Introduction
Osteoarthritis (OA) is a joint disease that is
commonly reported in the elderly. It is characterized by
progressive destruction of articular cartilage, inflammation of
synovium, exposure and sclerosis of subchondral bone, and
osteophyte formation (1–5). The morbidity of OA is >10%
(6,7). The aim of nonsurgical treatment is
mainly to alleviate the symptoms including pain and turgescence,
but not to prevent the pathogenesis, i.e. to cure cartilage
degradation (8–10).
Although the main etiological factor that
antagonizes OA remains to be elucidated, it is presumed that an
imbalance between anabolic and catabolic metabolism in articular
cartilage is an essential event in the progression of OA, which
results in pervasive cartilage damage (2).
Previous reports have suggested that a high
concentration of proinflammatory cytokine interleukin-1β (IL-1β)
was detected in the synovial fluid of patients with arthritis and
rat models (11), and that
neutralizing IL-1β using antibodies may ameliorate the degeneration
and destruction of articular cartilage (12). However, the high cost limits the
application of this method. In addition, gene expression of IL-1β
in the peripheral blood mononuclear cells is upregulated in
patients with OA (13), implying
that IL-1β may serve a vital role in synovial inflammation and
cartilage degradation. The IL-1β-induced gene expression of matrix
metalloproteinases (MMPs) and a disintegrin and metalloproteinase
with thrombospondin motifs (ADAMTS) in chondrocytes is regulated
predominantly at the transcriptional level, particularly by
mitogen-activated protein kinase (MAPK) signaling pathways
(14). The attenuation of one or
more of these signals was considered to reveal a potential target
for OA therapy and, therefore, meriting further investigation
(15). However, the precise
mechanism has yet to be fully elucidated.
The degradation process of articular cartilage is
mainly regulated by the activation of enzymes belonging to the MMP
and aggrecanase families (15). A
number of previous studies have illuminated the roles of MMPs in
collagen catabolism (16,17). For example, MMP-3 and MMP-9 are
considered to downregulate the extracellular matrix (ECM) in
cartilage (resolving the degenerated collagen) irreversibly
(17). Collagen type X (col X) is
an indicator of cartilage degradation in this pathogenesis
(18,19). Aggrecanase1 was initially
identified in 1999 (20),
followed by aggrecanase2 (21).
They were subsequently termed ADAMTS-4 and ADAMTS-5. It is reported
that ADAMTS5 was the primary ‘aggrecanase’ responsible for aggrecan
degradation in a murine model of OA (22); however, ADAMTS4 appeared not to be
associated with OA pathogenesis (23). As with other ADAMTS, including
ADAMTS-1, ADAMTS-8 and ADAMTS-15, they were of less interest on
account of their poor distribution in degradation cartilage tissues
in OA (24–26). However, the role of aggrecanase,
which belongs to the ADAMTS family, has yet to be elucidated. A
previous study suggested that ADAMTS-5 was upregulated in arthritis
and served a therapeutic role in OA treatment; silencing ADAMTS-5,
alone or combined with ADAMTS-4, which may merit further
investigation (27). In addition
to ADAMTS, cyclooxygenase (COX)-2 also serves a vital role in
cartilage metabolism and inflammation associated with OA. It is
widely used in medical reagents, including Celebrex, for its
anti-inflammatory action (10,28,29).
Significant interest has emerged in epigenetic
disease therapy. For example, inhibition of histone deacetylase
(HDAC) activity has been a promising treatment strategy for
leukemia and cancer (30). The
regulation by HDAC in certain cardiovascular diseases has also been
demonstrated in previous studies (31,32). Furthermore, several studies have
demonstrated that HDAC inhibitors regulate inflammatory responses
(33–36). As they are able to suppress the
expression of MMPs to suppress tumor metastasis (37), they are also used in OA treatment
(38). Despite extensive
investigations, no acknowledged therapy target for HDACs has been
reported. Little is known of the effects and underlying mechanism
of HDAC on OA pathogenesis (38–41).
As the excessive expression of ADAMTS-4 and ADAMTS-5
and production of col X and COX-2 are implicated in the
pathogenesis of OA, their inhibition may be useful for OA
treatment. However, inhibiting any one of them may lead to unknown
side effects or may not be effective (42). The present study assessed the
effect of two types of HDAC inhibitors [Trichostatin A (TSA) and
PCI-34051] and specific small interfering RNAs (siRNAs) on the
expression of OA-related genes and activation of MAPK pathways in
IL-1β-induced rat articular chondrocytes (rACs) to investigate the
underlying mechanisms of OA pathogenesis.
Materials and methods
Isolation and cultivation of rACs
Animal experiments were approved by the Animal Care
and Use Committee for Teaching and Research of Tongji Medical
College, Huazhong University of Science and Technology (Wuhan,
China).
Male Sprague-Dawley rats (n=12; weight, 50–60 g;
age, 4 weeks; Tongji Medical College) were dissected following
sacrifice and bilateral knee articular cartilage specimens were
extracted and cut into 1-mm3 pieces. Cells were isolated
using a sequential proteinase and collagenase digestion technique,
as described in a previous study (27). The cartilage sections were treated
with 0.25% trypsin without EDTA (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) for 30 min at 37°C and 0.1% collagenase II
(Gibco; Thermo Fisher Scientific, Inc.) overnight at 37°C.
Subsequently, the cells were resuspended and cultured in Dulbecco’s
modified Eagle’s medium (DMEM)/F12 (1:1; Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.), containing 100 U/ml
penicillin G sodium and 100 µg/ml streptomycin sulfate
(Gibco; Thermo Fisher Scientific, Inc.). The cells were expanded in
monolayer culture in the medium in an incubator (Thermo Fisher
Scientific, Inc.) at 37°C under a humidified atmosphere containing
5% CO2, with an exchange of the medium every 3 days. Up
to ~90% confluence, the chondrocytes were passaged at a ratio of
1:3 using 0.25% trypsin containing 0.05% EDTA. Chondrocytes of the
third passage were seeded in 6-well or 60-mm plates at a density of
105 cells/cm2 prior to treatment to avoid
phenotype loss.
Generation of an in vitro OA model and
investigation of the expression of HDACs and activation of
MAPKs
Cells were incubated in serum-free DMEM/F12 medium
overnight at 37°C prior to treatment with different reagents.
Recombinant rat IL-1β (Peprotech, Inc., Rocky Hill, NJ, USA) was
resolved in ultrapure water and stored at −20°C prior to use.
IL-1β-induced rACs were used to generate an in
vitro OA model, as described in a previous study (43). The chondrocytes were seeded in
six-well plates at a density mentioned earlier and treated with
2.5, 5, 10, 20 or 40 ng/ml IL-1β for 24 h in the serum-free medium
once reaching confluence, and untreated cells were used as a
control group (44–47).
The chondrocytes were harvested and the gene
expression of ADAMTS-4 and ADAMTS-5 for cartilage catabolism was
evaluated via reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) as detailed below. The expression of collagen
type X indicated cartilage degradation and the expression of COX-2
for inflammation were assayed by western blotting as detailed
below. Antibodies against col X were purchased from Abcam
(Cambridge, UK; cat. no. ab182563) and antibodies against COX-2
were purchased from Cell Signaling Technology, Inc. (Danvers, MA,
USA; cat. no. 12282).
Two groups, 10 ng/ml IL-1β and blank control groups,
were evaluated for the expression of HDACs by RT-qPCR as detailed
below. These groups were observed under an inverted microscope
(magnification, ×200) directly and following toluidine blue
staining (Wuhan Boster Biological Technology, Ltd., Wuhan, China),
according to the manufacturer’s protocols. Following treatment, the
cells were fixed with 4% paraformaldehyde for 30 min at room
temperature, then stained with toluidine blue for 5–10 min at room
temperature, followed by washing with PBS for 3–5 min several times
and finally observed under an inverted microscope (magnification,
×200) following air drying.
IL-1β-treated chondrocytes (10 ng/ml) were used to
set sequential time points of 5, 15 and 30 min, and 1, 2, 6 and 24
h to detect the regularity of the OA-related indicators and the
activating role of MAPK signaling pathways. The chondrocytes
without IL-1β were set as a blank control.
The OA-related indicators were analyzed using the
aforementioned grouping method. The activation of all three
signaling pathways was evaluated using western blotting as detailed
below. Antibodies against phosphorylated (p)-extracellular
signal-regulated kinase (ERK)-1/2, total (t)-ERK1/2, p-c-Jun
N-terminal kinase (JNK), t-JNK, p-p38 and t-p38 were purchased from
Cell Signaling Technology, Inc. (cat. nos. 5726, 9107, 4668, 9258,
9216 and 8690, respectively).
For subsequent experiments, all activators and
inhibitors were dissolved in dimethyl sulfoxide (DMSO,
Sigma-Aldrich, Ontario, Canada) and stored at −80°C prior to use.
For the kinase assays, rACs were pretreated with specific
inhibitors of ERK (FR180204 at 50 µM) and JNK (SP600125 at
10 µM; both from Selleck Chemicals, Houston, TX, USA) and
exposed to IL-1β, or treated with the specific activators of ERK
(ceramide C6 at 50 µM; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) or JNK (anisomycin at 1 µM; Selleck
Chemicals) alone. rACs treated with DMSO were used as a vehicle
control and with IL-1β were used as a positive control. The
expression of ADAMTS-4 and ADAMTS-5 was analyzed by RT-qPCR as
detailed below.
Investigation of the effect of HDAC
inhibitors or specific siRNAs in the in vitro OA model
Subsequently, two HDAC inhibitors (Selleck
Chemicals) were selected according to the results of the previous
step: TSA, a general HDAC inhibitor (only HDAC8 was not sensitive
to it) and PCI-34051, an HDAC8-specific inhibitor. They were
dissolved in DMSO and diluted to working concentrations of 10–500
µM (a further dilution of 1:1,000).
Inhibitors were initially used at various
concentrations for 24 h following a 6-h treatment with 10 ng/ml
IL-1β at 37°C to detect the effective concentration for regulating
the expression of ADAMTS-4 and ADAMTS-5. Then, 100 nM of both
inhibitors were used for 24-h pretreatment and 10 ng/ml IL-1β
treatment at 37°C was performed to detect the regulation of
OA-related genes, HDACs at 24 h and MAPK signaling pathway at 15
min.
Three pairs of RNA oligonucleotides specific for
HDAC4 and HDAC8 coding regions and non-targeting, scrambled siRNA
(siR-Ribo™ Negative Control, Standard; 5 nmol; cat. no.
siN05815122147-1-5) were designed and chemically synthesized
(Table I) by Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). Cells were seeded on six-wells at a
density of 5×105/well prior to transfection, then, 5
µl Lipofectamine® 3000 (Thermo Fisher Scientific,
Inc.) and 100 pmol siRNAs were diffused in Opti-MEM (Thermo Fisher
Scientific, Inc.) separately. A non-targeting, scrambled siRNA was
used as the negative control. Untreated cells were set as the blank
control. Following a 5-min incubation at room temperature, they
were mixed for a further 5-min incubation at room temperature. The
mixtures were placed into the medium and the medium was replaced
with a new one after 6 h at 37°C. Following 24 h of incubation in
total, the transfection efficiency was detected using Cy3 under a
fluorescence inverted microscope (magnification, ×200); the
wavelength for the excitation filter was 555–585 nm.
 | Table ISequences of siRNAs. |
Table I
Sequences of siRNAs.
| Gene | Sequence
number | Sense (5′-3′) | Anti-sense
(5′-3′) |
|---|
| HDAC4 | si1 |
GAAACUGGACAGUAAGAAA |
UUUCUUACUGUCCAGUUUC |
| si2 |
GCAAGAUCCUCAUCGUGGA |
UCCACGAUGAGGAUCUUGC |
| si3 |
GAAAAGGUUUUACAGCAAA |
UUUGCUGUAAAACCUUUUC |
| HDAC8 | si1 |
UGAUCAUCCGGACUCCAUA |
UAUGGAGUCCGGAUGAUCA |
| si2 |
GGUCCCGGUUUAUAUCUAU |
AUAGAUAUAAACCGGGACC |
| si3 |
GACGGUACUACAGUGUAA |
UUACACUGUAGUACCGUCC |
The cells were harvested following 24-h incubation
and the expression of HDAC4 and HDAC8 was assessed by western blot
analysis. Anti-HDAC4 antibodies were purchased from Cell Signaling
Technology, Inc. (cat. no. 5392) and anti-HDAC8 antibodies were
purchased from Abcam (cat. no. ab187139) and RT-qPCR, respectively
as detailed below.
The sequence of high efficiency was selected for
transfection and cells were treated with 10 ng/ml IL-1β at 37°C for
24 h. Subsequently, the expression of ADAMTS-4, ADAMTS-5 and HDAC
was examined by RT-qPCR as detailed below to confirm the effect of
HDACs on IL-1β treatment. The expression of col X and COX-2 was
analyzed by western blotting as detailed below separately.
Protein extraction and western
blotting
Following the aforementioned treatments, the total
protein was extracted from rACs (106 cells/well or
2×106 cells/plate) using radioimmunoprecipitation assay
buffer containing 1% phenylmethylsulfonyl fluoride and 1%
phosphatase inhibitor cocktail (optional, when protein was used for
phosphorylation detection; Wuhan Boster Biological Technology,
Ltd.), according to the manufacturer’s protocols. Chondrocytes were
initially placed on ice and washed with ice-cold PBS three times.
The extract was collected and centrifuged at 12,000 × g for 20 min
at 4°C. The concentration of total protein supernatant was
determined using a bicinchoninic acid kit (Wuhan Boster Biological
Technology, Ltd.). Then, 5X loading buffer (Wuhan Boster Biological
Technology, Ltd.) was added in proportion to the protein, mixed and
boiled for 5 min. Equal amounts of protein (20 µg per lane)
were resolved on 10% SDS-PAGE and the separated protein was
transferred by electroblotting onto 0.45-µm polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA). The
membranes were preincubated in blocking buffer [5% bovine serum
albumin (Wuhan Boster Biological Technology, Ltd.) in Tris-buffered
saline with Tween 20 (50 mM Tris, pH 7.6, 150 mM NaCl and 0.1%
Tween 20)] for 2 h at room temperature and then incubated at 4°C in
blocking buffer with specific primary antibodies at various
dilutions [t-p38, p-ERK, p-JNK, t-JNK, col X, COX-2 and HDAC4 at
1:1,000; p-p38 and t-ERK at 1:2,000; GAPDH at 1:10,000; HDAC8 at
1:50,000] overnight or for at least 16 h and subsequently incubated
with the secondary antibody (horseradish peroxidase-linked goat
anti-rabbit or goat anti-mouse at 1:5,000; Wuhan Boster Biological
Technology, Ltd.; cat. nos. BA1054 and BA1050.) for 2 h at room
temperature. Immunolabeling was visualized using enhanced
chemiluminescence reagent (Wuhan Boster Biological Technology,
Ltd.), and recorded and analyzed using the Bio-Rad ImageLab system
version 5.0 (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
RNA isolation and RT-qPCR analyses
Following the respective treatments, the total RNA
was extracted from rACs (106 cells/well or
2×106 cells/plate) using AZfresh™ Total RNA Extraction
Kit (MiniPrep; Azanno Biotech AB, Gothenburg, Sweden), according to
the manufacturer’s protocol. The concentration and purity of the
total RNA were assayed using a NanoDrop spectrometer (Thermo Fisher
Scientific, Inc.). Subsequently, 1 µg total RNA was used to
synthesize cDNA, using a ReverTra Ace qPCR RT Kit (cat. no.
FSQ-101; Toyobo Life Science, Osaka, Japan; Code No.FSQ-101)
according to the manufacturer’s protocol. Briefly, RNA samples were
mixed with the kit, incubated at 37°C for 15 min, and 99°C for 5
min. cDNA samples were diluted in diethylpyrocarbonate water and
stored at −20°C prior to use. cDNA was amplified and recorded using
SYBR® Green Realtime PCR Master Mix (cat. no. QPK-201;
Toyobo Life Science) on the iCycler real-time PCR instrument
(Bio-Rad Laboratories, Inc.). PCR analysis was performed using
gene-specific primers (Table
II). All primers were synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.). The expression of the gene of interest was
normalized to the housekeeping gene GAPDH, with data expressed as
relative to the corresponding control group. PCR reactions were
performed on a StepOnePlus Real-Time PCR System (Bio-Rad
Laboratories, Inc.) according to the manufacturer’s instructions.
The thermocycling profiles for PCR consisted of 30 cycles of 15 sec
at 94°C, 10 sec at 60°C and 40 sec at 72°C, followed by a final 8
min extension at 55°C. Each set of samples included a template-free
control. Data were analyzed using the 2−ΔΔCq method
(48).
 | Table IISequences of primers. |
Table II
Sequences of primers.
| Gene | Sequence |
|---|
| GAPDH | F:
5′-CTCATGACCACAGTCCATGC-3′ |
| R:
5′-TTCAGCTCTGGGATGACCTT-3′ |
| ADAMTS-5 | F:
5′-TGTGGTGCGCCAAGGCCAAA-3′ |
| R:
5′-CCCTGTGCAGTAGCGGCCAC-3′ |
| ADAMTS-4 | F:
5′-ACAATGGCTATGGACACTGCCTCT-3′ |
| R:
5′-TGTGGACAATGGCTTGAGTCAGGA-3′ |
| HDAC1 | F:
5′-TCCAAGTCCCTTACTTTCCC-3′ |
| R:
5′-CGCTGGTCCCTATCTAGTCC-3′ |
| HDAC2 | F:
5′-TAACTGTCAAAGGTCACGCTAA-3′ |
| R:
5′-ATACGTCCAACATCGAGCAA-3′ |
| HDAC3 | F:
5′-CCAGATCCGCCAGACCATC-3′ |
| R:
5′-GGCCTCGTCAGTCCTGTCATA-3′ |
| HDAC4 | F:
5′-ACTTTCCCGTGTCACTCCTG-3′ |
| R:
5′-TCTTCCTTCGTGGCTTTCC-3′ |
| HDAC5 | F:
5′-TCTTTGCATCGCTACGACAAT-3′ |
| R:
5′-ATCAAACCCAGCGGAGACTAG-3′ |
| HDAC6 | F:
5′-ATGCCGTGACTCCACTATCC-3′ |
| R:
5′-TGAACCGATCCACAGGTCTT-3′ |
| HDAC7 | F:
5′-GTGCCAAACCTTCATCTCCC-3′ |
| R:
5′-TCCCACCTTCCTATCCCTCA-3′ |
| HDAC8 | F:
5′-CTGGAAATCACGCCAAGC-3′ |
| R:
5′-TGTCTTCATTATGGGTTCTGC-3′ |
| HDAC9 | F:
5′-CTACCGTCAGGGAGGCTAA-3′ |
| R:
5′-TCCACCAGTCTGTGAATACGA-3′ |
| HDAC10 | F:
5′-GTGTTCTGTTTAGCCGATGTT-3′ |
| R:
5′-TCGCCAGTCGCTTGAAAT-3′ |
| HDAC11 | F:
5′-GGCAGCATAACCAAAGGC-3′ |
| R:
5′-GGAATACCCACATGGTAATGAA-3′ |
Statistical analysis
All experiments were performed in at least
triplicate using independent samples. All data were presented as
the mean ± standard deviation. Comparisons of multiple groups were
performed using one-way and two-way analysis of variance, followed
by pairwise comparisons with a Bonferroni post hoc test. P<0.05
was considered to indicate a statistically significant difference.
All data were analyzed using GraphPad Prism version 6.00 (GraphPad
Software, Inc., La Jolla, CA, USA), or SPSS version 20.0 (IBM
Corp., Armonk, NY, USA).
Results
Successful construction of an in vitro
model of OA
rACs were cultured with various concentrations of
IL-1β (2.5–40 ng/ml) for 24 h. The expression levels of OA-related
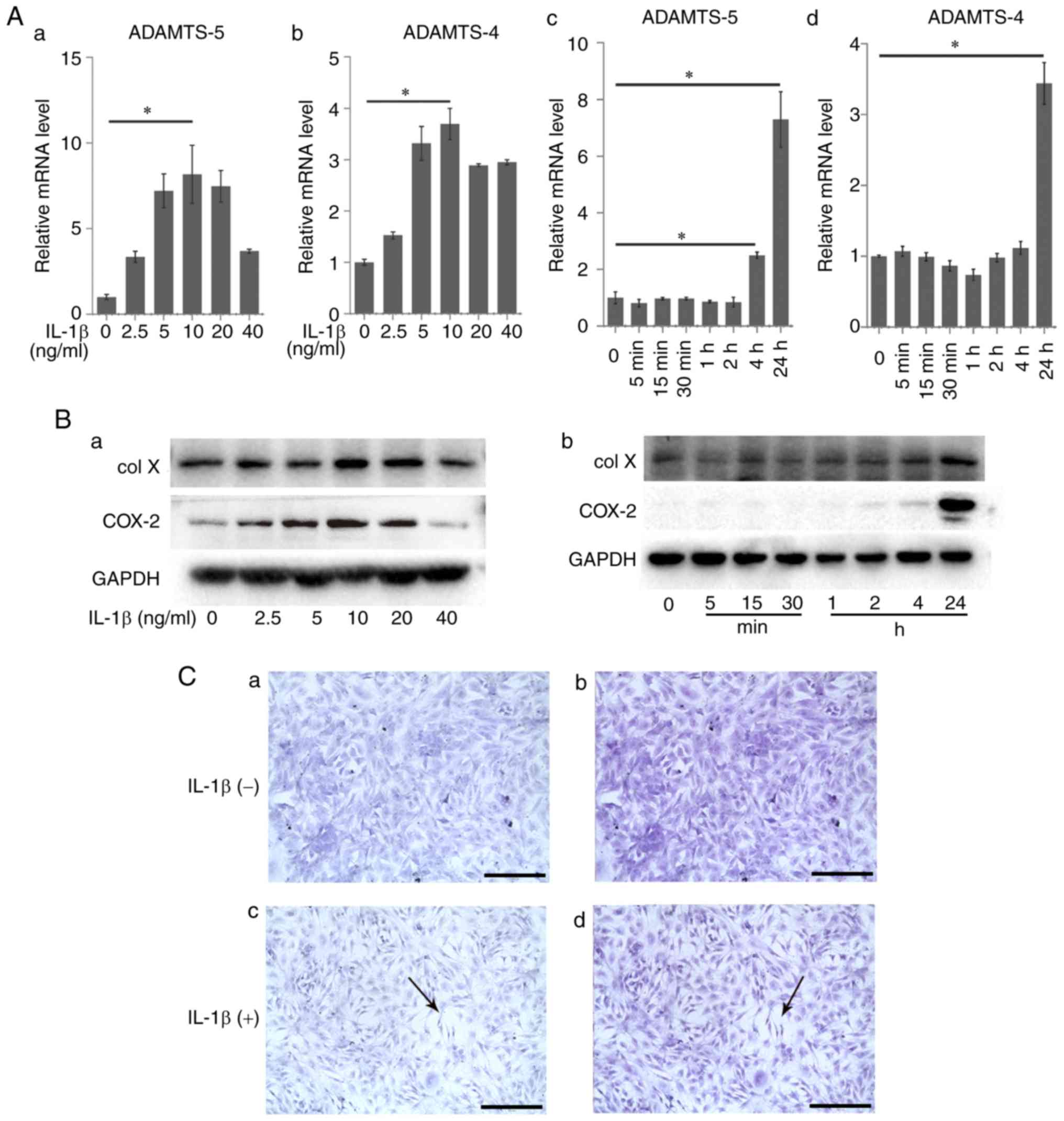
markers were measured via RT-qPCR (Fig. 1A) and western blot analysis
(Fig. 1B). The expression of
ADAMTS-5 (8.17±1.71) and ADAMTS-4 (3.69±0.31) increased
significantly following treatment with 10 ng/ml IL-1β compared with
the control group (1.00±0.16 and 1.00±0.06; P<0.05; Fig. 1A–a and –b). The expression of col
X and COX-2 also increased following IL-1β treatment (Fig. 1B–a). The expression of ADAMTS-5
was constant from the beginning of the experiment (1.00±0.21) to 2
h (0.85±0.17; P>0.05); then it significantly increased at 4 h
(2.50±0.12; P<0.05) and reached a peak at 24 h (7.29±0.98;
P<0.05; Fig. 1A–c). Other
indicators, ADAMTS-4, col X and COX-2, demonstrated similar effects
(Fig. 1A–d and B–b). The results
demonstrated that 10 ng/ml was the appropriate concentration for
further steps and the increase in OA-related markers was
time-dependent.
Morphological observation of rACs
Untreated rACs were small, exhibiting an identical
distribution, and were almost triangular or polygonal. The
secretory vesicles and organelles in the cytoplasm exhibited a
prolific distribution (Fig.
1C–a). Toluidine blue staining demonstrated that the ECM was
rich in proteoglycans and no abnormal cells were observed (Fig. 1C–b). ECM degradation, cell
shrinkage and intercellular broadening were observed following
IL-1β treatment. Most cells were elongated and acquired a narrow
triangular or spindle shape (Fig.
1C–c). Toluidine blue staining demonstrated morphological
changes including degradation of ECM and a decrease in the content
of proteoglycans (Fig. 1C–d).
IL-1β promotes the expression of ADAMTS-4
and ADAMTS-5 through the activation of MAPK signaling
The present study examined the effects of treatment
with IL-1β on the phosphorylation of 3 members of the MAPK family
in rACs. The phosphorylation of p38 did not demonstrate notable
activation following IL-1β treatment. However, ERK1/2 demonstrated
a rapid increase in the first 5 min, reaching a peak at 15 min and
then decreased gradually. Meanwhile, JNK demonstrated a rapid
increase at 15 min. remained constant at 30 min and then decreased
rapidly. Levels of the housekeeping gene GAPDH did not change
during this period (Fig. 2A).
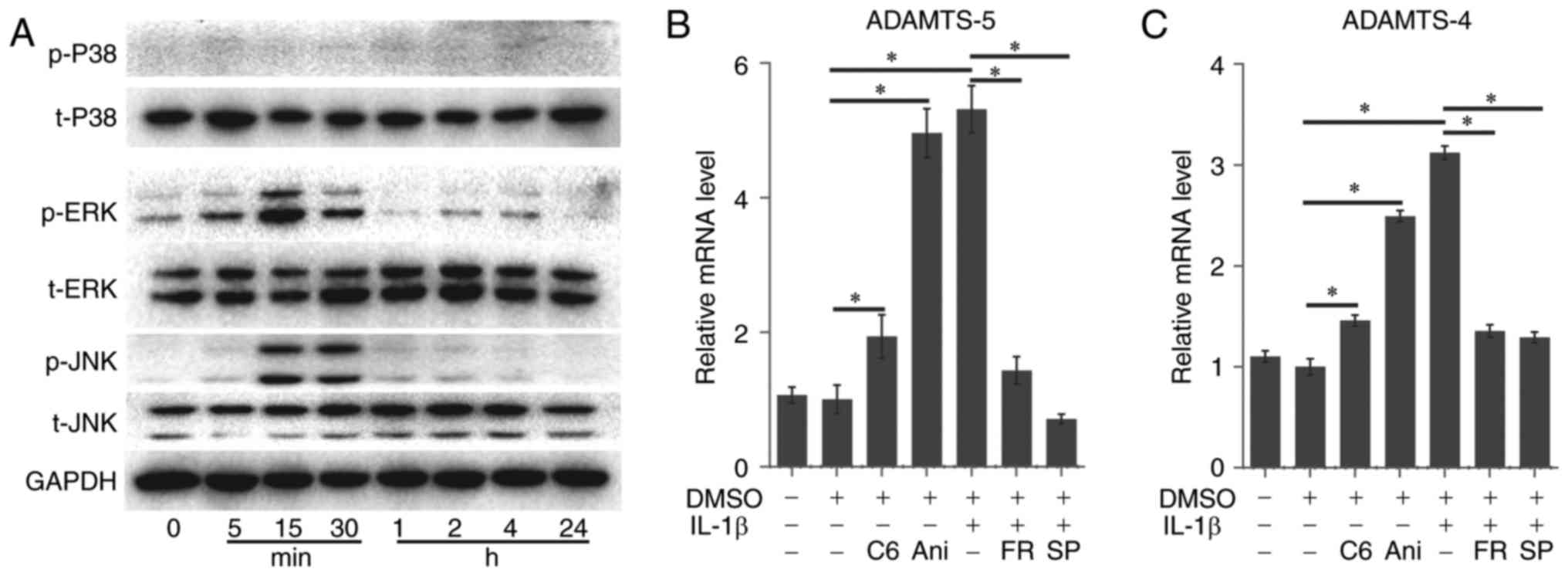 | Figure 2IL-1β promoted the expression of
ADAMTS-4 and ADAMTS-5 through the activation of MAPK signaling. (A)
Of the 3 MAPK isoforms, ERK1/2 and JNK were markedly phosphorylated
following treatment with IL-1β. The gene expression of (B) ADAMTS-5
and (C) ADAMTS-4 was facilitated by C6 (ERK activator) or Ani (JNK
activator). The IL-1β-induced gene expression of (B) ADAMTS-5 and
(C) ADAMTS-4 was inhibited by FR (ERK inhibitor) or SP (JNK
inhibitor). Data present the means ± standard deviation, n=3.
*P<0.05. IL, interleukin; ADAMTS, a disintegrin and
metalloproteinase with thrombospondin motifs; MAPK,
mitogen-activated protein kinase; ERK, extracellular
signal-regulated kinase; JNK, c-Jun N-terminal kinase; DMSO,
dimethyl sulfoxide; C6, Ceramide C6; Ani, Anisomycin; FR, FR180204;
SP, SP600125; p, phosphorylated; t, total. |
The expression of ADAMTS-5 (1.00±0.21 vs. 1.06±0.12)
and ADAMTS-4 (1.00±0.08 vs. 1.10±0.06) demonstrated no evident
change in the negative control (NC) group using DMSO alone to treat
the cartilage cells compared with the blank control group (Fig. 2B and C).
The specific activators of JNK (anisomycin, 1
µM) and ERK (ceramide C6, 50 µM) significantly
upregulated the expression of ADAMTS-5 (1.94±0.32 and 4.96±0.36 vs.
1.00±0.21; P<0.05) and ADAMTS-4 (1.46±0.05 and 2.49±0.06 vs.
1.00±0.21; P<0.05), respectively, compared with the NC group
(Fig. 2B and 2C).
The specific inhibitors of JNK (SP600125, 10 mM;
0.71±0.07 and 1.29±0.05) and ERK (FR180204, 50 µM; 1.43±0.21
and 1.35±0.06) also significantly blocked the IL-1β-induced
expression compared with the positive control group (5.31±0.35 and
3.12±0.07; P<0.05; Fig. 2B and
C).
However, the ERK activator-induced expression could
not reach the peak in the positive control group even at a higher
concentration, suggesting that ERK may serve a subordinate role in
the upregulation of ADAMTS-4 and ADAMTS-5.
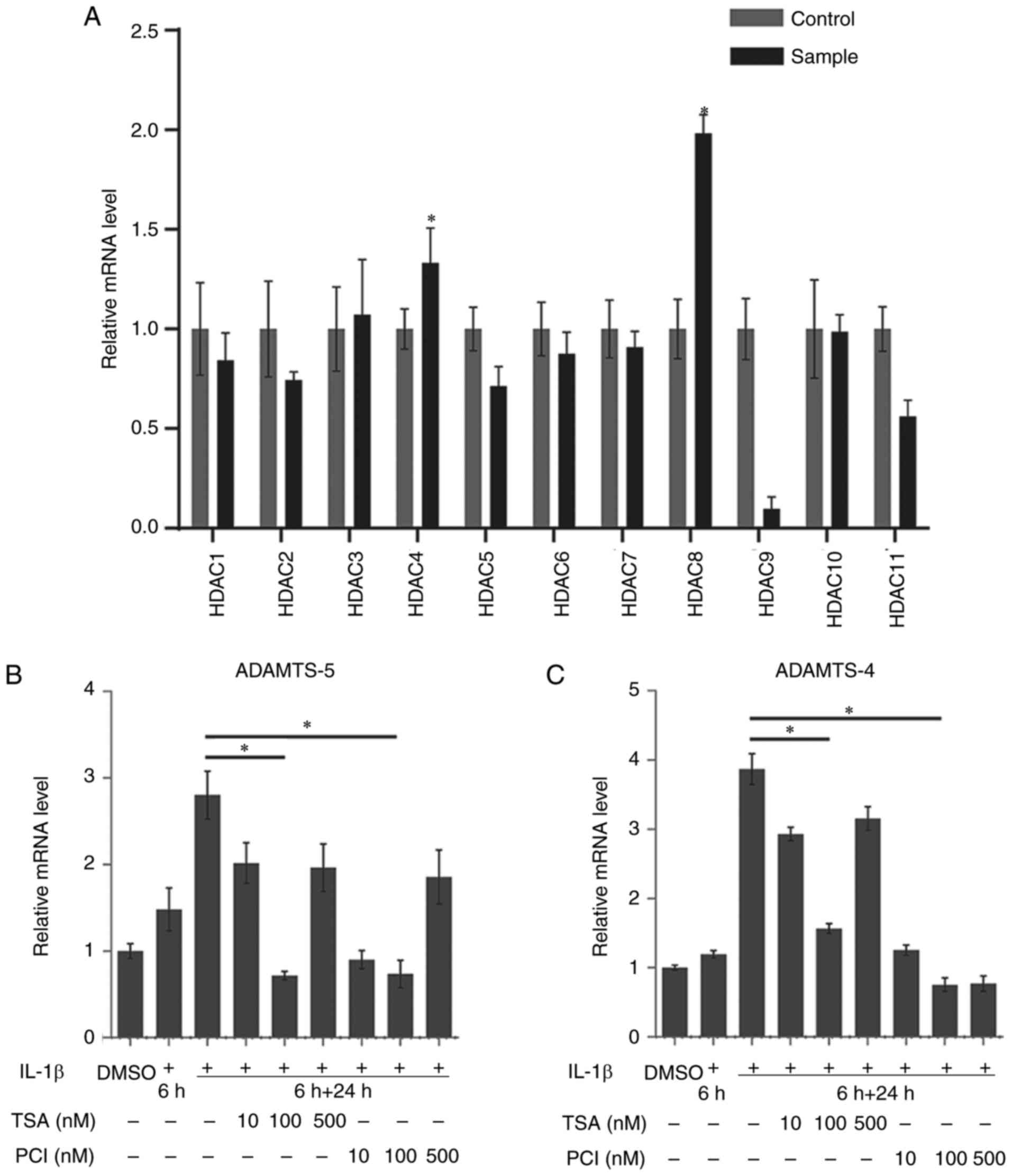
Inhibitory effect and mechanism of HDAC
in an in vitro OA model
The expression of HDAC1-11 mRNA was evaluated using
RT-qPCR. The expression of HDAC4 (1.33±0.17 vs. 1.00±0.10;
P<0.05) and HDAC8 (1.98±0.09 vs. 1.00±0.15; P<0.05) was
upregulated following IL-1β treatment (Fig. 3A). This suggested that HDAC4 and
HDAC8 may be associated with the upregulation of the expression of
ADAMTS-5 in IL-1β-treated chondrocytes.
In rACs stimulated with IL-1β (10 ng/ml) for 6 h and
treated or not treated with TSA or PCI-34051 for 24 h, the
expression of ADAMTS-5 and ADAMTS-4 were observed over 30 h. TSA at
a concentration of 100 nM (0.72±0.25) and PCI-34051 at a
concentration of 100 nM demonstrated a significant inhibitory
effect on the IL-1β-induced expression of ADAMTS-5 (2.80±0.28 and
0.74±0.16; P<0.05; Fig. 3B).
TSA at a concentration of 100 nM (1.57±0.07) and PCI-34051 at 100
nM (0.76±0.10) demonstrated a similar inhibitory effect on the
expression of ADAMTS-4 (3.87±0.22; P<0.05; Fig. 3C). These results demonstrated that
HDAC inhibitors were able to downregulate the expression of ADAMTS
in IL-1β-stimulated rACs and therefore a concentration of 100 nM
for TSA and PCI-34051 was selected for subsequent experiments.
TSA- or PCI-pretreated cells were exposed to IL-1β.
ERK phosphorylation demonstrated a marked decrease in the TSA group
compared with the positive control group, but no apparent change in
the PCI group was observed. However, JNK phosphorylation
demonstrated a marked decrease in the two groups, the latter
appearing to be more evident (Fig.
4A).
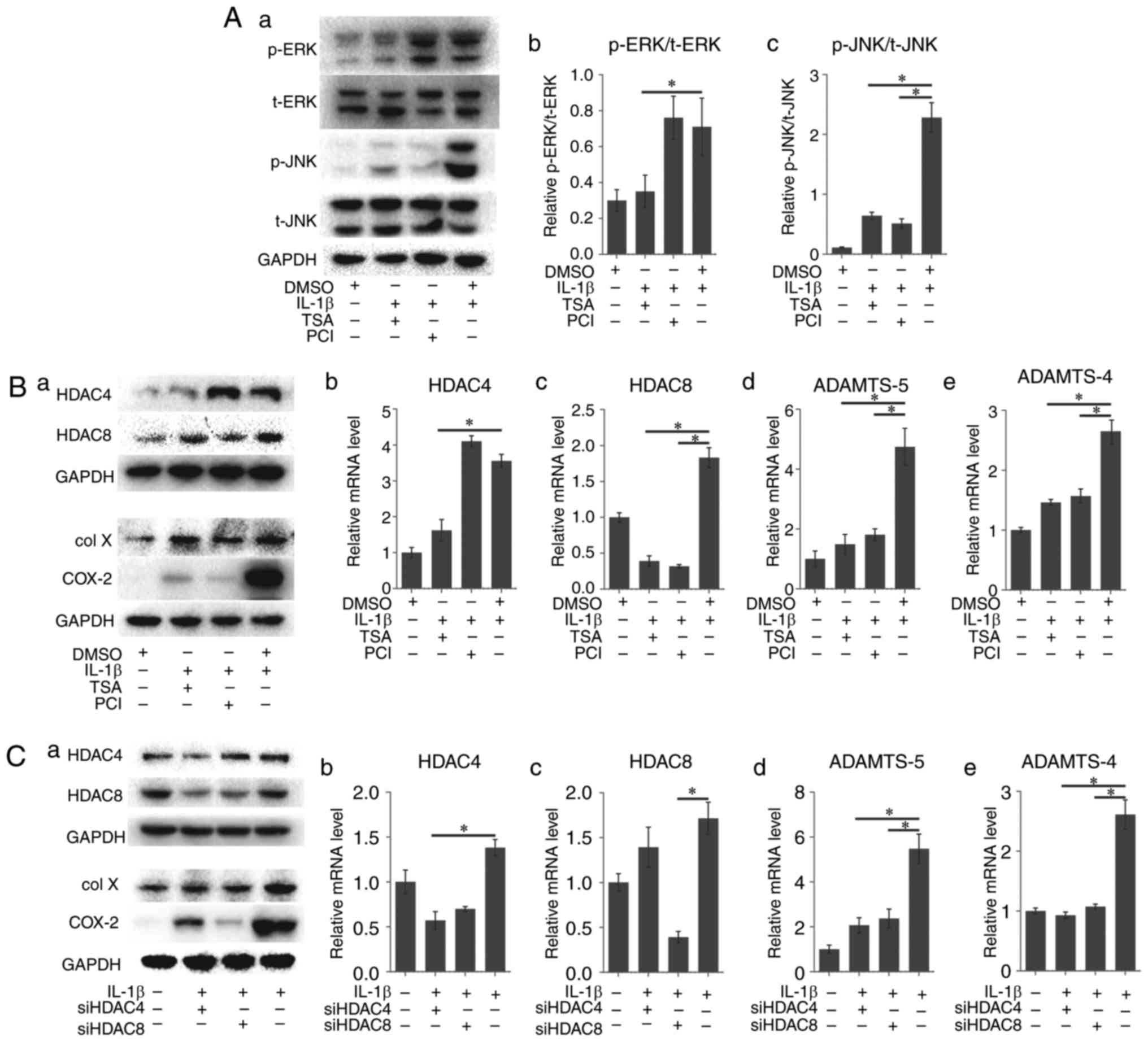 | Figure 4HDAC regulates the
osteoarthritis-related genes and protein expression of rACs via
ERK1/2 and JNK. (A) IL-1β-induced ERK1/2 phosphorylation was
blocked by TSA, and not blocked by PCI; JNK phosphorylation was
blocked by TSA and PCI. (B) HDAC inhibitors (TSA or PCI) suppressed
HDAC expression at both mRNA and protein level, and prevented the
upregulation of osteoarthritis-related genes (col X, COX2, ADAMTS-5
and ADAMTS-4) expression induced by IL-1β. Results are presented
with respect to GAPDH expression. (C) siRNAs (targeting HDAC4 or
HDAC8) suppressed HDAC expression at both mRNA and protein level,
and prevented the upregulation of osteoarthritis-related gene
expression induced by IL-1β. Results are expressed with respect to
GAPDH expression. Data represent the means ± standard deviation,
n=3. *P<0.05. HDAC, histone deacetylase; ERK,
extracellular signal-regulated kinase; JNK, c-Jun N-terminal
kinase; IL, interleukin; TSA, Trichostatin A; DMSO, dimethyl
sulfoxide; PCI, PCI-34051; col X, type X collagen; COX2,
cyclooxygenase; ADAMTS, a disintegrin and metalloproteinase with
thrombospondin motifs; siRNA, small interfering RNA; p,
phosphorylated; t, total. rACs, rat articular chondrocytes. |
The expression of col X and COX-2 as analyzed via
western blotting demonstrated downregulation compared with the
IL-1β-treated group; the PCI-pretreated group demonstrated a
stronger effect (Fig. 4B–a). The
expression of HDAC4 in the TSA group (1.62±0.30 vs. 3.56±0.18;
P<0.05; Fig. 4B–b) and that of
HDAC8 in the PCI group decreased significantly (0.32±0.02 vs.
1.83±0.14; P<0.05; Fig. 4B–c).
The expression of ADAMTS-5 was significantly lower in the
pretreated groups than in the positive control group (1.49±0.33 and
1.81±0.20 vs. 4.74±0.62, P<0.05; Fig. 4B–d), in addition to the expression
of ADAMTS-4 (1.46±0.05 and 1.57±0.12 vs. 2.65±0.20, P<0.05;
Fig. 4B–e). These results
demonstrated that HDAC inhibitors were able to block MAPK pathways
and downregulate OA-related markers at various levels.
The cells pretreated with specific siRNAs targeting
HDAC4 or HDAC8 transfection were stimulated with IL-1β, which
demonstrated a similar effect as that of HDAC inhibitors.
The expression of col X and COX-2 as analyzed by
western blotting demonstrated similar effects; the HDAC8 siRNA
pretreated group demonstrated a stronger inhibitory expression of
COX-2 (Fig. 4C–a). The expression
of HDAC4 decreased in the siHDAC4 group (0.57±0.10 vs. 1.38±0.09;
P<0.05; Fig. 4C–b) whereas
that of HDAC8 decreased in the siHDAC8 group (0.39±0.06 vs.
1.72±0.18; P<0.05; Fig. 4C–c).
The expression of ADAMTS-4 and ADAMTS-5 was similar to that in the
inhibitor-pretreated groups (P<0.05; Fig. 4C–d and C–e).
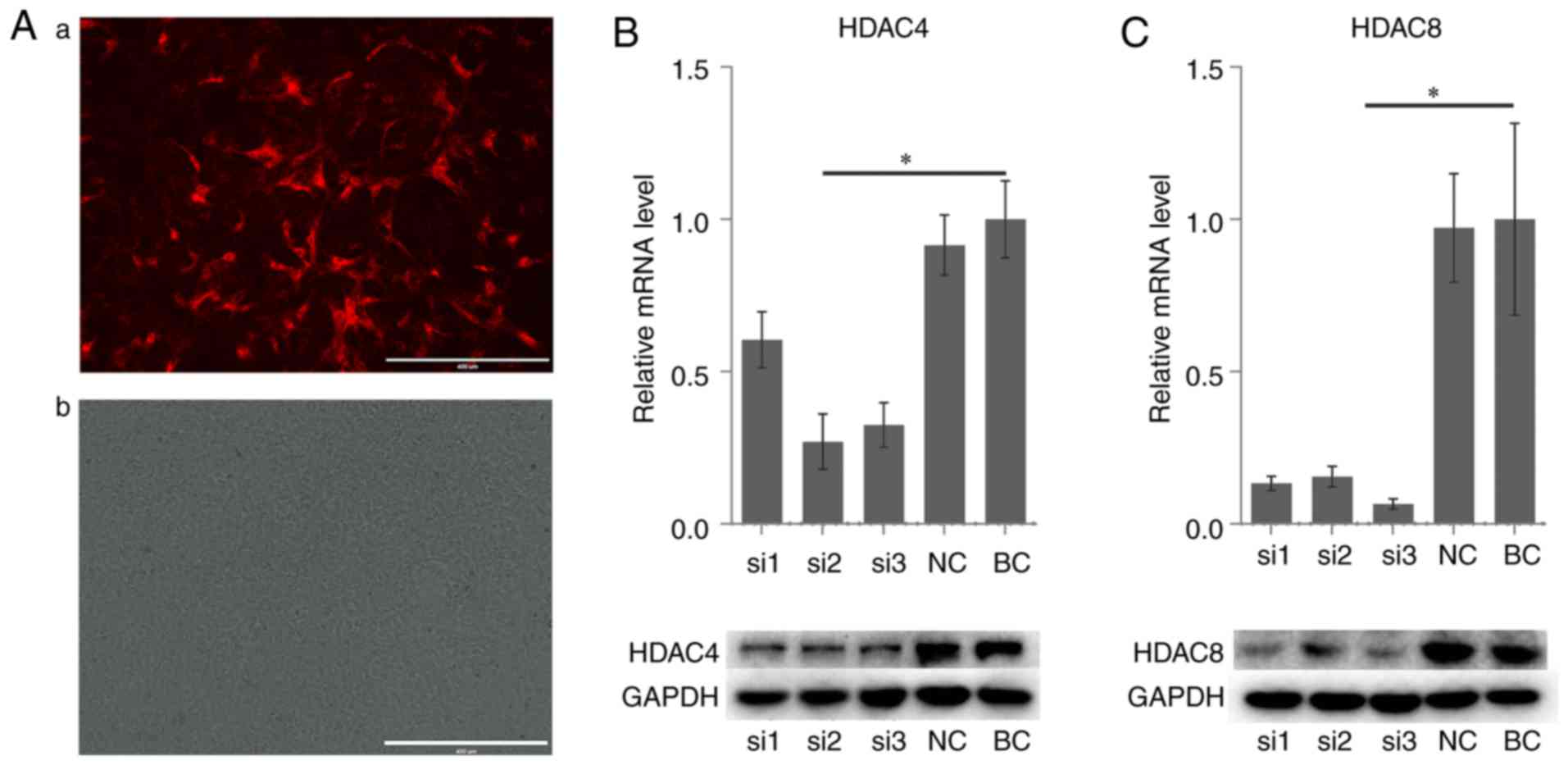
Effect of siRNA on HDAC4 and HDAC8 gene
expression in rACs
Subsequently, 3 pairs of siRNA for each gene were
used to silence the mRNA expression of HDAC4 and HDAC8 in the rACs.
A non-targeting, scrambled siRNA was used as the negative control.
The transfection efficiency was observed under a fluorescence
microscope at 24 h following transfection. The red fluorescence
cells were considered to be transfected successfully (Fig. 5A). All groups demonstrated a
downregulation in the mRNA and protein expression of HDAC4
(Fig. 5B) and HDAC8 (Fig. 5C). Appropriate sequences based on
the results were selected for subsequent experiments.
Discussion
The most characteristic feature of OA is the large
amount of proinflammatory cytokines in articular cartilage produced
by synovial cells and progressive damage of cartilage. As current
therapies for OA act only on symptoms and do not prevent the
pathogenesis of OA (29),
researchers have attempted to find effective agents that may
inhibit the degeneration and catabolism of articular cartilage. In
addition, no reliable indicator exists for the early diagnosis of
OA. Therefore, investigating the mechanism for diagnosis and remedy
is important. The modulation of synthesis or activity of cartilage
degradation-related genes including MMPs and ADAMTS has been one of
the major targets in this endeavor.
Previous researchers have focused on tissue
engineering to repair the damage of articular cartilage (49–52). However, this type of cartilage may
degrade under inflammatory conditions. Although certain stem
cell-based therapies may be useful for chondrogenic differentiation
under a stable environment (53–56), they may induce osteogenic
differentiation under an OA environment. Therefore, research on
anti-inflammatory agents is urgent (57).
Numerous studies have demonstrated that IL-1β may be
an important trigger in the progression of OA (13,58). A previous study also suggested
ADAMTS-5 as a downstream indicator for OA therapy and demonstrated
that a single dose of ADAMTS-5 siRNA lentivirus injection may
prevent cartilage degradation (27). Whether a combination with ADAMTS-4
silencing can achieve an improved effect has been explored
(59,60). As a single inhibitor for one type
of ADAMTS may be inefficient and possess a number of side effects,
the investigation of upstream regulation is crucial. An in
vitro model of OA was successfully constructed in the present
study and the IL-1β-induced increased expression of ADAMTS-4 and
ADAMTS-5, and overproduction of col X and COX-2, were observed in
cultured chondrocytes.
MAPK is a group of serine/threonine protein kinases
involved in the cellular signal transduction. The members of this
signaling pathway group include p38, JNK and ERK1/2 (61). Previous studies have demonstrated
the effect of MAPK signals on tumor metastasis (62,63), osteogenesis (64,65), or certain inflammatory diseases
(66). The IL-1β activation of
ADAMTS requires the integration of several signaling pathways and
activated transcription factors. The present study investigated the
regulation and mechanisms of cartilage inflammation and degradation
in an in vitro model. The effects of IL-1β-induced MAPK
activation in chondrocytes was also explored, and this demonstrated
that IL-1β-induced expression of ADAMTSs was mediated by the MAPK
family signal transduction molecules. Following treatment with
IL-1β, the phosphorylation of ERK and JNK occurred in chondrocytes
in the initial 30 min; the upregulation of the expression of
ADAMTS-4 and ADAMTS-5 occurred at a later time, suggesting a
chronological order. Notably, the ERK activator had less ability to
upregulate the expression of ADAMTS-4 and ADAMTS-5 compared with
IL-1β or JNK activator, suggesting that ERK may serve a subordinate
role in this process or have a synergistic effect with JNK. p38 was
not activated in the present study, unlike in the previous findings
of Saito et al (42). This
discrepancy may be due to cell line disparity.
Epigenetic therapy has been widely researched and
used in laboratory and clinical trials, mainly in tumor or
cardiovascular research and therapy. Several researchers have
investigated HDAC inhibitors in OA therapy, owing to their
anti-inflammatory properties (67). However, no clear consensus exists
as to which type(s) of HDAC serve(s) a decisive role in arthritis
pathogenesis and development. For instance, Saito et al
(42) used two types of HDAC
inhibitors to block the expression of ADAMTS-5 in mechanically
induced cartilage cells, which was presumed to be via class I HDACs
including HDAC1/2/3 or 8. Other researchers have explored the nexus
between HDAC4 and HDAC7 in human OA and proposed specific
mechanisms (68–70). HDAC inhibitor vorinostat can
inhibit the IL-1β-induced expression of MMPs and this effect is
mediated through the phosphorylation of p38 and ERK1/2 (2). These findings demonstrated the
therapeutic role and target in the OA treatment of HDACs. The close
link between ADAMTS or MMPs and HDAC has promoted the studies of
their effects on OA treatment.
The present study selected 2 types of inhibitors.
TSA is a broad-spectrum HDAC inhibitor, but HDAC8 was the only one
not affected by it. PCI-34051 was a selective HDAC inhibitor mainly
targeting HDAC8. It also had an effect on HDAC2. HDAC4 or HDAC8
demonstrated relatively high expression at an early stage in
IL-1β-treated rACs. Blocking HDAC4 or HDAC8 potently suppressed
ADAMTS-4 and ADAMTS-5 production and expression in IL-1β-stimulated
chondrocytes, suggesting that they may be a promising target for
diagnosis and therapy and may be associated with regulation.
As the inhibition of ADAMTS induced by IL-1β was
known to protect from degradation and alleviate the pathology
progression, the effect of inhibition of HDAC4 and HDAC8 on
IL-1β-induced inflammation was encouraging.
The effect of HDAC4 and HDAC8 on JNK and ERK1/2
activation in chondrocytes stimulated with IL-1β was also
investigated in the present study. The effect on p38 was not
studied as no activation in IL-1β-stimulated chondrocytes was
observed. Notably, TSA was revealed to inhibit the activation of
JNK and markedly inhibit the activation of ERK. However, PCI-34051
inhibited the activation of JNK but had almost no effect on the
activation of ERK. These results indicated the possible association
of JNK and ERK1/2 pathway with the selective inhibition via ADAMTS
production. HDAC4 and HDAC8 were associated with this process.
Considering the complex effect of inhibitors, specific siRNAs were
used to repeat the experiment and similar results were achieved,
confirming the regulatory effect of HDAC4 and HDAC8. As other
signaling pathways are also associated with the regulation of
OA-related gene expression, further studies are required to reveal
the precise mechanisms that underlie the regulation.
The inhibitors and siRNAs were revealed to exert a
wide range of effects on the expression of genes and proteins
associated with OA pathogenesis and correlated with the attenuation
of MAPK pathways in rat chondrocytes. The results of the present
study enhanced the understanding of the molecular mechanisms of OA
and suggested that they may be of potential value in treating and
preventing inflammation associated with OA. The lower expression of
HDAC9 and HDAC11 may negatively regulate the OA process, which
merits further studies. Therefore, the findings of the present
study may contribute to the development of anti-OA drugs and
diagnostic methods targeting the HDACs.
In conclusion, the present study demonstrated that
HDAC inhibitors or siRNAs inhibited the IL-1β-induced expression of
ADAMTS-4, ADAMTS-5, col X and COX2, all of which may serve a
pivotal role in the progression of OA, suggesting their inhibitory
effect on cartilage degradation. Therefore, HDAC inhibitors merit
consideration as therapeutic targets in treating and preventing
OA.
Furthermore, the study also demonstrated that this
effect was mediated through the regulation of the inhibition of
phosphorylation of JNK and ERK1/2 (Fig. 6). This suggested that HDAC4 and
HDAC8 may be of diagnostic significance in the early phase of
OA.
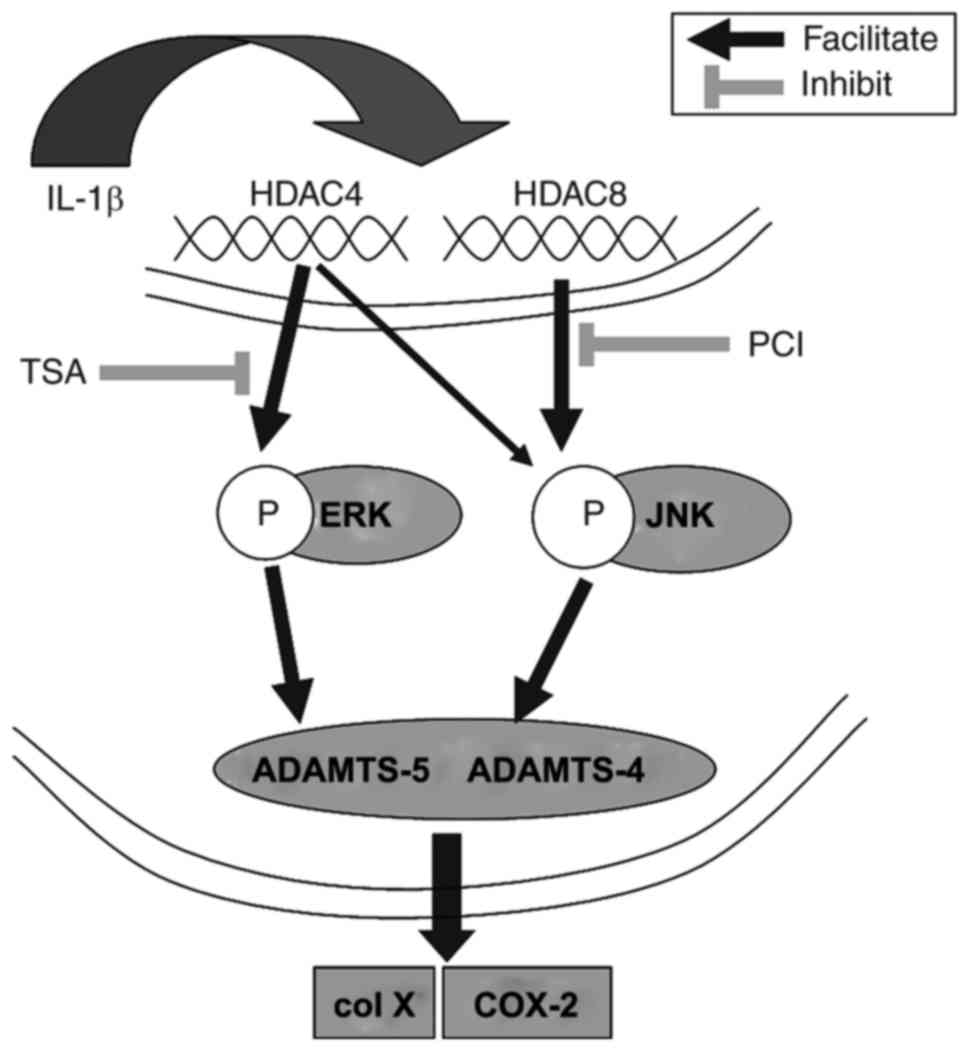 | Figure 6Possible signaling pathways
associated with ADAMTS, col X, and COX-2 regulation by IL-1β in
chondrocytes. ADAMTS, a disintegrin and metalloproteinase with
thrombospondin motifs; col X, type X collagen; COX2,
cyclooxygenase; IL, interleukin; HDAC, histone deacetylase; ERK,
extracellular signal-regulated kinase; JNK, c-Jun N-terminal
kinase; IL, interleukin; TSA, Trichostatin A; PCI, PCI-34051. |
Abbreviations:
|
ADAMTS
|
a disintegrin and metalloproteinase
with thrombospondin motifs
|
|
DMSO
|
dimethyl sulfoxide
|
|
ECM
|
extracellular matrix
|
|
IL-1β
|
interleukin-1β
|
|
MMP
|
matrix metalloproteinase
|
|
NC
|
negative control
|
|
OA
|
osteoarthritis
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TSA
|
Trichostatin A
|
|
siRNA
|
small interfering RNA
|
Acknowledgments
The present study was supported by the Fundamental
Research Funds for the Central Universities (grant no.
2017KFYXJJ104) and the Science and Technology Research Projects of
Wuhan (grant no. 2014060101010048).
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Hayami T, Pickarski M, Wesolowski GA,
McLane J, Bone A, Destefano J, Rodan GA and Duong LT: The role of
subchondral bone remodeling in osteoarthritis: Reduction of
cartilage degeneration and prevention of osteophyte formation by
alendronate in the rat anterior cruciate ligament transection
model. Arthritis Rheum. 50:1193–1206. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhong HM, Ding QH, Chen WP and Luo RB:
Vorinostat, a HDAC inhibitor, showed anti-osteoarthritic activities
through inhibition of iNOS and MMP expression, p38 and ERK
phosphorylation and blocking NF-κB nuclear translocation. Int
Immunopharmacol. 17:329–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tetsunaga T, Nishida K, Furumatsu T,
Naruse K, Hirohata S, Yoshida A, Saito T and Ozaki T: Regulation of
mechanical stress-induced MMP-13 and ADAMTS-5 expression by RUNX-2
transcriptional factor in SW1353 chondrocyte-like cells.
Osteoarthritis Cartilage. 19:222–232. 2011. View Article : Google Scholar
|
|
4
|
Arden N and Nevitt MC: Osteoarthritis.
Epidemiology Best Pract Res Clin Rheumatol. 20:3–25. 2006.
View Article : Google Scholar
|
|
5
|
Goldring MB and Goldring SR:
Osteoarthritis. J Cell Physiol. 213:626–634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cooper C, Javaid MK and Arden N:
Epidemiology of osteoarthritis. Atlas Osteoarthritis. 21–36.
2015.
|
|
7
|
Issa SN and Sharma L: Epidemiology of
osteoarthritis: An update. Curr Rheumatol Rep. 8:7–15. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dancevic CM and McCulloch DR: Current and
emerging therapeutic strategies for preventing inflammation and
aggrecanase-mediated cartilage destruction in arthritis. Arthritis
Res Ther. 16:4292014. View Article : Google Scholar
|
|
9
|
Krader CG: Guidance on non-surgical
management of knee osteoarthritis. Med Econ. 91:122014.PubMed/NCBI
|
|
10
|
McAlindon TE, Bannuru RR, Sullivan MC,
Arden NK, Berenbaum F, Bierma-Zeinstra SM, Hawker GA, Henrotin Y,
Hunter DJ, Kawaguchi H, et al: OARSI guidelines for the
non-surgical management of knee osteoarthritis. Osteoarthritis
Cartilage. 22:363–388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dodge GR and Poole AR: Immunohistochemical
detection and immunochemical analysis of type II collagen
degradation in human normal, rheumatoid, and osteoarthritic
articular cartilages and in explants of bovine articular cartilage
cultured with interleukin 1. J Clin Invest. 83:647–661. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Farrajota K, Cheng S, Martel-Pelletier J,
Afif H, Pelletier JP, Li X, Ranger P and Fahmi H: Inhibition of
interleukin-1beta-induced cyclooxygenase 2 expression in human
synovial fibroblasts by 15-deoxy-Delta12,14-prostaglandin J2
through a histone deacetylase-independent mechanism. Arthritis
Rheum. 52:94–104. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Attur M, Belitskaya-Lévy I, Oh C,
Krasnokutsky S, Greenberg J, Samuels J, Smiles S, Lee S, Patel J,
Al-Mussawir H, et al: Increased interleukin-1β gene expression in
peripheral blood leukocytes is associated with increased pain and
predicts risk for progression of symptomatic knee osteoarthritis.
Arthritis Rheum. 63:1908–1917. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chowdhury TT, Salter DM, Bader DL and Lee
DA: Signal transduction pathways involving p38 MAPK, JNK, NFkappaB
and AP-1 influences the response of chondrocytes cultured in
agarose constructs to IL-1beta and dynamic compression. Inflam Res.
57:306–313. 2008. View Article : Google Scholar
|
|
15
|
Lim H and Kim HP: Matrix
metalloproteinase-13 expression in IL-1β-treated chondrocytes by
activation of the p38 MAPK/c-Fos/AP-1 and JAK/STAT pathways. Arch
Pharm Res. 34:109–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mitchell PG, Magna HA, Reeves LM,
Lopresti-Morrow LL, Yocum SA, Rosner PJ, Geoghegan KF and Hambor
JE: Cloning, expression, and type II collagenolytic activity of
matrix metalloproteinase-13 from human osteoarthritic cartilage. J
Clin Invest. 97:761–768. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lauer-Fields JL, Juska D and Fields GB:
Matrix metalloproteinases and collagen catabolism. Biopolymers.
66:19–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qing C, Wei-ding C and Wei-min F:
Co-culture of chondrocytes and bone marrow mesenchymal stem cells
in vitro enhances the expression of cartilaginous extracellular
matrix components. Braz J Med Biol Res. 44:303–310. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zuo Q, Cui W, Liu F, Wang Q, Chen Z and
Fan W: Co-cultivated mesenchymal stem cells support chondrocytic
differentiation of articular chondrocytes. Int Orthop. 37:747–752.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bolton M: Purification and cloning of
aggrecanase-1. Arthritis Res Ther. 3:667251999.
|
|
21
|
Abbaszade I, Liu RQ, Yang F, Rosenfeld SA,
Ross OH, Link JR, Ellis DM, Tortorella MD, Pratta MA, Hollis JM, et
al: Cloning and characterization of ADAMTS11, an aggrecanase from
the ADAMTS family. J Biol Chem. 274:23443–23450. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Glasson SS, Askew R, Sheppard B, Carito B,
Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, et al:
Deletion of active ADAMTS5 prevents cartilage degradation in a
murine model of osteoarthritis. Nature. 434:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Glasson SS, Askew R, Sheppard B, Carito
BA, Blanchet T, Ma HL, Flannery CR, Kanki K, Wang E, Peluso D, et
al: Characterization of and osteoarthritis susceptibility in
ADAMTS-4-knockout mice. Arthritis Rheum. 50:2547–2558. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rodríguez-Manzaneque JC, Westling J, Thai
SN, Luque A, Knauper V, Murphy G, Sandy JD and Iruela-Arispe ML:
ADAMTS1 cleaves aggrecan at multiple sites and is differentially
inhibited by metalloproteinase inhibitors. Biochem Biophys Res
Commun. 293:501–508. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Collins-Racie LA, Flannery CR, Zeng W,
Corcoran C, Annis-Freeman B, Agostino MJ, Arai M, DiBlasio-Smith E,
Dorner AJ, Georgiadis KE, et al: ADAMTS-8 exhibits aggrecanase
activity and is expressed in human articular cartilage. Matrix
Biol. 23:219–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dancevic CM, Fraser FW, Smith AD, Stupka
N, Ward AC and Mcculloch DR: Biosynthesis and expression of a
disintegrin-like and metalloproteinase domain with thrombospondin-1
repeats-15: A novel versican-cleaving proteoglycanase. J Biol Chem.
288:37267–37276. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chu X, You H, Yuan X, Zhao W, Li W and Guo
X: Protective effect of lentivirus-mediated siRNA targeting
ADAMTS-5 on cartilage degradation in a rat model of osteoarthritis.
Int J Mol Med. 31:1222–1228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan HW, Liu GY, Zhao CF, Li XF and Yang
XY: Differential expression of COX-2 in osteoarthritis and
rheumatoid arthritis. Genet Mol Res. 14:12872–12879. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su SC, Tanimoto K, Tanne Y, Kunimatsu R,
Hirose N, Mitsuyoshi T, Okamoto Y and Tanne K: Celecoxib exerts
protective effects on extracellular matrix metabolism of mandibular
condylar chondrocytes under excessive mechanical stress.
Osteoarthritis Cartilage. 22:845–851. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Herold C, Ganslmayer M, Ocker M, Hermann
M, Geerts A, Hahn EG and Schuppan D: The histone-deacetylase
inhibitor Trichostatin A blocks proliferation and triggers
apoptotic programs in hepatoma cells. J Hepatol. 36:233–240. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen J, Lai J, Yang L, Ruan G, Chaugai S,
Ning Q, Chen C and Wang DW: Trimetazidine prevents
macrophage-mediated septic myocardial dysfunction via activation of
the histone deacetylase sirtuin 1. Br J Pharmacol. 173:545–561.
2016. View Article : Google Scholar
|
|
32
|
Yan M, Chen C, Gong W, Yin Z, Zhou L,
Chaugai S and Wang DW: miR-21-3p regulates cardiac hypertrophic
response by targeting histone deacetylase-8. Cardiovasc Res.
105:340–352. 2015. View Article : Google Scholar
|
|
33
|
Adcock IM: HDAC inhibitors as
anti-inflammatory agents. Brit J Pharmacol. 150:829–831. 2007.
View Article : Google Scholar
|
|
34
|
Halili MA, Andrews MR, Labzin LI, Schroder
K, Matthias G, Cao C, Lovelace E, Reid RC, Le GT, Hume DA, et al:
Differential effects of selective HDAC inhibitors on macrophage
inflammatory responses to the Toll-like receptor 4 agonist LPS. J
Leukocyte Biol. 87:1103–1114. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sweet MJ, Shakespear MR, Kamal NA and
Fairlie DP: HDAC inhibitors: Modulating leukocyte differentiation,
survival, proliferation and inflammation. Immunol Cell Biol.
90:14–22. 2012. View Article : Google Scholar
|
|
36
|
Shuttleworth SJ, Bailey SG and Townsend
PA: Histone deacetylase inhibitors: New promise in the treatment of
immune and inflammatory diseases. Curr Drug Targets. 11:1430–1438.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song C, Zhu S, Wu C and Kang J: Histone
deacetylase (HDAC) 10 Suppresses cervical cancer metastasis through
inhibition of matrix metalloproteinase (MMP) 2 and 9 Expression. J
Biol Chem. 288:28021–28033. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Young DA, Lakey RL, Pennington CJ, Jones
D, Kevorkian L, Edwards DR, Cawston TE and Clark IM: Histone
deacetylase inhibitors modulate metalloproteinase gene expression
in chondrocytes and block cartilage resorption. Arthritis Res Ther.
7:R503–R512. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chabane N, Zayed N, Afif H, Mfuna-Endam L,
Benderdour M, Boileau C, Martel-Pelletier J, Pelletier JP, Duval N
and Fahmi H: Histone deacetylase inhibitors suppress
interleukin-1beta-induced nitric oxide and prostaglandin E2
production in human chondrocytes. Osteoarthritis Cartilage.
16:1267–1274. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nasu Y, Nishida K, Miyazawa S, Komiyama T,
Kadota Y, Abe N, Yoshida A, Hirohata S, Ohtsuka A and Ozaki T:
Trichostatin A, a histone deacetylase inhibitor, suppresses
synovial inflammation and subsequent cartilage destruction in a
collagen antibody-induced arthritis mouse model. Osteoarthritis
Cartilage. 16:723–732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Song Y, Jacobi JL and Tuan RS:
Inhibition of histone deacetylases antagonized FGF2 and IL-1beta
effects on MMP expression in human articular chondrocytes. Growth
Factors. 27:40–49. 2009. View Article : Google Scholar
|
|
42
|
Saito T, Nishida K, Furumatsu T, Yoshida
A, Ozawa M and Ozaki T: Histone deacetylase inhibitors suppress
mechanical stress-induced expression of RUNX-2 and ADAMTS-5 through
the inhibition of the MAPK signaling pathway in cultured human
chondrocytes. Osteoarthritis Cartilage. 21:165–174. 2013.
View Article : Google Scholar
|
|
43
|
Koshy PJ, Lundy CJ, Rowan AD, Porter S,
Edwards DR, Hogan A, Clark IM and Cawston TE: The modulation of
matrix metalloproteinase and ADAM gene expression in human
chondrocytes by interleukin-1 and oncostatin M: A time-course study
using real-time quantitative reverse transcription-polymerase chain
reaction. Arthritis Rheum. 46:961–967. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tian Y, Yuan W, Fujita N, Wang J, Wang H,
Shapiro IM and Risbud MV: Inflammatory cytokines associated with
degenerative disc disease control aggrecanase-1 (ADAMTS-4)
expression in nucleus pulposus cells through MAPK and NF-κB. Am J
Pathol. 182:2310–2321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pan T, Chen R, Wu D, Cai N, Shi X, Li B
and Pan J: Alpha-Mangostin suppresses interleukin-1β-induced
apoptosis in rat chondrocytes by inhibiting the NF-κB signaling
pathway and delays the progression of osteoarthritis in a rat
model. Int Immunopharmacol. 52:156–162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Feng Z, Zheng W, Li X, Lin J, Xie C, Li H,
Cheng L, Wu A and Ni W: Cryptotanshinone protects against
IL-1β-induced inflammation in human osteoarthritis chondrocytes and
ameliorates the progression of osteoarthritis in mice. Int
Immunopharmacol. 50:161–167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang Q, Zheng G, Feng Z, Tong M, Xu J, Hu
Z, Shang P, Chen Y, Wang C, Lou Y, et al: Wogonoside inhibits IL-1β
induced catabolism and hypertrophy in mouse chondrocyte and
ameliorates murine osteoarthritis. Oncotarget. 8:61440–61456.
2017.PubMed/NCBI
|
|
48
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
49
|
Guo X, Chu X, Li W, Pan Q and You H:
Chondrogenic effect of precartilaginous stem cells following
NLS-TAT cell penetrating peptide-assisted transfection of
eukaryotic hTGFβ3. J Cell Biochem. 114:2588–2594. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kim YI, Ryu JS, Yeo JE, Choi YJ, Kim YS,
Ko K and Koh YG: Overexpression of TGF-β1 enhances chondrogenic
differentiation and proliferation of human synovium-derived stem
cells. Biochem Biophys Res Commun. 450:1593–1599. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pan Q, Li W, Yuan X, Rakhmanov Y, Wang P,
Lu R, Mao Z, Shang X and You H: Chondrogenic effect of cell-based
scaffold of self-assembling peptides/PLGA-PLL loading the hTGFβ3
plasmid DNA. J Mater Sci Mater Med. 27:192016. View Article : Google Scholar
|
|
52
|
You H, Chen A, Liu T, Wang M and Zhang G:
Construction of eukaryotic expression plasmid of hTGF-β3 and its
inducing effect on differentiation of precartilaginous stem cells
into chondroblasts. J Huazhong Univ Sci Technol Med Sci.
31:524–529. 2011. View Article : Google Scholar
|
|
53
|
Bouffi C, Thomas O, Bony C, Giteau A,
Venier-Julienne MC, Jorgensen C, Montero-Menei C and Noël D: The
role of pharmacologically active microcarriers releasing TGF-beta3
in cartilage formation in vivo by mesenchymal stem cells.
Biomaterials. 31:6485–6493. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ding R, Zhang Y, You HB, Li F and Sun K:
Transfecting human transformation growth factor beta 3 gene into
precartilaginous stem cells cultured in three-dimensional
self-assembled peptide nanofiber scaffold of KLD-12. J Clin
Rehabilitative Tissue Eng Res. 14:5339–5343. 2010.
|
|
55
|
Qi J, Chen A, You H, Li K, Zhang D and Guo
F: Proliferation and chondrogenic differentiation of CD105-positive
enriched rat synovium-derived mesenchymal stem cells in
three-dimensional porous scaffolds. Biomed Mater. 6:0150062011.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Danišovič L, Varga I and Polák S: Growth
factors and chondrogenic differentiation of mesenchymal stem cells.
Tissue Cell. 44:69–73. 2012. View Article : Google Scholar
|
|
57
|
Osterman C, McCarthy MB, Cote MP, Beitzel
K, Bradley J, Polkowski G and Mazzocca AD: Platelet-rich plasma
increases anti-inflammatory markers in a human coculture model for
osteoarthritis. Am J Sports Med. 43:1474–1484. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ying X, Chen X, Cheng S, Shen Y, Peng L
and Xu HZ: Piperine inhibits IL-β induced expression of
inflammatory mediators in human osteoarthritis chondrocyte. Int
Immunopharmacol. 17:293–299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Seki S, Asanuma-Abe Y, Masuda K, Kawaguchi
Y, Asanuma K, Muehleman C, Iwai A and Kimura T: Effect of small
interference RNA (siRNA) for ADAMTS5 on intervertebral disc
degeneration in the rabbit anular needle-puncture model. Arthritis
Res Ther. 11:R1662009. View
Article : Google Scholar : PubMed/NCBI
|
|
60
|
Song RH, Tortorella MD, Malfait AM, Alston
JT, Yang Z, Arner EC and Griggs DW: Aggrecan degradation in human
articular cartilage explants is mediated by both ADAMTS-4 and
ADAMTS-5. Arthritis Rheum. 56:575–585. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Seger R and Krebs EG: The MAPK signaling
cascade. FASEB J. 9:726–735. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fliedner SM, Engel T, Lendvai NK,
Shankavaram U, Nölting S, Wesley R, Elkahloun AG, Ungefroren H,
Oldoerp A, Lampert G, et al: Anti-cancer potential of MAPK pathway
inhibition in paragangliomas-effect of different statins on mouse
pheochromocytoma cells. PLoS One. 9:e977122014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ge C, Yang Q, Zhao G, Yu H, Kirkwood KL
and Franceschi RT: Interactions between extracellular
signal-regulated kinase 1/2 and p38 MAP kinase pathways in the
control of RUNX2 phosphorylation and transcriptional activity. J
Bone Miner Res. 27:538–551. 2012. View Article : Google Scholar
|
|
64
|
Li W, Li G, Zhang Y, Wei S, Song M, Wang
W, Yuan X, Wu H and Yang Y: Role of 2 × 7 receptor in the
differentiation of bone marrow stromal cells into osteoblasts and
adipocytes. Exp Cell Res. 339:367–379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li W, Wei S, Liu C, Song M, Wu H and Yang
Y: Regulation of the osteogenic and adipogenic differentiation of
bone marrow-derived stromal cells by extracellular uridine
triphosphate: The role of P2Y2 receptor and ERK1/2 signaling. Int J
Mol Med. 37:63–73. 2016. View Article : Google Scholar :
|
|
66
|
Ismail HM, Yamamoto K, Vincent TL, Nagase
H, Troeberg L and Saklatvala J: Interleukin-1 Acts via the JNK-2
signaling pathway to induce aggrecan degradation by human
chondrocytes. Arthritis Rheumatol. 67:1826–1836. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang JH, Shih KS, Wu YW, Wang AW and Yang
CR: Histone deacetylase inhibitors increase microRNA-146a
expression and enhance negative regulation of interleukin-1β
signaling in osteoarthritis fibroblast-like synoviocytes.
Osteoarthritis Cartilage. 21:1987–1996. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lu J, Sun Y, Ge Q, Teng H and Jiang Q:
Histone deacetylase 4 alters cartilage homeostasis in human
osteoarthritis. BMC Musculoskelet Disord. 15:4382014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Higashiyama R, Miyaki S, Yamashita S,
Yoshitaka T, Lindman G, Ito Y, Sasho T, Takahashi K, Lotz M and
Asahara H: Correlation between MMP-13 and HDAC7 expression in human
knee osteoarthritis. Mod Rheumatol. 20:11–17. 2010. View Article : Google Scholar :
|
|
70
|
Song J, Jin EH, Kim D, Kim KY, Chun CH and
Jin EJ: MicroRNA-222 regulates MMP-13 via targeting HDAC-4 during
osteoarthritis pathogenesis. BBA Clin. 3:79–89. 2015. View Article : Google Scholar : PubMed/NCBI
|