Introduction
Aortic aneurysm is a serious condition that results
from an atherosclerotic aorta and is a leading cause of mortality
in humans (1). Studies on the
genetic basis of familial aortic aneurysm have centered on the
relationship between the condition to systemic connective tissue
disorders such as Marfan syndrome (2) and Ehlers-Danlos syndrome (3). Although the molecular mechanism
underlying nonsyndromic aortic aneurysm is complex and has not been
determined definitively, several risk factors, including age,
arteriosclerosis, hypertension and inflammatory or autoimmune
diseases that affect the aorta, have been identified clinically
(4,5). In addition to these conventional
risk factors, recent studies have shown the importance of genetic
factors in the development of sporadic aortic aneurysm by revealing
a heritability of ~70% (6).
Genome-wide association studies (GWASs) have uncovered several loci
and genes that confer susceptibility to aortic aneurysm in European
ancestry populations (7–12), but genetic variants that
contribute to the development of this condition in Japanese
individuals have not been identified definitively.
Genetic variants previously associated with aortic
aneurysm typically have a minor allele frequency (MAF) of >10%
and a small individual effect size (7–12).
Given that these common variants explain only a small fraction of
disease heritability, low-frequency (MAF of 0.5–5%) or rare (MAF of
<0.5%) variants with a larger effect size may contribute to the
genetic architecture of aortic aneurysm (13).
In the present study, we performed exome-wide
association studies (EWASs) with the use of exome array-based
genotyping methods to identify single nucleotide polymorphisms
(SNPs) - in particular, low-frequency or rare coding variants with
a moderate to large effect size - that confer susceptibility to
aortic aneurysm in Japanese individuals. Given that most
low-frequency or rare variants were not included in the arrays of
previous GWASs, we used Illumina HumanExome-12 DNA Analysis
BeadChip or Infinium Exome-24 BeadChip arrays, which provide
coverage for functional SNPs including low-frequency or rare
variants in entire exons.
Materials and methods
Study population
A total of 8,782 Japanese individuals (456 patients
with aortic aneurysm, 8,326 controls) was examined. The subjects
were recruited from individuals who visited outpatient clinics of
or were admitted to participating hospitals (Gifu Prefectural
Tajimi Hospital, Tajimi; Gifu Prefectural General Medical Center,
Gifu; Japanese Red Cross Nagoya First Hospital, Nagoya; Inabe
General Hospital, Inabe; Hirosaki University Hospital and Hirosaki
Stroke and Rehabilitation Center, Hirosaki) either because they
were experiencing various symptoms or for an annual health checkup
between 2002 and 2014; from community-dwelling individuals
recruited to a population-based cohort study in Inabe between 2010
and 2014 or in Tokyo or Kusatsu between 2011 and 2015; or from
individuals who underwent autopsy at Tokyo Metropolitan Geriatric
Hospital from 1995 to 2012.
True aortic aneurysm was defined as a permanent
localized dilation of the aorta with a ≥50% increase in diameter
relative to the expected normal size of the artery or with a
diameter of >5 cm (14).
Dissecting aortic aneurysm was defined as separation of the aortic
wall layers with resulting true and false lumens or as intramural
hematoma (15). The subjects with
aortic aneurysm (279 with true aneurysm and 181 with dissecting
aneurysm (four had both conditions) were examined by chest and
abdominal X-ray and echocardiography followed by contrast
medium-enhanced computed tomography. Some subjects were also
examined by aortic angiography. Individuals with Marfan syndrome,
Ehlers-Danlos syndrome, bicuspid aortic valve disease, aortitis
syndrome, connective tissue disorder, congenital malformations of
the heart or vessels, pseudoaneurysm, or traumatic aneurysm were
excluded from the study. The control individuals had no history of
aortic, coronary, or peripheral arterial disease; ischemic or
hemorrhagic stroke; intracranial aneurysm; or other
atherosclerotic, thrombotic, embolic or hemorrhagic disorders.
Autopsy cases without aortic aneurysm were excluded from
controls.
The study protocol complied with the Declaration of
Helsinki and was approved by the Committees on the Ethics of Human
Research of Mie University Graduate School of Medicine, Hirosaki
University Graduate School of Medicine, Tokyo Metropolitan
Institute of Gerontology, and participating hospitals. Written
informed consent was obtained from each participant or from
families of the deceased subjects.
EWASs
Venous blood (5 or 7 ml) was collected into tubes
containing 50 mmol/l ethylenediaminetetraacetic acid (disodium
salt), peripheral blood leukocytes were isolated, and genomic DNA
was extracted from these cells either with the use of a DNA
extraction kit (Genomix supplied by Talent, Trieste, Italy, or
SMITEST EX-R&D supplied by Medical and Biological Laboratories,
Nagoya, Japan) or by standard protocols based on phenol-chloroform
extraction and spin columns. In autopsy cases, genomic DNA was
extracted from kidneys. EWASs were performed for the 456 subjects
with aortic aneurysm (or for the 279 subjects with true aneurysm or
181 subjects with dissecting aneurysm) and the 8,326 control
subjects with the use of a HumanExome-12 v1.1 or v1.2 DNA Analysis
BeadChip or an Infinium Exome-24 v1.0 BeadChip (Illumina, San
Diego, CA, USA). These exome arrays include putative functional
exonic variants selected from >12,000 individual exome or
whole-genome sequences. The exonic content consists of ~244,000
SNPs representing diverse populations, including European, African,
Chinese and Hispanic individuals (16). SNPs contained in only one of the
exome arrays (~3.6%) were excluded from analysis. We performed
quality control (17) as follows.
i) Genotyping data with a call rate of <97% were discarded, with
the mean call rate for the remaining data being 99.9%. ii) Gender
specification was checked for each sample, with samples for which
the gender designation in the clinical records was inconsistent
with genetic sex being discarded. iii) Duplicated samples and
cryptic relatedness were checked by calculation of identity by
descent; all pairs with DNA samples showing identity by descent of
>0.1875 were inspected and one sample from each pair was
excluded. iv) The frequency of heterozygosity of SNPs was
calculated for all samples, with those found to have extremely low
or high heterozygosity (>3 standard deviations from the mean)
being discarded. v) SNPs in sex chromosomes or mitochondrial DNA
were excluded from the analysis, as were nonpolymorphic SNPs or
SNPs with a MAF of <0.1%. vi) SNPs whose genotype distributions
in control individuals deviated significantly (P<0.001) from
Hardy-Weinberg equilibrium were excluded. vii) The genotype data
for each EWAS were examined for population stratification by
principal components analysis (18), and population outliers were
excluded from the analysis. A total of 41,432 SNPs passed quality
control and was subjected to analysis.
Statistical analysis
Quantitative data for characteristics of the study
subjects were compared between patients with aortic aneurysm and
control individuals with the unpaired Student's t test. Categorical
data were compared between the two groups with Fisher's exact test.
Allele frequencies were estimated by the gene counting method, and
Fisher's exact test was applied to identify departure from
Hardy-Weinberg equilibrium. Allele frequencies of SNPs were
compared between patients with aortic aneurysm and control subjects
with Fisher's exact test. Multivariable logistic regression
analysis was performed with aortic aneurysm as a dependent variable
and independent variables including age, gender (0, woman; 1, man),
the prevalence of hypertension (0, no history of this condition; 1,
positive history), and genotype of each SNP. Genotypes of SNPs were
assessed according to dominant [0, AA; 1, AB+BB (A, major allele;
B, minor allele)], recessive (0, AA+AB; 1, BB), and additive
genetic models, and the P-value, odds ratio, and 95% confidence
interval were calculated. Additive models comprised additive 1 (0,
AA; 1, AB; 0, BB) and additive 2 (0, AA; 0, AB; 1, BB) models,
which were analyzed simultaneously with a single statistical model.
To compensate for multiple comparisons of genotypes with aortic
aneurysm, we applied Bonferroni's correction for statistical
significance of association. Given that 41,432 SNPs were finally
examined, a P-value of <1.21×10−6 (0.05/41,432) was
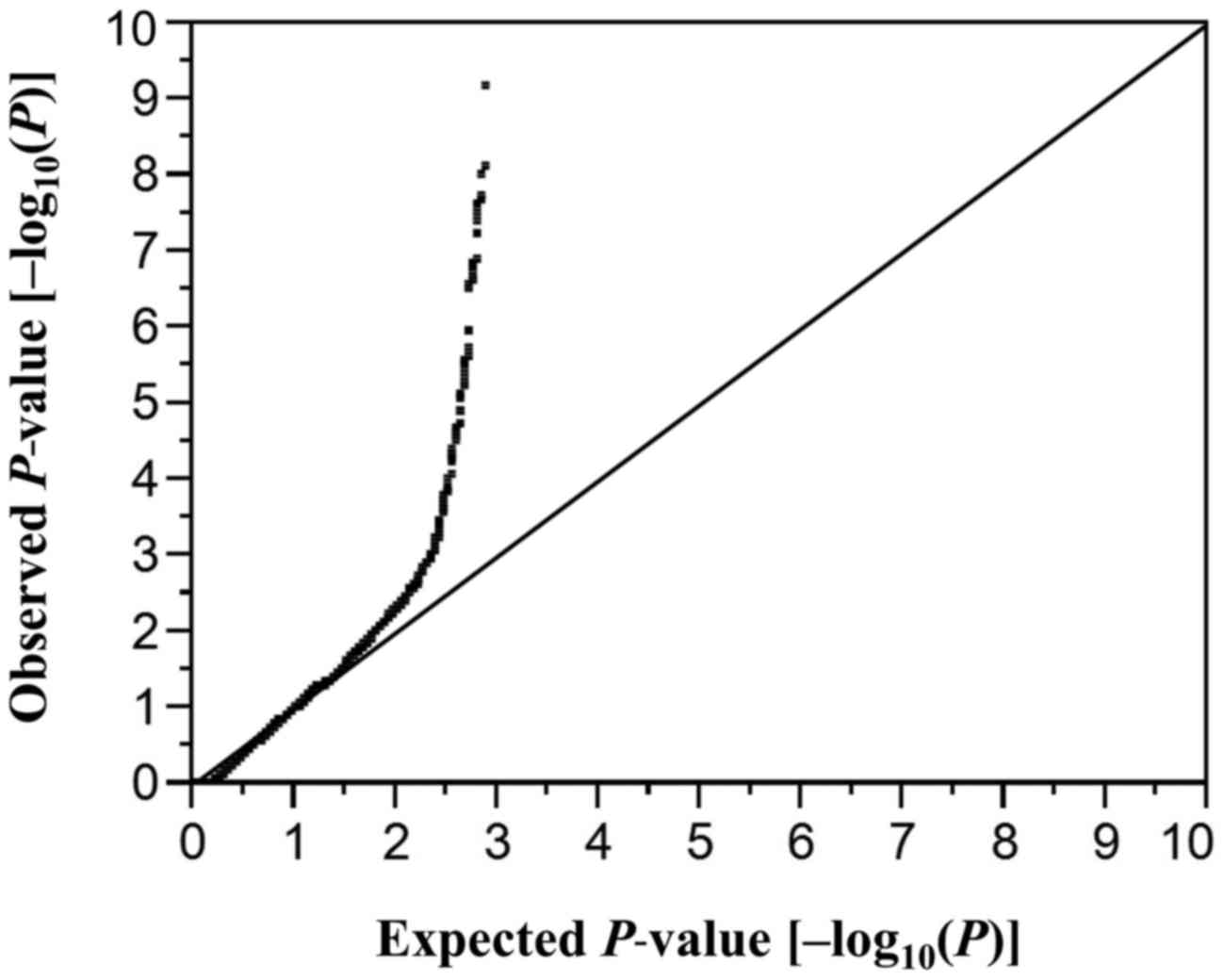
considered statistically significant. A quantile-quantile plot for
P-values of allele frequencies in the EWAS for aortic aneurysm is
shown in Fig. 1. The inflation
factor (λ) was 1.57. P-values for other comparisons were similarly
adjusted by Bonferroni's correction. Statistical tests were
performed with JMP Genomics version 6.0 software (SAS Institute,
Cary, NC, USA).
Results
Characteristics of the subjects
The characteristics of the subjects enrolled in the
study are shown in Table I. Age,
the frequency of males, and the prevalence of hypertension,
diabetes mellitus, dyslipidemia, chronic kidney disease and
hyperuricemia were significantly greater in patients with aortic
aneurysm than in control individuals.
 | Table ICharacteristics of the 8,782 study
subjects. |
Table I
Characteristics of the 8,782 study
subjects.
|
Characteristics | Aortic
aneurysm | Control | P-value |
|---|
| No. of
subjects | 456 | 8326 | |
| Age (years) | 74.7±13.5 | 57.3±13.5 | <0.0001 |
| Gender
(male/female, %) | 64.0/36.0 | 51.4/48.6 | <0.0001 |
| Body mass index
(kg/m2) | 23.2±3.5 | 23.1±3.5 | 0.7068 |
| Current or former
smoker (%) | 44.1 | 39.8 | 0.1777 |
| Hypertension
(%) | 94.5 | 40.7 | <0.0001 |
| Diabetes mellitus
(%) | 44.7 | 14.7 | <0.0001 |
| Dyslipidemia
(%) | 67.9 | 56.8 | 0.0006 |
| Chronic kidney
disease (%) | 41.9 | 17.5 | <0.0001 |
| Hyperuricemia
(%) | 25.7 | 16.1 | 0.0003 |
EWAS of aortic aneurysm
We examined the correlation of allele frequencies
for 41,432 SNPs that passed quality control to aortic aneurysm
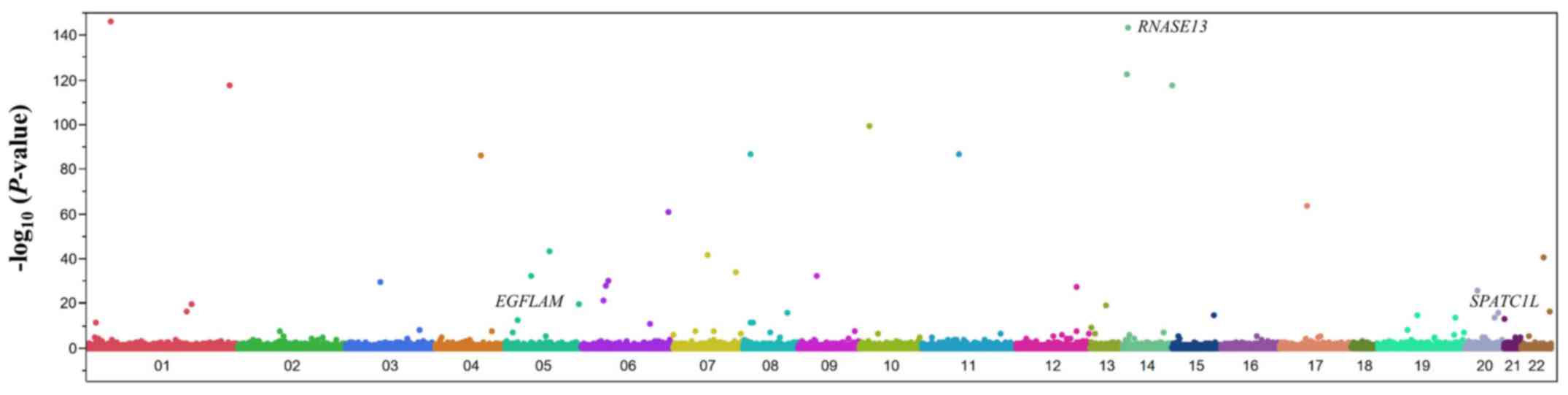
using the Fisher's exact test. A Manhattan plot for the EWAS of
aortic aneurysm is shown in Fig.
2. After Bonferroni's correction, 59 SNPs were found to be
significantly (P<1.21×10−6) associated with aortic
aneurysm (Table II). The
genotype distributions of these SNPs were in Hardy-Weinberg
equilibrium (P>0.001) both among patients with aortic aneurysm
and among the control individuals (data not shown).
 | Table IIThe 59 single nucleotide
polymorphisms (SNPs) significantly (P<1.21×10−6)
associated with aortic aneurysm in an exome-wide association study
(EWAS). |
Table II
The 59 single nucleotide
polymorphisms (SNPs) significantly (P<1.21×10−6)
associated with aortic aneurysm in an exome-wide association study
(EWAS).
| Gene | dbSNP | Nucleotide (amino
acid) substitutiona | Chromosome:
position | MAF (%) | P-value
(allele) | Allele OR |
|---|
|
CATSPER4 | rs11247866 | A/G (Q77R) | 1:26191303 | 0.4 |
9.82×10−147 | 1.81 |
| RNASE13 | rs143881017 | C/T (R140H) | 14:21033870 | 0.5 |
2.49×10−144 | 2.77 |
| RNASE10 | rs202109789 | G/A (G87S) | 14:20510730 | 0.2 |
4.32×10−123 | 0.47 |
| rs2582513 | A/G | 14:104948453 | 39.9 |
1.72×10−118 | 0.86 |
| HEATR1 | rs193150310 | T/A (V1975D) | 1:236554752 | 0.3 |
3.09×10−118 | 0.76 |
|
KIAA1217 | rs10828663 | G/A (A807T) | 10:24524525 | 10.4 |
2.68×10−100 | 1.00 |
| MTUS1 | rs3739407 | G/A (R148C) | 8:17755366 | 38.4 |
2.32×10−87 | 0.98 |
| OR5W2 | rs75634103 | G/A | 11:55914523 | 10.4 |
2.74×10−87 | 1.11 |
| ALPK1 | rs2074379 | A/G (I732M) | 4:112431743 | 32.0 |
1.03×10−86 | 1.11 |
| ATAD5 | rs11657270 | T/C (Y1419H) | 17:30887369 | 18.1 |
3.24×10−64 | 1.13 |
| ACAT2 | rs25683 | A/G (K211R) | 6:159775311 | 18.8 |
1.90×10−61 | 0.86 |
| ZNF474 | rs201335566 | G/A (R253Q) | 5:122152748 | 0.5 |
4.22×10−44 | 1.29 |
| ZNF804B | rs6963781 | A/G (M1105V) | 7:89336295 | 5.1 |
1.85×10−42 | 0.90 |
|
LOC100506679 | rs5751416 | G/A | 22:43036820 | 26.3 |
1.62×10−41 | 0.81 |
| SSPO | rs191064068 | G/A (R209H) | 7:149777738 | 1.1 |
8.55×10−35 | 1.13 |
|
ARHGEF28 | rs536568 | A/C | 5:73935841 | 45.8 |
3.61×10−33 | 1.03 |
| TMEM2 | rs142154818 | G/A (T1062M) | 9:71700645 | 1.0 |
6.41×10−33 | 1.90 |
| HLA-DMB | rs151719 | A/G | 6:32936123 | 25.7 |
1.11×10−30 | 1.03 |
| CCDC66 | rs61747994 | T/C (L802S) | 3:56619399 | 9.8 |
3.77×10−30 | 0.92 |
| rs3135365 | T/G | 6:32421478 | 18.9 |
1.62×10−28 | 0.84 |
| NAA25 | rs12231744 | C/T (R876K) | 12:112039251 | 35.1 |
5.02×10−28 | 1.07 |
|
RALGAPA2 | rs142962992 | G/C (E1676D) | 20:20505435 | 0.9 |
1.77×10−26 | 1.10 |
| NEU1 | rs13118 | T/A | 6:31859509 | 9.7 |
6.92×10−22 | 1.20 |
| AXDND1 | rs41267592 | C/T (T627M) | 1:179468524 | 0.3 |
1.42×10−20 | 0.63 |
| PHYKPL | rs146105181 | T/C (N88D) | 5:178230016 | 0.2 |
2.15×10−20 | 1.46 |
| PCDH8 | rs5030685 | A/G (V743A) | 13:52846209 | 0.3 |
9.92×10−20 | 2.72 |
| SELE | rs5361 | T/G (S149R) | 1:1169731919 | 3.3 |
2.49×10−17 | 0.95 |
| MOV10L1 | rs760749 | A/C (I454L) | 22:50117257 | 27.8 |
2.97×10−17 | 1.02 |
| HHLA1 | rs75623295 | C/G (T90R) | 8:132098893 | 2.9 |
1.23×10−16 | 0.80 |
| TUBB1 | rs6070697 | G/A (R307H) | 20:59024347 | 12.1 |
2.17×10−16 | 0.92 |
| ZNF708 | rs504280 | C/T (R66Q) | 19:21294577 | 7.4 |
2.16×10−15 | 0.96 |
| TICRR | rs79501973 | G/A (V1373I) | 15:89624427 | 14.7 |
2.51×10−15 | 0.97 |
| ADNP | rs148496595 | C/G (D924E) | 20:50891942 | 0.3 |
2.71×10−14 | 0.68 |
| FCAR | rs11666735 | G/A (D113N) | 19:54885501 | 3.2 |
3.10×10−14 | 1.03 |
| rs2823962 | G/A | 21:16673913 | 32.8 |
7.80×10−14 | 0.93 |
| EGFLAM | rs1465567 | T/C (W229R) | 5:38370435 | 25.1 |
5.92×10−13 | 1.19 |
| rs1480347 | G/A | 8:20489946 | 17.3 |
3.49×10−12 | 1.07 |
| UBE4B | rs180983516 | G/A (R331H) | 1:10106379 | 0.8 |
4.50×10−12 | 0.60 |
| rs448705 | A/G | 8:17837193 | 12.4 |
5.42×10−12 | 1.01 |
| rs11970286 | C/T | 6:118359211 | 17.3 |
1.38×10−11 | 0.90 |
| rs10047727 | T/C | 13:21743051 | 42.7 |
6.67×10−10 | 0.90 |
| rs507856 | C/T | 3:161736158 | 38.3 |
7.71×10−9 | 0.93 |
| SLC1A6 | rs7253812 | C/A | 19:14982691 | 26.7 |
9.43×10−9 | 0.99 |
| FGB | rs1800789 | G/A | 4:154561591 | 13.3 |
1.86×10−8 | 0.99 |
| SLC9A4 | rs1014286 | A/G (S784G) | 2:102532641 | 43.9 |
1.90×10−8 | 1.13 |
| HECTD4 | rs2074356 | C/T | 12:112207597 | 25.4 |
2.22×10−8 | 1.03 |
| PKD1L1 | rs66755489 | G/A (P2021L) | 7:47835032 | 2.9 |
2.50×10−8 | 1.16 |
| CAMSAP1 | rs201291561 | T/C (N1062S) | 9:135821476 | 0.2 |
3.04×10−8 | 1.14 |
| C7orf43 | rs3800952 | C/T (R353Q) | 7:100160331 | 6.3 |
4.08×10−8 | 1.08 |
| ZNF671 | rs3746207 | G/A (A149V) | 19:57721640 | 12.6 |
5.90×10−8 | 0.99 |
| RIN3 | rs7150931 | T/C | 14:92671696 | 46.2 |
1.26×10−7 | 1.03 |
| rs10805579 | G/A | 5:19127418 | 10.5 |
1.48×10−7 | 1.03 |
| rs12546220 | T/C | 8:69461493 | 29.1 |
1.82×10−7 | 0.96 |
| DRD2 | rs12363125 | C/T | 11:113415194 | 6.2 |
1.89×10−7 | 0.88 |
| MTUS2 | rs17571410 | G/A | 13:29007481 | 41.4 |
2.35×10−7 | 0.91 |
| GALNTL5 | rs11766982 | A/G | 7:151996417 | 27.6 |
2.56×10−7 | 1.05 |
| POLE | rs5745022 | C/T | 12:132632393 | 20.6 |
2.82×10−7 | 0.96 |
| CHAT | rs3810947 | A/G | 10:49613197 | 43.0 |
3.19×10−7 | 0.97 |
| LILRB5 | rs117421142 | A/G (I420T) | 19:54252383 | 1.0 |
1.16×10−6 | 1.29 |
Multivariable logistic regression
analysis of the correlation of SNPs to aortic aneurysm
The relation of the 59 identified SNPs to aortic
aneurysm was examined further by multivariable logistic regression
analysis with adjustment for age, gender and the prevalence of
hypertension. Although 8 SNPs were related (P<0.05) to aortic
aneurysm, no SNP was significantly [P<2.12×10−4
(0.05/236)] associated with this condition (Table III). We then examined the
correlation of the 8 identified SNPs to true or dissecting aortic
aneurysm separately. Five SNPs were related (P<0.05) to true
aortic aneurysm (Table IV),
among which rs1465567 [T/C (W229R)] of EGF-like, fibronectin type
III, and laminin G domains gene (EGFLAM) was significantly
[P<0.0016 (0.05/32)] associated with this condition, with the
minor C allele representing a risk factor. No SNP was found to be
related to dissecting aortic aneurysm (data not shown).
 | Table IIICorrelation of single nucleotide
polymorphisms (SNPs) to aortic aneurysm as determined by
multivariable logistic regression analysis. |
Table III
Correlation of single nucleotide
polymorphisms (SNPs) to aortic aneurysm as determined by
multivariable logistic regression analysis.
| SNP | | Dominant
| Recessive
| Additive 1
| Additive 2
|
|---|
| P-value | OR
(95% CI) | P-value | OR
(95% CI) | P-value | OR
(95% CI) | P-value (95% | OR
(95% CI) |
|---|
| rs143881017 | C/T (R140H) | 0.0208 | 3.00
(1.20–6.80) | 0.9665 | | 0.0207 | 3.00
(1.20–6.79) | 0.9667 | |
| rs5751416 | G/A | 0.0351 | 0.79
(0.63–0.98) | 0.2034 | | 0.0697 | | 0.1055 | |
| rs142154818 | G/A (T1062M) | 0.0486 | 1.94
(1.00–3.50) | 0.7044 | | 0.0452 | 1.97
(1.02–3.54) | 0.7078 | |
| rs13118 | T/A | 0.0415 | 1.32
(1.01–1.71) | 0.4182 | | 0.0245 | 1.37
(1.04–1.78) | 0.4879 | |
| rs5030685 | A/G (V743A) | 0.0293 | 2.94
(1.13–6.76) | 0.6067 | | 0.0227 | 3.12
(1.19–7.22) | 0.6090 | |
| rs1465567 | T/C (W229R) | 0.0004 | 1.49
(1.19–1.85) | 0.6932 | | 0.0004 | 1.51
(1.20–1.90) | 0.2807 | |
| rs7253812 | C/A | 0.0834 | | 0.0543 | | 0.0189 | 1.31
(1.05–1.65) | 0.1650 | |
| rs7150931 | T/C | 0.5033 | | 0.0224 | 1.36
(1.04–1.75) | 0.9225 | | 0.0586 | |
 | Table IVCorrelation of single nucleotide
polymorphisms (SNPs) to true aortic aneurysm as determined by
multivariable logistic regression analysis. |
Table IV
Correlation of single nucleotide
polymorphisms (SNPs) to true aortic aneurysm as determined by
multivariable logistic regression analysis.
| SNP | | Dominant
| Recessive
| Additive 1
| Additive 2
|
|---|
| P-value | OR
(95% CI) | P-value | OR
(95% CI) | P-value | OR
(95% CI) | P-value | OR
(95% CI) |
|---|
| rs143881017 | C/T (R140H) | 0.2899 | | ND | | 0.2899 | | ND | |
| rs5751416 | G/A | 0.0448 | 0.73
(0.54–0.99) | 0.1221 | | 0.1030 | | 0.0663 | |
| rs142154818 | G/A (T1062M) | 0.2214 | | 0.8209 | | 0.2154 | | 0.8231 | |
| rs13118 | T/A | 0.0253 | 1.51
(1.05–2.14) | 0.6076 | | 0.0167 | 1.57
(1.09–2.23) | 0.6967 | |
| rs5030685 | A/G (V743A) | 0.0779 | | 0.7314 | | 0.0667 | | 0.7333 | |
| rs1465567 | T/C (W229R) | 0.0014 | 1.63
(1.21–2.21) | 0.6670 | | 0.0014 | 1.66
(1.22–2.27) | 0.2911 | |
| rs7253812 | C/A | 0.0122 | 1.47
(1.09–1.99) | 0.2265 | | 0.0032 | 1.60
(1.17–2.18) | 0.5864 | |
| rs7150931 | T/C | 0.7329 | | 0.0431 1.45 | (1.01–2.04) | 0.2737 | | 0.2350 | |
EWASs of true or dissecting aortic
aneurysm
We next examined the relation of allele frequencies
for the total of 41,432 SNPs to true or dissecting aortic aneurysm
separately with the use of Fisher's exact test. After Bonferroni's
correction, 45 or 19 SNPs were found to be significantly
(P<1.21×10−6) associated with true (Table V) or dissecting (Table VI) aortic aneurysm, respectively.
The genotype distributions of these SNPs were in Hardy-Weinberg
equilibrium (P>0.001) both among patients with true or
dissecting aortic aneurysm and among control individuals (data not
shown).
 | Table VThe 45 single nucleotide
polymorphisms (SNPs) significantly (P<1.21×10−6)
associated with true aortic aneurysm in an exome-wide association
study (EWAS). |
Table V
The 45 single nucleotide
polymorphisms (SNPs) significantly (P<1.21×10−6)
associated with true aortic aneurysm in an exome-wide association
study (EWAS).
| Gene | dbSNP | Nucleotide (amino
acid) substitutiona | Chromosome:
position | MAF (%) | P-value
(allele) | Allele OR |
|---|
|
KIAA1217 | rs10828663 | G/A (A807T) | 10:24524525 | 10.4 |
6.87×10−94 | 1.14 |
| NRAP | rs79461687 | G/T (H1246Q) | 10:113606247 | 1.3 |
1.58×10−90 | 1.67 |
| OR5W2 | rs75634103 | G/A | 11:55914523 | 10.4 |
4.84×10−81 | 1.17 |
| ATAD5 | rs11657270 | T/C (Y1419H) | 17:30887369 | 18.1 |
9.29×10−70 | 0.98 |
| rs9683944 | A/G | 4:137512008 | 10.8 |
1.65×10−56 | 0.83 |
| TMPRSS3 | rs928302 | C/T (V53I) | 21:42389975 | 27.6 |
1.34×10−52 | 0.99 |
| ZNF804B | rs6963781 | A/G (M1105V) | 7:89336295 | 5.1 |
6.67×10−47 | 1.01 |
| ZNF474 | rs201335566 | G/A (R253Q) | 5:122152748 | 0.5 |
5.79×10−42 | 0.84 |
|
LOC100506679 | rs5751416 | G/A | 22:43036820 | 26.3 |
2.46×10−26 | 0.71 |
|
ARHGEF28 | rs536568 | A/C | 5:73935841 | 45.8 |
3.10×10−23 | 1.03 |
|
RALGAPA2 | rs142962992 | G/C (E1676D) | 20:20505435 | 0.9 |
8.85×10−23 | 0.72 |
| LYSMD1 | rs79024247 | G/T (Q150K) | 1:151160974 | 5.2 |
7.43×10−21 | 0.88 |
| CNGA1 | rs192912733 | C/T (R493Q) | 4:47937223 | 0.7 |
8.13×10−21 | 1.17 |
| MOV10L1 | rs760749 | A/C (I454L) | 22:50117257 | 27.8 |
1.23×10−20 | 1.18 |
| HLA-DMB | rs151719 | A/G | 6:32936123 | 25.7 |
1.96×10−19 | 1.17 |
| TUBB1 | rs6070697 | G/A (R307H) | 20:59024347 | 12.1 |
3.20×10−18 | 0.97 |
| rs3135365 | T/G | 6:32421478 | 18.9 |
3.94×10−17 | 0.83 |
| CCDC33 | rs1484214 | A/C | 15:74288732 | 49.3 |
6.60×10−17 | 1.09 |
| ZNF708 | rs504280 | C/T (R66Q) | 19:21294577 | 7.4 |
3.03×10−15 | 0.98 |
| rs2823962 | G/A | 21:16673913 | 32.8 |
3.36×10−14 | 0.97 |
| SGCZ | rs1037934 | G/A | 8:14399065 | 9.0 |
3.44×10−13 | 1.22 |
| DUOX1 | rs199549867 | A/T (R569S) | 15:45141997 | 0.2 |
1.58×10−10 | 0.81 |
| CTSC | rs3888798 | T/C (I453V) | 11:88294041 | 16.5 |
3.21×10−10 | 1.06 |
|
C10orf128 | rs118189413 | C/G (H67D) | 10:49166908 | 6.3 |
4.04×10−10 | 0.84 |
| OR5V1 | rs9405124 | A/G | 6:29401036 | 19.9 |
4.15×10−10 | 0.98 |
| UBAP2L | rs143080179 | T/C (S641P) | 1:154255163 | 1.3 |
4.20×10−10 | 0.66 |
| CAMSAP1 | rs201291561 | T/C (N1062S) | 9:135821476 | 0.2 |
5.67×10−10 | 1.87 |
| TICRR | rs79501973 | G/A (V1373I) | 15:89624427 | 14.7 |
7.76×10−10 | 1.04 |
| NCAM1 | rs7111410 | C/T | 11:113178565 | 15.9 |
2.63×10−9 | 0.80 |
| IRGQ | rs3817 | C/A | 19:43586043 | 47.7 |
3.30×10−9 | 0.92 |
| SSPO | rs191064068 | G/A (R209H) | 7:149777738 | 1.1 |
2.35×10−8 | 1.67 |
| SLC9A4 | rs1014286 | A/G (S784G) | 2:102532641 | 43.9 |
2.84×10−8 | 1.11 |
| rs2138852 | A/G | 17:29376331 | 2.5 |
2.92×10−8 | 0.77 |
| SPATC1L | rs113710653 | C/T (E231K) | 21:46161921 | 1.9 |
3.91×10−8 | 7.39 |
| DRD2 | rs12363125 | C/T | 11:113415194 | 6.2 |
4.99×10−8 | 0.87 |
| CCT5 | rs201280643 | C/G (S373C) | 5:10262584 | 0.4 |
7.04×10−8 | 0.99 |
| AXDND1 | rs41267592 | C/T (T627M) | 1:179468524 | 0.3 |
7.85×10−8 | 1.03 |
| rs507856 | C/T | 3:161736158 | 38.3 |
1.10×10−7 | 1.02 |
| rs962040 | A/G | 8:15454369 | 30.1 |
1.21×10−7 | 0.96 |
| rs11970286 | C/T | 6:118359211 | 17.3 |
1.60×10−7 | 0.83 |
| NINL | rs199671123 | C/T (A796T) | 20:25476905 | 0.2 |
2.45×10−7 | 0.85 |
| rs12531488 | C/T | 7:145194993 | 16.0 |
7.91×10−7 | 1.14 |
| MROH7 | rs143029488 | G/C (A1313P) | 1:54710152 | 0.8 |
1.05×10−6 | 1.34 |
| AFAP1 | rs28406288 | G/C (C403S) | 4:7800500 | 0.1 |
1.10×10−6 | ND |
| GALM | rs6741892 | A/T (N190Y) | 2:38689828 | 20.1 |
1.12×10−6 | 0.93 |
 | Table VIThe 19 single nucleotide
polymorphisms (SNPs) significantly (P<1.21×10−6)
associated with dissecting aortic aneurysm in an exome-wide
association study (EWAS). |
Table VI
The 19 single nucleotide
polymorphisms (SNPs) significantly (P<1.21×10−6)
associated with dissecting aortic aneurysm in an exome-wide
association study (EWAS).
| Gene | dbSNP | Nucleotide (amino
acid) substitutiona | Chromosome:
position | MAF (%) | P-value
(allele) | OR |
|---|
| ATXN7 | rs3774729 | G/A (V862M) | 3:63996406 | 46.8 |
3.81×10−35 | 0.91 |
|
KIAA1217 | rs10828663 | G/A (A807T) | 10:24524525 | 10.4 |
1.34×10−27 | 0.78 |
| RNASE13 | rs143881017 | C/T (R140H) | 14:21033870 | 0.5 |
5.12×10−26 | 4.48 |
| INPP5F | rs3736822 | A/G (I453V) | 10:119806397 | 1.7 |
8.50×10−23 | 0.62 |
| rs9683944 | A/G | 4:137512008 | 10.8 |
4.61×10−17 | 1.17 |
| rs9610342 | A/G | 22:35734530 | 30.7 |
8.20×10−17 | 0.79 |
| FAM98C | rs3745962 | C/A (T240K) | 19:38405604 | 16.7 |
8.99×10−15 | 0.96 |
| ZNF474 | rs201335566 | G/A (R253Q) | 5:122152748 | 0.5 |
1.02×10−12 | 1.95 |
| rs3135365 | T/G | 6:32421478 | 18.9 |
1.21×10−12 | 0.86 |
| DEPDC7 | rs34161108 | G/A (A192T) | 11:33027795 | 6.2 |
2.29×10−12 | 0.74 |
|
ARHGEF28 | rs536568 | A/C | 5:73935841 | 45.8 |
5.53×10−12 | 1.04 |
| RALGPS1 | rs57728614 | G/T (G383C) | 9:127196583 | 9.5 |
5.73×10−11 | 0.84 |
| AIM1L | rs34370465 | C/T (R847H) | 1:26344118 | 23.9 |
3.63×10−10 | 1.09 |
| ANXA7 | rs3750575 | C/T (R419Q) | 10:73378933 | 5.7 |
4.20×10−10 | 1.06 |
| AXDND1 | rs41267592 | C/T (T627M) | 1:179468524 | 0.3 |
1.77×10−8 | ND |
| rs2138852 | A/G | 17:29376331 | 2.5 |
1.49×10−7 | 1.32 |
| CHAT | rs78925077 | C/G (S119R) | 10:49622109 | 0.5 |
1.50×10−7 | 1.93 |
| GPR156 | rs902790 | A/T (E512D) | 3:120167929 | 4.8 |
2.24×10−7 | 0.71 |
| SELE | rs5361 | T/G (S149R) | 1:169731919 | 3.3 |
3.58×10−7 | 0.98 |
Multivariable logistic regression
analysis of the correlation of SNPs to true or dissecting aortic
aneurysm
The corrrelation of the 45 identified SNPs to true
aortic aneurysm was examined further by multivariable logistic
regression analysis with adjustment for age, gender and the
prevalence of hypertension. Among these SNPs, rs113710653 [C/T
(E231K)] of the spermatogenesis and centriole-associated 1-like
gene (SPATC1L) was significantly [P<2.78×10−4
(0.05/180)] associated with true aortic aneurysm, with the minor T
allele representing a risk factor for this condition (Table VII). The correlation of the 19
identified SNPs to dissecting aortic aneurysm was also further
examined by multivariable logistic regression analysis with
adjustment for age, gender and the prevalence of hypertension. The
SNP rs143881017 [C/T (R140H)] of the ribonuclease A family member
13 gene (RNASE13) was significantly [P<6.58×10−4
(0.05/76)] associated with dissecting aortic aneurysm, with the
minor T allele representing a risk factor for this condition
(Table VII).
 | Table VIIRelation of single nucleotide
polymorphisms (SNPs) to true or dissecting aortic aneurysm as
determined by multivariable logistic regression analysis. |
Table VII
Relation of single nucleotide
polymorphisms (SNPs) to true or dissecting aortic aneurysm as
determined by multivariable logistic regression analysis.
| SNP | Dominant
| Recessive
| Additive 1
| Additive 2
|
|---|
| P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) |
|---|
True aortic
aneurysm
rs113710653 C/T (E231K) | 0.0002 | 5.32
(2.33–11.14) | 0.9263 | | 0.0002 | 5.34
(2.34–11.18) | 0.9299 | |
Dissecting aortic
aneurysm
rs143881017 C/T (R140H) | 0.0006 | 5.77
(2.25–12.95) | 0.9615 | | 0.0006 | 5.48
(2.26–12.97) | 0.9621 | |
Correlation of SNPs to intermediate
phenotypes of aortic aneurysm
Finally, we examined the correlation of three SNPs
(rs1465567, rs113710653 and rs143881017) to intermediate phenotypes
(hypertension, diabetes mellitus, hypertriglyceridemia,
hypo-HDL-cholesterolemia, hyper-LDL-cholesterolemia, chronic kidney
disease, obesity and hyperuricemia) of aortic aneurysm. No SNP was
found to be significantly [P<0.0021 (0.05/24)] associated with
intermediate phenotypes (data not shown).
Discussion
True and dissecting aneurysms of the aorta develop
as a result of progressive weakening of the vessel wall. They are
associated with characteristic histological features including
medial degeneration, which involves degeneration and fragmentation
of elastic fibers as well as loss of smooth muscle cells and an
accumulation of basophilic ground substances (19). In the present study, we showed
that rs1465567 [T/C (W229R)] of EGFLAM and rs113710653 [C/T
(E231K)] of SPATC1L were significantly associated with true
aortic aneurysm, whereas rs143881017 [C/T (R140H)] of
RNASE13 was significantly associated with dissecting aortic
aneurysm, in Japanese individuals. The minor alleles of these SNPs
were all risk factors for these conditions.
The EGFLAM is located at chromosomal region
5p13.2-p13.1 (NCBI Gene, https://www.ncbi.nlm.nih.gov/gene) and is expressed in
various tissues and organs including vascular smooth muscle (The
Human Protein Atlas, http://www.protein-atlas.org). EGFLAM is an
extracellular matrix-like protein that colocalizes with both
dystrophin and dystroglycan to the synaptic cleft of the
photoreceptor ribbon synapse in the retina and which directly
interacts with dystroglycan. It plays an important role in
interactions between the photoreceptor ribbon synapse and bipolar
dendrites (20,21), and it is implicated in defective
photoreceptor synaptic function associated with congenital muscular
dystrophies such as muscle-eye-brain disease caused by defective
glycosylation of α-dystroglycan (22). A genome-wide pharmacogenomics
study identified EGFLAM as a potential susceptibility locus
for citalopram-induced side effects (23). We have now shown that rs1465567
[T/C (W229R)] of EGFLAM was significantly associated with
true aortic aneurysm, with the minor C allele representing a risk
factor for this condition, although the molecular mechanism
underlying this association remains unclear.
The SPATC1L is located at chromosomal region
21q22.3 (NCBI Gene) and is expressed in various tissues and organs
including vascular smooth muscle (The Human Protein Atlas). SPATC1L
is distributed in the cytoplasm, nucleus, and perinuclear region of
cells, and it translocates to the sites of cell-cell junctions in
response to stimulation of cells with the neuropeptide neurokinin A
(24). Expression of
SPATC1L was also found to modulate the response of cells to
N-methyl-N′-nitro-N-nitrosoguanidine and may
thereby protect cells from cell death induced by this DNA-damaging
agent (25). We demonstrated that
rs113710653 [C/T (E231K)] of SPATC1L was significantly
associated with true aortic aneurysm, with the minor T allele
representing a risk factor for this condition, although the
functional relevance of this association remains to be
elucidated.
RNASE13 is located at chromosomal region
14q11.2 (NCBI Gene) and is expressed at a high level in the
epididymis (The Human Protein Atlas). A GWAS showed that an SNP
(rs3748348) located in the vicinity of RNASE13 was
associated with executive functioning resilience (26). Gene-based analyses also revealed a
genome-wide significant association between RNASE13 and
executive functioning resilience (27). We now showed that rs143881017 [C/T
(R140H)] of RNASE13 was significantly associated with
dissecting aortic aneurysm, with the minor T allele representing a
risk factor for this condition, although the molecular mechanism
underpinning this association remains unknown.
Previous GWASs identified the SNPs: rs10757278 of
CDKN2BAS, rs7025486 of DAP2IP, rs1466535 of
LRP1, rs2118181 of FBN1, rs6511720 of LDLR and
rs599839 of SORT1 as susceptibility loci for aortic aneurysm
(7–12). The MAFs of these SNPs were
>10%, and the odds ratios were 0.8–1.8 (5.7–12.28). We now
identified three novel loci that may confer susceptibility to true
or dissecting aortic aneurysm, with the odds ratios (MAF, %) of
rs1465567 of EGFLAM, rs113710653 of SPATC1L, and
rs143881017 of RNASE13 being 1.63 (25.1%), 5.32 (1.9%),and
5.77 (0.5%), respectively. Although rs1465567 of EGFLAM was
a common variant with a small effect size, rs113710653 of
SPATC1L and rs143881017 of RNASE13 were low-frequency
variants with moderate to large effect sizes.
There are some limitations to the present study: i)
Given that the number of subjects with aortic aneurysm was
relatively small and the results of the study were not replicated,
our findings will require validation with other independent subject
panels or in other ethnic groups. ii) It is possible that rs1465567
of EGFLAM, rs113710653 of SPATC1L, or rs143881017 of
RNASE13 is in linkage disequilibrium with other
polymorphisms in the same gene or in other nearby genes that are
actually responsible for the development of true or dissecting
aneurysm. iii) The functional relevance of these SNPs to the
pathogenesis of true or dissecting aneurysm remains to be
elucidated.
In conclusion, rs1465567 of EGFLAM and
rs113710653 of SPATC1L may be susceptibility loci for true
aortic aneurysm and rs143881017 of RNASE13 may be such a
locus for dissecting aortic aneurysm in Japanese individuals.
Determination of genotypes for these SNPs may prove informative for
assessment of the genetic risk for these conditions in Japanese
individuals.
Acknowledgments
The present study was supported by CREST (H25-H30),
Japan Science and Technology Agency (to Y.Y., J.S. and I.T.) and by
Japan Society for the Promotion of Science KAKENHI grants
JP15H04772 (to Y.Y.), JP25242062 (to M.T.) and JP16H01872 (to
M.T.).
References
|
1
|
Chiesa R, Melissano G, Civilini E, de
Moura ML, Carozzo A and Zangrillo A: Ten years experience of
thoracic and thoracoabdominal aortic aneurysm surgical repair:
Lessons learned. Ann Vasc Surg. 18:514–520. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mizuguchi T, Collod-Beroud G, Akiyama T,
Abifadel M, Harada N, Morisaki T, Allard D, Varret M, Claustres M,
Morisaki H, et al: Heterozygous TGFBR2 mutations in Marfan
syndrome. Nat Genet. 36:855–860. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schwarze U, Schievink WI, Petty E, Jaff
MR, Babovic-Vuksanovic D, Cherry KJ, Pepin M and Byers PH:
Haploinsufficiency for one COL3A1 allele of type III procollagen
results in a phenotype similar to the vascular form of
Ehlers-Danlos syndrome, Ehlers-Danlos syndrome type IV. Am J Hum
Genet. 69:989–1001. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nordon IM, Hinchliffe RJ, Loftus IM and
Thompson MM: Pathophysiology and epidemiology of abdominal aortic
aneurysms. Nat Rev Cardiol. 8:92–102. 2011. View Article : Google Scholar
|
|
5
|
Ye Z, Austin E, Schaid DJ and Kullo IJ: A
multi-locus genetic risk score for abdominal aortic aneurysm.
Atherosclerosis. 246:274–279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harrison SC, Holmes MV, Agu O and
Humphries SE: Genome wide association studies of abdominal aortic
aneurysms - biological insights and potential translation
applications. Atherosclerosis. 217:47–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Helgadottir A, Thorleifsson G, Magnusson
KP, Grétarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT,
Rinkel GJ, Blankensteijn JD, Ronkainen A, et al: The same sequence
variant on 9p21 associates with myocardial infarction, abdominal
aortic aneurysm and intracranial aneurysm. Nat Genet. 40:217–224.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gretarsdottir S, Baas AF, Thorleifsson G,
Holm H, den Heijer M, de Vries JP, Kranendonk SE, Zeebregts CJ, van
Sterkenburg SM, Geelkerken RH, et al: Genome-wide association study
identifies a sequence variant within the DAB2IP gene conferring
susceptibility to abdominal aortic aneurysm. Nat Genet. 42:692–697.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bown MJ, Jones GT, Harrison SC, Wright BJ,
Bumpstead S, Baas AF, Gretarsdottir S, Badger SA, Bradley DT,
Burnand K, et al CARDIoGRAM Consortium; Global BPgen Consortium;
DIAGRAM Consortium; VRCNZ Consortium: Abdominal aortic aneurysm is
associated with a variant in low-density lipoprotein
receptor-related protein 1. Am J Hum Genet. 89:619–627. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
LeMaire SA, McDonald ML, Guo DC, Russell
L, Miller CC III, Johnson RJ, Bekheirnia MR, Franco LM, Nguyen M,
Pyeritz RE, et al: Genome-wide association study identifies a
susceptibility locus for thoracic aortic aneurysms and aortic
dissections spanning FBN1 at 15q21.1. Nat Genet. 43:996–1000. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bradley DT, Hughes AE, Badger SA, Jones
GT, Harrison SC, Wright BJ, Bumpstead S, Baas AF, Grétarsdóttir S,
Burnand K, et al: A variant in LDLR is associated with abdominal
aortic aneurysm. Circ Cardiovasc Genet. 6:498–504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jones GT, Bown MJ, Gretarsdottir S,
Romaine SP, Helgadottir A, Yu G, Tromp G, Norman PE, Jin C, Baas
AF, et al: A sequence variant associated with sortilin-1 (SORT1) on
1p13.3 is independently associated with abdominal aortic aneurysm.
Hum Mol Genet. 22:2941–2947. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Manolio TA, Collins FS, Cox NJ, Goldstein
DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR,
Chakravarti A, et al: Finding the missing heritability of complex
diseases. Nature. 461:747–753. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johnston KW, Rutherford RB, Tilson MD,
Shah DM, Hollier L and Stanley JC: Suggested standards for
reporting on arterial aneurysms. Subcommittee on Reporting
Standards for Arterial Aneurysms, Ad Hoc Committee on Reporting
Standards, Society for Vascular Surgery and North American Chapter,
International Society for Cardiovascular Surgery. J Vasc Surg.
13:452–458. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Olsson C, Thelin S, Ståhle E, Ekbom A and
Granath F: Thoracic aortic aneurysm and dissection: Increasing
prevalence and improved outcomes reported in a nationwide
population-based study of more than 14,000 cases from 1987 to 2002.
Circulation. 114:2611–2618. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grove ML, Yu B, Cochran BJ, Haritunians T,
Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, et
al: Best practices and joint calling of the HumanExome BeadChip:
The CHARGE Consortium. PLoS One. 8:e680952013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Anderson CA, Pettersson FH, Clarke GM,
Cardon LR, Morris AP and Zondervan KT: Data quality control in
genetic case-control association studies. Nat Protoc. 5:1564–1573.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Price AL, Patterson NJ, Plenge RM,
Weinblatt ME, Shadick NA and Reich D: Principal components analysis
corrects for stratification in genome-wide association studies. Nat
Genet. 38:904–909. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barbour JR, Spinale FG and Ikonomidis JS:
Proteinase systems and thoracic aortic aneurysm progression. J Surg
Res. 139:292–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sato S, Omori Y, Katoh K, Kondo M,
Kanagawa M, Miyata K, Funabiki K, Koyasu T, Kajimura N, Miyoshi T,
et al: Pikachurin, a dystroglycan ligand, is essential for
photoreceptor ribbon synapse formation. Nat Neurosci. 11:923–931.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanagawa M, Omori Y, Sato S, Kobayashi K,
Miyagoe-Suzuki Y, Takeda S, Endo T, Furukawa T and Toda T:
Post-translational maturation of dystroglycan is necessary for
pikachurin binding and ribbon synaptic localization. J Biol Chem.
285:31208–31216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu H, Li J, Zhang Z and Yu M: Pikachurin
interaction with dystroglycan is diminished by defective O-mannosyl
glycosylation in congenital muscular dystrophy models and rescued
by LARGE overexpression. Neurosci Lett. 489:10–15. 2011. View Article : Google Scholar :
|
|
23
|
Adkins DE, Clark SL, Åberg K, Hettema JM,
Bukszár J, McClay JL, Souza RP and van den Oord EJ: Genome-wide
pharmacogenomic study of citalopram-induced side effects in STAR*D.
Transl Psychiatry. 2:e1292012. View Article : Google Scholar
|
|
24
|
Lecat S, Matthes HW, Pepperkok R, Simpson
JC and Galzi JL: A fluorescent live imaging screening assay based
on translocation criteria identifies novel cytoplasmic proteins
implicated in G protein-coupled receptor signaling pathways. Mol
Cell Proteomics. 14:1385–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fry RC, Svensson JP, Valiathan C, Wang E,
Hogan BJ, Bhattacharya S, Bugni JM, Whittaker CA and Samson LD:
Genomic predictors of interindividual differences in response to
DNA damaging agents. Genes Dev. 22:2621–2626. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mukherjee S, Kim S, Gibbons LE, Nho K,
Risacher SL, Glymour MM, Habeck C, Lee GJ, Mormino E, Ertekin-Taner
N, et al: Alzheimer's Disease Neuroimaging Initiative: Genetic
architecture of resilience of executive functioning. Brain Imaging
Behav. 6:621–633. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mukherjee S, Kim S, Ramanan VK, Gibbons
LE, Nho K, Glymour MM, Ertekin-Taner N, Montine TJ, Saykin AJ and
Crane PK: Alzheimer's Disease Neuroimaging Initiative: Gene-based
GWAS and biological pathway analysis of the resilience of executive
functioning. Brain Imaging Behav. 8:110–118. 2014. View Article : Google Scholar :
|
|
28
|
van't Hof FN, Ruigrok YM, Lee CH, Ripke S,
Anderson G, de Andrade M, Baas AF, Blankensteijn JD, Böttinger EP,
Bown MJ, et al Aneurysm Consortium; Vascular Research Consortium of
New Zealand: Shared genetic risk factors of intracranial,
abdominal, and thoracic aneurysms. J Am Heart Assoc. 5:e0026032016.
View Article : Google Scholar
|