Introduction
Cervical cancer (CC) is one of the most familiar
cancers and a leading cause of cancer-associated mortality in women
(1). Although the pathological
examination of cervical cancer tissue and improved early diagnosis
have significantly reduced the morbidity and mortality of this
disease in the past decades (2,3),
it remains an urgent problem in most developing countries due to
poor medical services. Up to now, the common treatments for CC are
still surgical procedures and radiotherapy. In addition, patients
who experience metastasis or recurrence are remedied by
chemotherapeutics (4). Although
the certain molecular mechanisms of cervical carcinogenesis have
been investigated (5), the
explicit developmental process of cervical carcinogenesis is still
mostly indistinct.
MicroRNAs (miRNAs or miRs) include a number of
endogenous small non-coding RNAs (approximately 18–22 nucleotides)
that serve important roles in controlling gene-targeted expression
at the post-transcriptional level by degradation of mRNA or
inhibition of translation (6).
Previously, miRNAs have been reported as important regulators of
many physiological and pathological processes, including cell
proliferation, cell differentiation, cell migration and invasion,
cell apoptosis and tumor metastasis. miRNAs are reported to
regulate target gene expression by binding to their 3′-untranslated
regions (3′-UTRs) (7,8). Previous studies have indicated that
miR-378 is upregulated and seems to function as an oncogene in
ovarian cancer (9), liver cancer
(10), human breast cancer
(11), nasopharyngeal carcinoma
(12), acute myeloid leukemia
(13), NSC differentiation
(14), human colorectal cancer
(15), gastric cancer (16) and renal cell carcinoma (17). However, the underlying clinical
roles and characteristic molecular mechanism of miR-378 in CC
remain unclear.
Suppression of tumorigenicity 7-like (ST7L) was
identified based on its similarity to the ST7 tumor suppressor gene
(18). ST7L is downregulated in
different cancers, including gastric cancer (19), glioma (20), ovarian cancer (21) and hepatocellular carcinoma
(22). For example, Chen et
al (20) demonstrated that
the deletion of miR-24 suppressed β-catenin/Tcf-4 transcription
activity by targeting ST7L in glioma. In addition, Yang et
al (21) reported that
upregulated miR-23a promoted cell malignant phenotype by targeting
ST7L in epithelial ovarian cancer cells. Recently, Zhuang et
al (22) indicated that ST7L
could interact with the carboxyl terminal region of AKT and
suppress AKT/GSK3β/β-catenin pathway in HCC cells. However, the
significance of ST7L in CC and the underlying mechanism has not
been elucidated.
In the present study, the authors demonstrated that
miR-378 might function as an oncogene by promoting cell growth,
accelerating the cell cycle and inhibiting cell apoptosis by
directly downregulating ST7L in HeLa and SiHa cells. Overexpression
of the miR-378-activated Wnt/β-catenin pathway in CC. Collectively,
these findings may provide insight into tumorigenesis and a
potential biomarker for CC.
Materials and methods
Materials
A total of 27 pairs of human cervical tissue,
consisting of human CC and matched normal cervical tissue from the
same patient, were used in the study. Written informed consent was
obtained from all enrolled patients, and all relevant
investigations were performed according to the principles of the
Declaration of Helsinki. The samples were received from the
Department of Oncology, Xintai Affiliated Hospital of Taishan
Medical University, Taian, China. Total RNA was extracted from the
human samples and purified using the miRVana miRNA Isolation kit
(Ambion, Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's instructions. The study was
approved by the Ethical Review Committee of Xintai Affiliated
Hospital of Taishan Medical University (Ethics approval number:
20150023).
Cell lines and transfection assay
Normal human endocervical epithelial cell lines
(Endl/E6E7) was obtained from Shanghai Medical College, Fudan
University (Shanghai, China), which were cultured in KER-SFM medium
supplemented with 10% calf serum (Biological Industries, Carlsbad,
CA, USA) at 37°C with 5% CO2. Other cervical cancer
cells used in the present study were obtained from the American
Type Culture Collection (ATCC; Manassas, VA, USA) and were
cultivated in RPMI-1640 (Invitrogen, Thermo Fisher Scientific)
supplemented with 8% fetal calf serum (FCS; Biological Industries),
100 U/ml penicillin, and 100 µg/ml streptomycinat 37°C in a
5% CO2 constant temperature incubator. A transfection
assay preceded using Lipofectamine™ 2000 reagent (Invitrogen,
Thermo Fisher Scientific) according to the protocol supplied by the
manufacturer (Invitrogen).
Plasmid construction
For miR-378, an overexpression vector (pri-miR-378)
containing a miR-378 precursor region was amplified from the
genomic DNA and inserted into the vector of pcDNA3. Pri-miR-378-S,
5′-CGACGCGTCGGGCTGCG AGGAGTGAGCG-3′ and Pri-miR-718-AS,
5′-CCATCGATGGGAGTTCAAATGGCTTGCTCC-3′. The 2′-O-methyl-modified
miR-378 antisense oligonucleotide (ASO-miR-378) was commercially
synthesized as an inhibitor of miR-378. ASO-miR-378,
5′-CCUUCUGACUCCAAGUCCAGU-3′ and ASO-NC,
5′-CAGUACUGUAGUGUAGUACTT-3′. The segment of 3′-UTR of ST7L
containing the miR-378 targets was acquired by polymerase chain
reaction with gene-specific primers, and then cloned into
pcDNA3/enhanced green fluorescent protein (EGFP) following the stop
codon of luciferase with BamHI and HindIII sites.
ST7L-3′-UTR-S, 5′-CGGGATTCGGTCAAAGAGAAAGAACTCTAATGTCCAGCTGCTC
CATCGA-3′ and ST7L-3′-UTR-AS,
5′-CGGAATTCTCGATGGAGCAGCTGGACATTAGAGTTCTTTCTCTTTGACC-3′.
ST7L-3′-UTR-mut-S,
5′-CGGGATTCGGTCAAAGAGAAAGAAGTATACTGCTGACCTGCTCCATCGA-3′ and
ST7L-3′-UTR-mut-AS, 5′-CGGAATTCTCGATGGAGCA
GCTGGTCAGCAGTATACTTTCTCTTTGACC-3′. The full-length sequences of
human ST7L cDNA (NM_017744.4) were obtained by reverse
transcriptase-quantitative polymerase chain reaction (RT-qPCR) and
were cloned into pcDNA3 at KpnI and EcoRI sites. The
resulting plasmid was termed pST7L. ST7L-S,
5′-CAGGGGTACCGCCACCATGGCGGACCGTGGCGGCGTG-3′ and ST7L-AS,
5′-CCGGAATTC GCCAGAACTCAAACCTAGGTCTTC-3′.
RNA isolation and detection
Total RNA was extracted with TRIzol reagent
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according to the
manufacturer's instructions. The quality and integrity of acquired
total RNA was evaluated by NanoDrop 2000c; Thermo Fisher
Scientific, Inc. (Wilmington, DE, USA) and 1% gel electrophoresis,
respectively. For RT-qPCR, 2 µg total RNA was reverse
transcribed with miR-378, U6 RT primers, or oligo-dT with M-MLV
reverse transcriptase. RT-qPCR was performed with kits, and
produced the following reaction: 2 µl RT products, 5 pmol
forward primer, 5 pmol reverse primer, 7.5 µl 2X SYBR-Green
buffer and nuclease-free water to 15 µl. The primers used
were followed by: miR-378-RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACGCCTTCT-3′;
miR-378-forward, 5′-TGCGGACUGGACUUGGAGUC-3′; U6-RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGA-3′;
U6-qPCR-forward, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′; Oligo-dT,
5′-TTTTTTTTTTTTTTTTTT-3′; β-actin-qPCR-forward,
5′-CGTGACATTAAGGAGAAGCTG-3′ and reverse, 5′-CT
AGAAGCATTTGCGGTGGAC-3′; Universal reverse qPCR primer,
5′-CCAGTGCAGGGTCCGAGGT-3′; ST7L-qPCR-forward,
5′-CGCGGATCCCCTCTGTGTGT GTGTGTGTAAC-3′ and reverse,
5′-CCGGAATTCGCATTCCTGGGCAGGTCGGT-3′.
MTT assay
Cervical cells were seeded into the plates of a
96-well plate at 5,000 cells/well one day prior to transfection.
The HeLa and SiHa cells were transfected with pri-miR-378,
ASO-miR-378, or the respective control vectors. Cell viability at
24, 48 and 72 h post-transfection was determined by MTT assay. The
absorbance values at 490 nm were measured via the Quant Microplate
spectrophotometer (BioTek Instruments, Inc., Winooski, VT,
USA).
Colony formation assay
For the colony formation ability assay, the HeLa and
SiHa cells were counted at 24 h post-transfection and seeded into
24-well plates at 500 cells/well. Culture medium was replaced every
72 h. After approximately two weeks, cells were cleaned with 1X
phosphate-buffered saline (PBS), stained with common crystal violet
dye, and colonies containing >50 cells were counted.
Prediction of miRNA targets
The hypothetical targets of miR-378 were predicted
using TargetScan 7.1, RNAhybrid and microRNA.org.
EGFP reporter assay
The EGFP reporter plasmids with ST7L 3′-UTR or ST7L
3′-UTR-mut were transfected into HeLa and SiHa cells with
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific)
and RFP expressing plasmid was integrated as a transfection
efficiency control. Cells were lysed 48 h post-transfection, and
the intensities of EGFP and RFP fluorescence were determined with a
spectrophotometer.
Western blot assay
Cell extracts were cleaned with 1X PBS buffer,
prepared with RIPA buffer supplemented with cocktail, and protein
concentrations were quantified using the BCA protein assay kit
(Beyotime Institute of Biotechnology, Shanghai, China) according to
the manufacturer's protocols. Equal amounts of proteins were
separated by 10% SDS-PAGE and subsequently transferred to
polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica,
MA, USA). The membranes were then blocked with 5% non-fat milk in
Tris-buffered saline with Tween-20 (TBST) for ~2 h, then followed
by incubation with the primary antibodies against GAPDH (1:2,000;
WL01547; Wanlei Biotech Co., Ltd., Beijing, China) and ST7L
(1:1,000; 17567-1-AP; Proteintech Co., Ltd., Wuhan, China)
overnight at 4°C. After washing with TBST, the blots were incubated
with horseradish peroxidase (HRP) conjugated secondary antibody
(1:5,000; A0216; Beyotime Institute of Biotechnology) at 37°C for 1
h. Thereafter, the proteins of interest were visualized using
enhanced chemiluminescence (ECL; Wanlei Biotech) and densitometric
analysis was performed using Gel-Pro Analyzer system (Beijing Liuyi
Instrument Factory, Beijing, China). LabWorks™ Image Acquisition
and Analysis software UVP EC3 (UVP, LLC, Upland, CA, USA) were used
to quantify band intensities.
Cell cycle and apoptosis flow cytometric
analyses
At 48 h after transfection, transfected CC cells
were harvested by trypsinization and resuspended in cold PBS for
analysis. For the analysis of cell cycle, cells stained with
propidium iodide (PI) according to the manufacturer's manual. The
rate of cell apoptosis was detected using an Annexin V-FITC/PI
apoptosis detection kit (Nanjing Kaiji Biotechnology Development
Co., Ltd., Nanjing, China). These analyses were conducted according
to the protocol provided by Nanjing Kaiji Biotechnology
Development.
TOP/FOP flash reporter assays
To assay the transcriptional activity of Wnt
pathway, pri-miR-378, pST7L, or pri-miR-378 and pST7L treated cells
were co-transfected with either the Wnt signaling reporter TopFlash
or the negative control FopFlash according to the protocol (EMD
Millipore). HeLa cells were transiently transfected with either 2
µg pTopFlash (TCF reporter plasmid) or pFopFlash (mutant,
inactive TCF binding site) plasmids (EMD Millipore) and 0.5
µg pSV40-Renilla plasmid as an internal control
(Promega Corp., Madison, WI, USA) for 48 h. The Dual-Luciferase
reporter assay system (DLR™ E1910; Promega) was used to assay the
firefly and Renilla luciferase activity ratio.
Statistical analysis
Data are presented as the means ± standard deviation
(SD) from at least three independent experiments. Statistical
analyses were performed using Student's t-tests. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-378 is upregulated in human cervical
cancer and cervical cell lines
To investigate the role and clinical significance of
miR-378 in cervical cancer, the authors first detected the level of
miR-378 by RT-qPCR assay in 27 pairs of human cervical tumors and
matched normal cervical tissues. Compared with the corresponding
non-tumorous counterparts, the miR-378 expression level was
significantly upregulated in cervical tumor tissues (Fig. 1A). In addition, RT-qPCR was also
used to examine the expression level of miR-378 in HeLa, C33A, SiHa
and CaSKi cells, and in End1/E6E7 cells, which is a human normal
cervical epithelium cell line. The results demonstrated that
miR-378 was obviously elevated in cervical cancer cells compared to
End1/E6E7 cells (Fig. 1B). These
results identified that miR-378 was upregulated in human cervical
cancer.
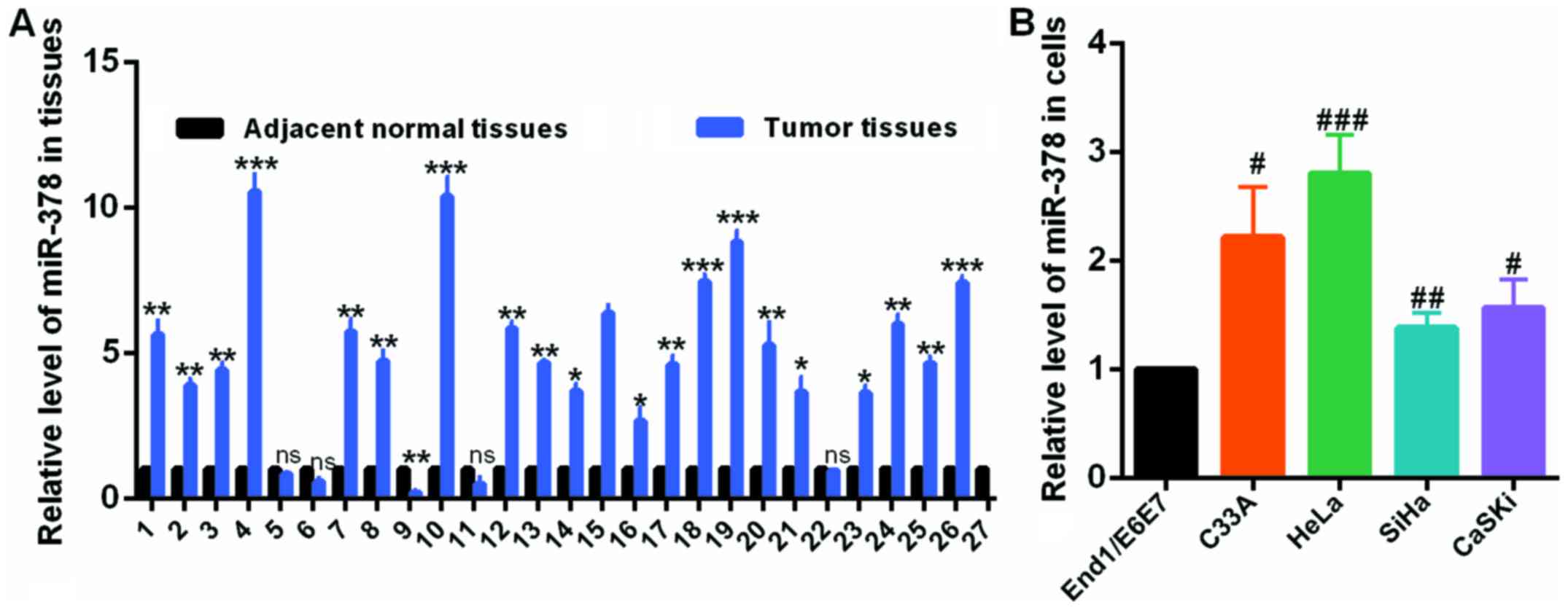 | Figure 1miR-378 was upregulated in CC tissues
and CC cells. (A) The levels of miR-378 in CC and normal tissues
were examined by RT-qPCR assay. (B) The levels of miR-378 in C33A,
HeLa, SiHa, CaSKi cells and End1/E6E7 cells were examined by
RT-qPCR assay. Data are presented as mean ± SD (n=3).
*P<0.05, **P<0.01,
***P<0.001 vs. the adjacent normal tissues;
#P<0.05, ##P<0.01,
###P<0.001 vs. End1/E6E7 cell line. CC, cervical
cancer; miR, microRNA; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction. |
miR-378 promotes cervical cancer cell
growth in vitro
The roles of miR-378 in cervical cancer cells were
evaluated by transfection of miR-378 overexpression or knockdown
plasmids in HeLa and SiHa cells (Fig.
2A). An MTT assay was then performed to determine cell
viability. Data indicated that transfection with pri-miR-378
significantly facilitated the proliferation of HeLa and SiHa cells;
on the contrary, ASO-miR-378 transfection in HeLa and SiHa cells
remarkably decreased cell viability compared to the controls
(Fig. 2B). Furthermore,
upregulation of miR-378 increased the relative colony formation
rate in HeLa and SiHa cells, whereas miR-378 downregulation
inhibited the colony formation ability of HeLa and SiHa cells
(Fig. 2C). These data suggested
that miR-378 served as an onco-miRNA in cervical cancer cells.
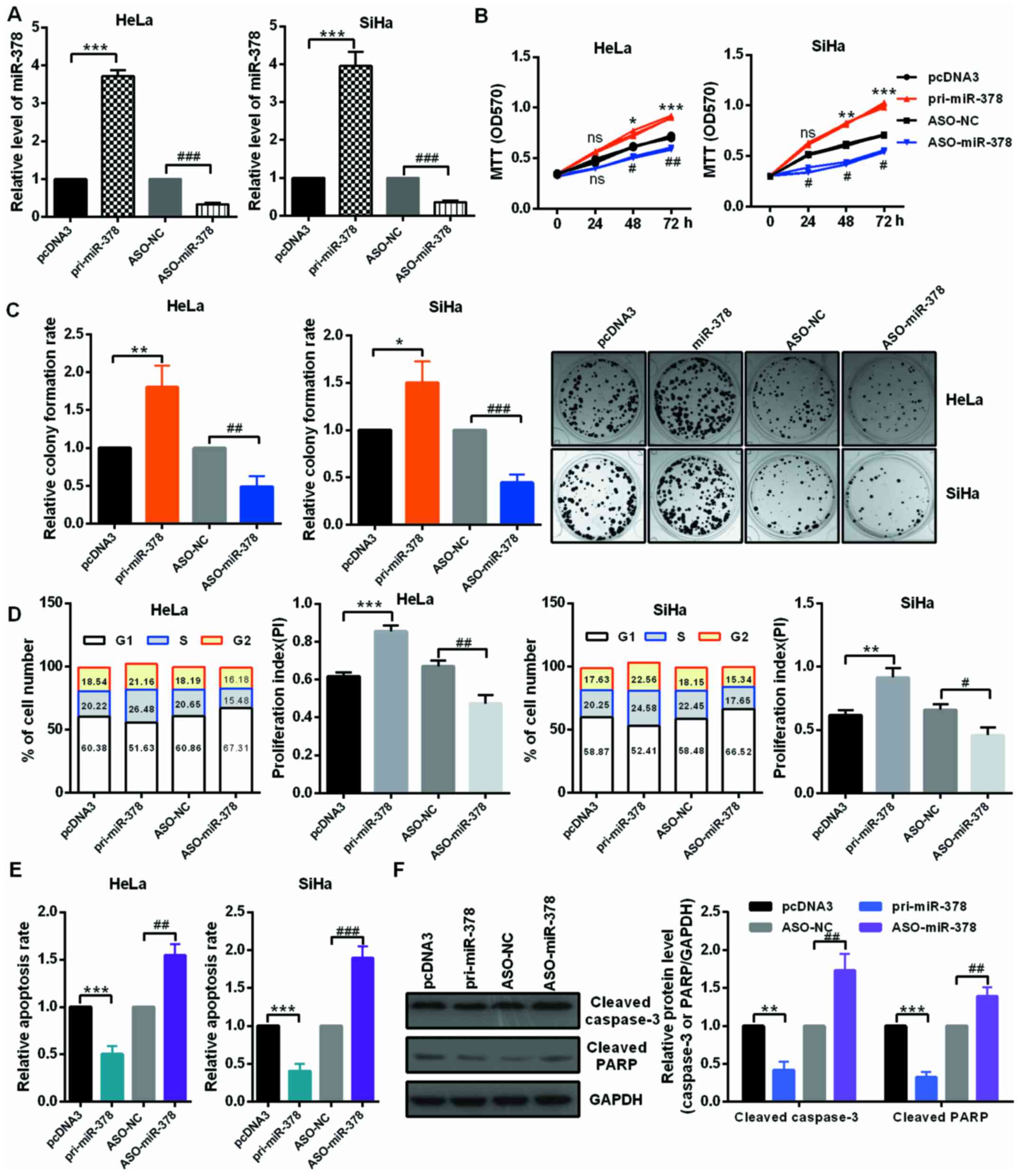 | Figure 2miR-378 served as an onco-miRNA in
cervical cancer cells. (A) The efficiency of miR-378 overexpression
and knockdown plasmids was confirmed by reverse
transcription-quantitative polymerase chain reaction assay. HeLa
and SiHa cells were transfected with the pri-miR-378 or ASO-miR-378
and the control groups, respectively. (B) The cell viability of
miR-378 on HeLa and SiHa cells were determined by MTT assay.
Overexpression of miR-378 promoted cell viability and knockdown of
miR-378 inhibited cell viability. (C) Relative colony formation
rate of HeLa and SiHa cells with indicated treatment was determined
by colony formation assay. Overexpression of miR-378 promoted
colony formation ability and knockdown of miR-378 inhibited colony
formation ability. Original magnification, ×1. (D) Flow cytometry
cell cycle assays demonstrated that miR-378 increased the number of
HeLa and SiHa cells in the S and G2 phases and decreased the number
of HeLa and SiHa cells in G1 phase. Overexpression of miR-378
enhanced the proliferation index. (E) Flow cytometry cell apoptosis
assays revealed that miR-378 overexpression suppressed the
apoptosis of HeLa and SiHa cells and miR-378 knockdown promoted
cell apoptosis. (F) The expression levels of cleaved PARP and
caspase-3 were examined by western blot assay in HeLa cells under
the described condition. Data are presented as mean ± SD (n=3).
*P<0.05, **P<0.01,
***P<0.001 vs. the pcDNA3 group;
#P<0.05, ##P<0.01,
###P<0.001 vs. the ASO-NC group. miR, microRNA; ASO,
antisense oligonucleotide; NC, negative control; ns, not
significant; pri, primary. |
miR-378 markedly accelerates cell cycle
progression and reduces cell apoptosis in CC cells
miR-378 significantly promoted the growth of HeLa
and SiHa cells, so it was speculated that miR-378 could accelerate
the cell cycle process in cervical cancer cells. The authors
reported that overexpression of miR-378 obviously decreased the
percentage of cells in the G1 phase and increased the percentage of
cells in the S and G2 phase in both HeLa and SiHa cells compared
with cells transfected with the controls; however, knockdown of
miR-378 increased the percentage of cells in the G1 phase and
decreased the percentage of cells in the S and G2 phase in HeLa and
SiHa cells by flow cytometry assay. In addition, cells treated with
pri-miR-378 markedly increased the proliferation index of HeLa and
SiHa cells, cells treated with ASO-miR-378 decreased the
proliferation index of HeLa and SiHa cells, respectively (Fig. 2D). Thus, the authors demonstrated
that miR-378 might promote the proliferation of cervical cancer
cells by accelerating cell cycle transition. To investigate whether
cell apoptosis took part in the miR-378-induced proliferative
effect, the relative cell apoptosis rates of HeLa and SiHa cells
were detected by flow cytometry cell apoptosis assay. Fig. 2E indicated that the relative
apoptotic rate of HeLa and SiHa cells was significantly lower in
the miR-378-treated group than that in the control groups. In
addition, the noted effect of miR-378 on apoptosis was confirmed,
as decreased activating cleavage of PARP and caspase-3 was induced
in the cervical cancer cells that overexpressed miR-378 (Fig. 2F).
miR-378 directly targets ST7L in CC
cells
miRNAs serve important roles in cell growth, cell
differentiation, cell apoptosis, cell cycle and other physiological
and pathological processes by binding to the 3′-UTR of target
genes, thus, regulating its expression of mRNA and protein level.
According to TargetScan 7.1, miRDB and microRNA.org,
ST7L is a predicted target gene of miR-378 (Fig. 3A). To verify that miR-378 can
directly target ST7L mRNA, EGFP reporter plasmids containing ST7L
wild-type or mutant 3′-UTR were constructed. In HeLa and SiHa
cells, the relative EGFP activity was significantly reduced in both
the pri-miR-378 and wild-type 3′-UTR of ST7L co-transfected group
and co-transfection with ASO-miR-378 and wild-type 3′-UTR of ST7L
increased the relative EGFP activity (Fig. 3B). However, miR-378 overexpression
or knockdown did not play a role in EGFP activity regulated by the
3′-UTR of ST7L mutation (Fig.
3C). In addition, significant downregulation of ST7L was
observed at both mRNA and protein levels in HeLa and SiHa cells
expressing pri-miR-378 and the upregulation of ST7L at both mRNA
and protein levels in the cells expressing ASO-miR-378 (Fig. 3D and E).
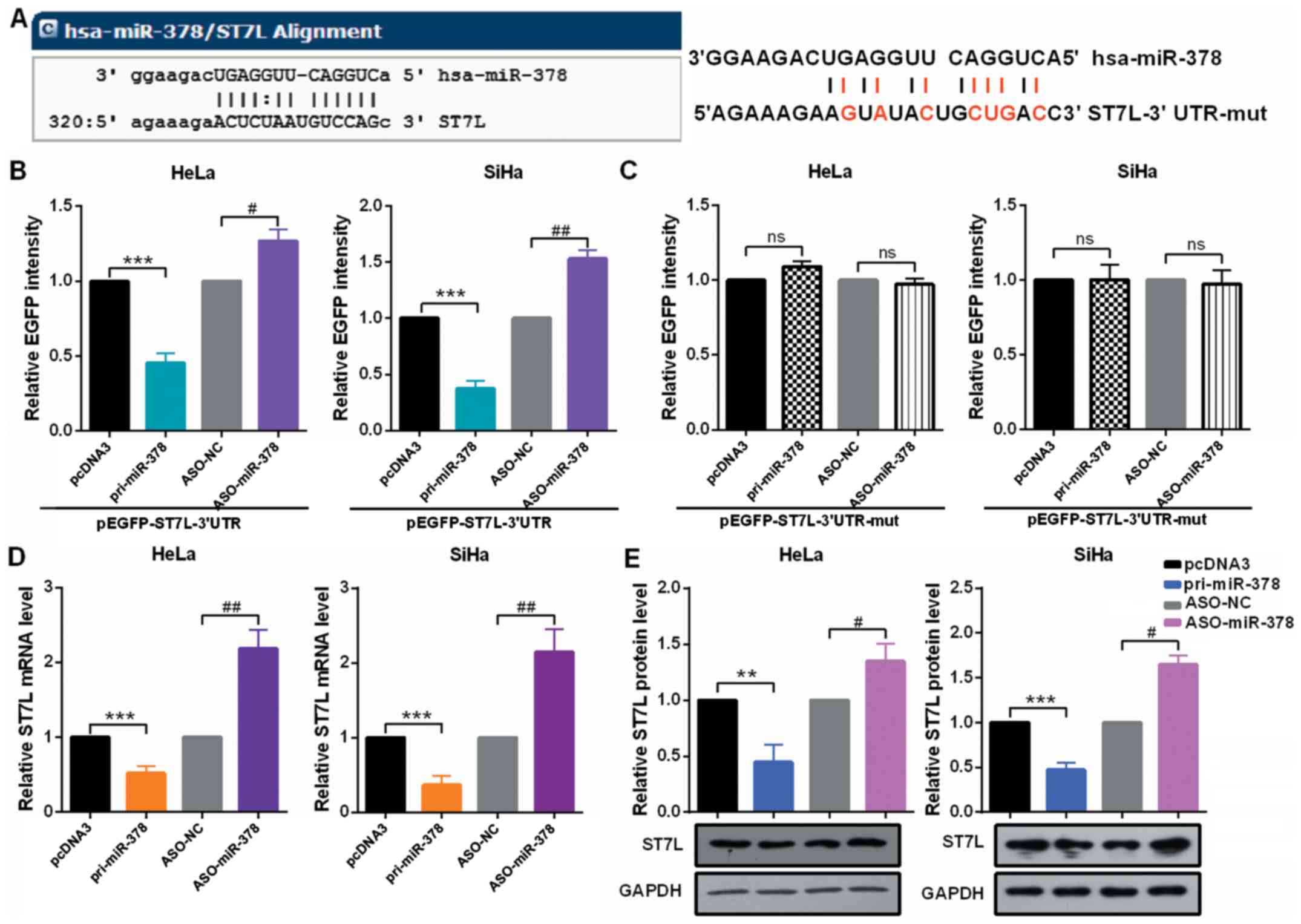 | Figure 3miR-378 directly targets ST7L in
cervical cancer cells. (A) The predicted miR-378 binding sites
using TargetScan 7.1 in ST7L mRNA 3′-UTR and the sites of ST7L mRNA
3′-UTR-mut is shown. (B and C) HeLa and SiHa cells were
co-transfected with pcDNA3/EGFP-ST7L 3′-UTR or 3′-UTR-mut and
pri-miR-378 or ASO-miR-378. EGFP intensity was determined by
spectrophotometer, and the value of the control group (pcDNA3 or
ASO-NC) was set to one. (D) HeLa and SiHa cells were transfected
with pri-miR-378 or ASO-miR-378. ST7L mRNA level in HeLa and SiHa
cells were measured by reverse transcription-quantitative
polymerase chain reaction assay. (E) ST7L protein level in HeLa and
SiHa cells transfected with pri-miR-378 or ASO-miR-378 and the
respective controls was determined by western blot assay. Data are
presented as mean ± SD (n=3). **P<0.01,
***P<0.001 vs. the pcDNA3 group;
#P<0.05, ##P<0.01 vs. the ASO-NC group.
ns, not significant; miR, microRNA; 3′-UTR, 3′-untranslated region;
mut, mutant; pri, primary ASO, antisense oligonucleotide; EGFP,
enhanced green fluorescent protein; NC, negative control. |
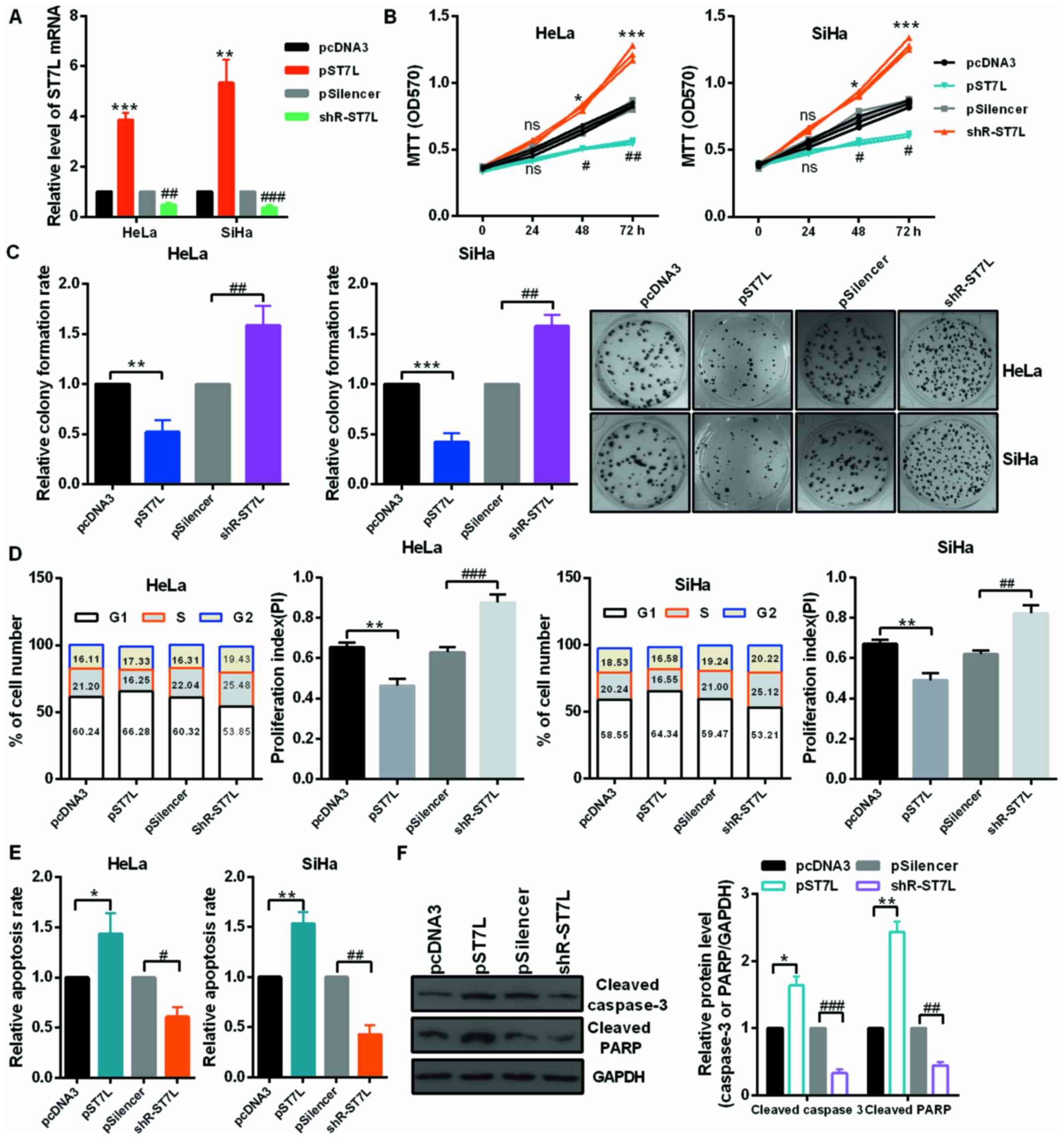
miR-378 promotes the malignancy of CC by
inhibiting ST7L expression
First, the authors detected the mRNA level of ST7L
by RT-qPCR in 27 pairs of cervical tumors and matched normal
cervical tissues and in cervical cancer cells and human normal
cervical epithelium cell. The mRNA level of ST7L was significantly
decreased in human cervical cancer tissues (Fig. 4A) and cervical cancer cells
(Fig. 4B) compared with the
control groups. As presented in Fig.
4C, the protein level of ST7L was lower in CC cell lines than
in End1/E6E7 cells by western blot assay. Moreover, the expression
level of ST7L was negatively correlated with the expression level
of miR-378 in tumor tissues (Fig.
4D). The overexpression or knockdown plasmid of ST7L was
effective in HeLa and SiHa cell lines (Fig. 5A). Then, authors performed an MTT
assay and colony formation assay using cells transfected with pST7L
or shR-ST7L and the control plasmids. At 24, 48 and 72 h
post-transfection, HeLa and SiHa cell viability were decreased by
overexpression of ST7L and increased by ST7L knockdown (Fig. 5B). The relative colony formation
rate was decreased by pST7L and increased by shR-ST7L compared to
the control groups (Fig. 5C). The
cell cycle assay demonstrated that ST7L overexpression increased
the number of HeLa and SiHa cells in the G1 phase, decreased the
number in the S phase (Fig. 5D),
and ST7L overexpression was observed to significantly promote cell
apoptosis in HeLa and SiHa cells (Fig. 5E). The noted effect of ST7L on
apoptosis was confirmed, because increased activating cleavage of
PARP and caspase-3 was induced in the cervical cancer cells that
overexpressed ST7L (Fig. 5F).
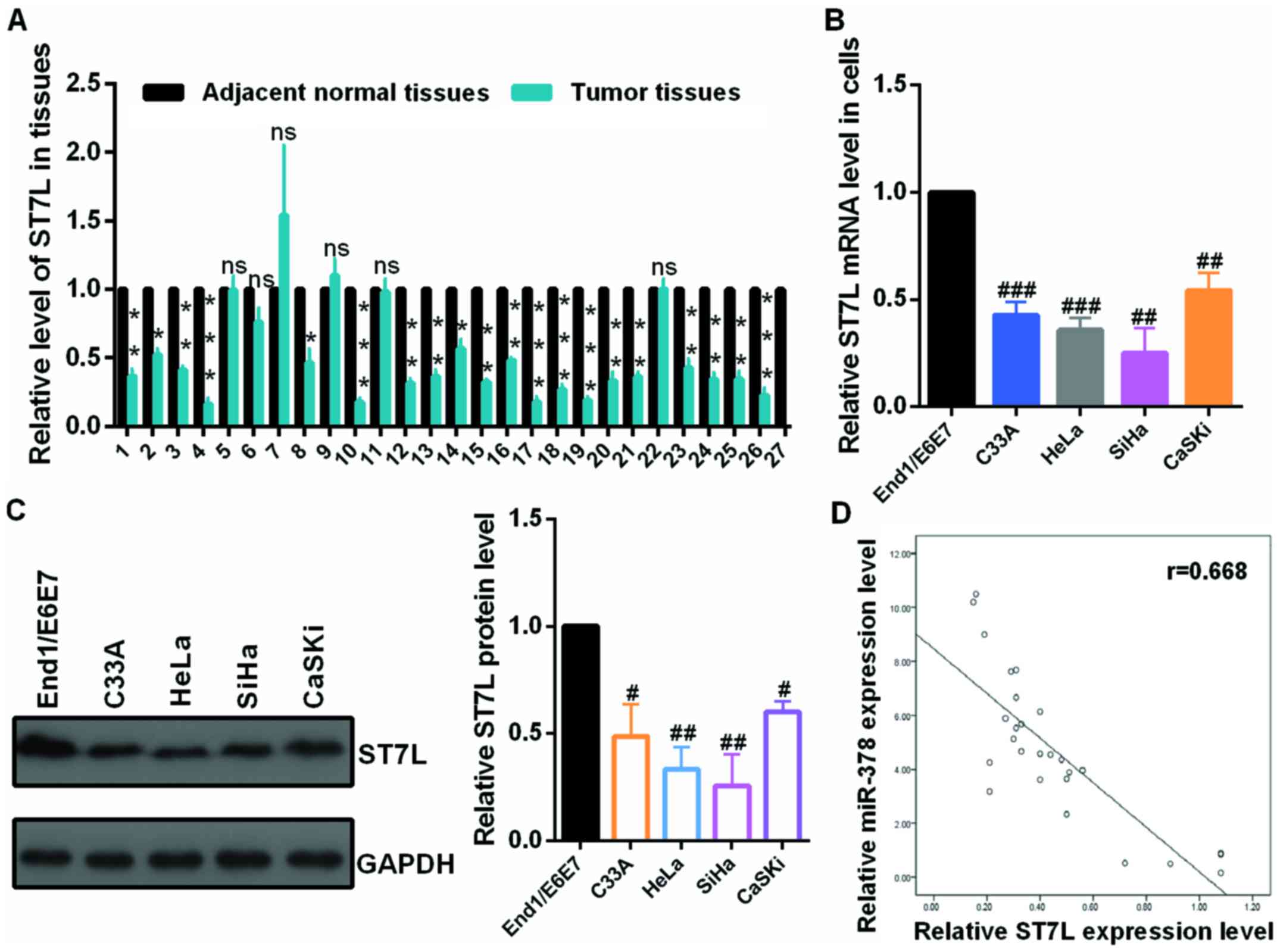 | Figure 4The mRNA level of ST7L was
downregulated in CC tissues and CC cells. (A) The mRNA levels of
ST7L in CC and normal tissues were examined by RT-qPCR assay. (B)
The mRNA levels of ST7L in C33A, HeLa, SiHa, CaSKi cells and
End1/E6E7 cells were examined by RT-qPCR assay. (C) The protein
levels of ST7L in C33A, HeLa, SiHa and CaSKi cells and End1/E6E7
cells were examined by western blot assay. (D) The correlation of
the expression of miR-378 and ST7L in tumor tissues was presented
using SPSS 19.0. Data are presented as mean ± SD (n=3).
*P<0.05, **P<0.01,
***P<0.001 vs. adjacent normal tissues;
#P<0.05, ##P<0.01,
###P<0.001 vs. the End1/E6E7 cells. CC, cervical
cancer; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; miR, microRNA. |
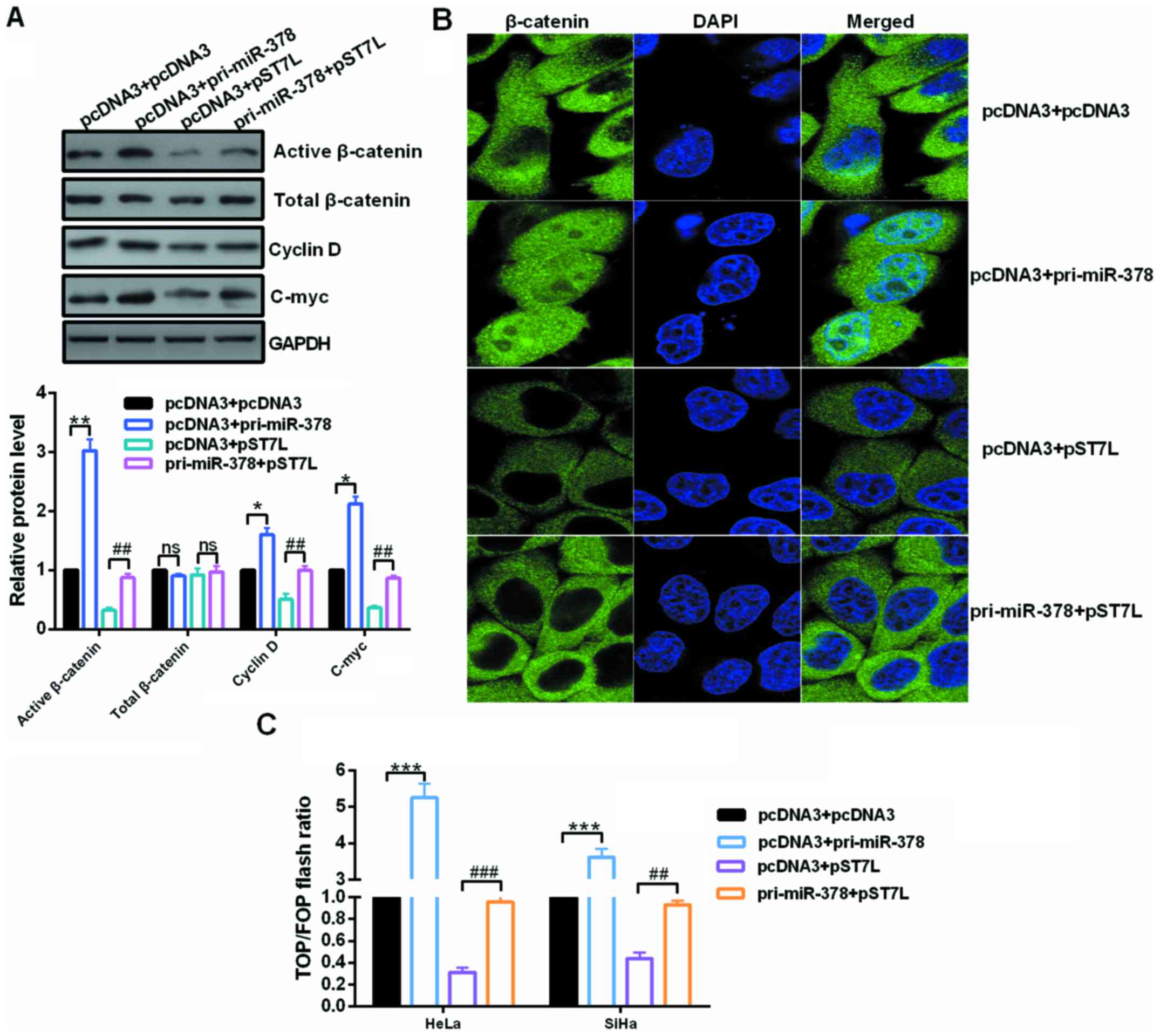
Upregulated miR-378 activates the
Wnt/β-catenin pathway in cervical cancer cells
Inactivation of Wnt/β-catenin signaling caused by
ST7L by repressing β-catenin expression could explain this
phenomenon to a certain extent in epithelial ovarian cancer
(21). To address these issues,
the expression of Wnt genes and the activity of β-catenin were
analyzed in the context of miR-378 overexpression or miR-378
overexpression in conjunction with pST7L in cervical cancer cells.
Western blot assays showed that the expression of activated
β-catenin, C-myc and cyclin D, which are the downstream effectors
of the Wnt/β-catenin pathway, were reduced by ST7L depletion and
significantly increased by miR-378 restoration (Fig. 6A). The localization of β-catenin
in HeLa cells was examined by immunofluorescence after transfection
with pri-miR-378 or pST7L or co-transfection with pri-miR-378 and
pST7L. The nuclear distribution of β-catenin was increased in
pri-miR-378-transfected cells and decreased in pST7L-transfected
cells (Fig. 6B). In addition,
TOP/FOP luciferase reporter assays were also performed, which is a
common Wnt pathway activation reporter assay, and the results
demonstrated that miR-378 enhanced the TOP/FOP flash ratio but ST7L
weakened it (Fig. 6C).
Discussion
Previously, miRNAs have appeared as a highlighted
class of gene regulators and dysregulation of miRNAs plays
important roles in the genesis and development of various cancers
including cervical cancer (23–25). Reports have indicated that miR-378
is overexpressed in many cancers (26). For example, upregulated miR-378
enhanced cell proliferation, cell migration and cell invasion in
HepG2 cells, and promoted the metastasis of the tumor cells to the
liver (10). In addition,
overexpression of miR-378 increased neural stem cell (NSC)
differentiation and reduced NSC proliferation, whereas suppression
of miR-378 led to decreased NSC differentiation and increased NSC
proliferation and miR-378 negatively regulated Tailless mRNA and
protein expression (14).
Furthermore, miR-378 levels significantly decreased in N2A cells
following oxygen-glucose deprivation treatment and overexpression
of miR-378 significantly enhanced cell viability, decreased cell
apoptosis and the immunoreactivity of cleaved-caspase-3 by
targeting the predicted 3′-UTR of caspase-3 gene in the ischemic
injury (27). However, Chen et
al (28) reported that
miR-378 suppresses cell growth by downregulating MAPK1 expression
in prostate cancer; and Li et al (29) demonstrated that the expression
level of miR-378 is obviously lower in glioma tissues compared with
normal brain tissues. In addition, Li et al (30) also concluded that miR-378 may
synergistically act with curcumin in inhibiting cell growth by
binding to the 3′-UTR of p38 in glioblastoma cells. These studies
suggested that miR-378 serves important roles in multiple types of
human cancer and may therefore be a therapeutic target in their
treatment. Thus, to investigate the role of miR-378 in various
cancers is a matter of great account. In the present study, miR-378
was significantly upregulated in both CC tissues and cell lines.
Functional assays demonstrated that miR-378 overexpression promoted
cell proliferation, accelerated the cell cycle process and reduced
apoptosis in cervical cancer cells. More importantly, miR-378
overexpression could activate the Wnt/β-catenin pathway in CC
cells. However, the mechanism that regulates the expression of
miR-378 in CC tissues and cells is not well understood; therefore,
the detail mechanism needs further investigation.
It is well known that miRNAs can play roles by
relying on the level of complementarities with the 3′-UTR of their
target mRNAs (31), the authors
next attempted to identify the potential targets of miR-378.
Bioinformatics software was used to predict the target genes of
miR-378, in the present study, ST7L was demonstrated to be a novel
candidate target of miR-378. Notably, the mRNA expression level of
ST7L was significantly decreased in HeLa, SiHa, C33A and Caski cell
lines, and CC tissues, which was inversely correlated with miR-378
level in CC tissues and cells. Furthermore, the EGFP reporter assay
presented that miR-378 binds directly to the 3′-UTR of the ST7L
mRNA in HeLa and SiHa cells. To examine the regulatory roles of
miR-378 on endogenous ST7L expression, RT-qPCR and western blot
analyses were utilized to measure ST7L expression at the mRNA and
protein levels in CC cells following transfection with pri-miR-378
or ASO-miR-miR-378. Results revealed that miR-378 overexpression
markedly reduced the expression of ST7L at mRNA and protein level
and miR-378 knockdown increased the expression of ST7L at mRNA and
protein level in HeLa and SiHa cells.
ST7L, also as known as ST7R, STLR and FAM4B, was
confirmed with its similarity to the ST7 tumor suppressor gene
found in the chromosome 7q31 region and clustered with the WNT2B
gene in a tail-to-tail manner in a chromosomal region known to be
deleted and rearranged in many cancers, including germ cell tumors,
breast cancer and non-small cell lung cancer, amongst others
(18,19). Furthermore, ST7L was reported to
inhibit the Wnt/β-catenin signaling pathway and act as a
tumor-suppressor gene in many cancers (20–22). However, the function and role of
ST7L in CC cells was unclear. The current results demonstrated that
ST7L was downregulated in CC tissues and cells. Functional assays
revealed that over-expression of ST7L suppressed cell proliferation
by arresting the cell cycle process and promoted cell apoptosis in
HeLa and SiHa cells. In addition, ST7L inhibited the Wnt/β-catenin
pathway in CC cells.
In conclusion, these reports confirmed that
upregulation of miR-378 is a common phenomenonin CC tissues and CC
cells, and the authors have identified that miR-378 takes effect in
regulating cell proliferation, cell cycle, cell apoptosis and the
Wnt/β-catenin pathway of CC cells. miR-378 inhibits ST7L expression
at both the mRNA and protein level by directly targeting the 3′-UTR
of the ST7L transcript. With its regulation of cervical cancer cell
malignant phenotype, knockdown of miR-378 functions as a novel
therapeutic strategy in CC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bansal N, Herzog TJ, Shaw RE, Burke WM,
Deutsch I and Wright JD: Primary therapy for early-stage cervical
cancer: Radical hysterectomy vs radiation. Am J Obstet Gynecol.
201:485.e1–485.e9. 2009. View Article : Google Scholar
|
|
3
|
Kosmas C, Mylonakis N, Tsakonas G, Vorgias
G, Karvounis N, Tsavaris N, Daladimos T, Kalinoglou N, Malamos N,
Akrivos T, et al: Evaluation of the paclitaxel-ifosfamide-cisplatin
(TIP) combination in relapsed and/or metastatic cervical cancer. Br
J Cancer. 101:1059–1065. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Colombo N, Carinelli S, Colombo A, Marini
C, Rollo D and Sessa C: Cervical cancer: ESMO Clinical Practice
Guidelines for diagnosis, treatment and follow-up. Ann Oncol.
23(Suppl 7): vii27–vii32. 2012.PubMed/NCBI
|
|
5
|
Castellsagué X, Díaz M, de Sanjosé S,
Muñoz N, Herrero R, Franceschi S, Peeling RW, Ashley R, Smith JS,
Snijders PJ, et al International Agency for Research on Cancer
Multicenter Cervical Cancer Study Group: Worldwide human
papilloma-virus etiology of cervical adenocarcinoma and its
cofactors: Implications for screening and prevention. J Natl Cancer
Inst. 98:303–315. 2006. View Article : Google Scholar
|
|
6
|
Wang Z and Cai H, Lin L, Tang M and Cai H:
Upregulated expression of microRNA-214 is linked to tumor
progression and adverse prognosis in pediatric osteosarcoma.
Pediatr Blood Cancer. 61:206–210. 2014. View Article : Google Scholar
|
|
7
|
Tamura M, Uyama M, Sugiyama Y and Sato M:
Canonical Wnt signaling activates miR-34 expression during
osteoblastic differentiation. Mol Med Rep. 8:1807–1811. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Davis-Dusenbery BN and Hata A: Mechanisms
of control of microRNA biogenesis. J Biochem. 148:381–392.
2010.PubMed/NCBI
|
|
9
|
Chan JK, Kiet TK, Blansit K, Ramasubbaiah
R, Hilton JF, Kapp DS and Matei D: MiR-378 as a biomarker for
response to anti-angiogenic treatment in ovarian cancer. Gynecol
Oncol. 133:568–574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma J, Lin J, Qian J, Qian W, Yin J, Yang
B, Tang Q, Chen X, Wen X, Guo H, et al: MiR-378 promotes the
migration of liver cancer cells by down-regulating Fus expression.
Cell Physiol Biochem. 34:2266–2274. 2014. View Article : Google Scholar
|
|
11
|
Browne G, Dragon JA, Hong D, Messier TL,
Gordon JA, Farina NH, Boyd JR, VanOudenhove JJ, Perez AW, Zaidi SK,
et al: MicroRNA-378-mediated suppression of Runx1 alleviates the
aggressive phenotype of triple-negative MDA-MB-231 human breast
cancer cells. Tumour Biol. 37:8825–8839. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang
L, Li G, Chen HH and Li XP: MicroRNA-378 functions as an onco-miR
in nasopharyngeal carcinoma by repressing TOB2 expression. Int J
Oncol. 44:1215–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qian J, Lin J, Qian W, Ma JC, Qian SX, Li
Y, Yang J, Li JY, Wang CZ, Chai HY, et al: Overexpression of
miR-378 is frequent and may affect treatment outcomes in patients
with acute myeloid leukemia. Leuk Res. 37:765–768. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang Y, Liu X and Wang Y: MicroRNA-378
regulates neural stem cell proliferation and differentiation in
vitro by modulating Tailless expression. Biochem Biophys Res
Commun. 466:214–220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng J, Xie Z, Cheng L, Zhang Y, Chen J,
Yu H, Li Z and Kang H: Paired design study by real-time PCR:
miR-378* and miR-145 are potent early diagnostic biomarkers of
human colorectal cancer. BMC Cancer. 15:1582015. View Article : Google Scholar
|
|
16
|
Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang
W, Ma Y and Xiao H: Genome-wide microRNA profiles identify miR-378
as a serum biomarker for early detection of gastric cancer. Cancer
Lett. 316:196–203. 2012. View Article : Google Scholar
|
|
17
|
Fedorko M, Stanik M, Iliev R,
Redova-Lojova M, Machackova T, Svoboda M, Pacik D, Dolezel J and
Slaby O: Combination of MiR-378 and MiR-210 serum levels enables
sensitive detection of renal cell carcinoma. Int J Mol Sci.
16:23382–23389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Katoh M: Molecular cloning and
characterization of ST7R (ST7-like, ST7L) on human chromosome 1p13,
a novel gene homologous to tumor suppressor gene ST7 on human
chromosome 7q31. Int J Oncol. 20:1247–1253. 2002.PubMed/NCBI
|
|
19
|
Kirikoshi H and Katoh M: Expression of
ST7R (ST7-like, ST7L) in normal tissues and cancer. Int J Oncol.
21:193–196. 2002.PubMed/NCBI
|
|
20
|
Chen L, Zhang A, Li Y, Zhang K, Han L, Du
W, Yan W, Li R, Wang Y, Wang K, et al: MiR-24 regulates the
proliferation and invasion of glioma by ST7L via β-catenin/Tcf-4
signaling. Cancer Lett. 329:174–180. 2013. View Article : Google Scholar
|
|
21
|
Yang Z, Wang XL, Bai R, Liu WY, Li X, Liu
M and Tang H: miR-23a promotes IKKα expression but suppresses ST7L
expression to contribute to the malignancy of epithelial ovarian
cancer cells. Br J Cancer. 115:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhuang L, Wang X, Wang Z, Ma X, Han B, Zou
H, Wu Z, Dong S, Qu Z, Zang Y, et al: MicroRNA-23b functions as an
oncogene and activates AKT/GSK3β/β-catenin signaling by targeting
ST7L in hepatocellular carcinoma. Cell Death Dis. 8:e28042017.
View Article : Google Scholar
|
|
23
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park H, Lee MJ, Jeong JY, Choi MC, Jung
SG, Joo WD, Lee C and An HJ: Dysregulated microRNA expression in
adenocarcinoma of the uterine cervix: Clinical impact of
miR-363-3p. Gynecol Oncol. 135:565–572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: Key players in the immune system, differentiation,
tumorigenesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li ZZ, Shen LF, Li YY, Chen P and Chen LZ:
Clinical utility of microRNA-378 as early diagnostic biomarker of
human cancers: A meta-analysis of diagnostic test. Oncotarget.
7:58569–58578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang N, Zhong J, Han S, Li Y, Yin Y and
Li J: MicroRNA-378 alleviates cerebral ischemic injury by
negatively regulating apoptosis executioner caspase-3. Int J Mol
Sci. 17:E14272016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen QG, Zhou W, Han T, Du SQ, Li ZH,
Zhang Z, Shan GY and Kong CZ: MiR-378 suppresses prostate cancer
cell growth through downregulation of MAPK1 in vitro and in vivo.
Tumour Biol. 37:2095–2103. 2016. View Article : Google Scholar
|
|
29
|
Li B, Wang Y, Li S, He H, Sun F, Wang C,
Lu Y, Wang X and Tao B: Decreased expression of miR-378 correlates
with tumor invasiveness and poor prognosis of patients with glioma.
Int J Clin Exp Pathol. 8:7016–7021. 2015.PubMed/NCBI
|
|
30
|
Li W, Yang W, Liu Y, Chen S, Chin S, Qi X,
Zhao Y, Liu H, Wang J, Mei X, et al: MicroRNA-378 enhances
inhibitory effect of curcumin on glioblastoma. Oncotarget. May
16–2017.Epub ahead of prin. View Article : Google Scholar
|
|
31
|
Farazi TA, Juranek SA and Tuschl T: The
growing catalog of small RNAs and their association with distinct
Argonaute/Piwi family members. Development. 135:1201–1214. 2008.
View Article : Google Scholar : PubMed/NCBI
|