Introduction
The formation of atherosclerotic plaque is the
pathological basis of coronary heart disease (CHD), vascular
complication of hypertension and restenosis after angioplasty or
bypass (1). The phenotypic
transition of vascular smooth muscle cells (VSMCs) plays an
important role in the formation and development of atherosclerosis
(AS) (1). Under normal
physiological condition, the mature VSMCs show differentiated
phenotype, and mainly perform contractile function and the
proliferation and migration are inefficient (2). They can synthezise and express some
contractile markers specific to smooth muscle, including α-SMA,
SM22α and α-SM-actin (3). Unlike
most mature cells such as skeletal muscle cells and cardiomyocytes,
VSMCs are not terminally differentiated cells and have remarkable
plasticity (2,4). VSMCs were able to convert from the
differentiated phenotype to dedifferentiated phenotype which can
ensure rapid adaptation to various environmental stimuli (4). The dedifferentiated phenotype was
characterized by the increased ability of proliferation/migration
and the reduction of contractile ability (5). Though an increasing number of
studies have reported various factors and mechanisms that may
control VSMCs phenotype modulation, further efforts are still
required to elucidate the detailed mechanism and biological
importance of the phenotype modulation of VSMCs.
Bcl-2-associated athanogene 3 (BAG3) is a member of
BAG family and plays an important role in diverse cellular
behaviors including cell apoptosis, autophagy, proliferation,
adhesion, migration and differentiation (6–9).
Accumulating evidence indicates that BAG3 is associated with
various cardiovascular diseases such as myocardial hypertrophy,
dilated cardiomyopathy, Takotsubo cardiomyopathy and chronic heart
failure (10–13). Normal tissues seldom express BAG3,
except for cardiomyocytes and skeletal muscle cells, but its
expression is induced upon exposure to various stressful stimuli
(14). However, there is no
previous investigation of BAG3 function in the phenotype modulation
of VSMCs.
Tumor necrosis factor-related apoptosis inducing
ligand (TRAIL), a member of tumor necrosis factor (TNF) family,
belongs to type II membrane protein (15). TRAIL can interact with five
different receptors, DR4 (TRAIL-R1), DR45 (TRAIL-R2), DcR1
(TRAIL-R3), DcR2 (TRAIL-R4), and OPG. After binding with death
receptors DR4 and DR5, TRAIL can trigger cell apoptosis. Due to
lack of the intracellular death domain, DcR1 and DcR2 are
considered as decoy receptors to protect normal cells from
apoptosis. OPG can work in bone tissue metabolism (16–18). Chiappetta et al reported
that BAG3 could downmodulate the apoptotic response to TRAIL in
human neoplastic thyroid cells (19). However, the relationship between
BAG3 and TRAIL and their effects the proliferation and migration in
VSMCs are rarely reported.
The present study analyzed BAG3 expression in VSMCs
and investigated its role in the phenotypic transition of VSMCs.
Moreover, we determined whether BAG3 could promote the
proliferation and migration of VSMCs via TRAIL.
Materials and methods
Ethics statement
Animals used in our study were treated in accordance
with the National Institutes of Health (NIH) Guide for the Care and
Use of Laboratory Animals. The procedures was in accordance with
the ethical standards of the Committee on Animal Experimentation of
China Medical University (project identification code,
SCXK-2013-0001).
Materials
Real-Time PCR system was purchased from Applied
Biosystems (Foster City, CA, USA). Click-iT Nascent RNA Capture kit
was purchased from Invitrogen Life Technologies (Carlsbad, CA,
USA). Western blotting related equipment was purchased from
Invitrogen Life Technologies. EdU Alexa Fluor 555 Imaging kit was
purchased from Invitrogen Life Technologies™. The real-time cell
analyzer (RTCA) xCELLigence system was purchased from ACEA
Biosciences (San Diego, CA, USA). Fluorescence microscope
(CKX41-F32FL) was purchased from Olympus (Tokyo, Japan). MicroChemi
4.2 was purchased from DNR Bio-Imaging Systems, Ltd. (Jerusalem,
Israel). Smart Blotter SB-10 was purchased from Wealtech (Reno, NV,
USA). Transwell related equipment was purchased from BD Biosciences
(Franklin Lakes, NJ, USA). Microplate reader was purchased from
Bio-Rad Laboratories, Inc. (Hercules, CA, USA). Antibodies for
α-SM-actin, BAG3, DcR1, DcR2, DR4, DR5, OPG and glyceraldehyde
3-phosphate dehydrogenase (GAPDH) were all purchased from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Lentiviral vectors
were purchased from GeneChem (Shanghai, China).
Isolation and culture of primary
VSMCs
Neonatal rats 1–2 days old were sacrificed by
cervical dislocation and disinfected with 75% alcohol, then moved
into the clean bench. The thoracic aorta was excised and the
inner/outer layers of blood vessel were removed. And then primary
neonatal rat VSMCs were isolated as previous described by us
(20). VSMCs were cultured in
complete medium including 10% fetal bovine serum (FBS) and
penicillin-streptomycin (100 U/ml-100 µg/ml) at 37°C, 5%
CO2, and with humidified atmosphere as previously
described (20,21). Media were changed every other day.
After primary cells reached 80–90% confluence, 0.25% trypsin was
added into the culture plate for digestion. Thirty seconds later,
serum-containing medium was used to terminate the digestion.
Subsequently, a part of the cells were moved into a new culture
dish. After attachment, cells were incubated with complete medium,
which was passage 2 of VSMCs. With these methods, cells between
passages 1–8 were obtained and applied for the next
experiments.
Transduction of BAG3 and TRAIL to primary
rat VSMCs using lentiviral vectors
The lentiviral plasmids labelled by green
fluorescent protein (GFP) were used in the knockdown experiments,
which contained short hairpin RNA (shRNA) against rat TRAIL or
control shRNA. There were five shRNA oligonucleotides specific for
rat TRAIL, i.e., TRAIL #1, 2, 3, 4 and 5. Furthermore, the
lentiviral plasmids labelled by GFP containing BAG3 cDNA were used
in the overexpression experiments. Transfection of shRNA
oligonucleotide was performed with Lipofectamine 2000 (Invitrogen
Life Technologies) according to the manufacturer's recommendations.
Transduced cells were cultured for 2 days and then proteins were
extracted and analyzed by western blotting.
Cell proliferation assay
Cells were seeded into 6-well plates at a density of
1×104 cells/well and cultured with 10% FBS. The medium
was changed every 2 days or as necessary. Cell number at the
indicated time-point was determined by counting using a
haemocytometer.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 15 min
at room temperature. After washed with phosphate-buffered saline
(PBS) three times, cells were permeabilized with 0.25% Triton X-100
in PBS for 5 min. After blocked with 1% BSA, cells were incubated
with first antibody (1:200) in PBS overnight at 4°C. After washed
with PBS three times, cells were incubated with secondary antibody
for 1 h in the dark. After washed with PBS three times, cells were
incubated with 0.1% DAPI for 5 min at room temperature. Finally
cells were analyzed under a fluorescent microscope.
Western blotting
Western blotting was performed as our previous
studies indicated (20,22). Briefly, cells were solubilized in
a lysis buffer for 30 min, and then total protein concentrations
were measured by a BCA Protein Assay kit. After heat denaturation,
the samples were analyzed on a 12 or 14% Tris-glycine gradient gel,
and then transferred to PVDF membranes and blocked with 5% non-fat
milk in Tris-buffered saline (TBS) for 1.5 h at the room
temperature. The membranes were incubated with primary antibody
overnight at 4°C. After washing three times with TBS, the membranes
were incubated with secondary antibodies for 1.5 h at room
temperature. After washing steps, immunoreactive binding were
detected with enhanced chemiluminescence (ECL) (Amersham
Biosciences, Piscataway, NJ, USA). The band intensity was
quantified using ImageJ 1.47 software and GAPDH as a control.
Dot blot
The secreted proteins in the media were extracted
and quantified similarly to those in western blotting. Then 100
µl sample volume was blotted onto a nitrocellulose membrane
and blocked with 5% non-fat milk for 1.5 h at the room temperature.
Subsequently, the membranes were incubated with first antibody at
1:1,000 dilution overnight at 4°C. After washing three times with
TBS, the membranes were incubated with secondary antibodies for 1 h
at room temperature. After washing three times with TBS,
immunoreactive binding was detected with ECL detection reagent with
MicroChemi 4.2.
MTT assay
MTT assay was used to determined cell viability.
VSMCs were seeded in a 96-well plate at a density of
4×103 cells/well. After cultured 48 h, cells were
incubated with MTT solution (final concentration, 5 mg/ml) for 4 h
at 37°C. Then the culture media containing MTT were removed and
replaced with 100 µl DMSO. Then the plate was gently rotated
on a linear and orbital shaker for 5 min to completely dissolve the
precipitation. The absorbance was measured with microplate reader
at 570 nm. The percentage of cell viability was calculated
according to the following formula: cell viability (%) = optical
density (OD) of the treatment group/OD of the control group
×100%.
RTCA proliferation assays
To analyze the cell proliferation continuously over
time, growth curve assays were performed in RTCA in quadruplicate
with the xCELLigence system according to the methods described
(23). Briefly, 5,000 cells/well
were seeded in RTCA E-plates. After the chambers were set up, the
RTCA E-plate was put into xCELLigence instrument at 37°C, 5%
CO2 incubator and cell index was recorded every 15 min.
The shift of the electrical impedance was expressed as the cell
index, which was a parameter of cell viability.
EdU incorporation analysis
As described by our previous studies (20,24) and the manufacturer's instructions,
DNA synthesis rate in VSMCs was determined by EdU incorporation
analysis using Click-iT™ EdU Alexa Fluor 555 Imaging kit. Briefly,
the cells were incubated with EdU-labeling solution for 8 h at
37°C, and then fixed with 4% cold formaldehyde for 30 min at room
temperature. After permeabilization with 1% Triton X-100, the cells
were reacted with Click-iT reaction cocktails (Invitrogen Life
Technologies) for 30 min. Subsequently, the DNA contents of the
cells were stained with Hoechst 33342 for 30 min. Finally,
EdU-labeled cells were counted using ImageJ 1.47 software and
normalized to the total number of Hoechst-stained cells. At least
500 cells in each experiment were counted, EdU-positive cells were
expressed as a percentage of the total cells.
Wound healing assay
Cells at 80–90% confluence were wounded with a 200
µl pipette tip and incubated with SFM or CM for 24 h.
Multiple views of the leading edge of the scratch were photographed
under a microscope at 0 and 24 h. The experiments were performed
three times independently.
Transwell migration assay
For the Transwell migration assay (8-µm
pores; BD Biosciences), cells were seeded at density of
2×106 cells in the upper chamber. The lower chamber was
filled with SFM or CM. After cultured for 24 h, cells on the upper
chamber were removed by gentle abrasion with a cotton bud, and the
cells on the lower chamber were fixed and stained with Hoechst
33342. Three experiments were performed independently. The cells
that passed through the filter were photographed under fluorescence
microscope with ultraviolet light. Hoechst-labeled cells in five
representative microscopic fields were counted using ImageJ 1.47
software.
RNA extraction and real-time
(RT)-PCR
The total RNA was extracted by Qiagen RNeasy Mini
kit (Qiagen, Berlin, Germany). After the determination of
concentration, the synthesis of cDNA was performed. With the primer
design software we synthesized the sense and antisense primers of
each fragment: TRAIL sense primer,
5′-tcgtgatcttcacagtgctcctgcagtc-3′ and antisense primer,
5′-tctaacgagctgacggagttgccacttg-3′; DcR1 sense primer,
5′-gattacaccaacgcttccaac-3′ and antisense primer,
5′-gctggtgttcattgtctcttc-3′; DcR2 sense primer,
5′-ttcttgcctgctatgtacag-3′ and antisense primer,
5′-aggatggtggtcactgtctc-3′; DR4 sense primer,
5′-gagtacatctaggtgcgttcctgg-3′ and antisense primer,
5′-agagccccacactttgctgg-3′; DR5 sense primer,
5′-tagcactcactggaatgacc-3′ and antisense primer,
5′-gtggacacattcgatgtcac-3′; OPG sense primer,
5′-gcctaactggcttagtgtcttg-3′ and antisense primer,
5′-ccaatgtgccgctgcagctg-3′; GAPDH sense primer,
5′-cgtcccgtagacaaaatggt-3′ and anti-sense primer,
5′-ttgatgttagtggggtctcg-3′. Quantitative RT-PCR was run and
analyzed with the 7500 RT-PCR system (Applied Biosystems). Results
were normalized against those of GAPDH and presented as arbitrary
unit.
Label and capture nascent RNA
Click-iT Nascent RNA Capture kit was used to detect
newly synthesized RNA according to the manufacturer's instructions.
5-Ethymyl uridine (EU) is an alkyne-modified uridine analog and it
could be efficiently and naturally incorporated into the nascent
RNA. Cells were incubated in 0.2 mM of EU for 4 h and total RNA
labeled with EU was isolated using TRIzol reagent (Invitrogen Life
Technologies). Then EU-labeled RNA was biotinylated in a Click-iT
reaction buffer with 0.5 mM of biotin azide and subsequently
captured on streptavidin magnetic beads.
Statistical analysis
All data were obtained from at least three
individual experiments. Continuous variables were expressed as the
mean ± SD and tested by one-way analysis of variance (ANOVA) or
Student's t-test. Categorical variables were expressed as
percentage and tested by Chi-square test. All the statistical
analyses were performed using SPSS statistical software for
Windows, version 17.0 (SPSS, Inc., Chicago, IL, USA) and P-values
<0.05 were considered statistically significant.
Results
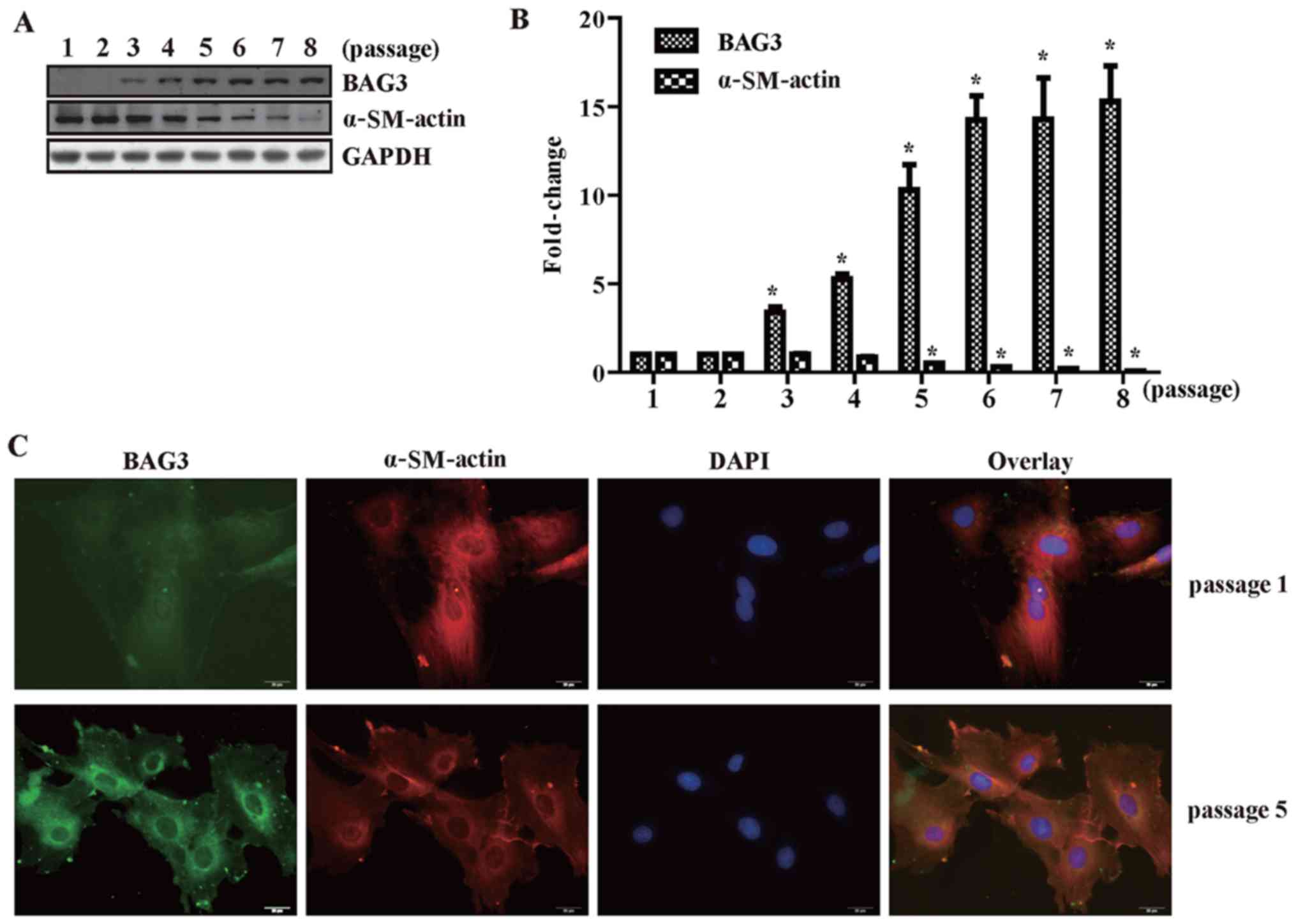
The expression of BAG3 increases with
continued passages in cultured primary VSMCs
BAG3 is rarely expressed in primary VSMCs, which
were mainly in contractile phenotype characterized by expression of
α-SM-actin. We also found that BAG3 expression was negatively
related with VSMC marker protein α-SM-actin. BAG3 expression
increased and α-SM-actin expression decreased with continued
passages in cultured primary VSMCs (Fig. 1A and B). This was furthermore
confirmed by the results of immunofluorescence staining (Fig. 1C). Passage 1 (differentiated
phenotype) and passage 5 (undifferentiated phenotype) were selected
to conduct the immunofluorescence staining assay. The fluorescence
intensity of BAG3 increased observably in VSMCs of passage 5
compared with that in passage 1 (Fig.
1C). On the contrary, the fluorescence intensity of α-SM-actin
decreased observably in VSMCs of passage 5 compared with that in
passage 1 (Fig. 1C).
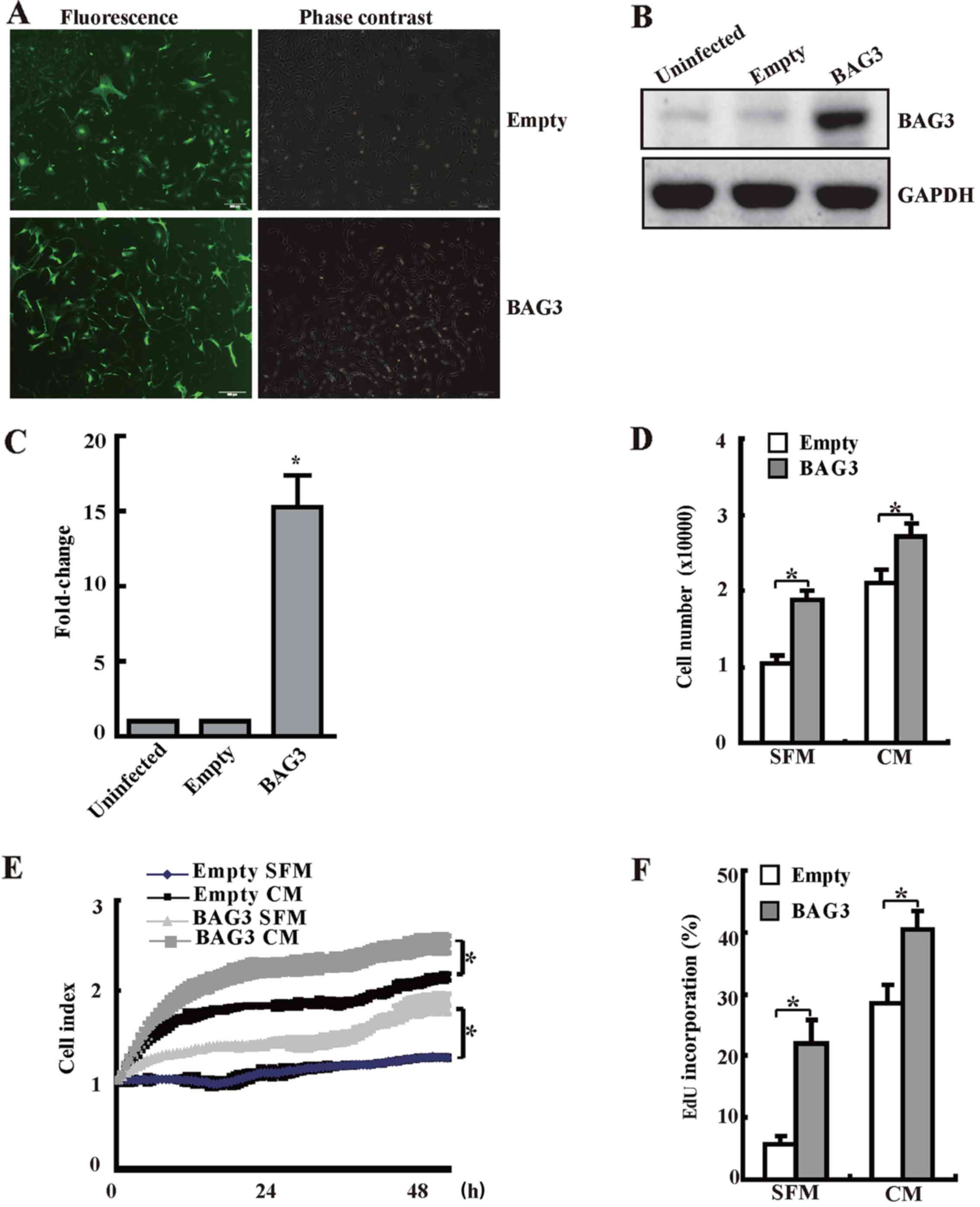
BAG3 promotes the proliferation of
primary rat VSMCs
We have generated lentiviral vectors to overexpress
BAG3 in order to explore the role of BAG3 in proliferation of
primary rat VSMCs. Measurement of GFP+ cells under
fluorescence microscopy demonstrated that transduction efficiency
by lentiviral vectors at 100 multiplicity of infection (MOI) was
80–90% (Fig. 2A). Western
blotting confirmed that BAG3 was successfully expressed in VSMCs
infected with BAG3-containg lentivirus (Fig. 2B and C). Importantly, results from
cell counting (Fig. 2D), RTCA
(Fig. 2E) and EdU staining
(Fig. 2F) consistently
demonstrated that forced overexpression of BAG3 could significantly
promote the proliferation of primary rat VSMCs in both SFM group
(serum free medium) and CM group (complete medium with 10%
FBS).
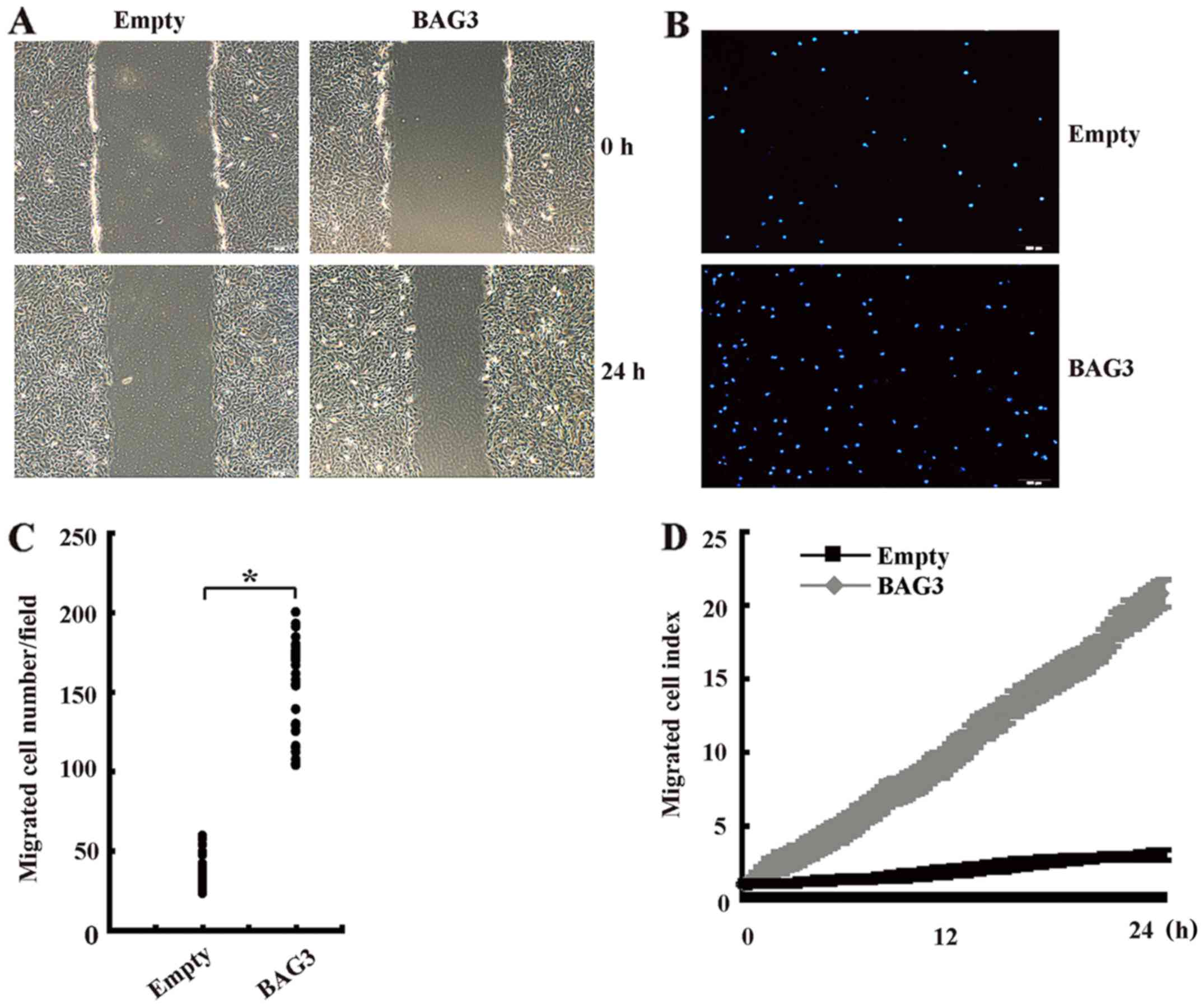
BAG3 promotes the migration of primary
rat VSMCs
Wound healing assay showed that BAG3 significantly
increased the migration of primary rat VSMCs at 24 h compared with
control group (Fig. 3A).
Transwell assay demonstrated that the number of invaded cells in
BAG3-overexpression VSMCs group was significantly higher than those
in the control group (Fig. 3B and
C). RTCA also confirmed that BAG3 promoted migration of primary
rat VSMCs in a time-dependent manner (Fig. 3D).
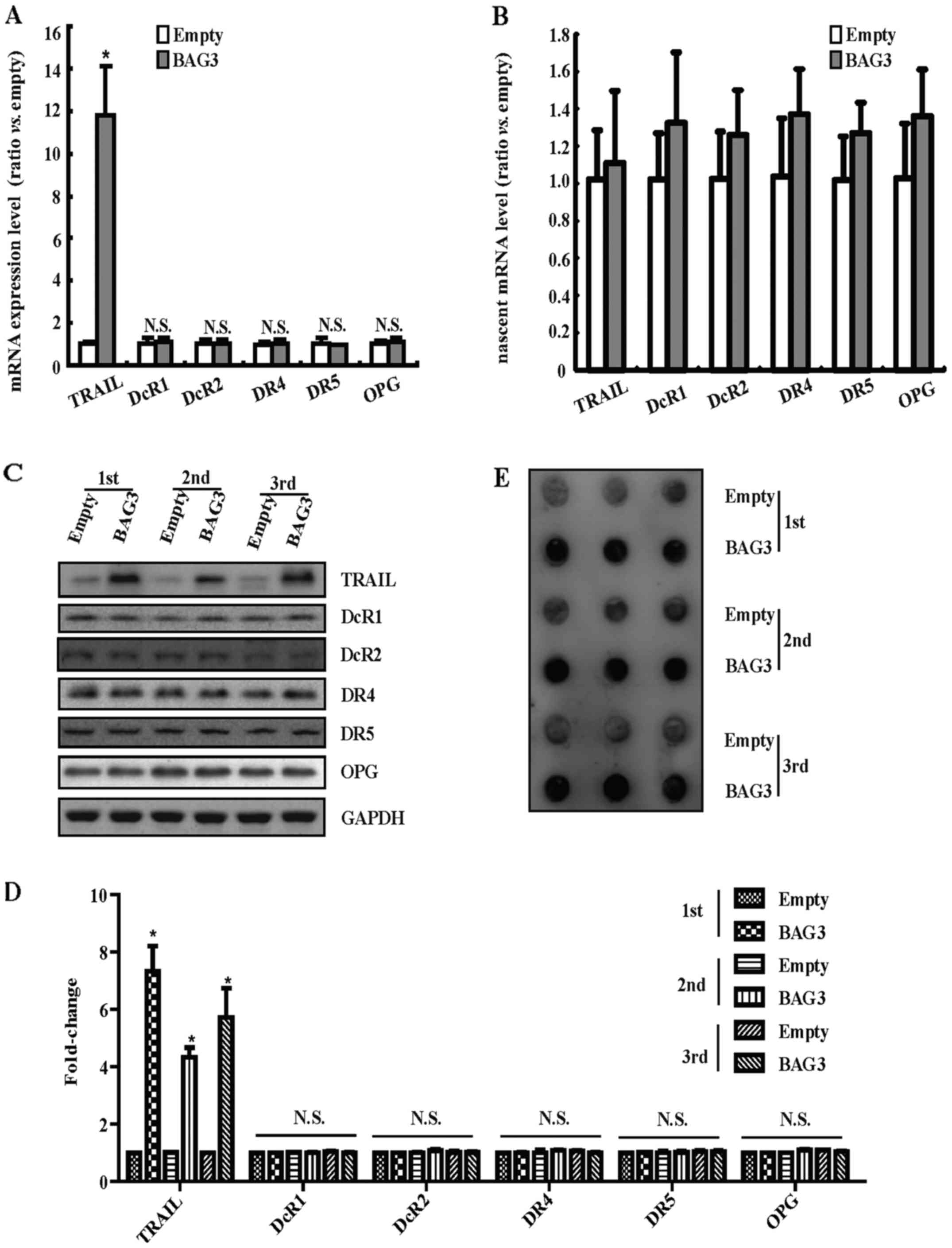
TRAIL was implicated in the proliferation
mediated by BAG3 in primary rat VSMCs
We wanted to clarify the mechanism underlying the
proliferation and migration of primary rat VSMCs promoted by BAG3
overexpression. The results of RT-PCR demonstrated that BAG3
overexpression could significantly increase the mRNA level of
TRAIL, but had no obvious effects on the receptors of TRAIL
including DcR1, DcR2, DR4, DR5 and OPG (Fig. 4A). The increase in mRNA level
could result from the increase of mRNA synthesis or decrease of
mRNA degradation. Next, TRAIL gene transcription was analyzed using
Click-iT Nascent RNA Capture kit to label and isolate newly
synthesized RNA. The results of RT-PCR demonstrated that
BAG3-overexpression did not alter the level of newly synthesized
TRAIL mRNA (Fig. 4B).
Consistently, western blotting demonstrated that BAG3
overexpression could promote the expression of TRAIL protein while
have no effects on the receptors of TRAIL (Fig. 4C and D). Furthermore, dot blot
demonstrated that BAG3-overexpression could increase the level of
secreted TRAIL protein in the cultured media (Fig. 4E).
BAG3 promotes the proliferation of
primary rat VSMCs via TRAIL
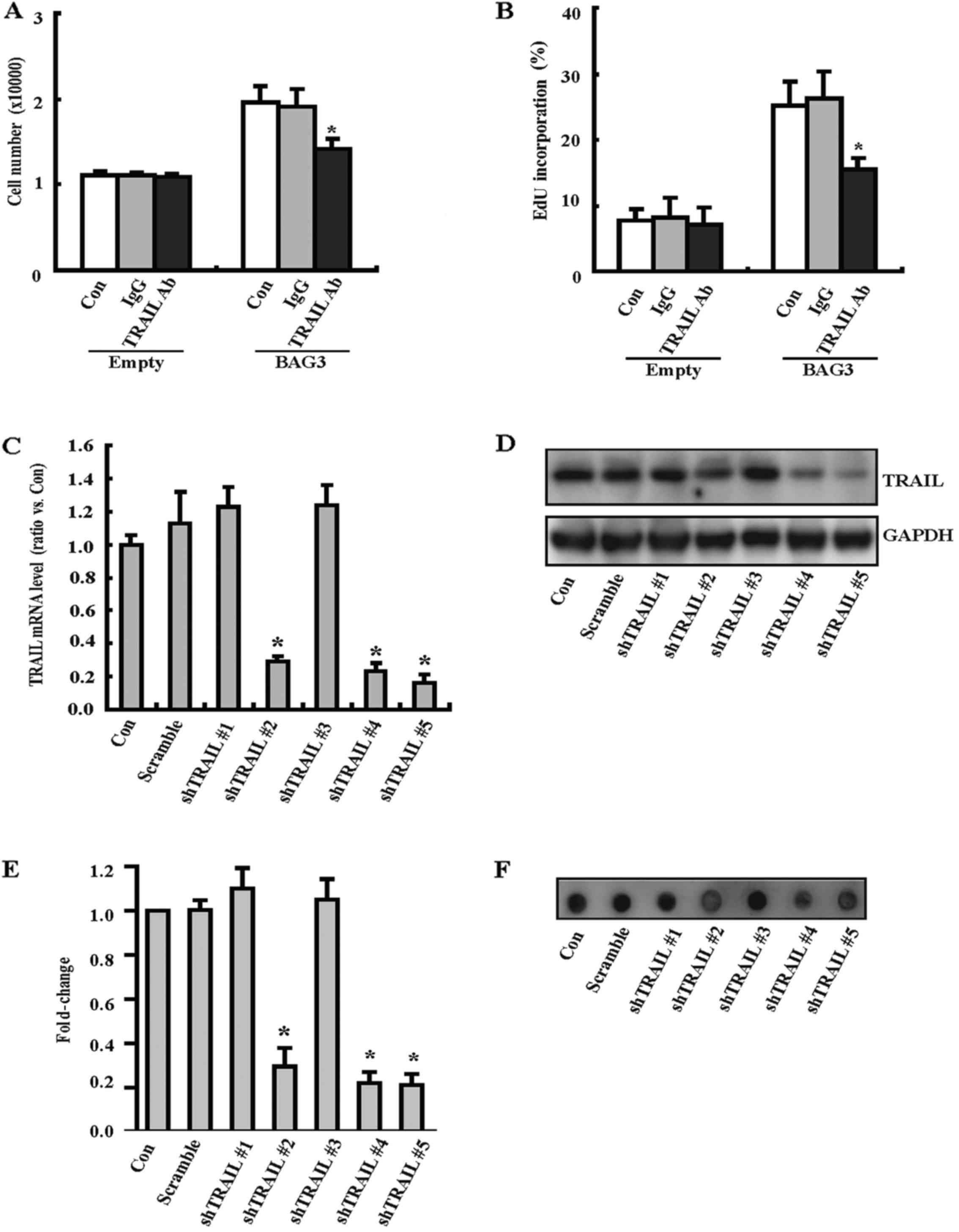
To investigate the potential involvement of TRAIL in
the proliferation of VSMCs induced by BAG3, a neutralizing antibody
against TRAIL (TRAIL Ab) was used. Cells counting demonstrated that
BAG3 overexpression could significantly promote the proliferation
of VSMCs, and this biological process was attenuated by TRAIL Ab
(Fig. 5A). EdU staining also
confirmed that TRAIL Ab could decrease EdU incorporation rate of
VSMCs induced by BAG3 overexpression (Fig. 5B). To further investigate the
involvement of TRAIL, we generated lentiviral vectors containing
shRNAs against TRAIL (shTRAIL) to knock down TRAIL expression in
primary rat VSMCs. RT-PCR demonstrated that three shRNAs
(shTRAIL#2, shTRAIL#4 and shTRAIL#5) significantly decreased the
mRNA level of TRAIL in VSMCs (Fig.
5C). Western blotting demonstrated that shTRAIL#2, shTRAIL#4
and shTRAIL#5 significantly decreased the protein expression of
TRAIL in VSMCs (Fig. 5D and E).
Dot blot exhibited that shTRAIL#2, shTRAIL#4 and shTRAIL#5
significantly decreased the secreted protein of TRAIL in culture
medium (Fig. 5F). Importantly,
cell counting (Fig. 5G) and EdU
staining (Fig. 5H) consistently
demonstrated that shTAIL#2, shTRAIL#4 and shTRAIL#5 could decrease
the proliferation of VSMCs.
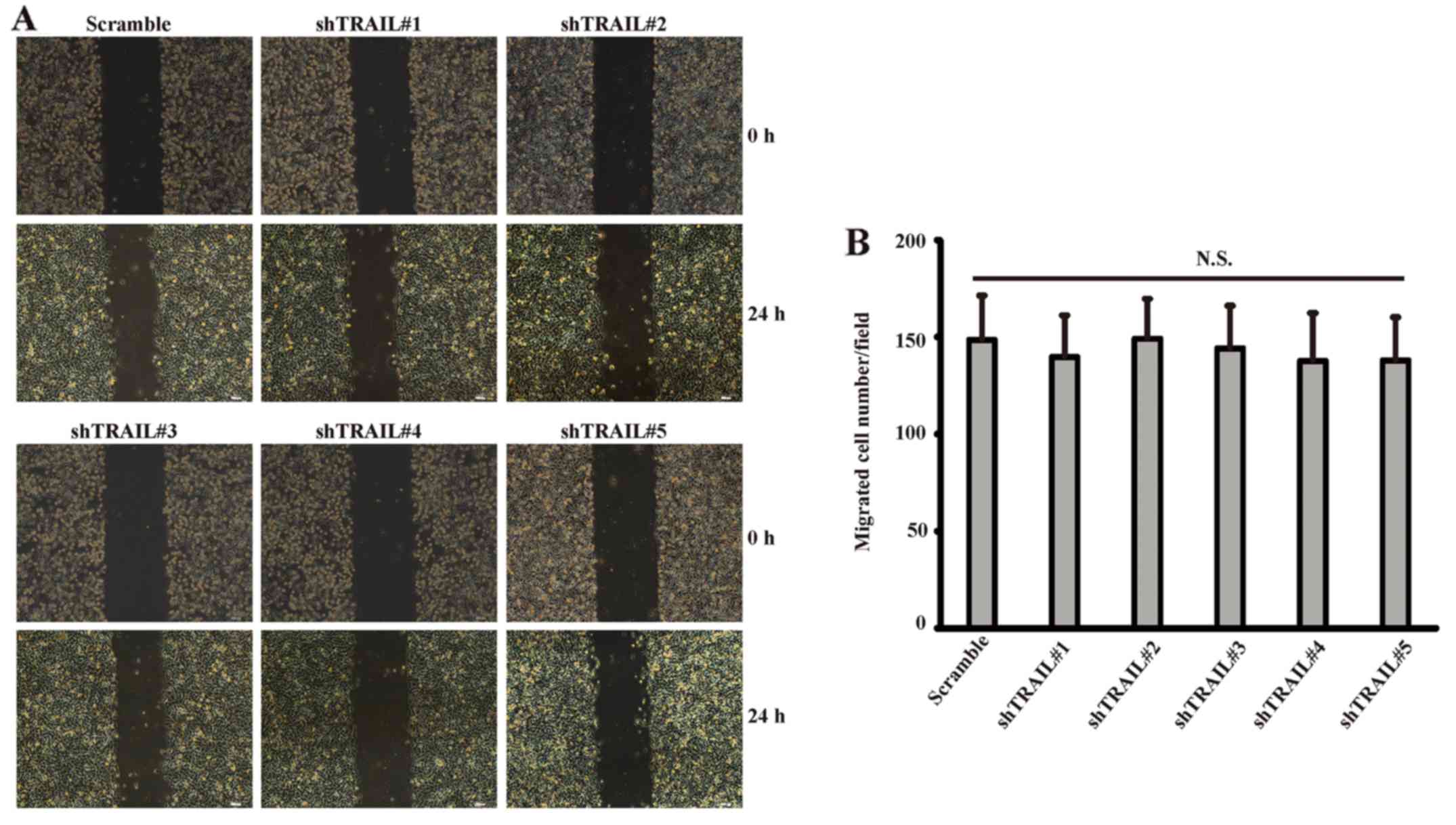
Knockdown of TRAIL exerted no obvious
influence on the migration of primary rat VSMCs
We then investigated the potential involvement of
TRAIL in the migration of primary rat VSMCs. Wound healing assay
(Fig. 6A) demonstrated that
knockdown of TRAIL per se exerted no obvious influence on the
migration of VSMCs. Transwell assay also confirmed that the number
of migrated cells did not have significant statistical differences
between knockdown group and control group (Fig. 6B).
Discussion
The present study demonstrated a novel role for BAG3
in regulating the phenotypic transformation in primary rat VSMCs.
We found that BAG3 expression increased with continued passages in
cultured primary rat VSMCs and promoted the proliferation and
migration of primary rat VSMCs. Furthermore, we demonstrated that
BAG3 promoted the proliferation of primary rat VSMCs via TRAIL.
Several lines of evidence suggested that the
expression of BAG3 was elevated in various tumors including
glioblastoma, acute lymphoblastic leukemia and prostate carcinoma
(25–27). However, normal tissues seldom
express BAG3, except for cardiomyocytes and skeletal muscle cells
(14). The expression of BAG3 can
be induced by a variety of stimuli such as the early growth
response gene-1 (Egr-1) (28),
heat shock factor 1 (HSF1) (29),
proteasome inhibitors (30),
heavy metals as well as heat stress (31,32). Previous studies indicated that
BAG3 is involved in various CVD such as myocardial hypertrophy,
dilated cardiomyopathy, Takotsubo cardiomyopathy and chronic heart
failure (10–13). In the present study, we first
confirmed that the expression of BAG3 increased with continued
passages in cultured primary rat VSMCs.
In this study, we also demonstrated for the first
time that overexpression of BAG3 could promote the proliferation
and migration of primary rat VSMCs. BAG3 contains three main
protein binding motifs that can mediate potential interactions with
chaperons and/or other proteins, a WW domain at the N-terminal, a
proline-rich region (PXXP) in the central region, two IPV motifs
(Ile-Pro-Val) between the WW domain and the PXXP region (33). Previous study found that in Cos7
cells, BAG3 could promote the movement and adhesion by binding to
the PPDY structure of the guanine nucleotide exchange factor 2
(PDZ-GEF2) through WW domain (34). In MDA435 human breast cancer
cells, BAG3 overexpression contributed to the decrease of
migration, which is related to PXXP domain (35). Meng et al reported silence
of BAG3 can elevate migratory capacity by activating the
transcription of ZEB1 in thyroid cancer cells (36). We speculated BAG3 may play
different roles in different cells. It is well known that cancer
cells are characterized by infinite proliferation, migration and
invasion, and similar changes occurred in VSMCs during the
formation of atheromatous plaque. Therefore, we hypothesized BAG3
may be a common signaling molecule of the two pathological
processes. However, further efforts are still required to elucidate
which domain played the main role.
Both BAG3 and TRAIL are involved in apoptosis and
immune responses to human immunodeficiency virus (HIV)-1. Existing
evidence showed that BAG3 acted synergistically with Bcl-2 and Bax
in antagonizing the intrinsic apoptosis induced by TRAIL. In the
host cell of HIV-1 infection, BAG3 and Bcl-2 resist apoptosis
induced by TRAIL together, which may be an important factor for the
chronicity of HIV infection (37,38). Furthermore, Chiappetta et
al demonstrated silencing BAG3 could downgrade the apoptotic
response to TRAIL in human neoplastic thyroid cells (19). However, the association between
BAG3 and TRAIL and their role in the phenotypic transformation of
VSMCs has not been fully clarified. The present study demonstrated
for the first time that BAG3 upregulation could promote the
expression of TRAIL in primary rat VSMCs, which may be realized by
decreasing the degradation of TRAIL mRNA. Furthermore, the present
study also demonstrated BAG3 promotes the proliferation of primary
rat VSMCs via TRAIL. BAG3 is an upstream signal-regulated molecule
of TRAIL. We originally predicted that knockdown of TRAIL may
affect BAG3-induced migration. However, no effect was observed in
migration assay, which indicated that other signal-regulated
molecules may exist.
The present study demonstrated for the first time
that BAG3 promotes the proliferation and migration in primary rat
VSMCs. BAG3 promoted VSMCs proliferation via the expression of
TRAIL. The result revealed incomplete mechanism of atherosclerotic
plaque and provided a possible drug target.
Acknowledgments
Not applicable.
Notes
[1]
Funding
This study was supported by National Natural Science
Foundation of China (grant nos. 81470417 and 81670231).
[2] Availability
of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
[3] Authors'
contributions
YF and YS made substantial contributions to
conception and design. YF, YeC, SC, YL, YiC, GS, SY, NY and CL
cooperated with each other to perform the experiments in this
manuscript. YF and YeC were major contributors in writing the
manuscript. YS gave the final approval of the version to be
published. All authors read and approved the final manuscript.
[4] Ethics
approval and consent to participate
Not applicable.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Doran AC, Meller N and McNamara CA: Role
of smooth muscle cells in the initiation and early progression of
atherosclerosis. Arterioscler Thromb Vasc Biol. 28:812–819. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rzucidlo EM, Martin KA and Powell RJ:
Regulation of vascular smooth muscle cell differentiation. J Vasc
Surg. 45(Suppl A): 25–32. 2007. View Article : Google Scholar
|
|
3
|
Owens GK: Regulation of differentiation of
vascular smooth muscle cells. Physiol Rev. 75:487–517. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Owens GK, Kumar MS and Wamhoff BR:
Molecular regulation of vascular smooth muscle cell differentiation
in development and disease. Physiol Rev. 84:767–801. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Orr AW, Hastings NE, Blackman BR and
Wamhoff BR: Complex regulation and function of the inflammatory
smooth muscle cell phenotype in atherosclerosis. J Vasc Res.
47:168–180. 2010. View Article : Google Scholar :
|
|
6
|
Iwasaki M, Homma S, Hishiya A, Dolezal SJ,
Reed JC and Takayama S: BAG3 regulates motility and adhesion of
epithelial cancer cells. Cancer Res. 67:10252–10259. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bloemberg D, McDonald E, Dulay D and
Quadrilatero J: Autophagy is altered in skeletal and cardiac muscle
of spontaneously hypertensive rats. Acta Physiol (Oxf).
210:381–391. 2014. View Article : Google Scholar
|
|
8
|
Liu BQ, Du ZX, Zong ZH, Li C, Li N, Zhang
Q, Kong DH and Wang HQ: BAG3-dependent noncanonical autophagy
induced by proteasome inhibition in HepG2 cells. Autophagy.
9:905–916. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rosati A, Graziano V, De Laurenzi V,
Pascale M and Turco MC: BAG3: a multifaceted protein that regulates
major cell pathways. Cell Death Dis. 2:e1412011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Falco A, Festa M, Basile A, Rosati A,
Pascale M, Florenzano F, Nori SL, Nicolin V, Di Benedetto M,
Vecchione ML, et al: BAG3 controls angiogenesis through regulation
of ERK phosphorylation. Oncogene. 31:5153–5161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Norton N, Li D, Rieder MJ, Siegfried JD,
Rampersaud E, Züchner S, Mangos S, Gonzalez-Quintana J, Wang L,
McGee S, et al: Genome-wide studies of copy number variation and
exome sequencing identify rare variants in BAG3 as a cause of
dilated cardiomyopathy. Am J Hum Genet. 88:273–282. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Citro R, d'Avenia M, De Marco M, Giudice
R, Mirra M, Ravera A, Silverio A, Farina R, Silvestri F, Gravina P,
et al: Polymorphisms of the antiapoptotic protein bag3 may play a
role in the pathogenesis of tako-tsubo cardiomyopathy. Int J
Cardiol. 168:1663–1665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Marco M, Falco A, Basile A, Rosati A,
Festa M, d'Avenia M, Pascale M, Dal Piaz F, Bisogni R, Barcaroli D,
et al: Detection of soluble BAG3 and anti-BAG3 antibodies in
patients with chronic heart failure. Cell Death Dis. 4:e4952013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hishiya A, Kitazawa T and Takayama S: BAG3
and Hsc70 interact with actin capping protein CapZ to maintain
myofibrillar integrity under mechanical stress. Circ Res.
107:1220–1231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA,
et al: Identification and characterization of a new member of the
TNF family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan G, O'Rourke K, Chinnaiyan AM, Gentz R,
Ebner R, Ni J and Dixit VM: The receptor for the cytotoxic ligand
TRAIL. Science. 276:111–113. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pan G, Ni J, Wei YF, Yu G, Gentz R and
Dixit VM: An antagonist decoy receptor and a death
domain-containing receptor for TRAIL. Science. 277:815–818. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Emery JG, McDonnell P, Burke MB, Deen KC,
Lyn S, Silverman C, Dul E, Appelbaum ER, Eichman C, DiPrinzio R, et
al: Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J
Biol Chem. 273:14363–14367. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiappetta G, Ammirante M, Basile A,
Rosati A, Festa M, Monaco M, Vuttariello E, Pasquinelli R, Arra C,
Zerilli M, et al: The antiapoptotic protein BAG3 is expressed in
thyroid carcinomas and modulates apoptosis mediated by tumor
necrosis factor-related apoptosis-inducing ligand. J Clin
Endocrinol Metab. 92:1159–1163. 2007. View Article : Google Scholar
|
|
20
|
Ma M, Guo X, Chang Y, Li C, Meng X, Li S,
Du ZX, Wang HQ and Sun Y: Advanced glycation end products promote
proliferation and suppress autophagy via reduction of cathepsin D
in rat vascular smooth muscle cells. Mol Cell Biochem. 403:73–83.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salabei JK, Cummins TD, Singh M, Jones SP,
Bhatnagar A and Hill BG: PDGF-mediated autophagy regulates vascular
smooth muscle cell phenotype and resistance to oxidative stress.
Biochem J. 451:375–388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang Y, Li Y, Ye N, Guo X, Li Z, Sun G
and Sun Y: Atorvastatin inhibits the apoptosis of human umbilical
vein endothelial cells induced by angiotensin II via the
lysosomal-mitochondrial axis. Apoptosis. 21:977–996. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ke N, Wang X, Xu X and Abassi YA: The
xCELLigence system for real-time and label-free monitoring of cell
viability. Methods Mol Biol. 740:33–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen S, Liu B, Kong D, Li S, Li C, Wang H
and Sun Y: Atorvastatin calcium inhibits phenotypic modulation of
PDGF-BB-induced VSMCs via down-regulation the Akt signaling
pathway. PLoS One. 10:e01225772015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gentilella A and Khalili K: BAG3
expression in glioblastoma cells promotes accumulation of
ubiquitinated clients in an Hsp70-dependent manner. J Biol Chem.
286:9205–9215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu H, Wu W, Fu Y, Shen W, Miao K, Hong M,
Xu W, Young KH, Liu P and Li J: Overexpressed BAG3 is a potential
therapeutic target in chronic lymphocytic leukemia. Ann Hematol.
93:425–435. 2014. View Article : Google Scholar
|
|
27
|
Staibano S, Mascolo M, Di Benedetto M,
Vecchione ML, Ilardi G, Di Lorenzo G, Autorino R, Salerno V, Morena
A, Rocco A, et al: BAG3 protein delocalisation in prostate
carcinoma. Tumour Biol. 31:461–469. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gentilella A, Passiatore G, Deshmane S,
Turco MC and Khalili K: Activation of BAG3 by Egr-1 in response to
FGF-2 in neuroblastoma cells. Oncogene. 27:5011–5018. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Franceschelli S, Rosati A, Lerose R, De
Nicola S, Turco MC and Pascale M: Bag3 gene expression is regulated
by heat shock factor 1. J Cell Physiol. 215:575–577. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du ZX, Meng X, Zhang HY, Guan Y and Wang
HQ: Caspase-dependent cleavage of BAG3 in proteasome
inhibitors-induced apoptosis in thyroid cancer cells. Biochem
Biophys Res Commun. 369:894–898. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liao Q, Ozawa F, Friess H, Zimmermann A,
Takayama S, Reed JC, Kleeff J and Büchler MW: The anti-apoptotic
protein BAG-3 is overexpressed in pancreatic cancer and induced by
heat stress in pancreatic cancer cell lines. FEBS Lett.
503:151–157. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pagliuca MG, Lerose R, Cigliano S and
Leone A: Regulation by heavy metals and temperature of the human
BAG-3 gene, a modulator of Hsp70 activity. FEBS Lett. 541:11–15.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gamerdinger M, Carra S and Behl C:
Emerging roles of molecular chaperones and co-chaperones in
selective autophagy: focus on BAG proteins. J Mol Med (Berl).
89:1175–1182. 2011. View Article : Google Scholar
|
|
34
|
Iwasaki M, Tanaka R, Hishiya A, Homma S,
Reed JC and Takayama S: BAG3 directly associates with guanine
nucleotide exchange factor of Rap1, PDZGEF2, and regulates cell
adhesion. Biochem Biophys Res Commun. 400:413–418. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kassis JN, Virador VM, Guancial EA, Kimm
D, Ho AS, Mishra M, Chuang EY, Cook J, Gius D and Kohn EC: Genomic
and phenotypic analysis reveals a key role for CCN1 (CYR61) in
BAG3-modulated adhesion and invasion. J Pathol. 218:495–504. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Meng X, Kong DH, Li N, Zong ZH, Liu BQ, Du
ZX, Guan Y, Cao L and Wang HQ: Knockdown of BAG3 induces
epithelial-mesenchymal transition in thyroid cancer cells through
ZEB1 activation. Cell Death Dis. 5:e10922014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang Y, Erdmann N, Zhao J and Zheng J:
The signaling and apoptotic effects of TNF-related
apoptosis-inducing ligand in HIV-1 associated dementia. Neurotox
Res. 8:135–148. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cummins NW and Badley AD: Mechanisms of
HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis.
1:e992010. View Article : Google Scholar : PubMed/NCBI
|