Exp Cell Res. 316:2760–2778. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hall A: Rho GTPases and the actin
cytoskeleton. Science. 279:509–514. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jaffe AB and Hall A: Rho GTPases:
Biochemistry and biology. Annu Rev Cell Dev Biol. 21:247–269. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li E, Stupack D, Bokoch GM and Nemerow GR:
Adenovirus endocytosis requires actin cytoskeleton reorganization
mediated by Rho family GTPases. J Virol. 72:8806–8812.
1998.PubMed/NCBI
|
|
64
|
Schwamborn JC and Püschel AW: The
sequential activity of the GTPases Rap1B and Cdc42 determines
neuronal polarity. Nat Neurosci. 7:923–929. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Huang EJ and Reichardt LF: Trk receptors:
Roles in neuronal signal transduction. Annu Rev Biochem.
72:609–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen TJ, Gehler S, Shaw AE, Bamburg JR and
Letourneau PC: Cdc42 participates in the regulation of ADF/cofilin
and retinal growth cone filopodia by brain derived neurotrophic
factor. J Neurobiol. 66:103–114. 2006. View Article : Google Scholar
|
|
67
|
Shen W, Wu B, Zhang Z, Dou Y, Rao ZR, Chen
YR and Duan S: Activity-induced rapid synaptic maturation mediated
by presynaptic cdc42 signaling. Neuron. 50:401–414. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Franke K, Otto W, Johannes S, Baumgart J,
Nitsch R and Schumacher S: miR-124-regulated RhoG reduces neuronal
process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO
J. 31:2908–2921. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mojtahedi S, Kordi MR, Hosseini SE, Omran
SF and Soleimani M: Effect of treadmill running on the expression
of genes that are involved in neuronal differentiation in the
hippo-campus of adult male rats. Cell Biol Int. 37:276–283. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hing H, Xiao J, Harden N, Lim L and
Zipursky SL: Pak functions downstream of Dock to regulate
photoreceptor axon guidance in Drosophila. Cell. 97:853–863. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Schimmang T, Minichiello L, Vazquez E, San
Jose I, Giraldez F, Klein R and Represa J: Developing inner ear
sensory neurons require TrkB and TrkC receptors for innervation of
their peripheral targets. Development. 121:3381–3391.
1995.PubMed/NCBI
|
|
72
|
Schimmang T, Tan J, Müller M, Zimmermann
U, Rohbock K, Kôpschall I, Limberger A, Minichiello L and Knipper
M: Lack of Bdnf and TrkB signalling in the postnatal cochlea leads
to a spatial reshaping of innervation along the tonotopic axis and
hearing loss. Development. 130:4741–4750. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Brors D, Aletsee C, Dazert S, Huverstuhl
J, Ryan AF and Bodmer D: Clostridium difficile toxin B, an
inhibitor of the small GTPases Rho, Rac and Cdc42, influences
spiral ganglion neurite outgrowth. Acta Otolaryngol. 123:20–25.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Mullen LM, Pak KK, Chavez E, Kondo K,
Brand Y and Ryan AF: Ras/p38 and PI3K/Akt but not Mek/Erk signaling
mediate BDNF-induced neurite formation on neonatal cochlear spiral
ganglion explants. Brain Res. 1430:25–34. 2012. View Article : Google Scholar
|