Introduction
Breast cancer is the most common type of cancer
affecting women and a leading cause of mortality for females
world-wide (1,2). However, several studies have
reported that microRNAs (miRNAs or miRs) are involved in regulating
gene expression, as well as regulating diverse physiological and
pathological processes (3,4).
miRNAs are short, highly conserved small non-coding RNA molecules
of 19–25 nucleotides in length that regulate gene expression at the
post-transcriptional level. By targeting complementary binding
sites within the 3′ untranslated region (3′UTR) of target messenger
RNAs (mRNAs), they impair or inhibit translation and promote
degradation (5–8). Similarly, miR-30a is situated on
chromosome 6q.13 and is produced by an intronic transcriptional
unit (9,10). Two mature forms of miR-30a exist,
miR-30a-3p and miR-30a-5p. miR-30a is deregulated in several
malignant tumors, such as breast cancer (11), hepatocellular cancer (12), colon cancer (13), nasopharyngeal carcinoma (14), prostate cancer (15), endometrial cancer (16) and cutaneous squamous cell
carcinoma (17). Moreover,
miR-30a has been shown to be a potential prognostic marker in
breast cancer (18).
The Notch signaling pathway is highly conserved and
plays an important role in intercellular signaling and
developmental processes; it includes NOTCH ligands (JAG1, JAG2,
DLL1, DLL3 and DLL4) (19,20),
Notch receptors (Notch1, −2, −3 and −4), and the downstream target
genes, hairy and enhancer of split-1 (HES1) and cyclin D1, and
FADD-like apoptosis regulator (CFLAR) (21–23). Previous studies have confirmed
that the overexpression of Notch1 is associated with cancer,
particularly breast cancer, cancer cell proliferation and
apoptosis, as well as the promotion of migration and invasion
(24,25).
However, the function and mechanisms of action of
miR-30a in breast cancer are not yet fully understdood, nor is its
association with the Notch1 target gene. Thus, in this study, we
aimed to shed light into this matter. We propose a logical
hypothesis: miR-30a may mediate Notch1, and thus regulating its
biological function.
Materials and methods
Cell culture
The human breast cancer cell lines, MCF-7 and
MDA-MB-231, were purchased from the American Type Culture
Collection (ATCC, Rockville, MD, USA). All cell lines were cultured
in RPMI-1640 (Keygen, Nanjing China), supplemented with 10% fetal
bovine serum (FBS; Gibco-Life Technologies, Carlsbad, CA, USA) and
kept in a humidified atmosphere containing 5% CO2 at
37°C.
Cell transfection
Human hsa-miR-30a mimic and hsa-miR negative control
were purchased from Genepharma (Shanghai, China). Small interfering
RNA (siRNA) against human Notch1 (si-Notch1), negative control
siRNA (NC-siRNA) and si-h-GAPDH were purchased from RiboBio Co.,
Guangzhou, China. At 24 h prior to transfection, the MCF-7 and
MDA-MB-231 cells were plated in 6-well plates (2.5×105
cell/well), and then transfected with miR-30a mimic (at 100 nM
final concentration; Genepharma), or miR negative control (100 nM;
Genepharma) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA,
USA). NC-siRNA at 100 nM, si-Notch1 or si-h-GAPDH were delivered
into the cells following the manufacturer's instructions. The cells
transfected with the miRNA mimic or siRNA were harvested 12–48 h
post-transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen). RT-PCR was carried out using the
TaqMan® MicroRNA Reverse Transcription kit (Applied
Biosystems; Life Technologies, Carlsbad, CA, USA). Mature miRNA was
spotted using the TaqMan® MicroRNA assay kit (assay ID:
miR-30a: 000417 and RNU6B: 001093) (Applied Biosystems; Life
Technologies). All procedures were performed according to the
manufacturer's instructions. The primers used were as follows:
Notch1 forward, 5′-CACTGTGGGCGGGTCC-3′ and reverse,
5′-GTTGTATTGGTTCGGCACCAT-3′. Relative expression levels were
calculated using the ΔΔCt method and normalized to RNU6B expression
(fold difference relative to RNU6B). The quantitative analysis of
the changes in expression levels were calculated using the ABI 7300
real-time PCR machine (Applied Biosystems). All reverse
transcription and PCR assays were carried out in triplicate.
Cell proliferation assay
At 24 h following the transfection of miRNA mimics,
the cells were seeded into 96-well plates (5×103
cells/well). Subsequently, 10 µl of cell counting kit-8
assay solution (CCK-8; Dojindo, Kumamoto Prefecture, Kyushu, Japan)
were added to the cultured cells in 100 µl of culture medium
and incubated in a humidified atmosphere for 1 h at 37°C; each
experiment was performed in triplicate. The absorbance was measured
at 570 nm using CliniBio 128 (ASYS-Hitech, Eugendorf, Austria).
Cell apoptosis assay
The MCF-7 and MDA-MB-231 cells were transfected with
miR-30a mimic or negative control for 48 h. Subsequently, the cells
were trypsinized and collected by centrifugation at 1,000 rpm for 5
min. The cell pellets were rinsed twice with ice-cold
phosphate-buffered saline (PBS). Apoptosis was determined by dual
staining using the FITC Annexin V apoptosis detection kit I (BD
Biosciences, Franklin Lakes, NJ, USA) according to the
manufacturer's instructions. The experiment was carried out in
triplicate.
Cell migration assays
To examine cell migration, tissue culture inserts
(6.5 mm diameter) with an 8.0 µm pore size (Transwell;
Corning, Inc., Corning, NY, USA) were placed into the wells of
24-well culture plates, separating the upper and the lower
chambers. The cells transfected with either miR-30a mimics or
negative control miRNA were cultured in 6-well plates, and the
cells were starved for 24 h. The cells (1×105
cells/well) suspended in 200 µl RPMI-1640 serum-free medium
were added to the upper chamber and 600 µl RPMI-1640 medium
supplemented with 20% FBS was placed into the lower chamber.
Following 24 h of incubation in a humidified atmosphere, the cells
that migrated through the Transwell membrane were fixed in 4%
paraformaldehyde, stained with crystal violet (Beyotime, Shanghai,
China), and 4 random fields were counted under a microscope
(magnification, ×100; CFM-500; Carl Zeiss, Göttingen, Germany).
Cell invasion assay
Cell invasion was carried out using
Matrigel® Basement Membrane Matrix (Corning, Inc.). The
cells transfected with either miR-30a mimics or NC miRNA were
cultured in 6-well plates and then starved for 24 h. The cells
(1×105 cells/well) suspended in 200 µl RPMI-1640
serum-free medium were added to the upper chamber and 600 µl
RPMI-1640 medium supplemented with 20% FBS was placed into the
lower chamber. Following 24 h of incubation, the cells that had
invaded the Matrigel membrane were fixed in 4% paraformaldehyde,
stained with crystal violet, and 4 random fields counted under a
microscope (magnification, ×100). The experiment was performed in
triplicate.
miRNA target gene identification
The prediction of miRNA target genes was carried out
using TargetScan (http://www.targetscan.org), Pictar (http://pictar.mdc-berlin.de), microRNA (http://www.microrna.org).
Dual luciferase activity assay
The wild-type and mutant seed region of Notch1
containing the putative target site for Pre-miR-30a was synthesized
and cloned into the PGL3-promoter vector (Life Technologies). The
MCF-7 cells (5×104 cells/well) were incubated in 24-well
plates. miR-30a mimic or control mimic, PGL3-Notch1 3′UTR-WT vector
or PGL3-Notch1 3′UTR-MUT vector containing Firefly luciferase
reporter gene and 3′UTR of Notch1 gene (Promega, Madison, WI, USA)
were co-transfected into the cells using Lipofectamine 2000
(Invitrogen). After 48 h of transfection, luciferase activity was
analyzed by using the Dual Luciferase Reporter assay system
(Promega) and normalized to Renilla luciferase activity.
Western blot analysis
Total protein was extracted and lysed using RIPA
buffer (Beyotime). Equal amounts of protein were separated by
NuPAGE® LDS Sample Buffer (4X) and NuPAGE®
Reducing Agent (10X) (both from Life Technologies). A total of 20
µl protein from each sample was used for western blot
analysis. Initially, protein samples were separated using a 5% MOPS
SDS running buffer (20X) (Life Technologies), and they were
subsequently transferred onto polyvinylidene difluoride membranes
(Sigma, Deisenhofen, Germany). The membranes were then washed with
TBST, blocked with western blocking buffer (Beyotime) and incubated
with primary antibodies against human Notch1 (1:2,000; ab8925;
Abcam, Cambridge, MA, USA) overnight at 4°C. The membranes were
then washed with TBST and incubated with the goat anti-rabbit IgG
peroxidase-conjugated secondary antibody (1:5,000; BS10650;
Bioworld, Nanjing, China). The protein band was detected by
chemiluminescence with Pierce ECL kits (Millipore, Billerica, MA,
USA). β-actin (1:5,000; AP0060; Bioworld) was used as an internal
loading control to normalize the expression patterns of each
sample. Three separate experiments were performed and only
representative images are shown.
Patients and samples
Samples were acquired from patients (n=20) who were
diagnosed at the Jiangsu Cancer Hospital, Affiliated Hospital of
Nanjing Medical University, from 2012–2015 (Table I). All the patients provided
written informed consent and the study was approved by the Ethics
Committee of Jiangsu Cancer Hospital (Nanjing, China).
 | Table IClinicopathological characteristics
of 20 patients with breast cancer. |
Table I
Clinicopathological characteristics
of 20 patients with breast cancer.
| Clinicopathologic
parameters | No. of
patients |
|---|
| Age | 32–60 |
| Tumor diameter | |
| T1 | 3 |
| T2 | 15 |
| T3 | 2 |
| Lymph node | |
| Negative | 4 |
| Positive | 16 |
|
Histological-pathological types | |
| Invasive ductal
carcinoma | 18 |
| Invasive lobular
carcinoma | 2 |
| Clinical stage | |
| I–II | 6 |
| III | 14 |
| Hormone
receptor | |
| Negative | 7 |
| Positive | 13 |
| Her-2 | |
| Negative | 9 |
| Positive | 11 |
In vivo tumorigenicity
Female BALB/c nude mice were purchased from the
Experimental Animal Center of the Academy of Military Sciences,
China (serial no. 0027549). Female BALB/c nude mice (6–8 weeks old)
were separately caged in a standard environment. MDA-MB-231 cells
transfected with either the negative control or miR-30a mimic
(1×107 cells, 0.1 ml) were injected subcutaneously into
the right axillary lymph nodes of 6–8-week-old BALB/c nude mice (5
mice/group). Tumor growth rate was evaluated by measuring tumor
diameters and the tumor growth curve was recorded accordingly. Both
the maximum (L) and minimum (W) length of the tumor were recorded
using a slide caliper, and the tumor volume was calculated as ½LW2.
After 36 days, all the mice were sacrificed. All animal experiments
were approved by the Institutional Review Board of Nanjing
University of Chinese Medicine (Nanjing, China).
Statistical analysis
All experiments were performed in triplicate and
representative data are shown from 3 separate experiments.
Statistical analysis was performed using a t-test or one-way ANOVA
and Spearman's rank test using SPSS 16.0 software. A value of
p<0.05 was considered to indicate a statistically significant
difference.
Results
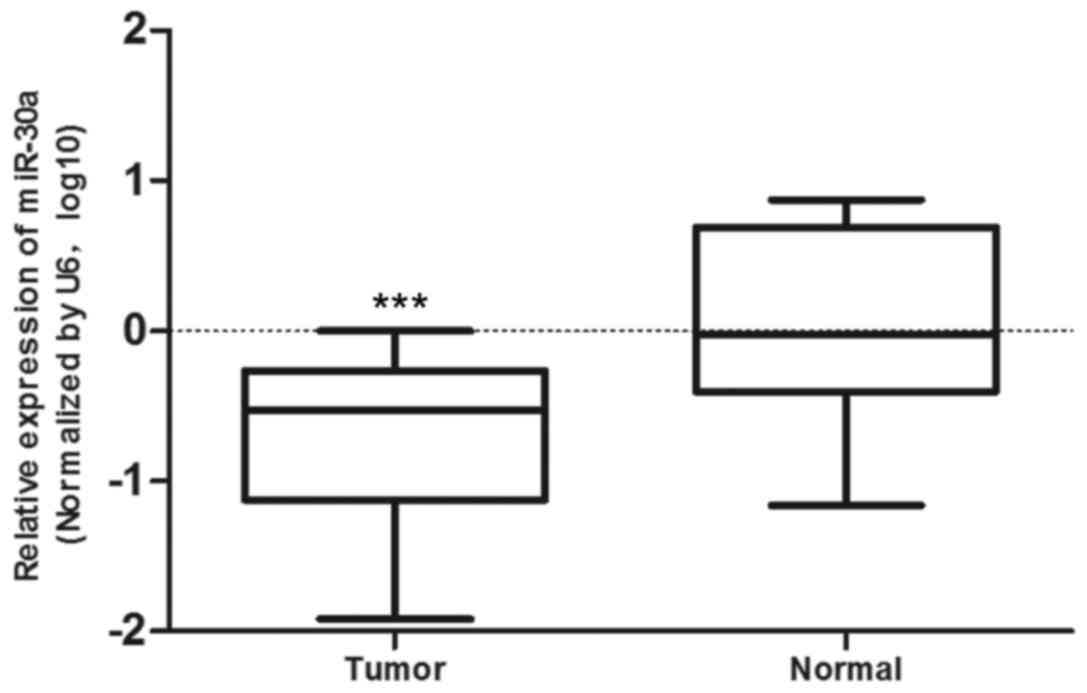
miR-30a is frequently downregulated in
breast cancer tissues
To research the expression of miR-30a in breast
cancer tissues and cell lines, we first compared the level of
miR-30a expression in 20 paired breast tumor tissues and adjacent
normal tissues by RT-qPCR (Table
I); From the results, we confirmed that miR-30a expression was
significantly downregulated in the tumor tissue compared to the
adjacent normal tissue (p<0.0005, means ± SD) (Fig. 1).
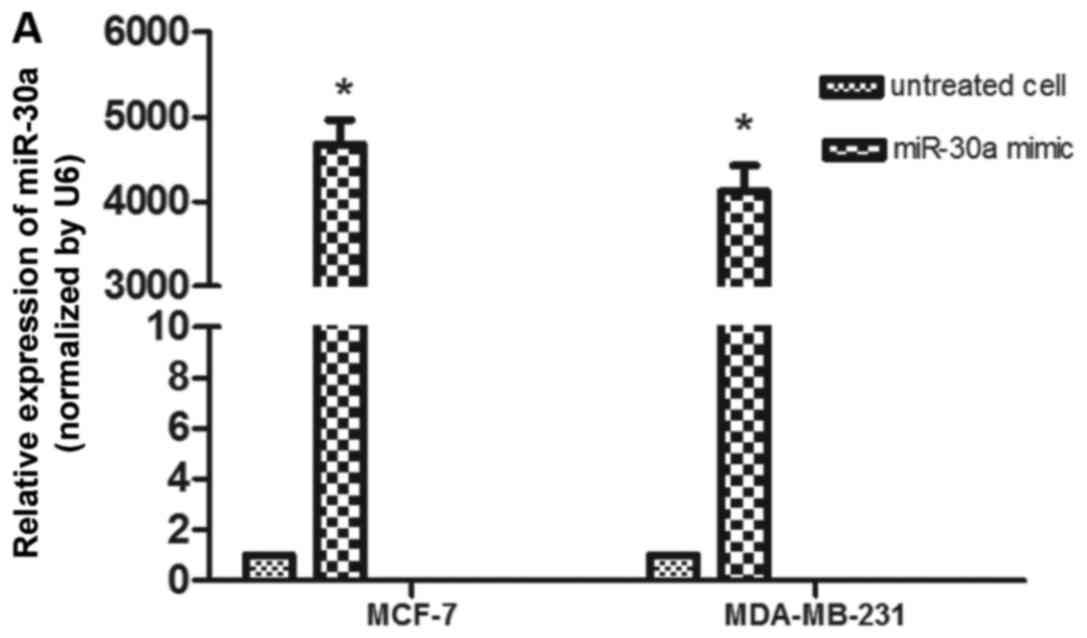
miR-30a inhibits the viability of breast
cancer cells in vitro and in vivo
The frequent downregulation of miR-30a in breast
cancer tissues indicated that miR-30a may play a role in the
development of breast cancer. In order to determine this, miR-30a
mimic and negative control were successfully transfected into MCF-7
and MDA-MB-231 cells. At 48 h after transfection, we performed
RT-qPCR to determine the expression level of miR-30a (Fig. 2A), and then a CCK-8 assay was
performed. As shown in Fig. 2B and
C, these assays confirm that the MCF-7 and MDA-MB-231 cells
transfected with miR-30a mimic exhibited a significant decrease in
cell proliferation compared to the cells transfected with the
negative control. To further confirm the growth inhibitory effect
of miR-30a on breast cancer cells in vivo, a xenograft tumor
growth assay was performed. We found that the tumors from the mice
injected with the miR-30a mimic-transfected cells were much smaller
in size (Fig. 2F). In addition,
as shown by the growth curve of the subcutaneous tumors, the tumors
from the mice injected with the miR-30a mimic-transfected cells had
a significantly lower growth rate compared to those from mice
injected with the negative control-transfected cells (Fig. 2D). The tumor volume was also
significantly lower in the nude mice injected with miR-30a
mimic-transfected cells as compared to that of those injected with
negative control-transfected cells (p<0.01; Fig. 2E).
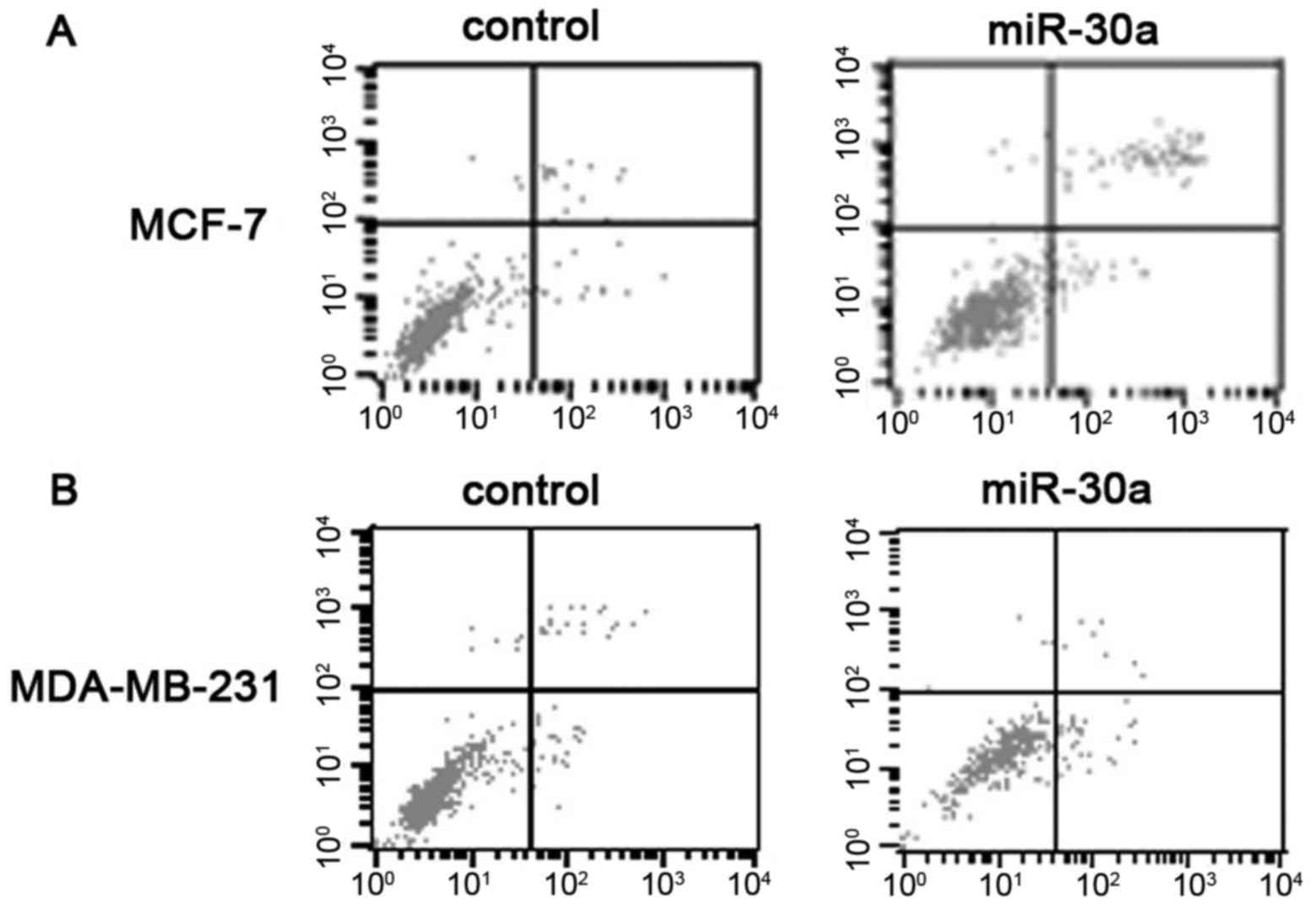
miR-30a induces the apoptosis of MCF-7
and MDA-MB-231 cells
We used the rate of cellular apoptosis to determine
the connection between miR-30a and apoptosis. In the MCF-7 cells,
the upregulation of miR-30a increased apoptosis, as compared with
the negative controls (Fig. 3A and
C). Similarly, as shown in Fig.
3B in the MDA-MB-231 cells, miR-30a also increased apoptosis,
as compared with the negative controls (Fig. 3B and C; Data represent the average
of apoptotic cells, respectively p<0.05, mean ± SD).
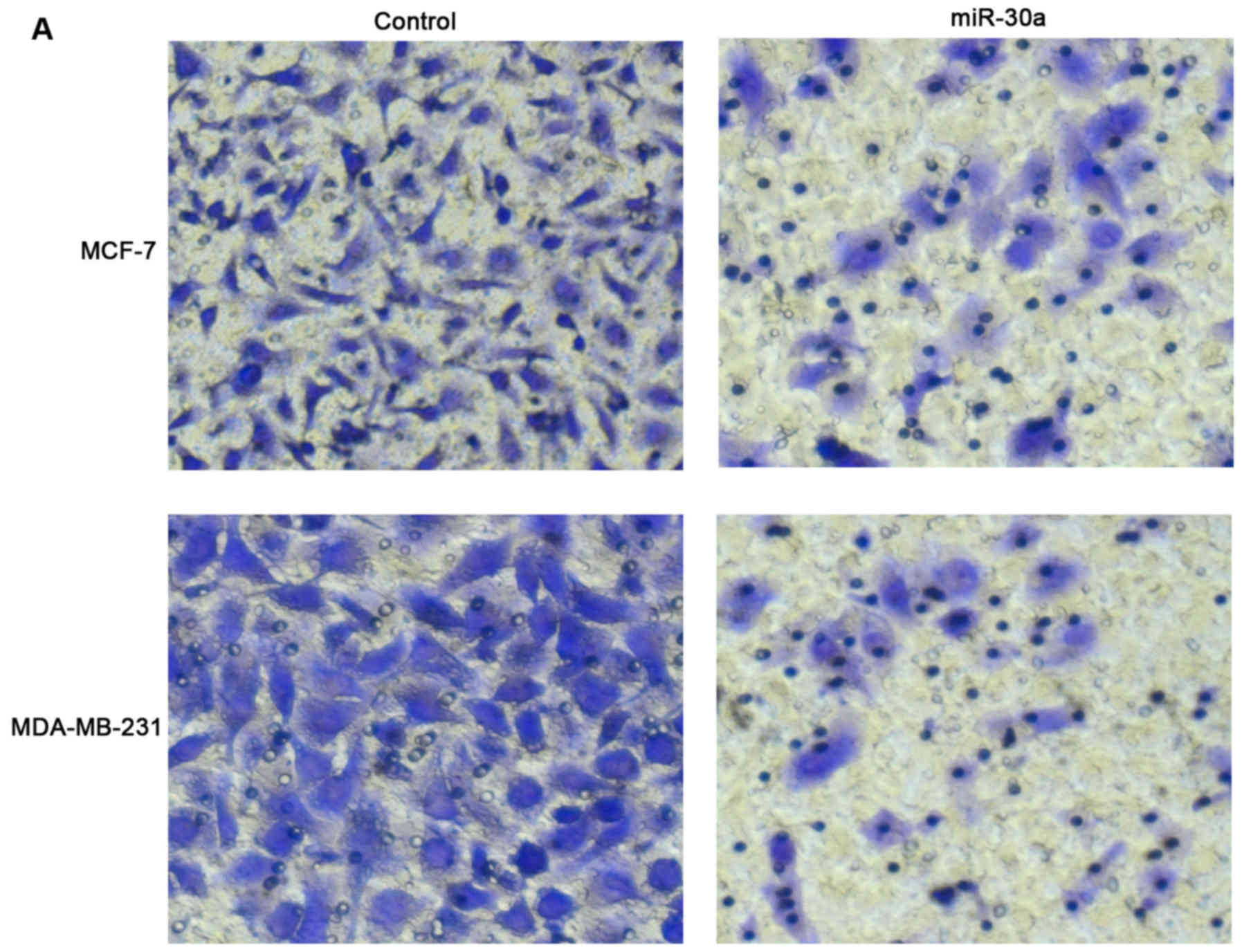
miR-30a inhibits breast cancer cell
migration and invasion in vitro
To verify the effects of miR-30a on the metastatic
ability of breast cancer cells, we transfected the MCF-7 and
MDA-MB-231 cells with miR-30a mimic and observed a significant
decrease in the number of migrated cells compared with the negative
control (Fig. 4A and C). We also
examinedthe effect of miR-30a on the invasion of breast cancer
cells. The MCF-7 and MDA-MB-231 cells transfected with miR-30a
mimic exhibited a markedly inhibited invasive ability (Fig. 4B and D).
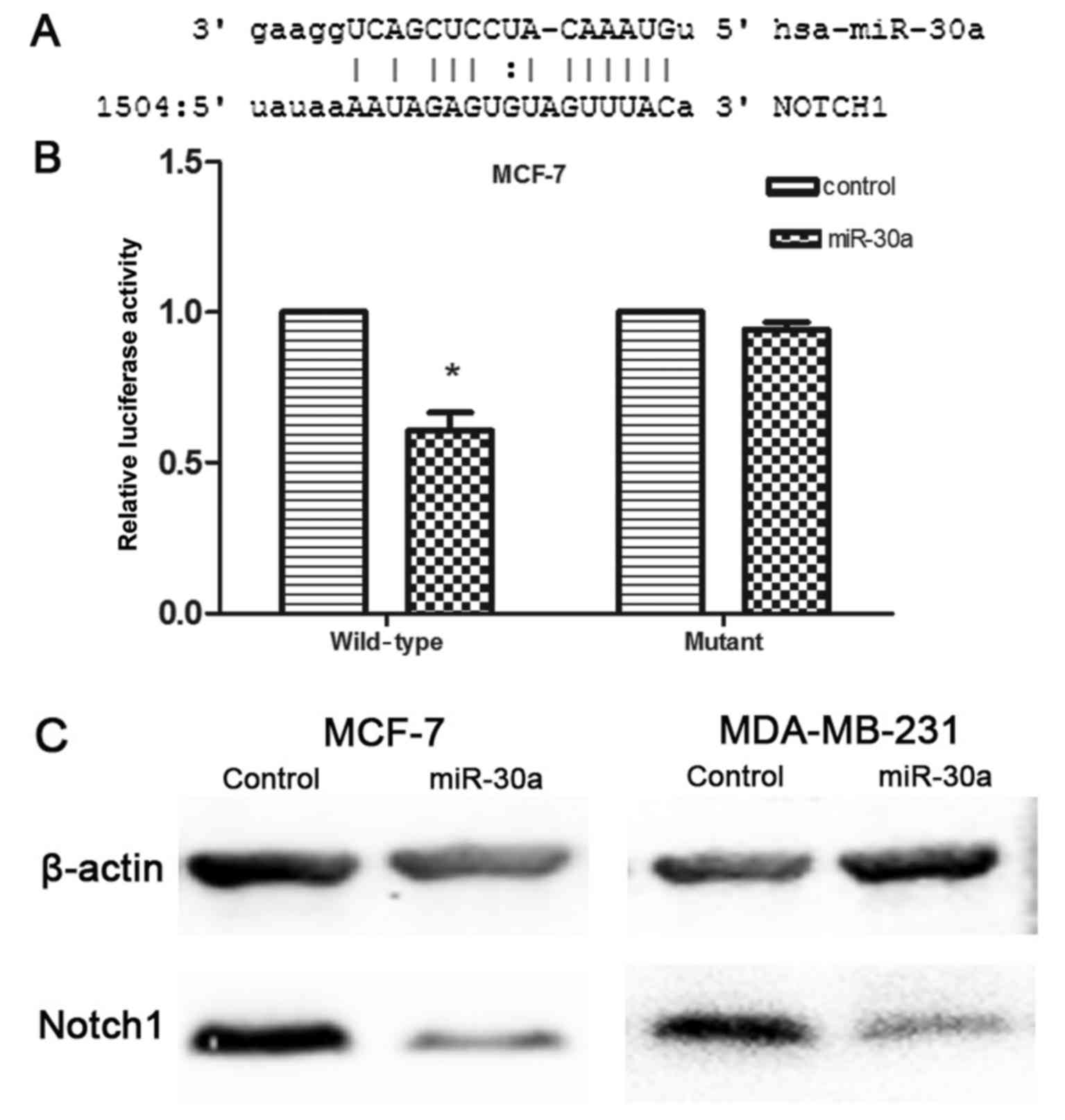
Notch1 is a direct target gene of miR-30a
in breast cancer
To explore the mechanisms through which miR-30a
executes its functions in breast cancer cells, we wished to
determine the potential target of miR-30a in breast cancer. We
found that Notch1 was a potential target of miR-30a, utilizing
three independently bioinformatic algorithms (TargetScan, Pictar
and microRNA). Thus, we examined the expression of Notch1 in the
cells transfected with miR-30a mimic or negative control. The
overexpression of miR-30a significantly decreased the protein
expression of Notch1, as shown by western blot analysis (Fig. 5C). This indirectly confirmed that
the expression level of Notch1 could be regulated by miR-30a.
Additionally, computational analysis revealed that the 3′UTR of
Notch1 contains a conserved binding site for miR-30a (Fig. 5A). To further confirm that miR-30a
directly and negatively regulates Notch1 expression, we constructed
luciferase reporter vectors that contained wild-type (Wt) and
mutant (Mt) miR-30a target sequences of the Notch1-3′UTR.
Co-transfection experiments in the breast cancer cells revealed
that miR-30a significantly inhibited the luciferase activity of Wt
Notch1-3′UTR reporter gene (p<0.05), but it failed to inhibit
the Mt Notch1-3′UTR reporter gene (Fig. 5B). Thus, we verified that Notch1
was a direct target of miR-30a in breast cancer cells.
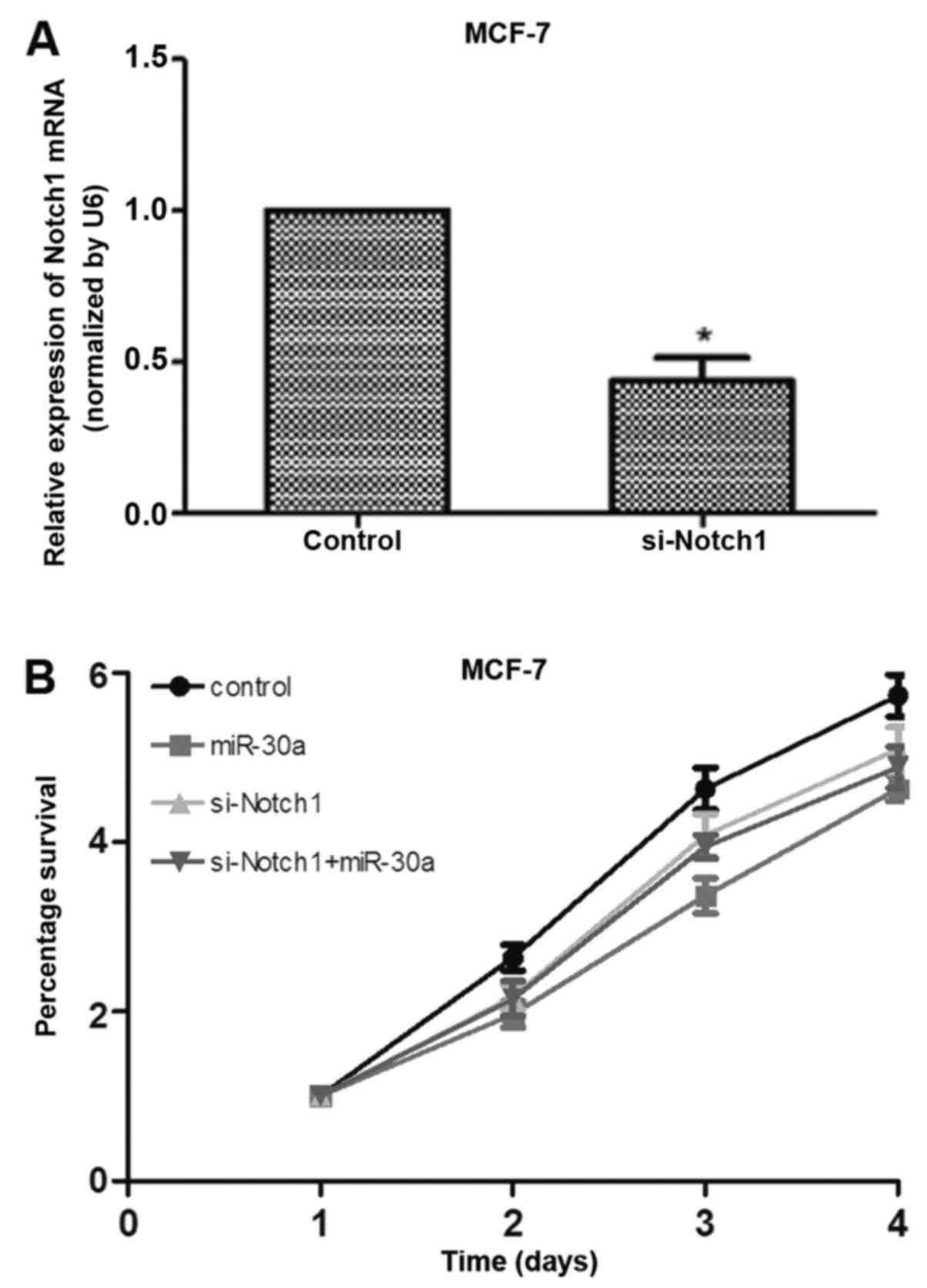
Notch1 knockdown significantly inhibits
breast cancer cell proliferation
To confirm that Notch1 directly regulates the
development of breast cancer, si-Notch1 was successfully
transfected into MCF-7 cells (Fig.
6A). The transfection of si-Notch1 significantly inhibited
breast cancer cell proliferation; however, compared with the cells
transfected with si-Notch1 only, cell proliferation did not
obviously change in the MCF-7 cells transfected with both si-Notch1
and miR-30a mimic, which implies that miR-30a inhibited cell
proliferation via Notch1 (Fig.
6B).
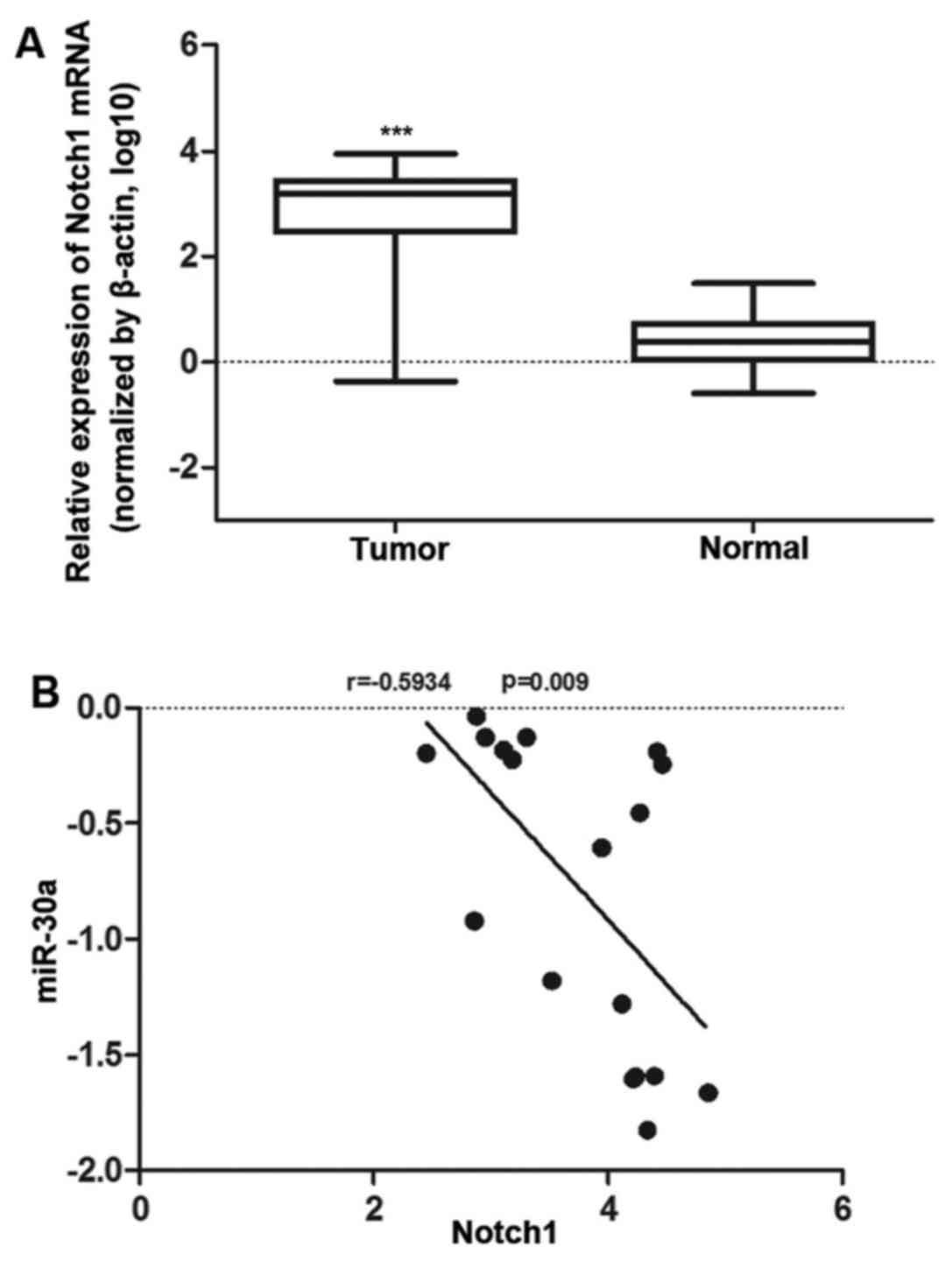
Notch1 inversely correlates with miR-30a
expression in vivo
To further confirm the correlation between Notch1
and miR-30a in tissues, we compared the level of Notch1 expression
and miR-30a expression in 20 paired breast tumor tissues and
adjacent normal tissues by RT-qPCR (Table I). The Notch1 expression levels
were significantly increased compared to the adjacent normal
tissues in breast cancer cells (Fig.
7B). Additionally, miR-30a expression was significantly
downregulated compared to the adjacent normal tissues (Fig. 1). Based on the mRNA expression of
Notch1 and miR-30a in breast tumor tissues, we revealed that there
was a significant inverse correlation, as calculated by linear
regression analysis (Spearman's rank test, r=−0.5934 p=0.009)
(Fig. 7C). These data further
confirmed that miR-30a targets Notch1.
Discussion
Accumulating studies have confirmed that miRNAs are
aberrantly expressed in a variety of human cancers; similarly, they
are involved in maintaining the balance of gene regulating networks
that control cell expression (11–14). Some studies have reported that
miRNAs not only play an important role in epigenetic changes, but
also effectively regulate protein expression by binding with target
mRNA 3′UTRs (26,27); moreover, aberrant miRNA expression
in tumors is involved in tumor growth, apoptosis and migration and
invasion, which may affect treatment and prognosis (11,14). miR-30a is situated on chromosome
6q.13, which has been confirmed to be a genomic fragile region due
to the loss of heterozygosity in breast cancer (28,29) and it is produced by an intronic
transcriptional unit (10).
miR-30a is deregulated in some malignant tumors, such as breast
cancer (11), hepatocellular
cancer (12), colon cancer
(13), nasopharyngeal carcinoma
(14), prostate cancer (15), endometrial cancer (16) and cutaneous squamous cell
carcinoma (17). Furthermore,
miR-30a has been shown to be significantly associated with the
prognosis of cancer (30,31).
The Notch signaling pathway is a highly conserved
cell signaling system in most multicellular organisms. Notch1 is
associated with cell transformation (32), cell cycle, progenitor/stem cell
maintenance and is aberrantly activated in breast cancer (33,34). Notch plays a key role in breast
cancer proliferation, apoptosis, differentiation and tumor
advance.
Our study confirmed the assumption that miR-30a
inhibits the development of breast cancer. Based on normal tissue
data, miR-30a exhibited a reduced expression in breast tumor
tissues compared with adjacent normal tissues, suggesting the
dysregulation of miR-30a in breast cancer tumorigenesis. This
suggested that miR-30a plays a tumor-suppressive role in breast
cancer. According to the cell proliferation assay, we discovered
that miR-30a significantly inhibited breast cancer cell growth.
Moreover, in nude mice, breast cancer cells over-expressing miR-30a
exhibited a significantly lower proliferation than the control
cancer cells. We thus hypothesized that the downregulation of
miR-30a may be associated with the clinical progression of breast
cancer, and miR-30a has been confirmed to inhibit proliferation in
a variety of tumor types, such as thyroid cancer, colon cancer
(13), liver cancer (12) and chronic myelogenous leukemia
(35). Furthermore, according to
flow cytometry, we also found that miR-30a overexpression induced
breast cancer cell apoptosis, and these results also suggested that
miR-30a may function as a tumor suppressor in breast cancer. It has
also been confirmed that miR-30a has the ability to inhibit the
invasion and metastasis of hepatocellular carcinoma via the
MTDH/PTEN/AKT pathway (36). In
this study, we demonstrated that miR-30a suppressed the metastatic
and invasive ability of MCF-7 and MDA-MB-231 breast cancer
cells.
Based on the databases for miRNA target prediction,
many miR-30a targets are common targets to other members of the
miR-30 family as they share the same seed sequence. To verify this,
we examined the effect of miR-30a on the translational regulation
of Notch1; that is to say, Notch1 serves as a critical effector of
miR-30a. From the dual-luciferase reporter assay, we found that
miR-30a significantly inhibited the luciferase activity of
Luc-Notch1-3′UTR by targeting the 3′UTR of Notch1 mRNA. However,
the dysregulated activity of Notch signaling is associated with
tumor proliferation, differentiation, apoptosis, invasion and
metastasis (37). In ths study,
transfection with si-Notch1 significantly inhibited breast cancer
cell proliferation; however, the overexpression of miR-30a did not
significantly inhibit cell proliferation in the MCF-7 cells
compared with the cells transfected with si-Notch1 only, which
implies that miR-30a inhibited cell proliferation via Notch1
(Fig. 6B). From the tissue data,
we found that Notch1 expression was significantly upregulated in
breast cancer tissues, which inversely correlated with miR-30a
(r=−0.5934, p=0.009), indicating a novel mechanism for the
regulation of Notch1 expression involving the miR-30a inhibition of
Notch1 expression at the translational level.
In conclusion, in this study, we found that miR-30a
is downregulation in breast cancer, and that it negatively
correlates with Notch1 expression, which is upregulated in breast
cancer. Our data suggest that miR-30a inhibited the proliferation,
invasion and metastasis, and induced the apoptosis of breast cancer
cells by targeting Notch1. These findings reveal the importance of
the miR-30a/Notch1 axis in breast cancer development and
progression.
Acknowledgments
This study was supported by grants from the National
High Technology Research and Development Program of China (no.
2014AA020604), the National Natural Science Foundation of China
(no. 81272470), the National Key Clinical Specialist Construction
Programs of China [no. 2013 (544)], the Major Program of Natural
Science Foundation of Jiangsu Province (no. BL2014090) and the
Natural Science Foundation of Jiangsu Province (no.
BK20151579).
References
|
1
|
Krishnan K, Steptoe AL, Martin HC,
Pattabiraman DR, Nones K, Waddell N, Mariasegaram M, Simpson PT,
Lakhani SR, Vlassov A, et al: miR-139-5p is a regulator of
metastatic pathways in breast cancer. RNA. 19:1767–1780. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Q, Kang X, Yang B, Wang J and Yang
F: Antiangiogenic effect of capecitabine combined with ginsenoside
Rg3 on breast cancer in mice. Cancer Biother Radiopharm.
23:647–653. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brown RH: Medicine. A reinnervating
microRNA. Science. 326:1494–1495. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo H, Hu X, Ge S, Qian G and Zhang J:
Regulation of RAP1B by miR-139 suppresses human colorectal
carcinoma cell proliferation. Int J Biochem Cell Biol.
44:1465–1472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen WX, Hu Q, Qiu MT, Zhong SL, Xu JJ,
Tang JH and Zhao JH: miR-221/222: Promising biomarkers for breast
cancer. Tumour Biol. 34:1361–1370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang J, Zhang H, Chen L, Sun DW, Mao C,
Chen W, Wu JZ, Zhong SL, Zhao JH and Tang JH: β-elemene reverses
chemoresistance of breast cancer via regulating MDR-related
microRNA expression. Cell Physiol Biochem. 34:2027–2037. 2014.
View Article : Google Scholar
|
|
7
|
Ratert N, Meyer HA, Jung M, Lioudmer P,
Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert
S, et al: miRNA profiling identifies candidate mirnas for bladder
cancer diagnosis and clinical outcome. J Mol Diagn. 15:695–705.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jia AY, Castillo-Martin M,
Domingo-Domenech J, Bonal DM, Sánchez-Carbayo M, Silva JM and
Cordon-Cardo C: A common MicroRNA signature consisting of miR-133a,
miR-139-3p, and miR-142-3p clusters bladder carcinoma in situ with
normal umbrella cells. Am J Pathol. 182:1171–1179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang R, Liang L, Luo D, Feng Z, Huang Q,
He R, Gan T, Yang L and Chen G: Downregulation of miR-30a is
associated with poor prognosis in lung cancer. Med Sci Monit.
21:2514–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J,
Cheng L, Fan Z, Yuan B, Tian P, et al: miR-30a suppresses breast
cancer cell proliferation and migration by targeting Eya2. Biochem
Biophys Res Commun. 445:314–319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai H, Kang B, Zuo D and Zuo G: Effect of
miR-30a-5p on the proliferation, apoptosis, invasion and migration
of SMCC-7721 human hepatocellular carcinoma cells. Zhonghua Gan
Zang Bing Za Zhi. 22:915–920. 2014.In Chinese.
|
|
13
|
Zhang Q, Tang Q, Qin D, Yu L, Huang R, Lv
G, Zou Z, Jiang XC, Zou C, Liu W, et al: Role of microRNA 30a
targeting insulin receptor substrate 2 in colorectal tumorigenesis.
Mol Cell Biol. 35:988–1000. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang HY, Li YY, Fu S, Wang XP, Huang MY,
Zhang X, Shao Q, Deng L, Zeng MS, Zeng YX, et al: MicroRNA-30a
promotes invasiveness and metastasis in vitro and in vivo through
epithelial-mesenchymal transition and results in poor survival of
nasopharyngeal carcinoma patients. Exp Biol Med (Maywood).
239:891–898. 2014. View Article : Google Scholar
|
|
15
|
Katz B, Reis ST, Viana NI, Morais DR,
Moura CM, Dip N, Silva IA, Iscaife A, Srougi M and Leite KR:
Comprehensive study of gene and microRNA expression related to
epithelialmesenchymal transition in prostate cancer. PLoS One.
9:e1137002014. View Article : Google Scholar
|
|
16
|
Tsukamoto O, Miura K, Mishima H, Abe S,
Kaneuchi M, Higashijima A, Miura S, Kinoshita A, Yoshiura K and
Masuzaki H: Identification of endometrioid endometrial
carcinoma-associated microRNAs in tissue and plasma. Gynecol Oncol.
132:715–721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sand M, Skrygan M, Georgas D, Sand D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Microarray analysis of
microRNA expression in cutaneous squamous cell carcinoma. J
Dermatol Sci. 68:119–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng CW, Wang HW, Chang CW, Chu HW, Chen
CY, Yu JC, Chao JI, Liu HF, Ding SL and Shen CY: MicroRNA-30a
inhibits cell migration and invasion by downregulating vimentin
expression and is a potential prognostic marker in breast cancer.
Breast Cancer Res Treat. 134:1081–1093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brennan K and Brown AM: Is there a role
for Notch signalling in human breast cancer? Breast Cancer Res.
5:69–75. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mittal S, Subramanyam D, Dey D, Kumar RV
and Rangarajan A: Cooperation of Notch and Ras/MAPK signaling
pathways in human breast carcinogenesis. Mol Cancer. 8:1282009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Won HY, Lee JY, Shin DH, Park JH, Nam JS,
Kim HC and Kong G: Loss of Mel-18 enhances breast cancer stem cell
activity and tumorigenicity through activating Notch signaling
mediated by the Wnt/TCF pathway. FASEB J. 26:5002–5013. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dunwoodie SL, Henrique D, Harrison SM and
Beddington RS: Mouse Dll3: A novel divergent Delta gene which may
complement the function of other Delta homologues during early
pattern formation in the mouse embryo. Development. 124:3065–3076.
1997.PubMed/NCBI
|
|
23
|
Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu
CW, Wang K, Zheng S, Ng SS, Chan FK, et al: microRNA-139-5p exerts
tumor suppressor function by targeting NOTCH1 in colorectal cancer.
Mol Cancer. 13:1242014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sharma A, Paranjape AN, Rangarajan A and
Dighe RR: A monoclonal antibody against human Notch1 ligand-binding
domain depletes subpopulation of putative breast cancer stem-like
cells. Mol Cancer Ther. 11:77–86. 2012. View Article : Google Scholar
|
|
25
|
Al-Hussaini H, Subramanyam D, Reedijk M
and Sridhar SS: Notch signaling pathway as a therapeutic target in
breast cancer. Mol Cancer Ther. 10:9–15. 2011. View Article : Google Scholar
|
|
26
|
Bao W, Fu HJ, Xie QS, Wang L, Zhang R, Guo
ZY, Zhao J, Meng YL, Ren XL, Wang T, et al: HER2 interacts with
CD44 to upregulate CXCR4 via epigenetic silencing of microRNA-139
in gastric cancer cells. Gastroenterology. 141:2076–2087. 2011.
View Article : Google Scholar
|
|
27
|
Au SL, Wong CC, Lee JM, Fan DN, Tsang FH,
Ng IO and Wong CM: Enhancer of zeste homolog 2 epigenetically
silences multiple tumor suppressor microRNAs to promote liver
cancer metastasis. Hepatology. 56:622–631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chappell SA, Walsh T, Walker RA and Shaw
JA: Loss of heterozygosity at chromosome 6q in preinvasive and
early invasive breast carcinomas. Br J Cancer. 75:1324–1329. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Noviello C, Courjal F and Theillet C: Loss
of heterozygosity on the long arm of chromosome 6 in breast cancer:
Possibly four regions of deletion. Clin Cancer Res. 2:1601–1606.
1996.PubMed/NCBI
|
|
30
|
Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu
Z, Hu G and Yang Q: MicroRNA-30a suppresses breast tumor growth and
metastasis by targeting metadherin. Oncogene. 33:3119–3128. 2014.
View Article : Google Scholar
|
|
31
|
Jiang D, Zheng X, Shan W and Shan Y: The
overexpression of miR-30a affects cell proliferation of
chondrosarcoma via targeting Runx2. Tumour Biol. 37:5933–5940.
2016. View Article : Google Scholar
|
|
32
|
Ronchini C and Capobianco AJ: Notch(ic)-ER
chimeras display hormone-dependent transformation, nuclear
accumulation, phosphorylation and CBF1 activation. Oncogene.
19:3914–3924. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bolós V, Mira E, Martínez-Poveda B, Luxán
G, Cañamero M, Martínez-A C, Mañes S and de la Pompa JL: Notch
activation stimulates migration of breast cancer cells and promotes
tumor growth. Breast Cancer Res. 15:R542013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Speiser J, Foreman K, Drinka E, Godellas
C, Perez C, Salhadar A, Erşahin Ç and Rajan P: Notch-1 and Notch-4
biomarker expression in triple-negative breast cancer. Int J Surg
Pathol. 20:139–145. 2012. View Article : Google Scholar
|
|
35
|
Liu Y, Song Y, Ma W, Zheng W and Yin H:
Decreased microRNA-30a levels are associated with enhanced ABL1 and
BCR-ABL1 expression in chronic myeloid leukemia. Leuk Res.
37:349–356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li WF, Dai H, Ou Q, Zuo GQ and Liu CA:
Overexpression of microRNA-30a-5p inhibits liver cancer cell
proliferation and induces apoptosis by targeting MTDH/PTEN/AKT
pathway. Tumour Biol. 37:5885–5895. 2016. View Article : Google Scholar
|
|
37
|
Song M, Yin Y, Zhang J, Zhang B, Bian Z,
Quan C, Zhou L, Hu Y, Wang Q, Ni S, et al: miR-139-5p inhibits
migration and invasion of colorectal cancer by downregulating AMFR
and NOTCH1. Protein Cell. 5:851–861. 2014. View Article : Google Scholar : PubMed/NCBI
|