Introduction
The ubiquitin-specific proteases (USPs) represent
the most evolutionarily conserved and well represented
deubiquitinating enzymes (DUBs) (1). They contain conserved Cys, His and
Asp/Asn domains in their amino acid sequ ences (2), which include the residues critical
for catalysis. Accordingly, USPs have the ability to remove
ubiquitin from a ubiquitinated substrate, thereby regulating the
localization and activity of this substrate (2–5).
The ability of USPs to cleave model ubiquitin fusion protein
substrates was previously investigated by co-expression in
Escherichia coli (E. coli) (6). USP enzymes are characterized by the
presence of two conserved active site domains, and several have
been shown to cleave ubiquitin from model substrates, such as
ubiquitin-β-galactosidase (Ub-Met-β-gal) and pGEX-Ub52 [glutathione
S-transferase (GST)-Ub52] (7), by
hydrolyzing the bond between the C-terminal double glycine of
ubiquitin and the linking methionine residue (6). However, ubiquitin C-terminal
7-amido-4-methylcoumarin (Ub-AMC) and its analogs, due to their
high fluorescence sensitivity and hydrolysis efficiency, are often
used to quantitatively determine the activities of DUBs without
considering their substrate specificities (8,9).
USP13, also named isopeptidase T-3, which was first
identified by Timms et al (10), is a well-characterized member of
USPs (11). USP13 shares a 54.8%
identity with its isoform USP5 (12,13). Both proteins contain 4
ubiquitin-associated (UBA) domains and an ubiquitin-specific
processing protease (UBP) domain. The UBP domain of USP13 harbors a
catalytic site, a zinc finger (ZnF) domain, and 2 UBA domains.
Domains such as UBA and ZnF play very important roles in USP13
(14). The UBA domain usually
facilitates substrate binding onto the enzymes, while the ZnF
domain may be an activator to stimulate the hydrolysis reaction or
a regulator to orchestrate other biological functions (15). Although USP13 contains all 4
putative ubiquitin-binding domains, it has not been shown to
hydrolyze polyubiquitin (16),
but rather serves as a protease for interferon-stimulated gene 15
(ISG15), whereby it reduces ISGylation (16). A previous study indicated that a
few residues in USP13-ZnF vary, particularly in the L2-loop region,
in which R221 and Y261 in USP5-ZnF are the key residues for
ubiquitin binding and catalytic activation (13). The ZnF domain of USP13 cannot bind
with ubiquitin, so that USP13 loses its ability to be activated by
free ubiquitin (14). However,
USP13 exhibits a weak deubiquitinating activity for Lys63-linked
polyubiquitin (K63-polyUb) in a non-activating manner, and it
cannot be activated by the reaction product (14). Nevertheless, in a recent study,
the ability of USP13 to bind free ubiquitin and cleave
ubiquitin-AFC (Ub-AFC) was tested in vitro, and USP13 was
almost completely inactive and could not efficiently cleave K48- or
K63-linked di-ubiquitin (17).
Thus, the catalytic function and the regulatory mechanism of USP13
remain to be defined.
In addition to studies on the enzymatic activity of
USP13, elucidating the substrate specificities of USP13 is vital to
understanding its biological function. It has recently been
demonstrated that USP13 can regulate Siah2 ligase stability and
activity via non-catalytic ubiquitin-binding domains (18). It can also deubiquitinate signal
transducer and activator of transcription 1 (STAT1) and increase
STAT1 protein stability (19),
reduce ubiquitination and degradation of Sky2 (20), and also appears to be responsible
for the protein stability of microphthalmia-associated
transcription factor (MITF), which is essential for the development
of melanocytes (21). Liu et
al also supported the existence of an interactive regulatory
association between USP10 and 13 with Vps34 complexes (22). Furthermore, USP13 was identified
as the first deubiquitylase that reverses phosphatase and tensin
homolog (PTEN) polyubiquitylation and stabilizes PTEN protein in
human breast cancer cells (23).
Recently, a new study found that USP13 interacted with PTEN in
fibroblasts and regulated PTEN ubiquitination and degradation
(24). Another study revealed an
unexpected interaction between USP13 and ERAD-specific ubiquitin
ligase gp78, thereby promoting ERAD (17). In addition to these findings,
further research is required to identify additional substrates of
USP13.
This study confirmed that USP13 proteins have no
detectable DUB activity detected by USP cleavage assay using two
different types of model substrates (GST-Ub52 and Ub-Met-β-gal). In
addition, a proteomics approach was taken by using two-dimensional
fluorescence difference gel electrophoresis (2D-DIGE) to detect
cellular proteins with significantly altered expression levels in
293T cells following the overexpression of USP13. As a specificity
control, this study also aimed to examine the effect of a
catalytically inactive mutant of USP13. To establish such a mutant,
the amino acid sequence of USP13 was aligned to USP5, a DUB with a
well-defined cysteine catalytic motif, and thereby cysteine 345 was
identified as the corresponding catalytically critical residue,
mutating it to serine (2).
Several proteins were detected and identified using liquid
chromatography-mass spectroscopy (LC-MS)/MS. Among the identified
proteins, the altered expression level of vinculin was associated
with the DUB activity of USP13, which was confirmed by western blot
analysis and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR).
Materials and methods
Plasmid construction and site-directed
mutagenesis
The plasmid pGEX-USP13 (wild-type), which expresses
the GST-USP13 fusion protein, was constructed by inserting the DNA
fragment encoding the corresponding open reading frames between the
SalI and NotI (both from New England Biolabs,
Hitchin, UK) sites of plasmid pGEX-6p-1. A catalytically inactive
version of USP13, pGEX-USP13 C345S (cysteine 345 mutated to
serine), was then constructed by site-directed mutagenesis with the
following primer sequences: 5′-AGCTATCTCAGCTCTGTCATG-3′ and
5′-GCTGTTGCCCAGGTTCTTCAG-3′. A pAC-T7-USP13 plasmid was produced by
inserting the comp lete coding sequence of USP13 into the pAC-T7
plasmid at the BamHI site. The pAC-T7 plasmid was produced
according to the method described in the study by Zhang et
al (7). To generate the
plasmids pEGFP-USP13 and pEGFP-USP13 C345S, the pGEX-USP13 and
pGEX-USP13 C345S vectors were digested with SalI and
NotI (both from New England Biolabs), respectively, and then
inserted into the pEGFP-C1 vector, which was linearized by
EcoRI (New England Biolabs).
Expression and USP cleavage assay
E. coli DH5α harboring pGEX-USP13 was
incubated in Luria-Bertani broth medium at 37 or 15°C overnight to
express GST-USP13 fusion proteins. GST fusion proteins were
purified using GSH-Sepharose resin (Sangon Biotech Co., Ltd.,
Shanghai, China) and detected using 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
The USP cleavage assays used Ub-Met-β-gal and
GST-Ub52 as model substrates, as previously described (7). For cleavage of the GST-Ub52
substrate, E. coli strain BL21 (DE3) cells harboring
pGEX-Ub52 were further transformed with pACT7-USP13 or pAC-T7-USP46
[plasmids expressing USP46 were used as positive controls (25)]. The protein expression was induced
by isopropyl β-D-1-thiogalactopyranoside (IPTG). Soluble protein
extracts were prepared after sonicating the cell lysate. GST fusion
proteins and their cleavage products were purified using
GSH-Sepharose resin (Sangon Biotech Co., Ltd.) and detected by 10%
SDS-PAGE. The USP cleavage assay using Ub-Met-β-gal as a model
substrate has been previously described (26). E. coli BL21 (DE3) bacteria
harboring pGEX-USP13 or pGEX-USP46 [plasmids expressing USP46 were
used as positive controls (27)]
were transformed with pAC-M-β-gal, AmpR CmR colonies were grown and
induced with IPTG, and total protein extracts were analyzed by west
ern blotting with anti-β-gal antibody (Z3781; Promega, Madison, WI,
USA). Total protein extracts were examined by western blot analysis
with anti-T7 purified polyclonal antibody (AB3790; Millipore,
Billerica, MA, USA) and anti-GST antibody (ab111947; Abcam,
Cambridge, UK) to detect the expression of USPs.
Cell culture and transient
transfection
The 293T cell line was purchased from the American
Type Culture Collection (ATCC, Manassas, VA, USA). The cell line
was seeded in a 6-well plate at a concentration of 4×105
cells/2 ml and cultured in Dulbecco's modified Eagle's medium
(DMEM) suppl emented with 10% fetal calf serum (FCS) (both from
Gibco Life Technologies, Grand Island, NY, USA) and 1%
penicillin-streptomycin at 37°C in a 5% CO2 atmosphere.
After 24 h, the medium was replaced with fresh medium without
antibiotics. The purified recombinant plasmids, pEGFP-C1,
pEGFP-USP13 and pEGFP-USP13 C345S, were transfected into the cells
using Lipofectamine 2000 transfection reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer's
instructions. Following incubating for 24 h, the cells were
collected by centrifugation (1,000 × g for 5 min at 4°C).
2D-DIGE
Protein isolation and sample
preparation
The proteins were extracted from the cells
transfected with the pEGFP-USP13, pEGFP-USP13 C345S or pEGFP-C1
plasmids using SDS extraction buffer (100 mM Tris-HCl, pH 8.0, 2%
SDS, 1% β-mercaptoethanol, 5 mM ethylene glycol tetraacetic acid,
10 mM ethylenediaminetetraacetic acid, 1 μM E-64, 1
μM bestatin, 1 μM pepstatin, 2 μM leupeptin, 1
mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail
and phosphatase inhibitor). The supernatants of the cell lysates
were extracted using an equivalent volume of ice-cold
Tris-saturated phenol (pH 8.0), and then precipitated overnight
with 5 volumes of chilled 0.1 M ammonium acetate in methanol at
−20°C. After rinsing twice with ice-cold ammonium acetate in
methanol, followed by 2 rinses with chilled methanol, the pellets
were solubilized in DIGE buffer [7M urea, 2M thiourea, 4%
3-[(3-(cholamidopropyl) dimethylammonio]-1-propanesulfonate
(CHAPS), pH 8.8]. The total protein concentrations were measured
using the Bradford method (28)
and adjusted to 5 μg/μl.
Protein labeling
2D-DIGE was performed following the method described
in the study by Tang (29). For
protein labeling, 50 pmol of CyDye was mixed with 10 μl of
protein sample and combined with Cy3- and Cy5-labeled control and
treated samples. Total protein extracts of USP13 were labeled with
Cy5 in the treated samples, whereas the EGFP proteins were labeled
with Cy3 in the control samples. The proteins were then pooled
together with the treated samples prior to 2D-DIGE. The samples
were incubated on ice for at least 2 h in the dark. The labeling
reaction was terminated by the addition 1 μl of 10 mM
lysine. For analytical 2D-DIGE analysis, 20 μl of combined
Cy3- or Cy5-labeled protein was mixed with 2D-DIGE buffer (7M urea,
2M thiourea, 4% CHAPS, 20 mM dithiothreitol, 0.5% IPG buffer) and
used for isoelectric focusing (IEF). IEF was performed on 24-cm IPG
strips, pH 4.0–7.0. The second-dimension electrophoresis was
performed using 10% SDS-polyacrylamide gels. The 2D gels were then
scanned on a Typhoon 9410 imager and DIGE images were analyzed
using DeCyder 6.5 software (both from GE Healthcare Life Sciences,
Logan, UT, USA), and spot detection was performed using the
differential in-gel analysis module and the DeCyder biological
variation analysis module. Protein spots displaying an average
≥1.1-fold change in abundance between the control and treated
samples with a value of P<0.05 were selected for picking on an
Ettan Spot Picker (GE Healthcare Life Sciences). For each
treatment, images from at least 3 biological repeats were used for
the statistical analysis of protein abundance.
LC-MS/MS
Protein spots were excised from the gels and sent to
the Department of Neuroplasticity of Shinshu University Graduate
School of Medicine in Japan for LC-MS/MS analysis.
Mass spectra were subjected to Mascot search engine
for protein identification. Database searching was performed using
the Mascot (http://www.matrixscience.com) program.
RT-qPCR
Total RNA from the cells transfected with
pEGFP-USP13, pEGFP-USP13 C345S, or pEGFP-C1 was extracted using
TRIzol reagent (Invitrogen Life Technologies) and
reverse-transcribed into cDNA using a RevertAid First Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
PCR quantification involved the use of TransStart Top Green qPCR
SuperMix (TransGen Biotech Co., Ltd., Beijing, China) with primer
sequences for vinculin (5′-AATGAGAGGGCAGTGTTTCC-3′ and
5′-GGATGTTCAGGCAAGGTTCT-3′). The mRNA levels of vinculin, as well
as those of the internal standard β-actin (Sangon Biotech Co.,
Ltd.), were measured by qPCR in triplicate using a Rotor Gene 6000
real-time detection system (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Western blot analysis
Total protein extracts of the cells transfected with
pEGFP-USP13, pEGFP-USP13 C345S or pEGFP-C1 were examined by western
blot analysis with vinculin antibody (OAAN00512; AVIVA Systems
Biology, San Diego, CA, USA) to detect the expression of vinculin.
With β-actin being an internal reference gene with anti-β-actin
(Proteintech Group, Inc., Chicago, IL, USA), the relative intensity
of the resulting bands was analyzed using Odyssey V3.0 software
(Odyssey, Lincoln, NE, USA).
Statistical analysis
All statistical analyses were performed using SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA). Statistical
analysis was performed using the Student's t-test and one-way
analysis of variance (ANOVA). A P-value <0.05 was considered to
indicate a statistically significant difference.
Results
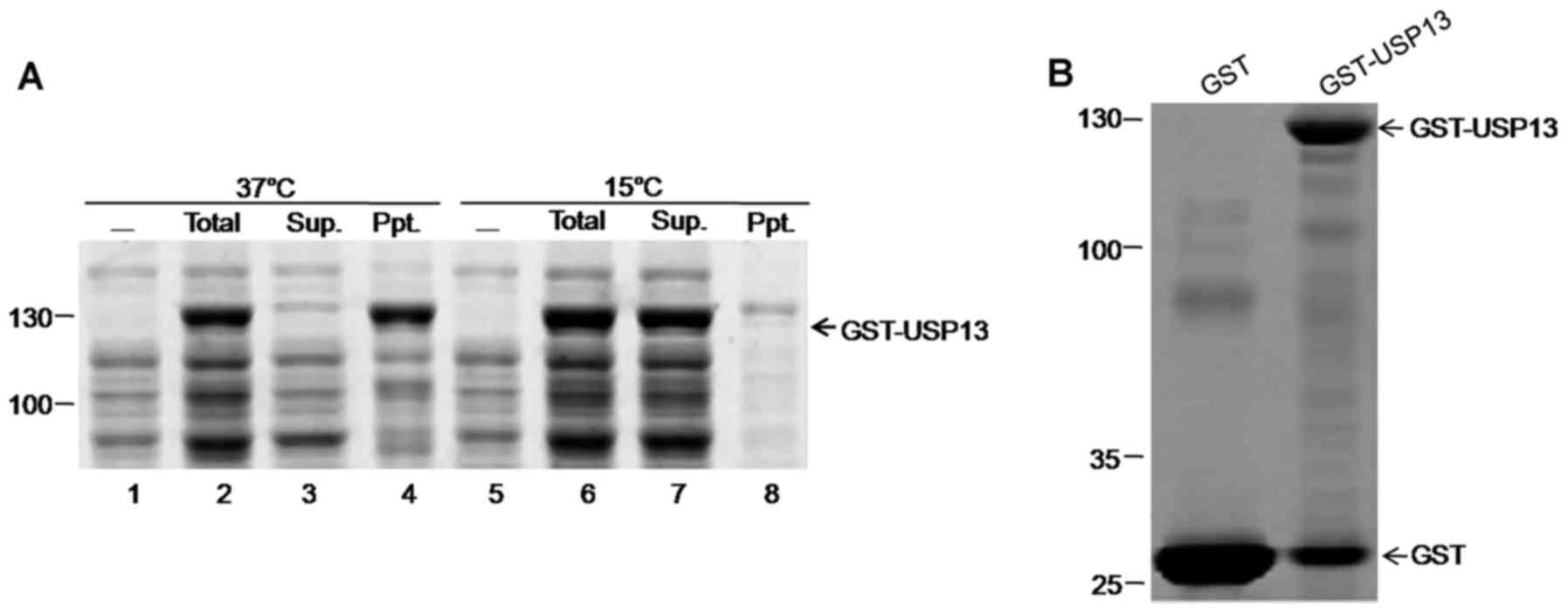
Low temperature improves the solubility
of GST-USP13 recombinant proteins
The nucleotide sequence of homo USP13 has been
submitted to GenBank under the accession no. HM138083. GST-USP13
fusion proteins of approximately 120 kDa were expressed in E.
coli BL21 and detected by 10% SDS-PAGE. Homo USP13 protein
fused with GST was expressed in E. coli strain BL21 and
induced by IPTG at 37 and 15°C. When induced at 37°C for 4 h, a
major part of GST-USP13 precipitated; on the contrary, GST-USP13
fusion protein induced with IPTG at 15°C mostly existed in the
supernatant (Fig. 1), thereby
facilitating the purification of the fusion proteins for further
analyses. GST fusion proteins were purified by GSH-Sepharose resin
(Sangon Biotech Co., Ltd.) (Fig.
1B) and used to measure the hydrolytic activities of
recombinant GST-fused USP13 (100 nM) using Ub-AMC as a substrate
(data not shown).
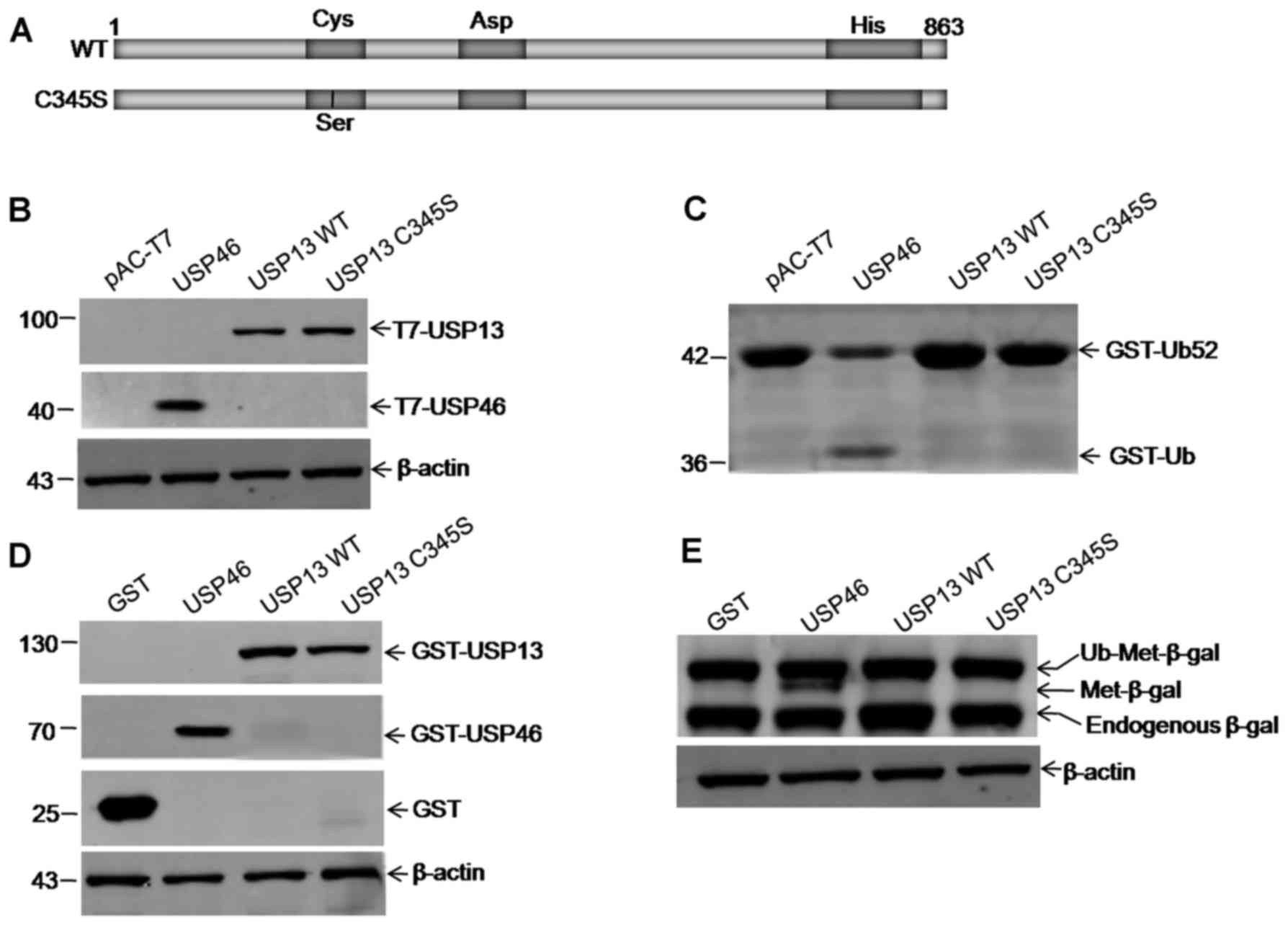
USP13 exhibits no detectable hydrolytic
activity for model substrates
To determine whether USP13 has DUB activity, a USP
cleavage assay was performed using GST-Ub52 as a model substrate.
USP46 cleaved GST-Ub52 efficiently to yield a product of the
expected size of 36 kDa as a positive control (Fig. 2C). Although western blot analysis
with anti-T7 antibody can detect the expression of T7-USP13
(Fig. 2B), the hydrolytic
activity of USP13 for GST-Ub52, as well as the empty vector alone
and the C345S mutant, was not detected (Fig. 2C). The hydrolytic activities of
recombinant GST-fused USP13 were further examined using
Ub-Met-β-gal as a substrate, and USP13 exhibited no detectable
activity either (Fig. 2D and E).
The hydrolytic activities of recombinant GST-fused USP13 (100 nM)
were also measured using Ub-AMC as a substrate, and similar results
were obtained (data not shown).
Screened proteins associated with the
overexpression of USP13 by 2D-DIGE and LC-MS/MS
To determine whether USP13 alters cellular protein
expression, a proteomic approach by 2D-DIGE was taken to detect
cellular proteins with significantly altered expression levels in
293T cell lines following the overexpression of USP13. 293T cells
were transfected with an expression vector for USP13, USP13 C345S
or an empty vector. Total protein extracts were labeled with Cy5 or
Cy3 and then pooled together with the treated samples prior to
2D-DIGE. DeCyder analysis of these replicates detected
approximately 2,500 spots in at least 2 of the 3 gels. For each
treatment, images from at least 3 biological repeats were used for
the statistical analysis of protein abundance.
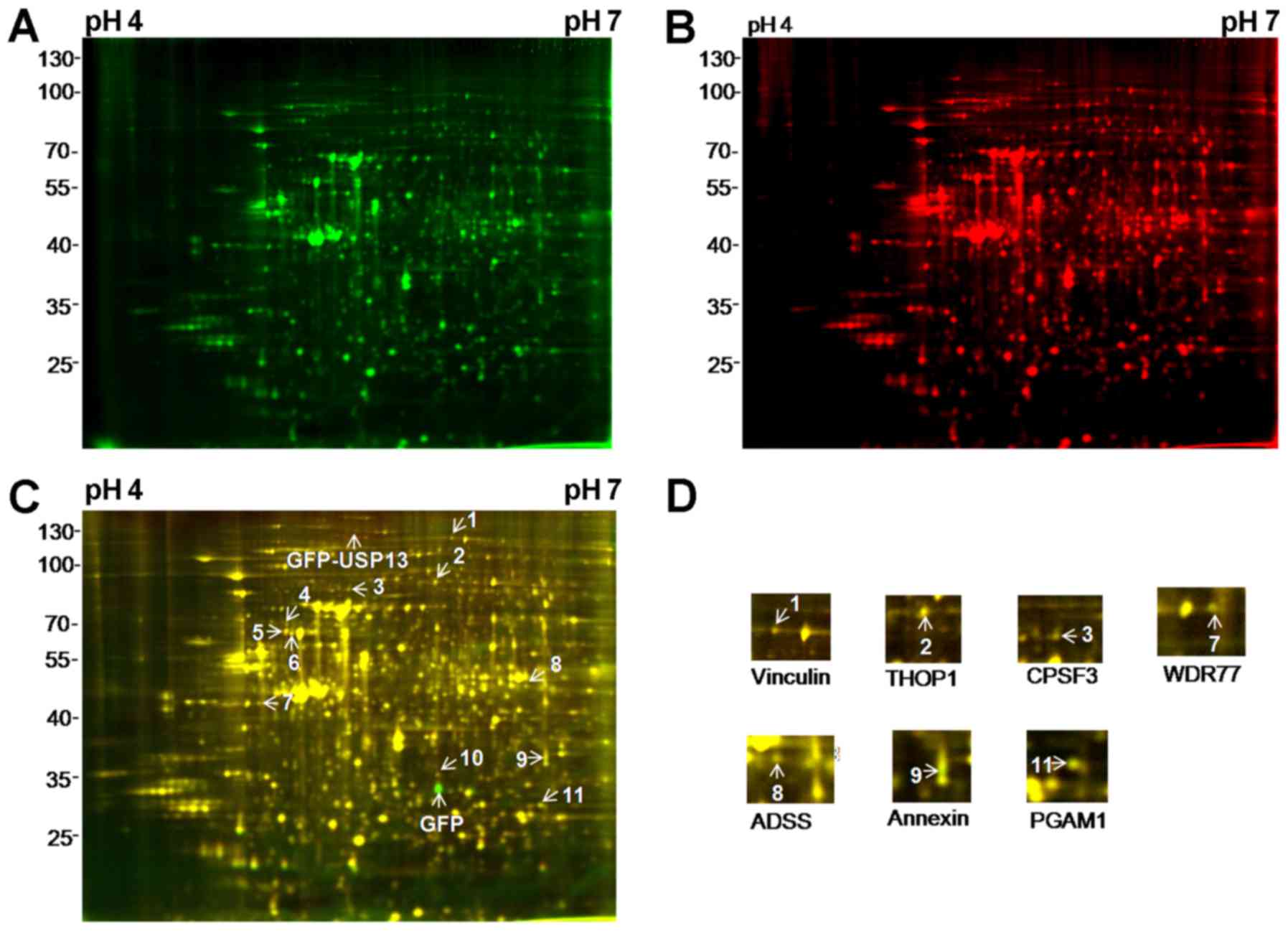
A total of 11 spots displayed an average ≥1.1-fold
change in abundance with a value of P<0.05 following the
overexpression of USP13 compared with that of EGFP (Fig. 3A–C). The arrowheads and numbers
indicate the spots with an increased and decreased expression. A
total of 7 of the 11 species were identified using LC-MS/MS. These
proteins included 4 proteins with an increased expression, namely
vinculin, thimet oligopeptidase 1 (THOP1), cleavage and
polyadenylation specific factor 3 (CPSF3) and methylosome protein
50 (WDR77), and 3 proteins with a decreased expression, namely
adenylosuccinate synthetase (ADSS), annexin and phosphoglycerate
mutase (PGAM), as shown in Fig.
3D and Table I. With regard
to the remaining four spots, spots 4 and 10 were identified as SH3
domain containing RING finger protein 3 and T complex protein 1
subunit α (TCP1), respectively, with isoelectric points of 9.99 and
7.67 (Table I). These isoelectric
points were beyond the range of the IPG strip used. Spots 5 and 6
could not be identified (Table
I). The reasons for the failure to identify the other species
may include problems in peptide elution, low amounts of protein, or
scoring below the threshold in the Mascot search engine.
 | Table ISummary of the proteins identified by
LC-MS/MS. |
Table I
Summary of the proteins identified by
LC-MS/MS.
| Master no.a | Accession no. | Protein name | Theoretical
| Protein score | Protein matched
name peptides | Protein sqeCover
(%) | Avg ratioa | P-valueb |
|---|
| Size (Da) | pI |
|---|
| 1/I | P18206 | Vinculin | 116,722.62 | 5.90 | 5,615.65 | 81 | 84.02 | 1.23/1.21 | 0.008/0.010 |
| 2 | P52888 | Thimet
Oligopeptidase (THOP1) | 78,840.00 | 5.87 | 1,693.12 | 13 | 37.48 | 1.21 | 0.008 |
| 3 | G5E9W3 | Cleavage and
polyadenylation specific factor 3 (CPSF3) | 77,486.24 | 5.42 | 1,053.86 | 19 | 38.31 | 1.16 | 0.008 |
| 4 | Q5T6W5 | SH3 domain
containing RING finger protein 3 | 93,973.91 | 9.99 | 1,580.02 | 11 | 25.81 | 1.15 | 0.019 |
| 5/IIc | B3GQS7 | | | | | | | 1.32/1.31 | 0.017/0.011 |
| 6/IIIc | F5GWR2 | | | | | | | 1.16/1.15 | 0.02/0.024 |
| 7 | Q9BQA1 | Methylosome protein
50 (WDR77) | 36,724.50 | 4.95 | 2,015.87 | 20 | 61.25 | 1.17 | 0.013 |
| 8 | F8W9D6 | Adenylosuccinate
synthetase (ADSS) | 48,338.13 | 6.37 12,416.72 | | 39 | 89.20 | −1.16 | 0.016 |
| 9 | E7EW96 | Annexin | 38,714.27 | 6.86 | 5,310.28 | 16 | 79.14 | −1.2 | 0.014 |
| 10 | P17987 | T complex protein 1
subunit α | 43,885.80 | 7.67 | 751.85 | 17 | 44.11 | −1.15 | 0.004 |
| 11 | Q6P6D7 | Phosphoglycerate
mutase (PGAM) | 28,776.89 | 6.46 39,293.07 | | 13 | 55.12 | −1.23 | 0.010 |
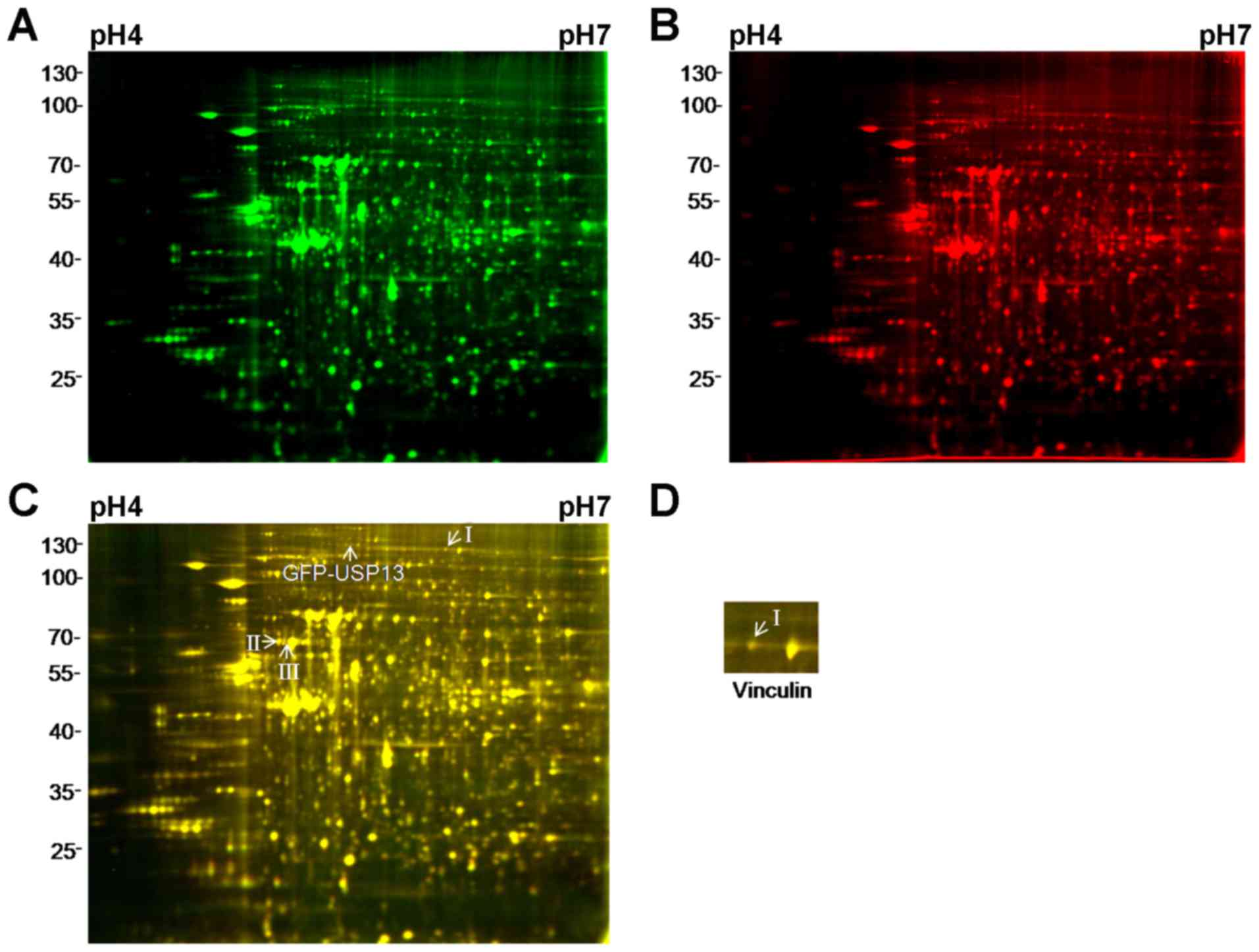
To investigate whether USP13 DUB activity affects
the expression of these proteins, the USP13 C345S proteins were
labeled with Cy3 in the control samples and pooled together with
the treated samples prior to 2D-DIGE. USP13 C345S completely
abolished its DUB activity (14).
Three of the 11 spots still displayed an average ≥1.1-fold increase
in abundance with P<0.05 (Fig.
4A–C). Following LC-MS/MS analysis, only vinculin exhibited an
increased expression as a result of the expression of USP13
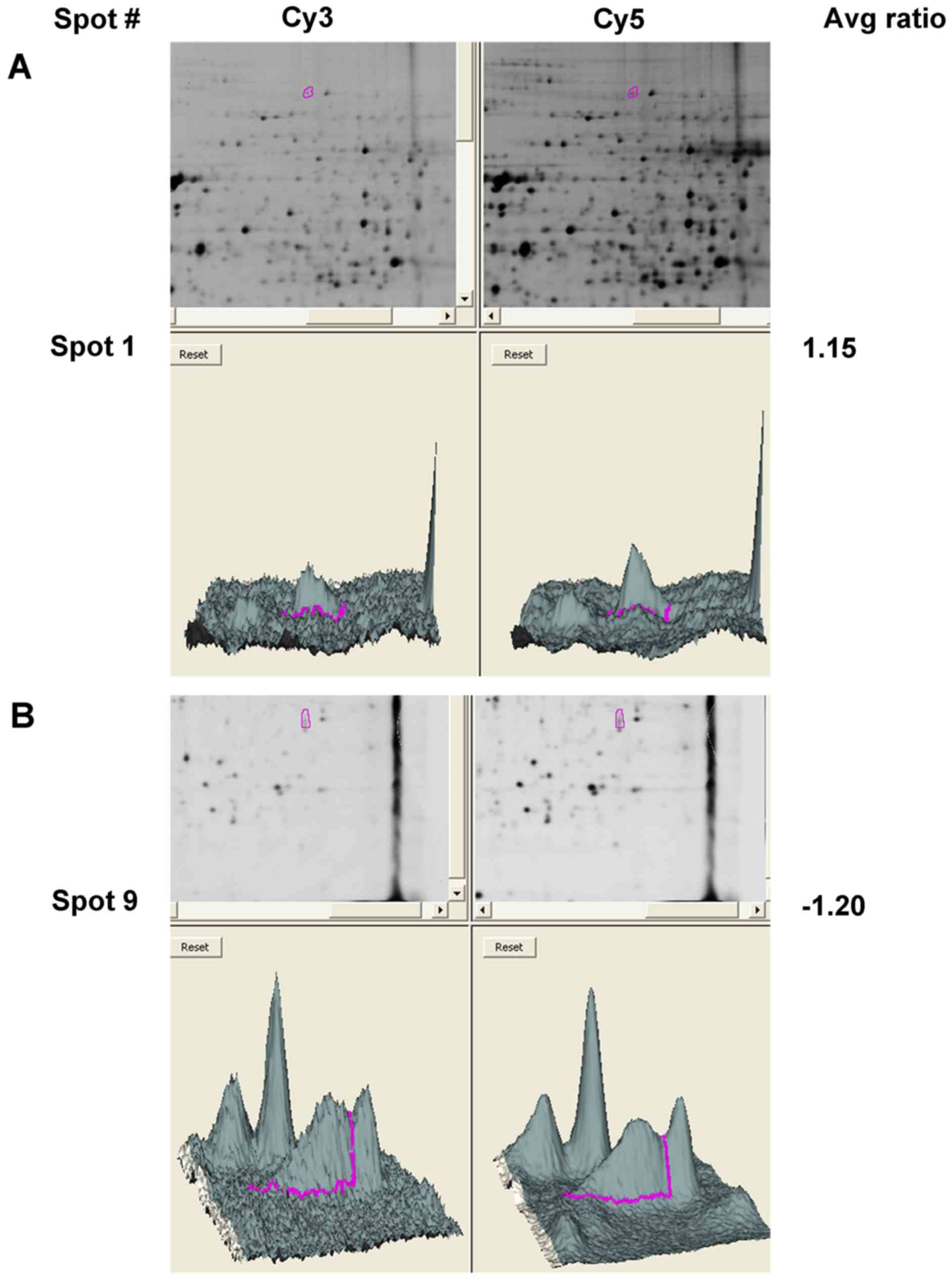
(Fig. 4D). Fig. 5 shows the three-dimensional plots
of spot density that increased in amount in 293T cell lines
expressing USP13 and USP13 C345S, where the z-axis (vertical
dimension) shows the fluorescence signal intensity in that channel.
These results suggest that the regulation of vinculin protein
levels by USP13 may depend on the enzymatic activity of USP13.
USP13 upregulates vinculin protein
levels, but not mRNA levels
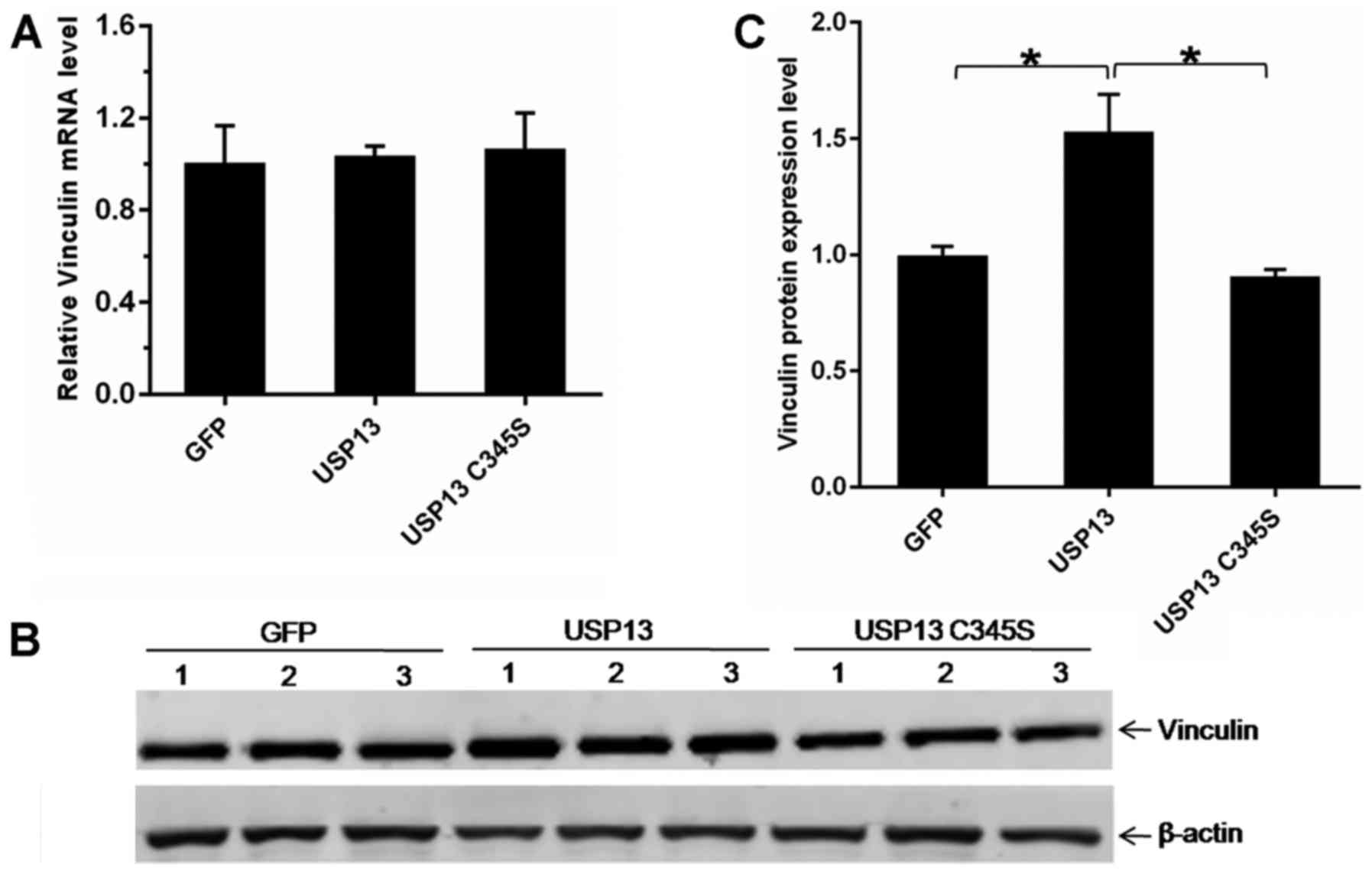
To further validate the upregulation of vinculin
protein levels by USP13, western blot analysis and RT-qPCR analysis
were performed. the 293T cells were transfected with an expression
vector for USP13, USP13 C345S, or an empty vector (pEGFP-C1). At 24
h following transfection, total protein extracts were examined by
western blot analysis to detect the expression of endogenous
vinculin. The results from 3 independent experiments revealed that
the overexpression of USP13, but not the enzyme-dead mutant USP13
C345S and the empty vector, led to a prominent increase in the
protein levels of vinculin (Fig. 6B
and C). On the contrary, the vinculin transcript levels were
not significantly altered in the cells following the overexpression
of USP13 (Fig. 6A). These results
suggest that USP13 upregulates vinculin protein levels, but not the
mRNA levels, through an enzymatically mediated mechanism.
Discussion
In the present study, the USP13 gene was
identified from 293T cells, and the solubility of GST-USP13
recombinant protein was found to be improved significantly at a
lower temperature (15°C). The USP cleavage assay using GST-Ub52 and
Ub-Met-β-gal as model substrates in vitro also revealed that
USP13 had no detectable DUB activity. In addition, a proteomics
approach was taken by using 2D-DIGE to detect cellular proteins
with significantly altered expression levels in 293T cells
following the overexpression of USP13 or its C345S mutant (the
catalytically inactive form) for the first time, to the best of our
knowledge. Several proteins were detected and identified using
LC-MS/MS. Among the identified proteins, the vinculin protein level
was upregulated by USP13 through an enzymatically mediated
mechanism, which was confirmed by western blot analysis and
RT-qPCR.
Attempts were made to purify and improve the
solubility of the fusion protein GST-USP13 for further analyses.
Commonly, lowering the temperature improves the solubility of
recombinant proteins (30–32)
as cell processes that involve transcription, translation and cell
division slow down (33). In
addition, due to reducing the activities of most proteases at a
lower temperature, recombinant proteins GST-USP13 are protected
from attack by other proteases that cause protein degradation and
formation of inclusion bodies (21). As shown in Fig. 1, when the GST-USP13 fusion
proteins were expressed at 15°C, the solubility of GST-USP13
recombinant protein improved significantly.
Two types of model substrates, GST-Ub52 and
Ub-Met-β-gal, were used in this study to detect the DUB acti vity
of USP13 for the first time (Fig. 2B
and D). Neither USP13 nor C345S mutant was found to have
detectable activity. GST-Ub52 and Ub-Met-β-gal, the general model
substrates, are often used to detect the activity of DUBs (7,25,27). The expression levels of USP13 are
different for the two different types of substrates due to the
different expression vectors. Thus, two different types of model
substrates were used to detect the DUB activity of USP13. The
present findings were in partial agreement with previous studies.
Catic et al (16)
discovered that USP13 reacts with ISG15VS rather than with UbVME,
which provided the first biochemical evidence for the protease
activity of USP13. In a recent study, USP13 was found to possess a
very weak deubiquitinating acti vity using Ub-AMC as a substrate,
but a high concentration of GST-USP13 (500 nM) is required
(14). However, the hydrolytic
activities of recombinant GST-fused USP13 (100 nM) were also
measured using Ub-AMC as a substrate, and no deubiquitinating
activity was detected for USP13 (data not shown). The reasons for
the failure may include fusion protein cleavage, low protein
concentration or low protein purity.
The types and lengths of ubiquitinated proteins
varied among species (34).
Therefore, each DUB member may have different substrate
specificity, but this specificity cannot be determined by using
model substrates.
In addition to studies on the enzymatic activity of
USP13, elucidating the substrate specificities of USP13 is vital to
understanding the biological function of the protein. The 2D-DIGE
technology is now recognized as a very powerful tool to address
changes related to a variety of effects on the cellular proteome.
The method allows detecting not only the changes in protein
quantity but also post-translational modifications that change the
charge or size of the protein (35). When coupled to LC-MS/MS, 2D-DIGE
can be used to identify proteins that change in structure or
concentration under specific conditions. This technology seems
applicable to identifying putative new substrates for USP13, as
USP13 generally upregulates substrate protein levels by stabilizing
proteins via deubiquitination. In this study, 2D-DIGE and LC-MS/MS
were used to identify cellular proteins with significantly altered
expression levels in 293T cell lines after the expression of USP13.
Several proteins displayed altered expression levels after the
overexpression of USP13 compared with that of EGFP (Fig. 3). However, when comparing the
samples after the overexpression of USP13 and USP13 C345S, only
vinculin exhibited increased expression levels (Fig. 4). These results suggest that the
regulation of the remaining proteins whose expression level altered
after the overexpression of USP13, such as THOP1, CPSF3 and Annexin
by USP13 may not require USP13 deubiquitinase activity. This is
simlar to the effect of USP13 on Siah2, which is not mediated by
its isopeptidase activity (18).
Future studies are warranted to determine the molecular mechanisms
through which USP13 regulates the levels of these proteins.
Although the 2D-DIGE technology allows the detection
of very small changes in spot intensity, it has some limitations.
First, some overlapping of protein spots due to similarities in
molecular mass and pI is possible. Second, the 2D-DIGE
approach allows the detection of proteins that are expressed at
relatively high levels. Hence, potentially important substrate
proteins that are expressed at levels below the detection limit of
2D-DIGE may be missed. These limitations may explain why the
present study failed to identify the known USP13 substrates MITF
(21) and PTEN (23,17–19). For example, MITF is a member of
the basic helix-loop-helix-leucine zipper transcription factor
family and a key regulator of development and survival for
melanocytes as well as melanomas (36). MITF is usually highly expressed in
the melanoma cell lines (21).
In this study, 2D-DIGE was used to explore whether
vinculin is upregulated by USP13. Vinculin is a 116-kDa
cytoskeletal protein that is involved in the linkage of integrin
adhesion molecules to the actin cytoskeleton (37). In a previous study, UCH-L1, a DUB
that removes Ub from Ub precursor proteins and hydrolyzes Ub
carboxyl-terminal esters and amides (38), was found to be involved in
regulating focal adhesions and adherens junctions, which is
supported by co-immunoprecipitation with key components of these
complexes, including FAK, paxillin, p120-catenin, β-catenin, and
vinculin (39). Furthermore, vinc
ulin is regularly co-precipitated by the wild-type and mutant
UCH-L1 after mild cross-linking. However, the deubiquitylase that
regulates vinculin polyubiquitylation and protein stability has not
yet been reported. The present study revealed that vinculin is
upregulated following the overexpression of USP13 and thereby
provides a molecular clue as to whether USP13 is responsible for
the deubiquitination of vinculin. Future studies are warranted to
determine the ubiquitination of vinculin in 293T cell lines and
whether USP13 deubiquitinates vinculin polyubiquitylation and
stabilizes vinculin protein.
However, regardless of the aforementioned
shortcomings, it is believed that the 2D-DIGE approach provides a
useful avenue to identify new USP substrates, provided that
sufficient follow-up validation studies are performed.
Acknowledgments
This study was supported by the Natural Science
Foundation of Hebei Province (no. H2015206406). The authors highly
appreciate Dr Man Li for contributing to mass spectrometry
analysis.
References
|
1
|
Kim YK, Kim YS, Yoo KJ, Lee HJ, Lee DR,
Yeo CY and Baek KH: The expression of Usp42 during embryogenesis
and spermatogenesis in mouse. Gene Expr Patterns. 7:143–148. 2007.
View Article : Google Scholar
|
|
2
|
Quesada V, Díaz-Perales A,
Gutiérrez-Fernández A, Garabaya C, Cal S and López-Otín C: Cloning
and enzymatic analysis of 22 novel human ubiquitin-specific
proteases. Biochem Biophys Res Commun. 314:54–62. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baek SH, Park KC, Lee JI II, Kim KI II,
Yoo YJ, Tanaka K, Baker RT and Chung CH: A novel family of
ubiquitin-specific proteases in chick skeletal muscle with distinct
N- and C-terminal extensions. Biochem J. 334:677–684. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
D'Andrea A and Pellman D: Deubiquitinating
enzymes: a new class of biological regulators. Crit Rev Biochem Mol
Biol. 33:337–352. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wing SS: Deubiquitinating enzymes - the
importance of driving in reverse along the ubiquitin-proteasome
pathway. Int J Biochem Cell Biol. 35:590–605. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Everett RD, Meredith M, Orr A, Cross A,
Kathoria M and Parkinson J: A novel ubiquitin-specific protease is
dynamically associated with the PML nuclear domain and binds to a
herpesvirus regulatory protein. EMBO J. 16:1519–1530. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang W, Tian QB, Li QK, Wang JM, Wang CN,
Liu T, Liu DW and Wang MW: Lysine 92 amino acid residue of USP46, a
gene associated with 'behavioral despair' in mice, influences the
deubiquitinating enzyme activity. PLoS One. 6:e262972011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dang LC, Melandri FD and Stein RL: Kinetic
and mechanistic studies on the hydrolysis of ubiquitin C-terminal
7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry.
37:1868–1879. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yin ST, Huang H, Zhang YH, Zhou ZR, Song
AX, Hong FS and Hu HY: A fluorescence assay for elucidating the
substrate specificities of deubiquitinating enzymes. Biochem
Biophys Res Commun. 416:76–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Timms KM, Ansari-Lari MA, Morris W, Brown
SN and Gibbs RA: The genomic organization of isopeptidase T-3
(ISOT-3), a new member of the ubiquitin specific protease family
(UBP). Gene. 217:101–106. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stein RL, Chen Z and Melandri F: Kinetic
studies of isopeptidase T: modulation of peptidase activity by
ubiquitin. Biochemistry. 34:12616–12623. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lacombe T and Gabriel JM: Further
characterization of the putative human isopeptidase T catalytic
site. FEBS Lett. 531:469–474. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reyes-Turcu FE, Horton JR, Mullally JE,
Heroux A, Cheng X and Wilkinson KD: The ubiquitin binding domain
ZnF UBP recognizes the C-terminal diglycine motif of unanchored
ubiquitin. Cell. 124:1197–1208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang YH, Zhou CJ, Zhou ZR, Song AX and Hu
HY: Domain analysis reveals that a deubiquitinating enzyme USP13
performs non-activating catalysis for Lys63-linked polyubiquitin.
PLoS One. 6:e293622011. View Article : Google Scholar
|
|
15
|
Bonnet J, Romier C, Tora L and Devys D:
Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem
Sci. 33:369–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Catic A, Fiebiger E, Korbel GA, Blom D,
Galardy PJ and Ploegh HL: Screen for ISG15-crossreactive
deubiquitinases. PLoS One. 2:e6792007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Soetandyo N, Lee JG, Liu L, Xu Y,
Clemons WM Jr and Ye Y: USP13 antagonizes gp78 to maintain
functionality of a chaperone in ER-associated degradation. eLife.
3:e013692014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scortegagna M, Subtil T, Qi J, Kim H, Zhao
W, Gu W, Kluger H and Ronai ZA: USP13 enzyme regulates Siah2 ligase
stability and activity via noncatalytic ubiquitin-binding domains.
J Biol Chem. 286:27333–27341. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yeh HM, Yu CY, Yang HC, Ko SH, Liao CL and
Lin YL: Ubiquitin-specific protease 13 regulates IFN signaling by
stabilizing STAT1. J Immunol. 191:3328–3336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen M, Gutierrez GJ and Ronai ZA:
Ubiquitin-recognition protein Ufd1 couples the endoplasmic
reticulum (ER) stress response to cell cycle control. Proc Natl
Acad Sci USA. 108:9119–9124. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao X, Fiske B, Kawakami A, Li J and
Fisher DE: Regulation of MITF stability by the USP13
deubiquitinase. Nat Commun. 2:4142011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L,
Cai Y, Norberg HV, Zhang T, Furuya T, et al: Beclin1 controls the
levels of p53 by regulating the deubiquitination activity of USP10
and USP13. Cell. 147:223–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Zhang P, Wei Y, Piao HL, Wang W,
Maddika S, Wang M, Chen D, Sun Y, Hung MC, et al: Deubiquitylation
and stabilization of PTEN by USP13. Nat Cell Biol. 15:1486–1494.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geng J, Huang X, Li Y, Xu X, Li S, Jiang
D, Liang J, Jiang D, Wang C and Dai H: Down-regulation of USP13
mediates phenotype transformation of fibroblasts in idiopathic
pulmonary fibrosis. Respir Res. 16:1242015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu YL, Zheng J, Tang LJ, Han W, Wang JM,
Liu DW and Tian QB: The deubiquitinating enzyme activity of USP22
is necessary for regulating HeLa cell growth. Gene. 572:49–56.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian QB, Okano A, Nakayama K, Miyazawa S,
Endo S and Suzuki T: A novel ubiquitin-specific protease, synUSP,
is localized at the post-synaptic density and post-synaptic lipid
raft. J Neurochem. 87:665–675. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang LJ, Li Y, Liu YL, Wang JM, Liu DW and
Tian QB: USP12 regulates cell cycle progression by involving c-Myc,
cyclin D2 and BMI-1. Gene. 578:92–99. 2016. View Article : Google Scholar
|
|
28
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang W: Quantitative analysis of plasma
membrane proteome using two-dimensional difference gel
electrophoresis. Methods Mol Biol. 876:67–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shirano Y and Shibata D: Low temperature
cultivation of Escherichia coli carrying a rice lipoxygenase L-2
cDNA produces a soluble and active enzyme at a high level. FEBS
Lett. 271:128–130. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kataeva I, Chang J, Xu H, Luan CH, Zhou J,
Uversky VN, Lin D, Horanyi P, Liu ZJ, Ljungdahl LG, et al:
Improving solubility of Shewanella oneidensis MR-1 and Clostridium
thermocellum JW-20 proteins expressed into Esherichia coli. J
Proteome Res. 4:1942–1951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Volontè F, Marinelli F, Gastaldo L, Sacchi
S, Pilone MS, Pollegioni L and Molla G: Optimization of
glutaryl-7-aminocephalosporanic acid acylase expression in E coli.
Protein Expr Purif. 61:131–137. 2008. View Article : Google Scholar
|
|
33
|
Chou CP: Engineering cell physiology to
enhance recombinant protein production in Escherichia coli. Appl
Microbiol Biotechnol. 76:521–532. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Larsen CN, Krantz BA and Wilkinson KD:
Substrate specificity of deubiquitinating enzymes: ubiquitin
C-terminal hydrolases. Biochemistry. 37:3358–3368. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kolkman A, Dirksen EH, Slijper M and Heck
AJ: Double standards in quantitative proteomics: direct comparative
assessment of difference in gel electrophoresis and metabolic
stable isotope labeling. Mol Cell Proteomics. 4:255–266. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Levy C, Khaled M and Fisher DE: MITF:
master regulator of melanocyte development and melanoma oncogene.
Trends Mol Med. 12:406–414. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ziegler WH, Liddington RC and Critchley
DR: The structure and regulation of vinculin. Trends Cell Biol.
16:453–460. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Das C, Hoang QQ, Kreinbring CA, Luchansky
SJ, Meray RK, Ray SS, Lansbury PT, Ringe D and Petsko GA:
Structural basis for conformational plasticity of the Parkinson's
disease-associated ubiquitin hydrolase UCH-L1. Proc Natl Acad Sci
USA. 103:4675–4680. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Frisan T, Coppotelli G, Dryselius R and
Masucci MG: Ubiquitin C-terminal hydrolase-L1 interacts with
adhesion complexes and promotes cell migration, survival, and
anchorage independent growth. FASEB J. 26:5060–5070. 2012.
View Article : Google Scholar : PubMed/NCBI
|