Introduction
The human lysyl oxidase-like 3 (LOXL3) gene
was originally identified by EST database searches, encoding a 753
amino acid-long polypeptide with a deduced molecular mass of 83 kDa
(1,2). LOXL3 belongs to an emerging family
of lysyl oxidases (LOX), each of which functions as a
copper-dependent amine oxidase for the formation of lysine-derived
cross-links found in collagen and elastin fibrils (3). All LOX paralogues (LOX, LOXL, LOXL2,
LOXL3 and LOXL4) contain a copper-binding domain, residues for
lysyl-tyrosyl quinone (LTQ) and a cytokine receptor-like (CRL)
domain in the C-terminus. In addition, four repeated copies of
scavenger receptor cysteine-rich (SRCR) domains are found in the
N-terminus of only LOXL2, LOXL3 and LOXL4, suggesting novel
functional roles assigned to these three members (4–6).
LOXL3 has been reported to be associated with
a diverse range of diseases in both humans and animals. LOXL3 was
reported to interact with Snail to downregulate the expression of
E-cadherin in carcinoma progression (7), to induce the autophagy of
chondrocytes in osteoarthritis (8) and to play an essential role in the
regulation of integrin signaling for correct positioning and
anchoring of myofibers (9).
Expression of LOXL3 was found to be significantly reduced at both
mRNA and protein levels in the vaginal tissues of patients with
pelvic organ prolapse (10). A
recent genetic study reported a homozygous missense mutation
(C676Y) in LOXL3 as the cause of Stickler syndrome, a
collagenopathy characterized by arthropathy and vitreoretinopathy
with high myopia and cleft palate, in a consanguineous Saudi family
(11). Several homozygous and
compound heterozygous frame-shift mutations of LOXL3 have
been identified in patients with early-onset high myopia (12). In animal studies,
Loxl3−/− mice showed perinatal lethality with
craniofacial and spinal defects (13), and knockdown of lox3b in
zebrafish resulted in craniofacial abnormalities (14).
Previously, we reported a variant of LOXL3,
termed LOXL3-sv1 (15),
which contained the characteristic C-terminal domains of the LOX
family but lacked the characteristic N-terminal region including
SRCR domains 1–3. The LOXL3-sv1 showed amine oxidase activity
toward different types of collagens with distinct substrate
specificity from LOXL3 (15). In
search of more human LOXL3 variants, we identified a novel
variant, termed LOXL3-sv2, which lacked the sequences
corresponding to exons 4 and 5 of LOXL3. The deduced amino
acid sequence of LOXL3-sv2 does not have the SRCR domain 2 but
contains all the other functional domains of LOXL3. The recombinant
LOXL3-sv2 protein showed β-aminopropionitrile (BAPN)-inhibitable
amine oxidase activity toward collagen type I. These findings
indicate that LOXL3 encodes at least three variants, LOXL3,
LOXL3-sv1 and LOXL3-sv2, each functioning as an amine oxidase
toward collagen.
Materials and methods
EST database search
The human LOXL3 (GenBank accession no.
NM_032603.3) cDNA sequence was used to search the human EST
database through the BLASTN program (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST). EST clones
showing significant sequence homology to human LOXL3 were
purchased from GE Dharmacon (Lafayette, CO, USA) and were
completely sequenced using an ABI 310 automated DNA sequencer.
RT-PCR analysis in human tissues
Human MTC (Multiple Tissue cDNA) Panels (Clontech
Laboratories, Inc., Mountain View, CA, USA) were PCR-amplified with
primers specific to LOXL3-sv1 (GenBank accession no.
DQ378059) and LOXL3-sv2 (GenBank accession no. NM_001289164)
using Ex-Taq polymerase (Takara Bio, Inc., Otsu, Japan). The
LOXL3-sv1-specific primers were: forward,
5′-TGGAAGTCAGGCTTCCTGAC-3′ and reverse, 5′-CTCAGAGCTTCTCGAGCACT-3′.
The LOXL3-sv2-specific primers were:
5′-GTGTGACTGAATGTGCCTCC-3′ and reverse, 5′-CAGTCATCCCCACAGATGAG-3′.
Both the LOXL3-sv1-specific and LOXL3-sv2-specific
primer sets were also designed to detect the LOXL3 mRNA. The
PCR conditions consisted of 30 cycles at 94°C for 45 sec, 56–58°C
for 45 sec, and 72°C for 45 sec with a predenaturation at 94°C for
5 min and a final extension at 72°C for 7 min. The amplified PCR
products were analyzed by electrophoresis on a 2% agarose gel.
GAPDH was used as an internal control. All RT-PCR analyses
were performed in the linear range of amplification, and
quantification was performed using Kodak Gel Logic Imaging System
(Eastman Kodak, Rochester, NY, USA).
Expression and purification of the
LOXL3-sv1 and LOXL3-sv2 proteins
For expression of the LOXL3-sv1 protein, the
pET21-LOXL3-sv1 construct that was previously reported by us
(15) was used. For expression of
LOXL3-sv2, an EST clone (clone ID no. 6650423; GE Dharmacon) was
used as a PCR template, and the entire coding region of
LOXL3-sv2 was PCR-amplified by PfuTurbo™ DNA polymerase
(Agilent Technologies, Santa Clara, CA, USA) according to the
manufacturer's instructions. The sequences of the oligonucleotide
primers used for the construction of the expression plasmids were
5′-GCGGCTAGCATGCGACCTGTCAGTGTCTG-3′ and
5′-GCGAAGCTTGATAATCTGGTTGCTGGTCTG-3′. A unique restriction site,
either NheI or HindIII, was introduced in each primer
for convenient subcloning into the pET21 vector. The PCR conditions
consisted of 30 cycles at 94°C for 60 sec, 58°C for 60 sec and 72°C
for 60 sec with a predenaturation at 94°C for 5 min and a final
extension at 72°C for 7 min. The sequence of the resulting
expression constructs was confi rmed by DNA-sequencing analysis
using a Cycle Sequencing Ready Reaction kit (Life Technologies,
Paisley, UK) according to the manufacturer's instructions. The
Escherichia coli (E. coli) strain BL21 (DE3)
(Novagen, Inc., Madison, WI, USA) was used for transformation of
the LOXL3-sv1 or LOXL3-sv2 expression construct. The recombinant
proteins were expressed, purified and refolded into an
enzymatically active form as previously reported (15).
Amine oxidase assays
The amine oxidase activity of the LOXL3-sv1 and
LOXL3-sv2 recombinant proteins were assessed using a
peroxidase-coupled fluorometric assay with the Amplex®
Red hydrogen peroxide assay kit (Molecular Probes, Eugene, OR, USA)
as previously described (16).
Each reaction contained 10 µg of the purified LOXL3-sv1 or
LOXL3-sv2 recombinant protein and 20 pmol of calfskin type I
collagen (Sigma-Aldrich, St. Louis, MO, USA) as a substrate in a
reaction volume of 200 µl. Parallel assays were performed in
the absence or presence of 1 mM BAPN for 30 min at 37°C.
Fluorescence was measured using a fluorescence spectrophotometer
(Molecular Devices, Sunnyvale, CA, USA) with excitation and
emission wavelengths of 500 nm and 650 nm, respectively. Total
amine oxidase activity was expressed as nM of
H2O2 produced per µg of the LOXL3-sv1
or LOXL3-sv2 recombinant protein, calculated by interpolation with
fluorescence values from an H2O2 calibration
curve.
Statistical analysis
Statistical analyses were performed by means of
one-way analysis of variance (ANOVA) followed by a
multiple-comparison Tukey's test. All statistical analyses were
performed using SPSS software version 12 (SPSS, Inc., Chicago, IL,
USA). Statistical significance was determined at P-values
<0.05.
Results
Distinct exon-intron structure of
LOXL3-sv2 from LOXL3 and LOXL3-sv1
Searches for novel variants of the human
LOXL3 gene resulted in the identification of ESTs (accession
nos. EL736465 and CN422297) in the GenBank database, which showed a
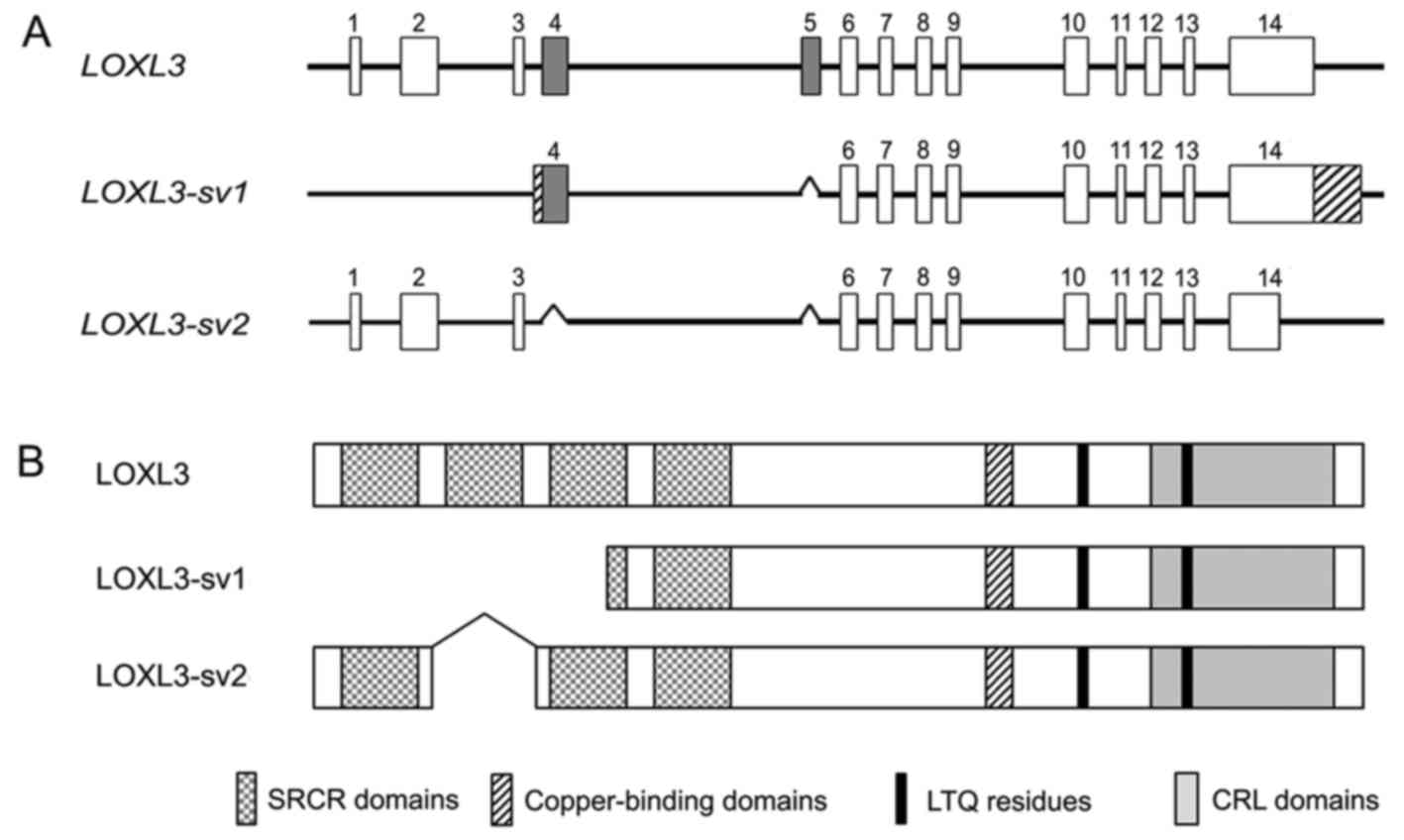
different exon-intron structure from LOXL3. Furthermore,
this novel variant, termed LOXL3-sv2, showed an exon-intron
structure distinct from the LOXL3-sv1 that we previously
reported (15), lacking exons 4
and 5 of LOXL3 (Fig. 1A).
The novel variant corresponded to the LOXL3 transcript variant 2
(LOXL3-v2) that was recently reported in the GenBank
(accession no. NM_001289164). However, there was a difference in
the length of the 3′-untranslated region (3′-UTR); our
LOXL3-sv2 contains 476 bp 3′-UTR, whereas the previously
reported LOXL3-v2 contains 1,348 bp 3′-UTR. LOXL3-sv2 also
corresponded to an unverified cDNA sequence of LOXL3 (accession no.
BC071865), but BC071865 contained an additional 30 bp sequence at
the 5′-end, originating from an unknown source.
The LOXL3-sv2 mRNA is at least 2,398 bp in
length, encoding a 608 amino acid-long polypeptide with a
calculated molecular mass of 67.4 kDa. The deletion of exons 4 and
5 do not change the open-reading frame of LOXL3 but results in
deletion of the SRCR domain 2 (Fig.
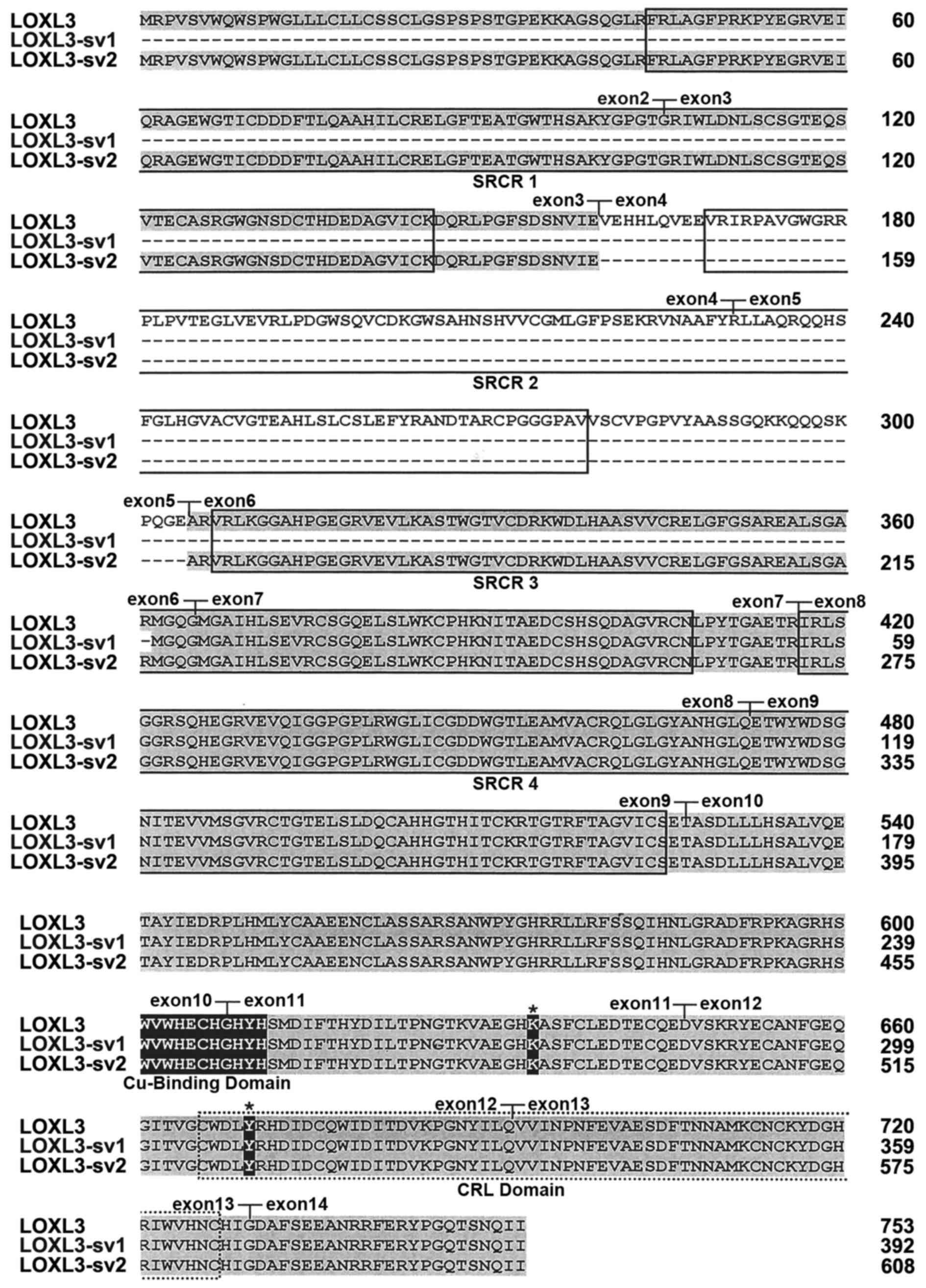
1B). The deduced amino acid sequence of LOXL3-sv2 contains the
characteristic C-terminal domains of the LOX family, including the
copper-binding domain, the residues for the LTQ cofactor, and the
CRL domain (Fig. 2).
Expression analysis of LOXL3, LOXL3-sv1
and LOXL3-sv2 in human tissues
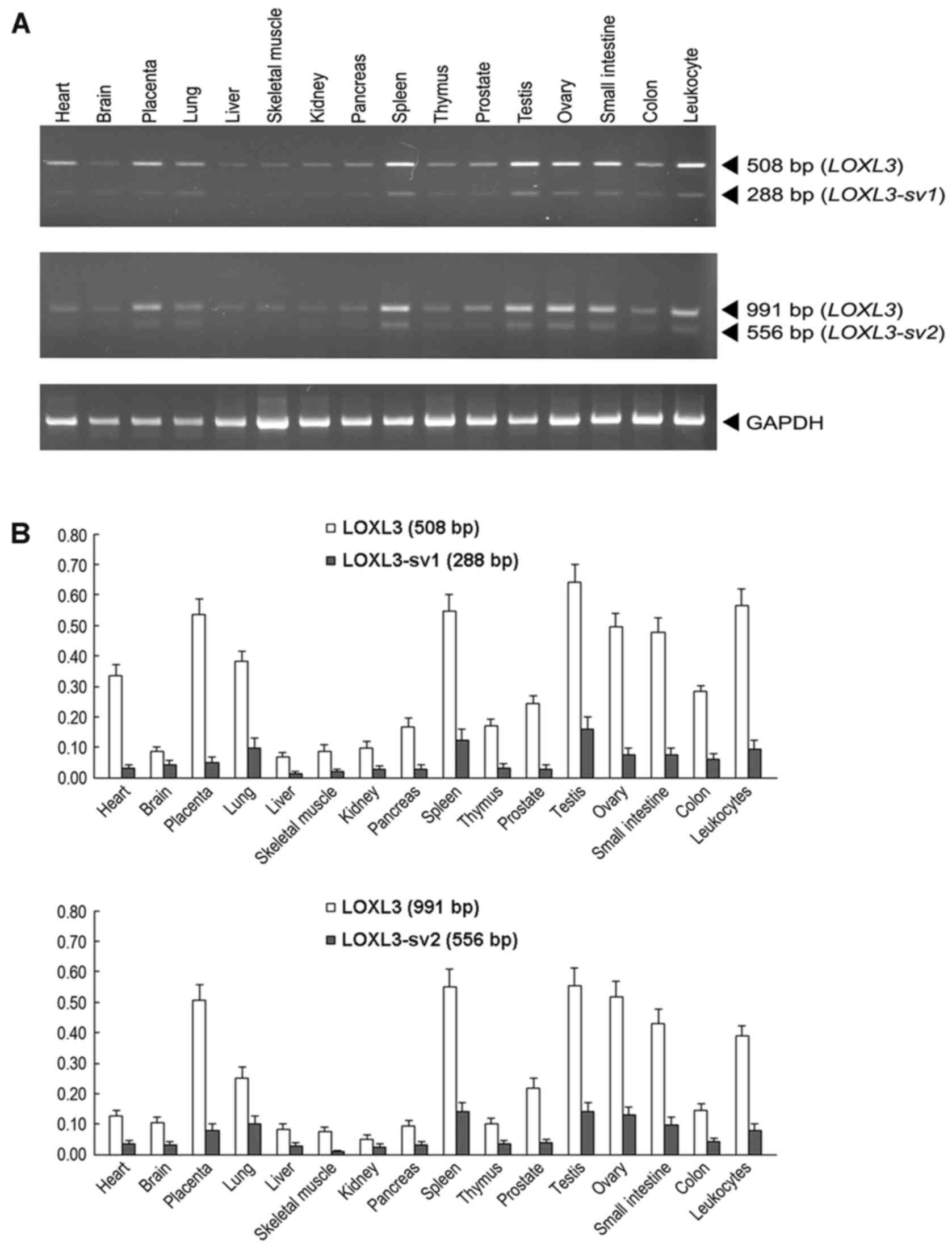
To compare the expression of LOXL3,
LOXL3-sv1 and LOXL3-sv2 in human tissues, RT-PCR
analyses were performed with poly(A)+ RNA isolated from
16 different human tissues. For specific detection of
LOXL3-sv1 and LOXL3-sv2 amplicons, the different
exonic structures of the LOXL3-sv1 and LOXL3-sv2
mRNAs were used to design the primers. Both the LOXL3-sv1-
and LOXL3-sv2-specific primer sets were also expected to
detect the LOXL3 mRNA that contains all the exonic
sequences. For the LOXL3-sv1-specific amplicon, the forward
primer was derived from exon 4, which was deleted in the
LOXL3-sv2 mRNA, and the reverse primer was derived from exon
6. This primer set was expected to amplify a 288 bp band from
nucleotide sequences 169–456 of LOXL3-sv1 and a 508 bp band
from nucleotide sequences 638–1,145 from LOXL3. For the
LOXL3-sv2-specific amplicon, the forward primer was derived
from exon 3, which was deleted in the LOXL3-sv1 mRNA, and
the reverse primers were derived from exon 8. This primer set was
expected to amplify a 556 bp band from nucleotide sequences
460–1,015 of LOXL3-sv2 and a 991 bp band from nucleotide
sequences 431–1,421 of LOXL3. In all tissues tested,
amplicons of expected sizes were detected for both LOXL3-sv1
and LOXL3-sv2, but the expression levels were significantly
lower than that of LOXL3 (Fig.
3). The expression patterns of LOXL3 and the two
variants were similar in all tissues tested (Fig. 3).
Amine oxidase activity of LOXL3-sv2
We previously reported that LOXL3-sv1 functions as
an amine oxidase (15). To
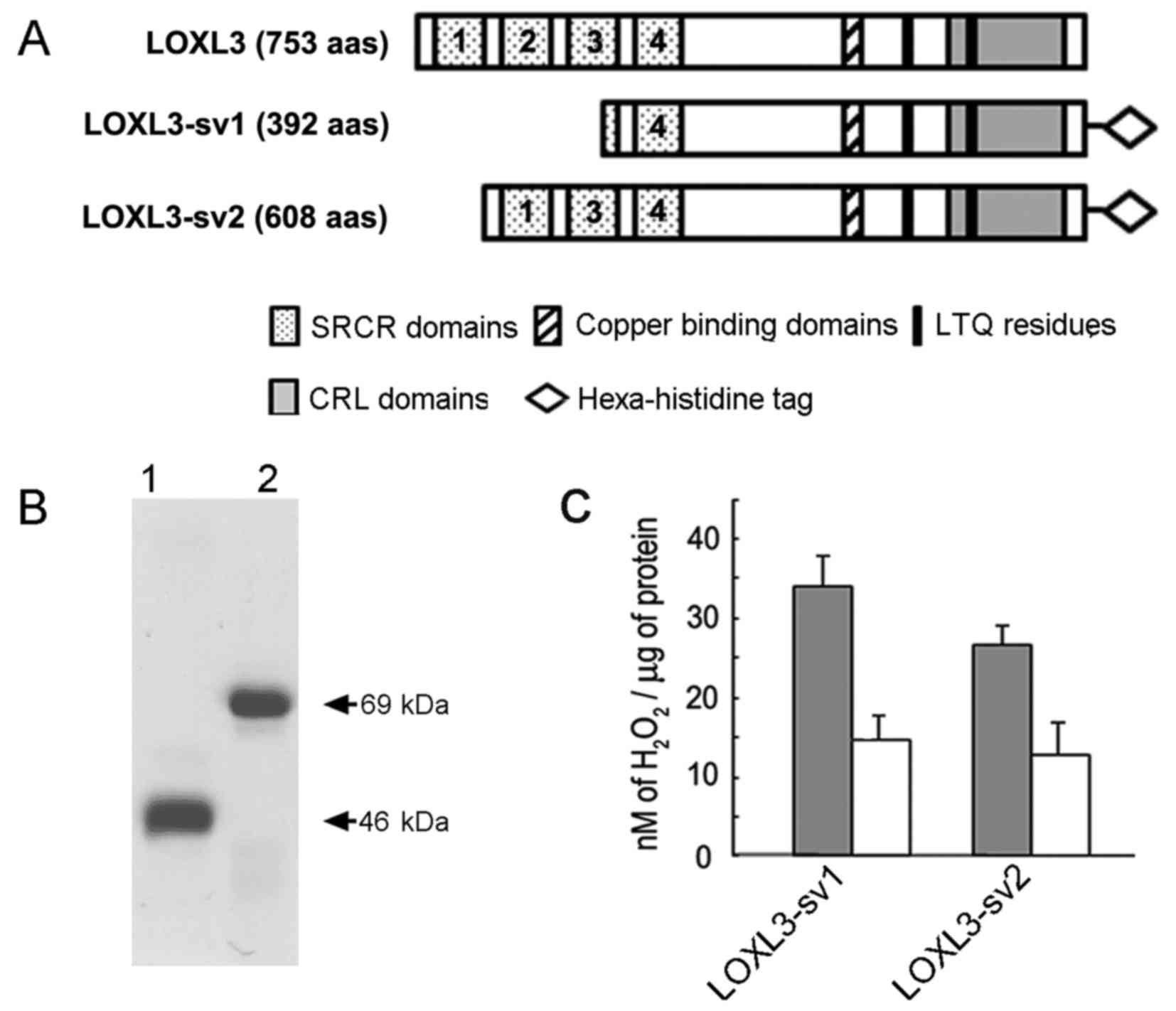
determine if LOXL3-sv2 functions as an amine oxidase, we expressed
and purified the LOXL3-sv2 protein in a hexa-histidine recombinant
form as previously described for LOXL3 and LOXL3-sv1 (15). The LOXL3-sv1 protein was also
expressed and purified in a hexa-histidine recombinant form for
comparison of the amine oxidase activity with LOXL3-sv2 (Fig. 4A). However, the full-length LOXL3
protein was not expressed in the E. coli overexpression
system, as we previously reported (15). It was probably due to the presence
of rare codons clustered in the N-terminus of LOXL3, such as CCC
(Pro11, Pro50, Pro53,
Pro173, Pro181, Pro183 and
Pro220) and AGG (Arg51, Arg192 and
Arg224) that are infrequently used in E. coli.
Clusters of rare codons were reported to reduce the expression
level in E. coli (17).
The apparent sizes of the purified recombinant proteins were in
agreement with the deduced molecular mass; 46 and 69 kDa for the
recombinant LOXL3-sv1 and LOXL3-sv2 proteins, respectively
(Fig. 4B).
The recombinant LOXL3-sv1 and LOXL3-sv2 proteins
were assessed for amine oxidase activity toward collagen type I
using a peroxidase-coupled fluorometric assay. Both the recombinant
LOXL3-sv1 and LOXL3-sv2 proteins showed significant levels of amine
oxidase activity toward collagen type I in a manner sensitive to
β-aminopropionitrile (BAPN), an irreversible inhibitor of the
LOX-derived amine oxidase activity (Fig. 4C). The LOXL3-sv1 protein showed
slightly higher amine oxidase activity than the LOXL3-sv2 protein;
however, the difference was not statistically significant (Fig. 4C). These results indicate that
LOXL3-sv2, as do LOXL3 and LOXL3-sv1, also functions as an amine
oxidase toward collagen type I, a physiological substrate of the
LOX family proteins.
Discussion
LOXL3-sv2, the newly identified variant of
LOXL3, encodes a 608 amino acid-long polypeptide with a
calculated molecular mass of 67.4 kDa. Previously, we reported an
approximately 67 kDa protein in several human tissues using a
polyclonal antibody against LOXL3 and proposed that the 67 kDa
protein could be derived from post-translational modification of
the full-length LOXL3 (15).
However, judging from the deduced molecular mass of LOXL3-sv2, the
previously reported 67 kDa band may correspond to a novel LOXL3-sv2
protein, although further detailed characterization is required. By
promoter analysis, we previously demonstrated that an alternative
promoter element present in the 5′-flanking intronic region of exon
4 of the LOXL3 gene regulates the transcriptional activation
of LOXL3-sv1. The promoter region responsible for expression
of the LOXL3 mRNA contained no TATA and CAAT boxes but
abundant CpG-rich sequences, as shown in a number of eukaryotic
housekeeping genes (18,19). In contrast, the
LOXL3-sv1-specific promoter contained a TATA box with a
number of potential transcription factor binding sites (15). The presence of exon 1 in the newly
identified LOXL3-sv2 mRNA indicates that the promoter
element located in the upstream region of exon 1 regulates the
transcriptional activation of LOXL3-sv2. Identification of
these LOXL3 variants suggest that at least three different
regulatory mechanisms exist for the transcription of the
LOXL3 gene, being responsible for the differential
expression of LOXL3, LOXL3-sv1 and
LOXL3-sv2.
The LOX family proteins have been proposed to exert
different functional roles through distinct substrate specificity
in the formation and maintenance of extracellular matrix. LOX and
LOXL1 are more likely involved in the formation of fibrillar
collagen fibers, such as types I-III, whereas LOXL2 is responsible
for cross-linking of collagen type IV associated with the basement
membranes (20). In
peroxidase-coupled fluorometric assays with different types of
collagens, LOXL3 showed higher amine oxidase activity toward
collagen type I, II and VIII, whereas LOXL3-sv1 showed higher amine
oxidase activity toward collagen types III and IV (15), further suggesting that the LOX
family proteins may play different functional roles through
distinct substrate specificity. In our amine oxidase assays, the
recombinant LOXL3-sv2 protein showed a significant level of amine
oxidase activity toward collagen type I in a BAPN-sensitive manner,
indicating that LOXL3-sv2 functions as an active amine oxidase,
although substrate specificity of LOXL3-sv2 has yet to be
determined. Considering the diverse structural divergence and
tissue distribution of different collagen types, detailed amine
oxidase assays and colocalization studies with different types of
collagens are required for further understanding of the functional
significance of this novel LOXL3 variant.
LOXL3 contains four repeated copies of SRCR domains
in the N-terminus, which are characterized by 6–8 cysteine residues
in 90–110 amino acid long regions. SRCR domains are found either on
the cell surface proteins or on secreted proteins, mediating
protein-protein interactions for cell adhesion and cell signaling
(21,22). The deduced amino acid sequence of
LOXL3-sv2 shows deletion of the SRCR domain 2 but contains all the
other SRCR domains as well as the characteristic C-terminal domains
of the full-length LOXL3. In contrast, LOXL3-sv1 does not have the
N-terminal SRCR domains 1–3 of LOXL3. Previously, using a series of
deletion constructs of the N-terminal SRCR domains, we showed that
the SRCR domains did not affect the amine oxidase activity of
LOXL4, a close paralogue of LOXL3 (23). In this study, the purified
recombinant proteins of both LOXL3-sv1 and LOXL3-sv2 showed
BAPN-inhibitable amine oxidase activities toward collagen type I,
further indicating that deletion of SRCR doma ins do not influence
the amine oxidase activity at least in in vitro assays.
Recently, LOXL2, another SRCR domain-containing LOX paralogue, was
shown to affect keratinocyte differentiation through the SRCR
domains but not by amine oxidase activity (24), suggesting unique functional roles
of the SRCR domains present in the LOXL proteins. The partial
absence of N-terminal SRCR domains in LOXL3-sv1 and LOXL3-sv2
suggests that these two variants may possess distinct interactive
profiles from LOXL3 in protein-protein interactions. Further
studies on the interactive properties of LOXL3, LOXL3-sv1 and
LOXL3-sv2 will be important for understanding the functional
differences of these three variants generated from the single gene
in the biogenesis of extracellular matrix proteins.
Acknowledgments
This study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF), funded by the Ministry of Education (no.
NRF-2015R1D1A3A01016577).
References
|
1
|
Mäki JM and Kivirikko KI: Cloning and
characterization of a fourth human lysyl oxidase isoenzyme. Biochem
J. 355:381–387. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jourdan-Le Saux C, Tomsche A, Ujfalusi A,
Jia L and Csiszar K: Central nervous system, uterus, heart, and
leukocyte expression of the LOXL3 gene, encoding a novel lysyl
oxidase-like protein. Genomics. 74:211–218. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith-Mungo LI and Kagan HM: Lysyl
oxidase: properties, regulation and multiple functions in biology.
Matrix Biol. 16:387–398. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim Y, Boyd CD and Csiszar K: A new gene
with sequence and structural similarity to the gene encoding human
lysyl oxidase. J Biol Chem. 270:7176–7182. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jourdan-Le Saux C, Tronecker H, Bogic L,
Bryant-Greenwood GD, Boyd CD and Csiszar K: The LOXL2 gene encodes
a new lysyl oxidase-like protein and is expr essed at high levels
in reproductive tissues. J Biol Chem. 274:12939–12944. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asuncion L, Fogelgren B, Fong KS, Fong SF,
Kim Y and Csiszar K: A novel human lysyl oxidase-like gene (LOXL4)
on chromosome 10q24 has an altered scavenger receptor cysteine rich
domain. Matrix Biol. 20:487–491. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peinado H, Del Carmen Iglesias-de la Cruz
M, Olmeda D, Csiszar K, Fong KS, Vega S, Nieto MA, Cano A and
Portillo F: A molecular role for lysyl oxidase-like 2 enzyme in
snail regulation and tumor progression. EMBO J. 24:3446–3458. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang ZM, Du SH, Huang LG, Li JH, Xiao L
and Tong P: Leptin promotes apoptosis and inhibits autophagy of
chondrocytes through upregulating lysyl oxidase-like 3 during
osteoarthritis pathogenesis. Osteoarthritis Cartilage.
24:1246–1253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kraft-Sheleg O, Zaffryar-Eilot S, Genin O,
Yaseen W, Soueid-Baumgarten S, Kessler O, Smolkin T, Akiri G,
Neufeld G, Cinnamon Y, et al: Localized LoxL3-dependent fibronectin
oxidation regulates myofiber stretch and inte grin-mediated
adhesion. Dev Cell. 36:550–561. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alarab M, Bortolini MA, Drutz H, Lye S and
Shynlova O: LOX family enzymes expression in vaginal tissue of
premenopausal women with severe pelvic organ prolapse. Int
Urogynecol J Pelvic Floor Dysfunct. 21:1397–1404. 2010. View Article : Google Scholar
|
|
11
|
Alzahrani F, Al Hazzaa SA, Tayeb H and
Alkuraya FS: LOXL3, encoding lysyl oxidase-like 3, is mutated in a
family with autosomal recessive Stickler syndrome. Hum Genet.
134:451–453. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Gao B, Xiao X, Li S, Jia X, Sun W,
Guo X and Zhang Q: Exome sequencing identified null mutations in
LOXL3 associated with early-onset high myopia. Mol Vis. 22:161–167.
2016.PubMed/NCBI
|
|
13
|
Zhang J, Yang R, Liu Z, Hou C, Zong W,
Zhang A, Sun X and Gao J: Loss of lysyl oxidase-like 3 causes cleft
palate and spinal deformity in mice. Hum Mol Genet. 24:6174–6185.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
van Boxtel AL, Gansner JM, Hakvoort HW,
Snell H, Legler J and Gitlin JD: Lysyl oxidase-like 3b is critical
for cartilage maturation during zebrafish craniofacial development.
Matrix Biol. 30:178–187. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee JE and Kim Y: A tissue-specific
variant of the human lysyl oxidase-like protein 3 (LOXL3) functions
as an amine oxidase with substrate specificity. J Biol Chem.
281:37282–37290. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Palamakumbura AH and Trackman PC: A
fluorometric assay for detection of lysyl oxidase enzyme activity
in biological samples. Anal Biochem. 300:245–251. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jonasson P, Liljeqvist S, Nygren PA and
Ståhl S: Genetic design for facilitated production and recovery of
recombinant proteins in Escherichia coli. Biotechnol Appl Biochem.
35:91–105. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
López-Maury L, Marguerat S and Bähler J:
Tuning gene expression to changing environments: from rapid
responses to evolutionary adaptation. Nat Rev Genet. 9:583–593.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang C, Bolotin E, Jiang T, Sladek FM and
Martinez E: Prevalence of the initiator over the TATA box in human
and yeast genes and identification of DNA motifs enriched in human
TATA-less core promoters. Gene. 389:52–65. 2007. View Article : Google Scholar :
|
|
20
|
Csiszar K: Lysyl oxidases: a novel
multifunctional amine oxidase family. Prog Nucleic Acid Res Mol
Biol. 70:1–32. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martínez VG, Moestrup SK, Holmskov U,
Mollenhauer J and Lozano F: The conserved scavenger receptor
cysteine-rich superfamily in therapy and diagnosis. Pharmacol Rev.
63:967–1000. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bowdish DM and Gordon S: Conserved domains
of the class A scavenger receptors: evolution and function. Immunol
Rev. 227:19–31. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim MS, Kim SS, Jung ST, Park JY, Yoo HW,
Ko J, Csiszar K, Choi SY and Kim Y: Expression and purification of
enzymatically active forms of the human lysyl oxidase-like protein
4. J Biol Chem. 278:52071–52074. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lugassy J, Zaffryar-Eilot S, Soueid S,
Mordoviz A, Smith V, Kessler O and Neufeld G: The enzymatic
activity of lysyl oxidas-like-2 (LOXL2) is not required for
LOXL2-induced inhibition of keratinocyte differentiation. J Biol
Chem. 287:3541–3549. 2012. View Article : Google Scholar :
|