Introduction
Collagen is a ubiquitous structural protein. There
are more than 20 different types of collagen, with specific
functions in each tissue (1,2).
These proteins have important roles in the maintenance of the
extracellular matrix environment (3-7).
Certain studies have demonstrated that collagen regulates cell
proliferation or apoptosis (8,9).
In this decade, collagens of marine origin (e.g., fish, sponges and
mollusks) have been considered a useful resource due to their high
availability (10-17). These collagens have been widely
used as functional foods or dietary supplements. Collagen has also
been used for skin substitutes and drug delivery vehicles (18-23).
Recently, collagen peptides (CPs), derived from
chemical and enzymatic collagen hydrolysis (24,25), have been increasingly used as
functional materials, due to their various bioactivities and high
bioavailability (26,27). Several studies have demonstrated
the beneficial effects of CPs. For instance, CPs derived from fish
skin were demonstrated to have several protective effects on skin
photo-aging and wound healing, as they improved moisture retention
and repaired endogenous collagen and elastin protein fibers
(28-32). Therefore, CPs are considered a
useful material for the development of cosmetics, pharmaceuticals
and medical products.
Previous studies by our group reported that tissue
from soft-shelled turtle, Pelodiscus sinensis, may be a
useful alternative source of collagen (33). Due its ability to induce
keratinocytes to enter the epithelial-mesenchymal transition (EMT),
which facilitates wound healing, this collagen may be a useful
component of pharmaceuticals and medical products (34). Furthermore, CPs from soft-shelled
turtle may have beneficial effects on skin. However, due to
differences in habitat environments, collagen from soft-shelled
turtle may differ greatly from collagen from mammalian or marine
sources, in terms of physicochemical properties, amino acid
composition and physiological functions. Therefore, further
research is required prior to the use of CPs derived from
soft-shelled turtle tissue in commercial products.
In the present study, a shotgun liquid
chromatography/mass spectrometry (LC/MS)-based global proteomic
analysis of human keratinocytes treated with CPs was performed to
examine the functional effects of CPs on human skin. A total of 211
differentially expressed proteins was identified in keratinocytes
treated with CPs compared with untreated keratinocytes. It was
investigated whether any of these proteins may be involved in the
induction of inflammatory factors in human skin.
Materials and methods
Chemicals and reagents
The highest-grade chemicals and reagents available
were purchased from Wako Pure Chemical Industries (Osaka, Japan).
Emperor tissue, a soft tissue in the region around the shell of
soft-shelled turtles (P. sinensis), was provided by
Shin-uoei, Inc. (Osaka, Japan).
Collagen extraction
Collagen extraction was performed as described in a
previous study (33). In brief,
emperor tissue was treated with 0.1 M formic acid at a ratio of
1:10 (w/v) for 24 h for demineralization. The sample was then
treated with 0.1 M NaOH at a ratio of 1:10 (w/v) for 3 days to
remove non-collagenous proteins, including endogenous proteases.
The NaOH solution was changed every day. Finally, the sample was
incubated with 0.03 M citric acid for 24 h. After the incubation,
the solution was centrifuged at 6,500 x g for 20 min at 4°C, and
the supernatant was collected. This collagen solution was used in
the subsequent experiments.
Tryptic digestion of collagen
The extracted collagen solution was subjected to
proteolytic activation with bovine pancreatic trypsin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in 100 mM ammonium
bicarbonate buffer (pH 8.0). The collagen was incubated with
trypsin at a trypsin/collagen ratio of 1:100 (w:w) at 37°C. At each
indicated time-point, reaction solutions were quickly removed and
heated to 100°C to terminate trypsin digestion.
Tricine-SDS-PAGE
The molecular weights of the tryptic digestion
products were determined with tricine-SDS-PAGE, as described
previously (35). For comparison,
molecular weight markers ranging from 3.5 to 42 kDa (Wako Pure
Chemical Industries) were used. The electrophoresed gel was stained
with Coomassie brilliant blue at room temperature for 1 h.
Matrix-assisted laser desorption-time of
flight/mass spec- trometry (MALDI-TOF/MS)
CPs were applied onto the MALDI target plate
(Shimadzu, Kyoto, Japan) with 10 mg/ml of α-cyano-4-hydroxy
cinnamic acid (Sigma-Aldrich; Merck KGaA) in 50% acetonitrile and
0.05% trifluoroacetic acid. The mass spectra of the CPs were
determined with an AXIMA Confidence (Shimadzu) in reflector mode.
Prior to acquiring the peptide mass spectrum of the sample, the
system was calibrated with a ProteoMass Peptide & Protein
MALDI-MS Calibration Kit (cat. no. MSCAL1-1KT; Bradykinin fragment
1-7, 757.3997; P14R, 1,533.8582; insulin oxidized
B-chain, 3,494.6513; Sigma-Aldrich; Merck KGaA).
Cell culture
HaCaT immortalized human keratinocytes were
purchased from CLS Cell Lines Service GmbH (Eppelheim, Germany).
The cells were cultured in RPMI-1640 medium supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) in an atmosphere containing 5% CO2 at 37°C.
Cell growth assay
Cells were plated at a density of 5×103
cells/well in a 96-well plate and grown in culture medium. The
medium was changed the next day, and different concentrations of
CPs were added. After 72-h treatments, the cells were incubated
with the WST-8 cell counting reagent (Wako Pure Chemical
Industries), and the optical density of the culture solution was
measured at 450 nm with an ELISA plate reader.
Protein preparation
HaCaT cells were plated in a 60-mm dish at a density
of 2×105 cells per dish and grown in culture medium. The
medium was changed the next day and CPs were added. After 72-h
treatments, the cells were solubilized in urea lysis buffer (7 M
urea, 2 M thiourea, 5% CHAPS, 1% Triton X-100). The protein
concentration was measured with the Bio-Rad Protein Assay (cat. no.
5000006JA; Bio-Rad Laboratories, Hercules, CA, USA).
In-solution trypsin digestion
The gel-free digestion method was applied as
described previously (36). In
brief, 10 μg protein extract from each sample was chemically
reduced by adding 45 mM dithiothreitol and 20 mM
tris(2-carboxyethyl)phos-phine. Subsequently, the protein was
alkylated with 100 mM iodoacetamide. After the alkylation, the
samples were digested with mass spectrometry grade trypsin gold
(Promega Corp., Madison, WI, USA) at 37°C for 24 h. Next, the
digests were purified with PepClean C-18 Spin Columns (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
protocol.
Liquid chromatography tandem MS
(LC-MS/MS) analysis for protein identification
Peptide samples (~2 μg) were injected into a
peptide L-trap column (Chemicals Evaluation and Research Institute,
Tokyo, Japan) with an HTC PAL autosampler (CTC Analytics, Zwingen,
Switzerland). The peptides were separated further in a Paradigm MS4
(AMR Inc., Tokyo, Japan) with a reverse-phase C18-column (L-column,
3-μm-diameter gel particles and 120 Å pore size, 0.2×150 mm,
Chemicals Evaluation and Research Institute). The mobile phase
consisted of 0.1% formic acid in water (solution A) and
acetonitrile (solution B). The column flow rate was 1 μl/min
with a concentration gradient of 5% B to 40% B over 120 min.
Gradient-eluted peptides were analyzed with an LTQ ion-trap mass
spectrometer (Thermo Fisher Scientific, Inc.). The results were
acquired in a data-dependent manner, where MS/MS fragmentation was
performed on the two most intense peaks of each full MS scan.
All MS/MS spectral data were entered into a search
for comparisons against the SwissProt Homo sapiens database
with the Mascot tool (version 2.4.01; Matrix Science, London, UK).
The search criteria were as follows: Enzyme, trypsin; with the
following allowances: Up to two missed cleavage peptides; mass
tolerance, ±2.0 kDa; MS/MS tolerance, ±0.8 kDa; and cysteine
carbamidomethylation and methionine oxidation modifications.
Semiquantitative analysis of identified
proteins
The fold-change in expression was calculated as the
log2 of the ratio of protein abundances (Rsc), evaluated
by spectral counting (37). For
comparison, the relative amounts of identified proteins were
calculated using the normalized spectral abundance factor (NSAF)
(38). Differential expression of
proteins were considered significant when the Rsc was >1 or
<-1, which corresponded to fold-changes of >2 or <0.5,
respectively.
Bioinformatics
The function of proteins that exhibited a
significant change in expression with CP treatment was
investigated. These sequences were processed by examining their
functional annotations in the Database for Annotation,
Visualization, and Integrated Discovery (DAVID) version 6.8
(http://david.abcc.ncifcrf.gov/home.jsp) (39-41).
Western blot analysis
Total protein (5 μg) that had been extracted
from CP-treated cells was added to each well of an SDS-PAGE gel and
electrophoresis was performed under reducing conditions. The
separated proteins were transferred to polyvinylidene fluoride
membranes (Merck KGaA) for 30 min at 15 V. After blocking in
TBS-Tween-20 (0.1%) buffer with 5% skimmed milk for 2 h at room
temperature, the membranes were incubated with an anti-calpain 1
antibody (1:1,000 dilution; cat. no. 2556; Cell Signaling
Technology, Inc., Beverly, MA, USA) at 4°C overnight. The membranes
were then washed and incubated with horseradish
peroxidase-conjugated anti-rabbit immunoglobulin (Ig)G antibody
(cat. no. A106PU; American Qualex, San Clemente, CA, USA) at room
temperature for 1 h. The blots were washed and visualized with
SuperSignal West Dura Extended Duration substrate (Thermo Fisher
Scientific, Inc.). The bands were analyzed with the myECL Imager
system (version 2.0; Thermo Fisher Scientific, Inc.). Next, the
membranes were stripped by Restore Western Blot Stripping buffer
(Thermo Fisher Scientific, Inc.), and the same membranes were
re-probed with an anti-β-actin antibody (1:5,000 dilution; cat. no.
sc-47778; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C
overnight, which served as the protein loading control. The
intensities of calpain-1 and β-actin were quantified with
myImageAnalysis software (version 2.0; Thermo Fisher Scientific,
Inc.). The relative quantities of calpain-1 over β-actin were used
to evaluate calpain-1 expression under different conditions. All
western blot analyses were performed as three independent
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HaCaT cells with the
GenElute Mammalian Total RNA Miniprep kit (cat. no. RTN70-1KT;
Sigma-Aldrich; Merck KGaA). Complementary (c)DNA was synthesized
with the High Capacity cDNA Reverse Transcription kit (cat. no.
4368814; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. To measure the expression levels of
interleukin (IL)-1α, IL-6, IL-8 and tumor necrosis factor (TNF)-α,
PCR amplification was performed in the 7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Primers and TaqMan
probes for detecting IL-1α (assay ID, Hs00174092_m1), IL-6 (assay
ID, Hs00985639_m1), IL-8 (assay ID, Hs00174103_m1), TNF-α (assay
ID, Hs01113624_g1) and 18S ribosomal (r)RNA (assay ID,
Hs03928990_g1) were supplied with the TaqMan Gene Expression Assay
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The relative
gene expression was calculated via the ΔΔCq method (42-46). The ΔΔCq method uses the normalized
ΔCq value of each sample, which was calculated with 18S rRNA as the
endogenous control gene. The ΔΔCq value is the difference between
treated and control samples. Finally, the fold-change was
determined as 2−ΔΔCq. Gene expression was evaluated in
triplicate.
Statistical analysis
All data are presented as the mean ± standard error
of the mean. The data were analyzed by one-way analysis of variance
followed by Dunnett's test or the unpaired Student's t-test for two
groups. P<0.05 was considered to indicate a signifi-cant
difference. Computations were performed with GraphPad Prism version
5.1 (GraphPad Software Inc., La Jolla, CA, USA).
Results
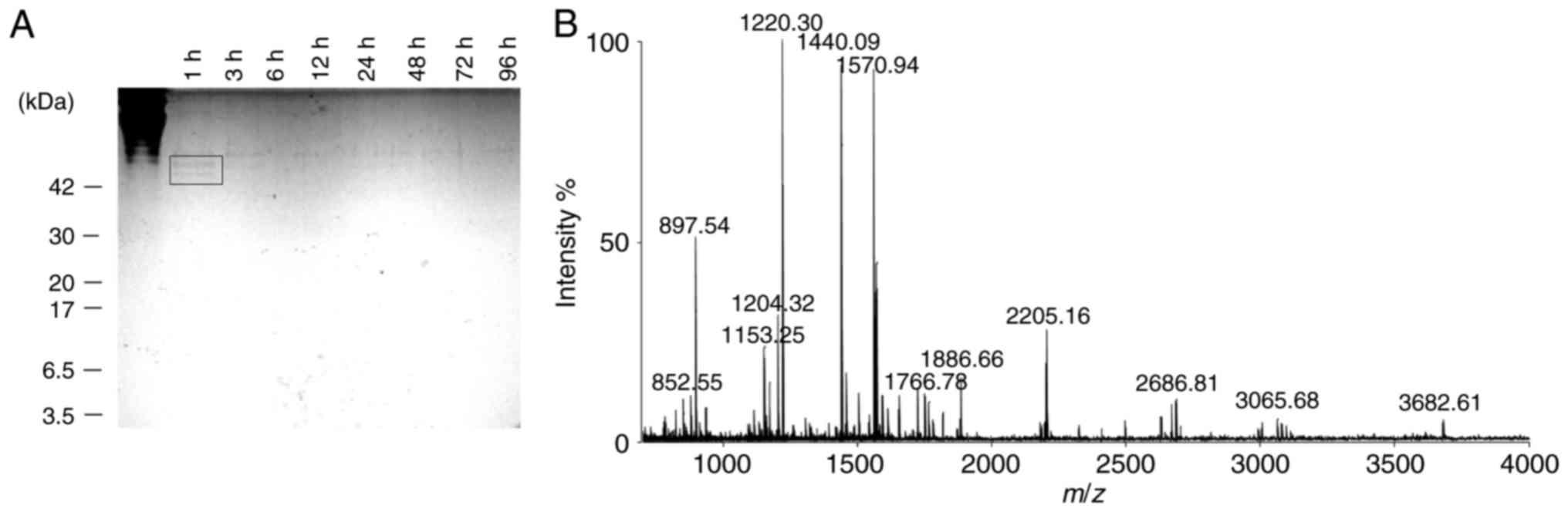
Tryptic digestion of collagen from
soft-shelled turtle
Collagen extracted from soft-shelled turtle was
digested with trypsin to obtain CPs with molecular weights of
<3.5 kDa. The collagen digestion was monitored by extracting
samples at different time-points. The samples were separated on a
15% tricine-SDS-PAGE gel (Fig.
1A). After 1 h of trypsin digestion, bands that corresponded to
collagen or CPs at around 42 kDa were observed (Fig. 1A; black square). After 96 h, these
bands completely disappeared (Fig.
1A). The molecular weight distribution of the digested CPs was
evaluated using MALDI-TOF/MS. The results indicated that the
collagen was digested to CPs with a molecular weight of <4.0
kDa, and most CPs had mass-to-charge (m/z) ratios of
800-2,500 (Fig. 1B).
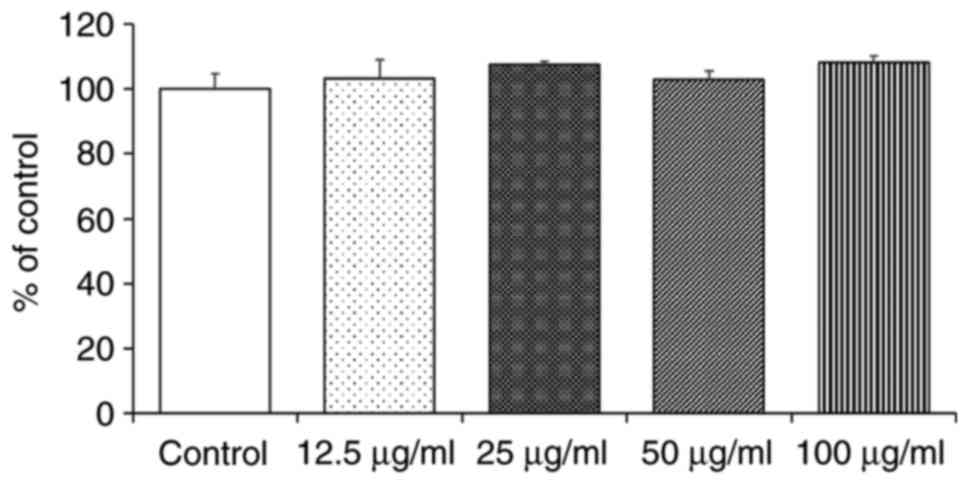
Cytotoxicity of CPs to HaCaT cells
To examine the possible cytotoxic effects of CPs on
HaCaT cells, it was assessed whether the cell growth rate was
affected when the cells were grown in culture medium containing CPs
at concentrations of 0.1-100 μg/ml. The results indicated
that CPs did not inhibit the HaCaT cell growth rate at any of the
tested concentrations (Fig. 2).
Therefore, the CPs were used at a concentration of 100 μg/ml
in the subsequent experiments.
Identification and semi-quantitative
comparison of differen- tially expressed proteins in CP-treated
HaCaT cells
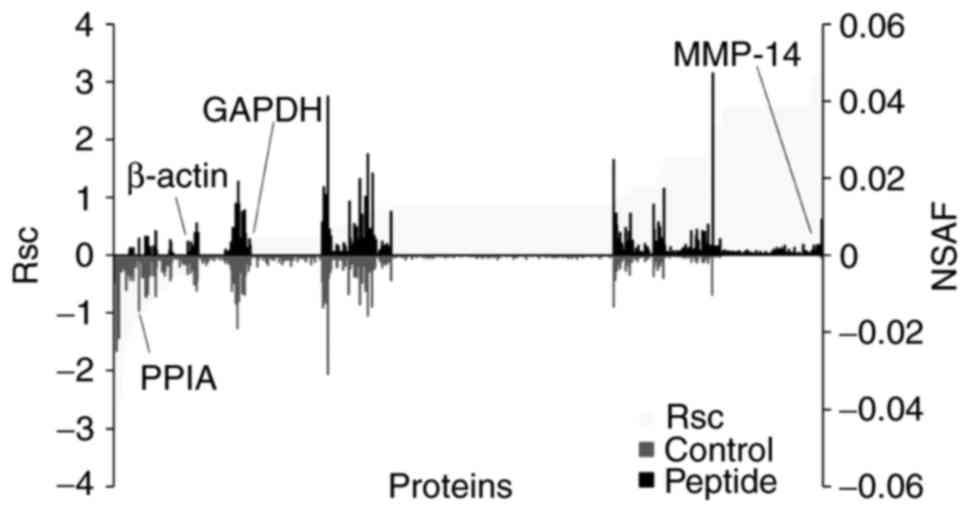
Next, the potential effect of CPs on cells in the
basal layer of the skin was investigated by treating HaCaT cells
with CPs. To determine the molecular profile of proteins that are
regulated by CPs, a shotgun proteomics approach was used. A
label-free semi-quantitative method based on spectral counting was
utilized to evaluate the proteins expressed in HaCaT cells. The Rsc
values were calculated for proteins that had been identified in
CP-treated HaCaT cells and untreated cells. A positive value
indicated increased expression by CP-treatment and a negative value
indicated reduced expression by CP-treatment (Fig. 3; light grey area). For each
protein that had been identified in CP-treated HaCaT cells and
untreated cells, the NSAF value was also calculated (Fig. 3; black bars; peptides, grey bar;
control). Proteins with a >1 and <-1 Rsc value were
considered candidate CP-regulated proteins.
Based on this semi-quantitative procedure, a total
of 211 proteins that were differentially expressed with CP
treatment were identified (Table
I). The expression of housekeeping proteins, including β-actin
and GAPDH, was not altered by CP treatment.
 | Table IProteins differentially expressed
(≥2-fold) after treatment with collagen peptides. |
Table I
Proteins differentially expressed
(≥2-fold) after treatment with collagen peptides.
| ID | Accession
number | Definition | Number of amino
acids | Fold change
(Rsc) |
|---|
| TBA1B_HUMAN | P68363 | Tubulin α-1B
chain | 451 | −2.937 |
| H2B1L_HUMAN | Q99880 | Histone H2B type
1-L | 126 | −2.890 |
| H2B1B_HUMAN | P33778 | Histone H2B type
1-B | 126 | −2.683 |
| H2B1K_HUMAN | O60814 | Histone H2B type
1-K | 126 | −2.683 |
| K2C75_HUMAN | O95678 | Keratin, type II
cytoskeletal 75 | 551 | −2.507 |
| K2C4_HUMAN | P19013 | Keratin, type II
cytoskeletal 4 | 534 | −2.443 |
| K2C79_HUMAN | Q5XKE5 | Keratin, type II
cytoskeletal 79 | 535 | −2.073 |
| ATPA_HUMAN | P25705 | ATP synthase
subunit α, mitochondrial | 553 | −2.073 |
| HS902_HUMAN | Q14568 | Putative heat shock
protein HSP 90-α A2 | 343 | −1.691 |
| HSP7C_HUMAN | P11142 | Heat shock cognate
71 kDa protein | 646 | −1.577 |
| H2A1B_HUMAN | P04908 | Histone H2A type
1-B/E | 130 | −1.577 |
| K2C6C_HUMAN | P48668 | Keratin, type II
cytoskeletal 6C | 564 | −1.409 |
| K2C5_HUMAN | P13647 | Keratin, type II
cytoskeletal 5 | 590 | −1.384 |
| TBA3C_HUMAN | Q13748 | Tubulin α-3C/D
chain | 450 | −1.383 |
| HS90B_HUMAN | P08238 | Heat shock protein
HSP 90-β | 724 | −1.343 |
| K1C19_HUMAN | P08727 | Keratin, type I
cytoskeletal 19 | 400 | −1.245 |
| TBA3E_HUMAN | Q6PEY2 | Tubulin α-3E
chain | 450 | −1.171 |
| TPIS_HUMAN | P60174 | Triosephosphate
isomerase | 286 | −1.171 |
| LMNA_HUMAN | P02545 | Prelamin-A/C | 664 | −1.171 |
| K2C3_HUMAN | P12035 | Keratin, type II
cytoskeletal 3 | 628 | −1.171 |
| PPIA_HUMAN | P62937 | Peptidyl-prolyl
cis-trans isomerase A | 165 | −1.125 |
| K2C73_HUMAN | Q86Y46 | Keratin, type II
cytoskeletal 73 | 540 | −1.005 |
| HS904_HUMAN | Q58FG1 | Putative heat shock
protein HSP 90-α A4 | 418 | −1.005 |
| RS12_HUMAN | P25398 | 40S ribosomal
protein S12 | 132 | −1.005 |
| H2A1A_HUMAN | Q96QV6 | Histone H2A type
1-A | 131 | −1.005 |
| FAS_HUMAN | P49327 | Fatty acid
synthase | 2,511 | −1.005 |
| K1C18_HUMAN | P05783 | Keratin, type I
cytoskeletal 18 | 430 | 1.009 |
| KRT84_HUMAN | Q9NSB2 | Keratin, type II
cuticular Hb4 | 600 | 1.029 |
| H2B1A_HUMAN | Q96A08 | Histone H2B type
1-A | 127 | 1.029 |
| PSA2_HUMAN | P25787 | Proteasome subunit
α type-2 | 234 | 1.029 |
| PHB2_HUMAN | Q99623 | Prohibitin-2 | 299 | 1.029 |
| ALBU_HUMAN | P02768 | Serum albumin | 609 | 1.029 |
| SPTN1_HUMAN | Q13813 | Spectrin α chain,
non-erythrocytic 1 | 2,472 | 1.029 |
| S10AE_HUMAN | Q9HCY8 | Protein
S100-A14 | 104 | 1.029 |
| HSPB1_HUMAN | P04792 | Heat shock protein
β-1 | 205 | 1.029 |
| PDIA3_HUMAN | P30101 | Protein
disulfide-isomerase A3 | 505 | 1.071 |
| 1433S_HUMAN | P31947 | 14-3-3 protein
σ | 248 | 1.071 |
| ACTG_HUMAN | P63261 | Actin, cytoplasmic
2 | 375 | 1.076 |
| ACTBM_HUMAN | Q9BYX7 | Putative
β-actin-like protein 3 | 375 | 1.165 |
| RSSA_HUMAN | P08865 | 40S ribosomal
protein SA | 295 | 1.165 |
| IASPP_HUMAN | Q8WUF5 | RelA-associated
inhibitor | 828 | 1.190 |
| FUMH_HUMAN | P07954 | Fumarate hydratase,
mitochondrial | 510 | 1.190 |
| HMGB1_HUMAN | P09429 | High mobility group
protein B1 | 215 | 1.190 |
| COR1B_HUMAN | Q9BR76 | Coronin-1B | 489 | 1.190 |
| COTL1_HUMAN | Q14019 | Coactosin-like
protein | 142 | 1.190 |
| LA_HUMAN | P05455 | Lupus La
protein | 408 | 1.190 |
| NCBP1_HUMAN | Q09161 | Nuclear cap-binding
protein subunit 1 | 790 | 1.190 |
| SQRD_HUMAN | Q9Y6N5 | Sulfide:Quinone
oxidoreductase, mitochondrial | 450 | 1.190 |
| TRAP1_HUMAN | Q12931 | Heat shock protein
75 kDa, mitochondrial | 704 | 1.190 |
| EZRI_HUMAN | P15311 | Ezrin | 586 | 1.190 |
| RL22_HUMAN | P35268 | 60S ribosomal
protein L22 | 128 | 1.190 |
| RS3_HUMAN | P23396 | 40S ribosomal
protein S3 | 243 | 1.190 |
| ARF4_HUMAN | P18085 | ADP-ribosylation
factor 4 | 180 | 1.190 |
| COR1C_HUMAN | Q9ULV4 | Coronin-1C | 474 | 1.190 |
| MARE1_HUMAN | Q15691 |
Microtubule-associated protein RP/EB
family member 1 | 268 | 1.190 |
| MYO1B_HUMAN | O43795 | Unconventional
myosin-Ib | 1,136 | 1.190 |
| FREM1_HUMAN | Q5H8C1 | FRAS1-related
extracellular matrix protein 1 | 2,179 | 1.190 |
| VPS35_HUMAN | Q96QK1 | Vacuolar protein
sorting-associated protein 35 | 796 | 1.190 |
| TBA1C_HUMAN | Q9BQE3 | Tubulin α-1C
chain | 449 | 1.242 |
| PRDX6_HUMAN | P30041 |
Peroxiredoxin-6 | 224 | 1.257 |
| SERPH_HUMAN | P50454 | Serpin H1 | 418 | 1.257 |
| AN32B_HUMAN | Q92688 | Acidic leucine-rich
nuclear phosphoprotein 32 family member B | 251 | 1.334 |
| CX6B1_HUMAN | P14854 | Cytochrome c
oxidase subunit 6B1 | 86 | 1.334 |
| ROA1_HUMAN | P09651 | Heterogeneous
nuclear ribonucleoprotein A1 | 372 | 1.334 |
| COX5A_HUMAN | P20674 | Cytochrome c
oxidase subunit 5A, mitochondrial | 150 | 1.417 |
| PSME1_HUMAN | Q06323 | Proteasome
activator complex subunit 1 | 249 | 1.417 |
| 4F2_HUMAN | P08195 | 4F2 cell-surface
antigen heavy chain | 630 | 1.417 |
| K1C9_HUMAN | P35527 | Keratin, type I
cytoskeletal 9 | 623 | 1.528 |
| LEG7_HUMAN | P47929 | Galectin-7 | 136 | 1.721 |
| EIF3M_HUMAN | Q7L2H7 | Eukaryotic
translation initiation factor 3 subunit M | 374 | 1.721 |
| HNRDL_HUMAN | O14979 | Heterogeneous
nuclear ribonucleoprotein D-like | 420 | 1.721 |
| TCPB_HUMAN | P78371 | T-complex protein 1
subunit β | 535 | 1.721 |
| CPNE3_HUMAN | O75131 | Copine-3 | 537 | 1.721 |
| DEST_HUMAN | P60981 | Destrin | 165 | 1.721 |
| TFR1_HUMAN | P02786 | Transferrin
receptor protein 1 | 760 | 1.721 |
| CNDP2_HUMAN | Q96KP4 | Cytosolic
non-specific dipeptidase | 475 | 1.721 |
| AHSA1_HUMAN | O95433 | Activator of 90 kDa
heat shock protein ATPase homolog 1 | 338 | 1.721 |
| DDB1_HUMAN | Q16531 | DNA damage-binding
protein 1 | 1,140 | 1.721 |
| ICAL_HUMAN | P20810 | Calpastatin | 708 | 1.721 |
| ASNA_HUMAN | O43681 | ATPase ASNA1 | 348 | 1.721 |
| CISY_HUMAN | O75390 | Citrate synthase,
mitochondrial | 466 | 1.721 |
| CSK23_HUMAN | Q8NEV1 | Casein kinase II
subunit α 3 | 391 | 1.721 |
| CNN2_HUMAN | Q99439 | Calponin-2 | 309 | 1.721 |
| SYYC_HUMAN | P54577 | Tyrosine-tRNA
ligase, cytoplasmic | 528 | 1.721 |
| SODM_HUMAN | P04179 | Superoxide
dismutase [Mn], mitochondrial | 222 | 1.721 |
| ACTY_HUMAN | P42025 | B-centractin | 376 | 1.721 |
| ML12A_HUMAN | P19105 | Myosin regulatory
light chain 12A | 171 | 1.721 |
| RL32_HUMAN | P62910 | 60S ribosomal
protein L32 | 135 | 1.721 |
| MYO1C_HUMAN | O00159 | Unconventional
myosin-Ic | 1,063 | 1.721 |
| FSCN1_HUMAN | Q16658 | Fascin | 493 | 1.722 |
| RL3_HUMAN | P39023 | 60S ribosomal
protein L3 | 403 | 1.722 |
| RPN1_HUMAN | P04843 |
Dolichyl-diphosphooligosaccharide-protein
glycosyltransferase subunit 1 | 607 | 1.722 |
| B2MG_HUMAN | P61769 |
B-2-microglobulin | 119 | 1.722 |
| RBP56_HUMAN | Q92804 | TATA-binding
protein-associated factor 2N | 592 | 1.722 |
| 2AAA_HUMAN | P30153 |
Serine/threonine-protein phosphatase 2A 65
kDa regulatory subunit A α isoform | 589 | 1.722 |
| ENOG_HUMAN | P09104 | Γ-enolase | 434 | 1.722 |
| RL27A_HUMAN | P46776 | 60S ribosomal
protein L27a | 148 | 1.722 |
| DCD_HUMAN | P81605 | Dermcidin | 110 | 1.722 |
| RS7_HUMAN | P62081 | 40S ribosomal
protein S7 | 194 | 1.722 |
| IQGA1_HUMAN | P46940 | Ras
GTPase-activating-like protein IQGAP1 | 1,657 | 1.723 |
| TERA_HUMAN | P55072 | Transitional
endoplasmic reticulum ATPase | 806 | 1.723 |
| SPTB2_HUMAN | Q01082 | Spectrin β chain,
non-erythrocytic 1 | 2,364 | 1.723 |
| RL18A_HUMAN | Q02543 | 60S ribosomal
protein L18a | 176 | 1.723 |
| CALX_HUMAN | P27824 | Calnexin | 592 | 1.723 |
| TBA4B_HUMAN | Q9H853 | Putative
tubulin-like protein α-4B | 241 | 1.724 |
| TCPE_HUMAN | P48643 | T-complex protein 1
subunit ε | 541 | 1.724 |
| K2C80_HUMAN | Q6KB66 | Keratin, type II
cytoskeletal 80 | 452 | 1.724 |
| HNRPK_HUMAN | P61978 | Heterogeneous
nuclear ribonucleoprotein K | 463 | 1.865 |
| VPP4_HUMAN | Q9HBG4 | V-type proton
ATPase 116 kDa subunit a isoform 4 | 840 | 2.029 |
| EF1G_HUMAN | P26641 | Elongation factor
1-γ | 437 | 2.110 |
| G6PI_HUMAN | P06744 | Glucose-6-phosphate
isomerase | 558 | 2.110 |
| H2B1M_HUMAN | Q99879 | Histone H2B type
1-M | 126 | 2.119 |
| RALY_HUMAN | Q9UKM9 | RNA-binding protein
Raly | 306 | 2.253 |
| TXTP_HUMAN | P53007 | Tricarboxylate
transport protein, mitochondrial | 311 | 2.253 |
| CAN1_HUMAN | P07384 | Calpain-1 catalytic
subunit | 714 | 2.253 |
| DHB12_HUMAN | Q53GQ0 | Estradiol
17-β-dehydrogenase 12 | 312 | 2.253 |
| DHX9_HUMAN | Q08211 | ATP-dependent RNA
helicase A | 1,270 | 2.253 |
| MYADM_HUMAN | Q96S97 | Myeloid-associated
differentiation marker | 322 | 2.253 |
| CUTA_HUMAN | O60888 | Protein CutA | 179 | 2.253 |
| CECR2_HUMAN | Q9BXF3 | Cat eye syndrome
critical region protein 2 | 1,484 | 2.253 |
| KRT81_HUMAN | Q14533 | Keratin, type II
cuticular Hb1 | 505 | 2.569 |
| GLOD4_HUMAN | Q9HC38 | Glyoxalase
domain-containing protein 4 | 313 | 2.569 |
| SURF4_HUMAN | O15260 | Surfeit locus
protein 4 | 269 | 2.569 |
| P4HA1_HUMAN | P13674 | Prolyl
4-hydroxylase subunit α-1 | 534 | 2.569 |
| ANX11_HUMAN | P50995 | Annexin A11 | 505 | 2.569 |
| CALU_HUMAN | O43852 | Calumenin | 315 | 2.569 |
| TFG_HUMAN | Q92734 | Protein TFG | 400 | 2.569 |
| ECHA_HUMAN | P40939 | Trifunctional
enzyme subunit α, mitochondrial | 763 | 2.569 |
| PA2G4_HUMAN | Q9UQ80 |
Proliferation-associated protein 2G4 | 394 | 2.569 |
| SF3A2_HUMAN | Q15428 | Splicing factor 3A
subunit 2 | 464 | 2.569 |
| SHLB2_HUMAN | Q9NR46 | Endophilin-B2 | 395 | 2.569 |
| MBOA7_HUMAN | Q96N66 | Lysophospholipid
acyltransferase 7 | 472 | 2.569 |
| AT1A1_HUMAN | P05023 |
Sodium/potassium-transporting ATPase
subunit α-1 | 1,023 | 2.569 |
| PPME1_HUMAN | Q9Y570 | Protein phosphatase
methylesterase 1 | 386 | 2.569 |
| IF4G2_HUMAN | P78344 | Eukaryotic
translation initiation factor 4 γ 2 | 907 | 2.569 |
| IPYR2_HUMAN | Q9H2U2 | Inorganic
pyrophosphatase 2, mitochondrial | 334 | 2.569 |
| CKAP4_HUMAN | Q07065 |
Cytoskeleton-associated protein 4 | 602 | 2.569 |
| COPB2_HUMAN | P35606 | Coatomer subunit
β | 906 | 2.569 |
| DLG1_HUMAN | Q12959 | Disks large homolog
1 | 904 | 2.569 |
| ATX2L_HUMAN | Q8WWM7 | Ataxin-2-like
protein | 1,075 | 2.569 |
| RMXL1_HUMAN | Q96E39 | RNA binding motif
protein, X-linked-like-1 | 390 | 2.569 |
| CERS2_HUMAN | Q96G23 | Ceramide synthase
2 | 380 | 2.569 |
| RM46_HUMAN | Q9H2W6 | 39S ribosomal
protein L46, mitochondrial | 279 | 2.569 |
| FDFT_HUMAN | P37268 | Squalene
synthase | 417 | 2.569 |
| CP26A_HUMAN | O43174 | Cytochrome P450
26A1 | 497 | 2.569 |
| EIF3A_HUMAN | Q14152 | Eukaryotic
translation initiation factor 3 subunit A | 1,382 | 2.569 |
| GLYM_HUMAN | P34897 | Serine
hydroxymethyltransferase, mitochondrial | 504 | 2.569 |
| ARHGJ_HUMAN | Q8IW93 | Rho guanine
nucleotide exchange factor 19 | 802 | 2.569 |
| GFPT1_HUMAN | Q06210 |
Glutamine-fructose-6-phosphate
aminotransferase[isomerizing] 1 | 699 | 2.569 |
| NOD2_HUMAN | Q9HC29 | Nucleotide-binding
oligomerization domain-containing protein 2 | 1,040 | 2.569 |
| NAGK_HUMAN | Q9UJ70 |
N-acetyl-D-glucosamine kinase | 344 | 2.569 |
| SYAC_HUMAN | P49588 | Alanine-tRNA
ligase, cytoplasmic | 968 | 2.569 |
| CLAP1_HUMAN | Q7Z460 | CLIP-associating
protein 1 | 1,538 | 2.569 |
| SPTN4_HUMAN | Q9H254 | Spectrin β chain,
non-erythrocytic 4 | 2,564 | 2.569 |
| MTU1_HUMAN | O75648 | Mitochondrial
tRNA-specific 2-thiouridylase 1 | 421 | 2.569 |
| PAOX_HUMAN | Q6QHF9 | Peroxisomal
N(1)-acetyl-spermine/spermidine
oxidase | 649 | 2.569 |
| MCM5_HUMAN | P33992 | DNA replication
licensing factor MCM5 | 734 | 2.569 |
| WNK3_HUMAN | Q9BYP7 |
Serine/threonine-protein kinase WNK3 | 1,800 | 2.569 |
| LIMK2_HUMAN | P53671 | LIM domain kinase
2 | 638 | 2.569 |
| VIPR2_HUMAN | P41587 | Vasoactive
intestinal polypeptide receptor 2 | 438 | 2.569 |
| DOS_HUMAN | Q8N350 | Protein Dos | 725 | 2.569 |
| TRDN_HUMAN | Q13061 | Triadin O | 729 | 2.569 |
| ZN318_HUMAN | Q5VUA4 | Zinc finger protein
318 | 2,279 | 2.569 |
| SUCO_HUMAN | Q9UBS9 | SUN
domain-containing ossification factor | 1,254 | 2.569 |
| 2AAB_HUMAN | P30154 |
Serine/threonine-protein phosphatase 2A 65
kDa regulatory subunit A β isoform | 601 | 2.569 |
| PCBP2_HUMAN | Q15366 | Poly(rC)-binding
protein 2 | 365 | 2.569 |
| 1433Z_HUMAN | P63104 | 14-3-3 protein
ζ/δ | 245 | 2.569 |
| MDHC_HUMAN | P40925 | Malate
dehydrogenase, cytoplasmic | 334 | 2.569 |
| DNM1L_HUMAN | O00429 | Dynamin-1-like
protein | 736 | 2.569 |
| ARL8A_HUMAN | Q96BM9 | ADP-ribosylation
factor-like protein 8A | 186 | 2.569 |
| DIAP1_HUMAN | O60610 | Protein diaphanous
homolog 1 | 1,272 | 2.569 |
| IF4H_HUMAN | Q15056 | Eukaryotic
translation initiation factor 4H | 248 | 2.569 |
| TEX35_HUMAN | Q5T0J7 | Testis-expressed
sequence 35 protein | 233 | 2.569 |
| GGCT_HUMAN | O75223 |
Γ-glutamylcyclotransferase | 188 | 2.569 |
| SF3A1_HUMAN | Q15459 | Splicing factor 3A
subunit 1 | 793 | 2.569 |
| ARPC2_HUMAN | O15144 | Actin-related
protein 2/3 complex subunit 2 | 300 | 2.569 |
| PP14B_HUMAN | Q96C90 | Protein phosphatase
1 regulatory subunit 14B | 147 | 2.569 |
| ARHGH_HUMAN | Q96PE2 | Rho guanine
nucleotide exchange factor 17 | 2,063 | 2.569 |
| SYSC_HUMAN | P49591 | Serine-tRNA ligase,
cytoplasmic | 514 | 2.569 |
| ALPK2_HUMAN | Q86TB3 | A-protein kinase
2 | 2,170 | 2.569 |
| SUSD3_HUMAN | Q96L08 | Sushi
domain-containing protein 3 | 255 | 2.569 |
| DP13B_HUMAN | Q8NEU8 | DCC-interacting
protein 13-β | 664 | 2.569 |
| XRCC6_HUMAN | P12956 | X-ray repair
cross-complementing protein 6 | 609 | 2.569 |
| HBS1L_HUMAN | Q9Y450 | HBS1-like
protein | 684 | 2.569 |
| RK_HUMAN | Q15835 | Rhodopsin
kinase | 563 | 2.569 |
| SPRY7_HUMAN | Q5W111 | SPRY
domain-containing protein 7 | 196 | 2.569 |
| BRSK2_HUMAN | Q8IWQ3 |
Serine/threonine-protein kinase BRSK2 | 736 | 2.569 |
| STAT1_HUMAN | P42224 | Signal transducer
and activator of transcription 1-α/β | 750 | 2.569 |
| ZEP1_HUMAN | P15822 | Zinc finger protein
40 | 2,718 | 2.569 |
| SOX4_HUMAN | Q06945 | Transcription
factor SOX-4 | 474 | 2.569 |
| REXO1_HUMAN | Q8N1G1 | RNA exonuclease 1
homolog | 1,221 | 2.569 |
| ZN521_HUMAN | Q96K83 | Zinc finger protein
521 | 1,311 | 2.569 |
| DLG5_HUMAN | Q8TDM6 | Disks large homolog
5 | 1,919 | 2.569 |
| TM155_HUMAN | Q4W5P6 | Protein
TMEM155 | 130 | 2.569 |
| ZYX_HUMAN | Q15942 | Zyxin | 572 | 2.569 |
| UBAC2_HUMAN | Q8NBM4 |
Ubiquitin-associated domain-containing
protein 2 | 344 | 2.569 |
| STPG2_HUMAN | Q8N412 | Sperm-tail PG-rich
repeat-containing protein 2 | 459 | 2.569 |
| K0556_HUMAN | O60303 | Uncharacterized
protein KIAA0556 | 1,618 | 2.569 |
| KLH11_HUMAN | Q9NVR0 | Kelch-like protein
11 | 708 | 2.569 |
| TTLL6_HUMAN | Q8N841 | Tubulin
polyglutamylase TTLL6 | 843 | 2.569 |
| CFA36_HUMAN | Q96G28 | Cilia- and
flagella-associated protein 36 | 342 | 2.569 |
| MMP14_HUMAN | P50281 | Matrix
metalloproteinase-14 | 582 | 3.101 |
| ADT1_HUMAN | P12235 | ADP/ATP translocase
1 | 298 | 3.101 |
| GPNMB_HUMAN | Q14956 | Transmembrane
glycoprotein NMB | 572 | 3.101 |
| PYGL_HUMAN | P06737 | Glycogen
phosphorylase, liver form | 847 | 3.101 |
| NIPS2_HUMAN | O75323 | Protein NipSnap
homolog 2 | 286 | 3.101 |
| ERP29_HUMAN | P30040 | Endoplasmic
reticulum resident protein 29 | 261 | 3.101 |
| ADT3_HUMAN | P12236 | ADP/ATP translocase
3 | 298 | 3.101 |
| LBN_HUMAN | Q86UK5 | Limbin | 1,308 | 3.101 |
| RAB1B_HUMAN | Q9H0U4 | Ras-related protein
Rab-1B | 201 | 3.201 |
| TF_HUMAN | P13726 | Tissue factor | 295 | 3.490 |
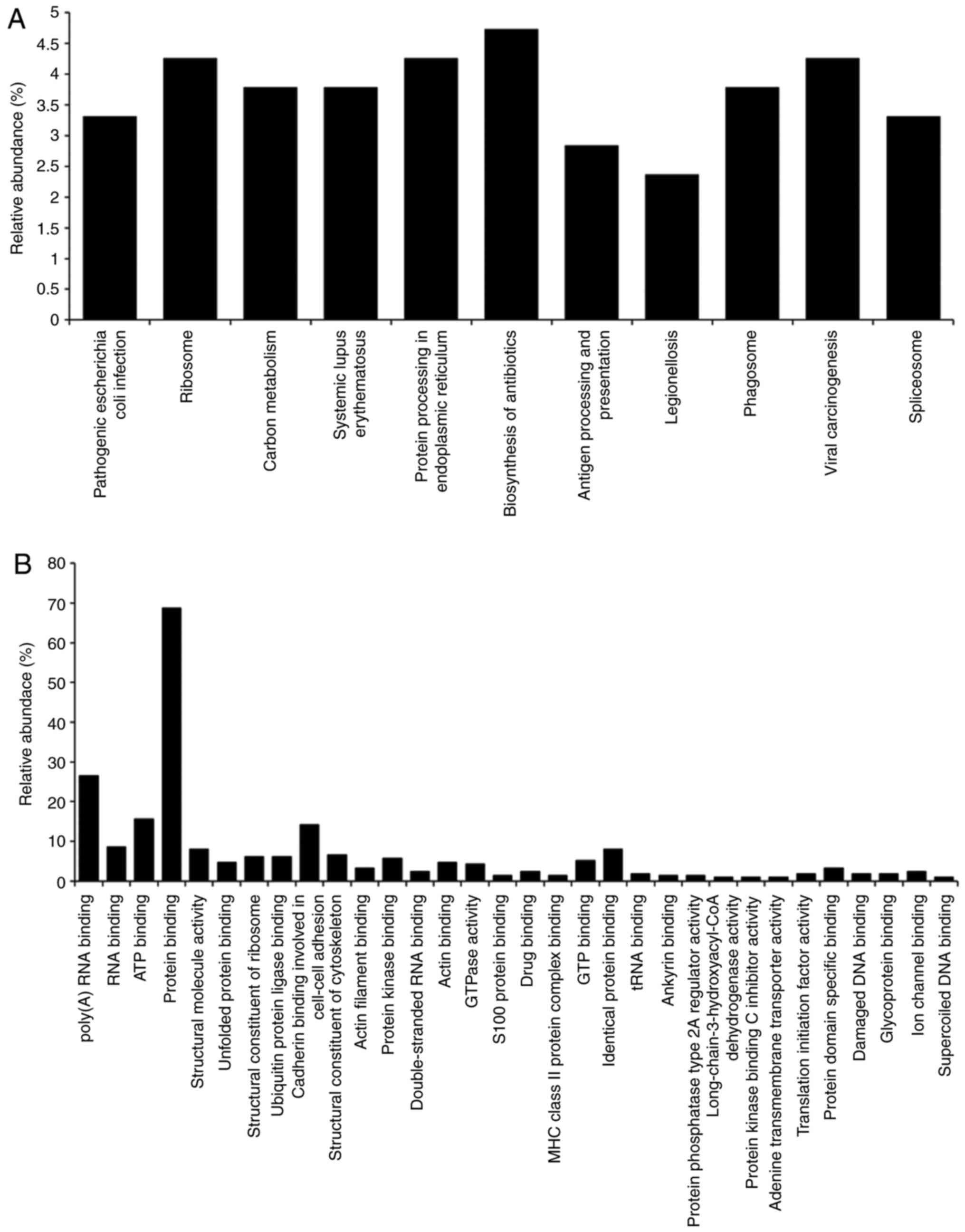
Functional annotation of proteins
regulated by CPs
A gene ontology (GO) analysis of the candidate
CP-regulated proteins was then performed. GO terms associated with
'pathway' (Fig. 4A) and
'molecular function' (Fig. 4B)
were searched for in DAVID. The search focused on proteins
classified as 'protein processing in endoplasmic reticulum'
(Table II).
 | Table IIDifferentially expressed proteins
categorized as 'protein processing in endoplasmic reticulum' in a
Gene Ontology analysis in human skin keratinocytes. |
Table II
Differentially expressed proteins
categorized as 'protein processing in endoplasmic reticulum' in a
Gene Ontology analysis in human skin keratinocytes.
| Accession
number | Description | Fold change
(Rsc) |
|---|
| P11142 | Heat shock cognate
71 kDa protein | −1.577 |
| P08238 | Heat shock protein
HSP 90-β | −1.343 |
| P30101 | Protein
disulfide-isomerase A3 | 1.071 |
| P04843 |
Dolichyl-diphosphooligosaccharide-protein
glycosyltransferase subunit 1 | 1.722 |
| P27824 C | alnexin | 1.723 |
| P55072 | Transitional
endoplasmic reticulum ATPase | 1.723 |
| P07384 | Calpain-1 catalytic
subunit | 2.253 |
| Q07065 |
Cytoskeleton-associated protein 4 | 2.569 |
| P30040 | Endoplasmic
reticulum resident protein 29 | 3.101 |
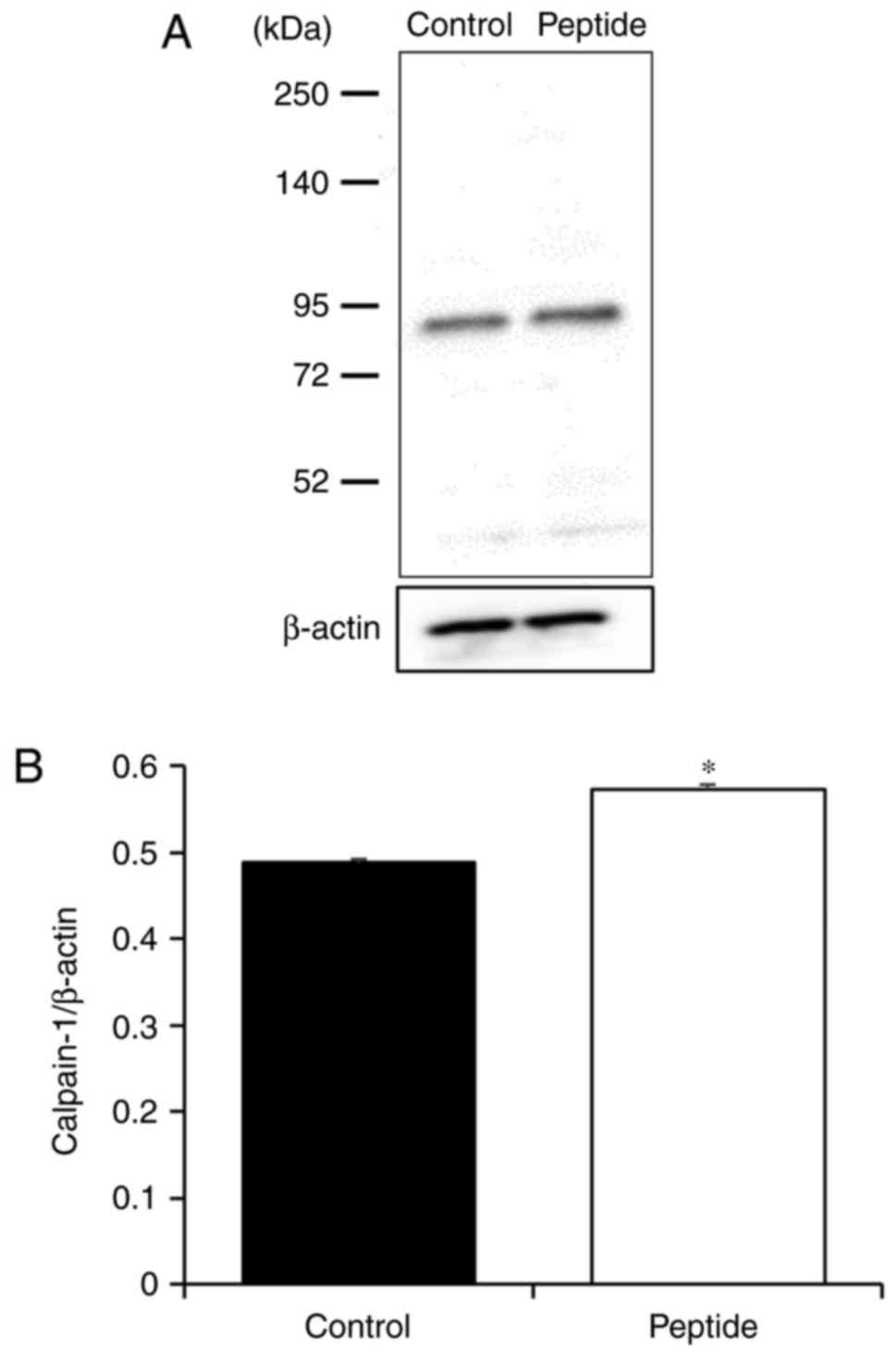
Effect of CP on calpain-1 expression in
HaCaT cells
To confirm that CP treatment altered caipain-1
expression, caipain-1 protein levels were examined in CP-treated
HaCaT cells. It was identified that caipain-1 expression was
signifi-cantly increased with CP treatment compared with that in
control cells (Fig. 5).
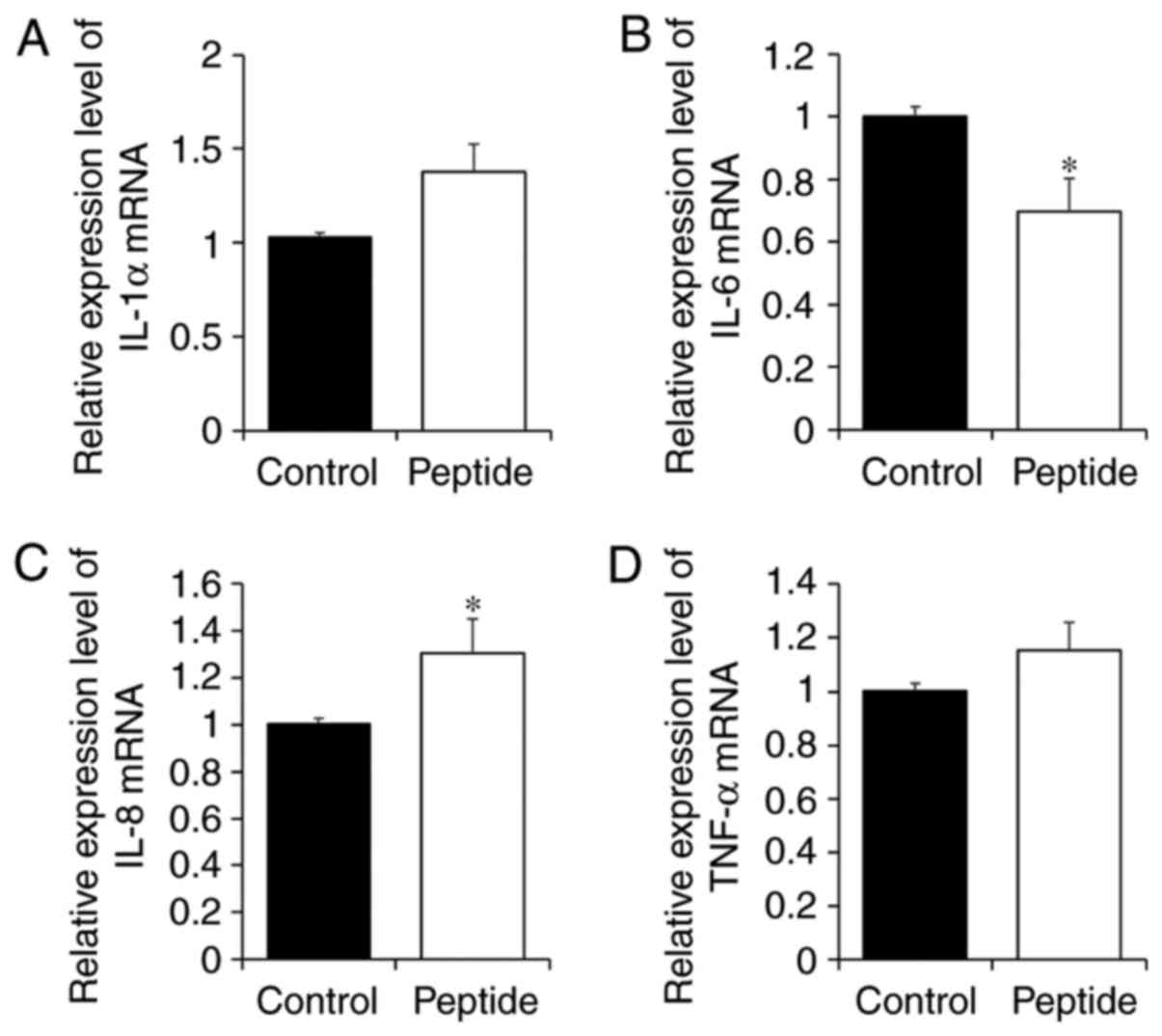
Effect of CP treatment on the expression
of inflammatory factors in HaCaT cells
To investigate whether the CP-induced increase in
calpain-1 expression was associated with the expression of
inflammatory cytokines in HaCaT cells, expression levels of IL-1α,
IL-6, IL-8 and TNF-α were examined (Fig. 6). Although CP-treated HaCaT cells
tended to exhibit increases in IL-1α expression (Fig. 6A), the change was not significant
(P=0.0781). However, IL-6 expression was significantly decreased in
HaCaT cells treated with CP, and by contrast, IL-8 expression was
significantly increased with CP treatment (Fig. 6B and C). Finally, CPs had no
effect on TNF-α expression (Fig.
6D).
Discussion
In the present study, a gel-free LC-MS-based
proteomics approach was used to examine the functional effects of
soft-shelled turtle CPs on human skin cells. A total of 211
proteins that exhibited >2-fold changes in expression after CP
treatment were successfully identified in HaCaT cells, based on a
semi-quantitative method of spectral counting. To examine the roles
of these identified proteins, a GO analysis was performed. The
study focused on the functions of proteins classified as 'protein
processing in endoplasmic reticulum', as they have important roles
in the synthesis of correctly folded proteins and in the
degradation of misfolded proteins. The function of calpain-1, which
is a member of this pathway, was also examined.
To validate the spectral counting results, a western
blot analysis was performed to examine whether calpain-1 expression
is increased in HaCaT cells with CP treatment. Calpain-1 is a
calcium-dependent intracellular cysteine protease (47,48). It has an important role in various
biological processes, including cell proliferation, cell migration,
apoptosis and cytoskeletal remodeling (49,50). Therefore, calpain is considered a
therapeutic target for disorders involving inflammation, wound
healing and tumor progression. A previous study reported that
downregulation of calpain-1 expression in IgE-activated mast cells
led to a reduced expression of cytokines, including IL-6 and TNF-α.
Thus, they concluded that calpain-1 may regulate IgE-mediated
allergic inflammation (51).
Another study indicated that downregulation of calpain-1 expression
in lung fibroblast cells also reduced the expression of cytokines,
including IL-6, IL-8, and TNF-α (52). Furthermore, calpain-1 knockout
mice exhibited impaired bactericidal activity in an acute bacterial
peritonitis model, due to a reduction in IL-1α production (53). Therefore, calpain-1 is considered
a key factor in the immune response through its regulation of
inflammation. Accordingly, it was hypothesized that CP-induced
increases in calpain-1 expression in HaCaT cells may affect the
expression of inflammatory cytokines in keratinocytes. The present
results support this hypothesis. Therefore, CP treatment may
regulate the immune system in the setting of skin wounds.
In the present GO analysis in the category molecular
function, several proteins classified as 'cadherin binding involved
in cell-cell adhesion' exhibited altered expression with CP
treatment. A previous study by our group reported that in HaCaT
cells, these proteins were changed by treatment with collagen
derived from soft-shelled turtle. Furthermore, it was observed that
changing the expression of these proteins enhanced the wound
healing properties of HaCaT cells by inducing EMT (34). In the present study, it was
revealed that CP treatment was associated with >2-fold increases
in ceramide synthase 2 expression in HaCaT cells compared with that
in control cells (Table I).
Ceramide is an epidermal sphingolipid that has important roles in
maintaining the skin barrier and supporting wound healing processes
(54-56). The present result is similar to
previous ones reported for collagen derived from soft-shelled
turtles and for CPs derived from other origins (25,31,32,57). It is therefore suggested that CPs
from soft-shelled turtles may facilitate wound healing.
Although the present study suggested that CPs from
soft-shelled turtles may be a useful material for pharmaceuticals
and medical products, it remains elusive whether these CPs affect
the wound healing processes in keratinocytes in vivo. Of
note, certain CP-regulated inflammatory cytokines including IL-6
and IL-8 were not been investigated to determine their potential
effects on the wound healing process in keratinocytes. Further
in vitro and in vivo studies are necessary to clarify
the effect of CPs from soft-shelled turtle on wound healing and the
role of CP-regulated inflammatory cytokines in keratinocytes. In
addition, it is also necessary to investigate the correlation
between the expression of calpain-1 and inflammatory cytokines in
CP-treated keratinocytes.
In conclusion, the present shotgun LC/MS-based
global proteomic analysis revealed that CP treatment regulated the
expression of inflammatory cytokines in HaCaT cells and induced the
expression of proteins associated with cell-cell adhesion and skin
barrier maintenance. Therefore, CPs from soft-shelled turtles may
provide significant benefits in maintaining the biological
environment of the skin. These peptides may be a useful material
for pharmaceuticals and medical products.
Funding
This study was supported in part by a Grant-in-Aid
for Scientific Research (C) from the Japanese Society for the
Promotion of Science to T.Y. (grant no. 15K09054).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TY and AT designed the study and analyzed the data.
TY and SN performed the experimental work. TY drafted the
manuscript. KM contributed conduct the literature review. AT
critically evaluated the study and final version of the manuscript.
All authors participated in discussion of the study and gave final
approval.
Ethical approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
MALDI-TOF/MS
|
matrix-assisted laser desorptiontime
of flight/mass spectrometry
|
|
LC-MS/MS
|
liquid chromatography tandem mass
spectrometry
|
|
NSAF
|
normalized spectral abundance
factor
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
IL
|
interleukin
|
|
TNF
|
tumor necrosis factor
|
Acknowledgments
The authors are grateful to Mr. Takashi Aboshi
(Shin-uoei, Inc.) for providing the soft-shelled turtle tissue used
in the present study.
References
|
1
|
Gelse K, Pöschl E and Aigner T:
Collagens-structure, function, and biosynthesis. Adv Drug Deliv
Rev. 55:1531–1546. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Myllyharju J and Kivirikko KI: Collagens,
modifying enzymes and their mutations in humans, flies and worms.
Trends Genet. 20:33–43. 2004. View Article : Google Scholar
|
|
3
|
Birk DE and Trelstad RL: Extracellular
compartments in tendon morphogenesis: Collagen fibril, bundle, and
macroaggregate formation. J Cell Biol. 103:231- 2401986. View Article : Google Scholar
|
|
4
|
Adachi E and Hayashi T: Anchoring of
epithelia to underlying connective tissue: Evidence of frayed ends
of collagen fibrils directly merging with meshwork of lamina densa.
J Electron Microsc (Tokyo). 43:264–271. 1994.
|
|
5
|
Park KH and Bae YH: Phenotype of
hepatocyte spheroids in Arg- GLY- Asp (RGD) containing a thermo-
reversible extracellular matrix. Biosci Biotechnol Biochem.
66:1473- 14782002. View Article : Google Scholar
|
|
6
|
Liu B, Weinzimer SA, Gibson TB,
Mascarenhas D and Cohen P: Type Ialpha collagen is an IGFBP- 3
binding protein. Growth Hormone IGF Res. 13:89- 972003. View Article : Google Scholar
|
|
7
|
Di Lullo GA, Sweeney SM, Korkko J,
Ala-Kokko L and San Antonio JD: Mapping the ligand-binding sites
and disease-associated mutations on the most abundant protein in
the human, type I collagen. J Biol Chem. 277:4223–4231. 2002.
View Article : Google Scholar
|
|
8
|
Saby C, Buache E, Brassart-Pasco S, El
Btaouri H, Courageot MP, Van Gulick L, Garnotel R, Jeannesson P and
Morjani H: Type I collagen aging impairs discoidin domain receptor
2-mediated tumor cell growth suppression. Oncotarget.
7:24908–24927. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maquoi E, Assent D, Detilleux J, Pequeux
C, Foidart JM and Noël A: MT1-MMP protects breast carcinoma cells
against type I collagen-induced apoptosis. Oncogene. 31:480–493.
2012. View Article : Google Scholar
|
|
10
|
Muralidharan N, Jeya Shakila R, Sukumar D
and Jeyasekaran G: Skin, bone and muscle collagen extraction from
the trash fish, leather jacket (Odonus niger) and their
characterization. J Food Sci Technol. 50:1106–1113. 2013.
View Article : Google Scholar :
|
|
11
|
Wang Y and Regenstein JM: Effect of EDTA,
HCl, and citric acid on Ca salt removal from Asian (silver) carp
scales prior to gelatin extraction. J Food Sci. 74:C426–C431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang C, Zhan CL, Cai QF, Du CH, Liu GM, Su
WJ and Cao MJ: Expression and characterization of common carp
(Cyprinus carpio) matrix metalloproteinase-2 and its activity
against type I collagen. J Biotechnol. 177:45- 522014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Benjakul S, Thiansilakul Y, Visessanguan
W, Roytrakul S, Kishimura H, Prodpran T and Meesane J: Extraction
and characterisation of pepsin-solubilised collagens from the skin
of bigeye snapper (Priacanthus tayenus and Priacanthus
macracanthus). J Sci Food Agric. 90:132–138. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nalinanon S, Benjakul S and Kishimura H:
Collagens from the skin of arabesque greenling (Pleurogrammus
azonus) solubilized with the aid of acetic acid and pepsin from
albacore tuna (Thunnus alalunga) stomach. J Sci Food Agric.
90:1492–1500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tziveleka LA, Ioannou E, Tsiourvas D,
Berillis P, Foufa E and Roussis V: Collagen from the marine sponges
Axinella cannabina and Suberites carnosus: Isolation and
morphological, biochemical, and biophysical characterization. Mar
Drugs. 15:E1522017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pallela R, Venkatesan J, Janapala VR and
Kim SK: Biophysicochemical evaluation of chitosan-
hydroxyap-atite-marine sponge collagen composite for bone tissue
engineering. J Biomed Mater Res A. 100:486- 4952012.
|
|
17
|
Coelho RCG, Marques ALP, Oliveira SM,
Diogo GS, Pirraco RP, Moreira-Silva J, Xavier JC, Reis RL, Silva TH
and Mano JF: Extraction and characterization of collagen from
Antarctic and Sub-Antarctic squid and its potential application in
hybrid scaffolds for tissue engineering. Mater Sci Eng C Mater Biol
Appl. 78:787–795. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gorell ES, Leung TH, Khuu P and Lane AT:
Purified type I collagen wound matrix improves chronic wound
healing in patients with recessive dystrophic epidermolysis
bullosa. Pediatr Dermat. 32:220–225. 2015. View Article : Google Scholar
|
|
19
|
Shevchenko RV, Sibbons PD, Sharpe JR and
James SE: Use of a novel porcine collagen paste as a dermal
substitute in full-thickness wounds. Wound Repair Regen.
16:198–207. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wollina U, Meseg A and Weber A: Use of a
collagen-elastin matrix for hard to treat soft tissue defects. Int
Wound J. 8:291–296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barhoumi A, Salvador-Culla B and Kohane
DS: NIR-triggered drug delivery by collagen-mediated second
harmonic generation. Adv Healthc Mater. 4:1159–1163. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wallace DG and Rosenblatt J: Collagen gel
systems for sustained delivery and tissue engineering. Adv Drug
Deliv Rev. 55:1631–1649. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Friess W: Collagen-biomaterial for drug
delivery. Eur J Pharm Biopharm. 45:113–136. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Song H, Zhang S, Zhang L and Li B: Effect
of orally administered collagen peptides from bovine bone on skin
aging in chronologically aged mice. Nutrients. 9:E12092017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu Z, Yang P, Zhou C, Li S and Hong P:
Marine collagen peptides from the skin of Nile Tilapia (Oreochromis
niloticus): Characterization and wound healing evaluation. Mar
Drugs. 15:E1022017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhuang Y, Hou H, Zhao X, Zhang Z and Li B:
Effects of collagen and collagen hydrolysate from jellyfish
(Rhopilema esculentum) on mice skin photoaging induced by UV
irradiation. J Food Sci. 74:H183- H1882009. View Article : Google Scholar
|
|
27
|
Zague V: A new view concerning the effects
of collagen hydroly-sate intake on skin properties. Arch Dermatol
Res. 300:479–483. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan J, Zhuang Y and Li B: Effects of
collagen and collagen hydrolysate from jellyfish umbrella on
histological and immunity changes of mice photoaging. Nutrients.
5:223–233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou H, Li B, Zhang Z, Xue C, Yu G, Wang J,
Bao Y, Bu L, Sun J, Peng Z and Su S: Moisture absorption and
retention properties, and activity in alleviating skin photodamage
of collagen polypeptide from marine fish skin. Food Chem. 135:1432-
14392012. View Article : Google Scholar
|
|
30
|
Song H, Meng M, Cheng X, Li B and Wang C:
The effect of collagen hydrolysates from silver carp
(Hypophthalmichthys molitrix) skin on UV-induced photoaging in
mice: Molecular weight affects skin repair. Food Funct.
8:1538–1546. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Z, Wang J, Ding Y, Dai X and Li Y:
Oral administration of marine collagen peptides from Chum Salmon
skin enhances cutaneous wound healing and angiogenesis in rats. J
Sci Food Agric. 91:2173–2179. 2011.PubMed/NCBI
|
|
32
|
Wang J, Xu M, Liang R, Zhao M, Zhang Z and
Li Y: Oral administration of marine collagen peptides prepared from
chum salmon (Oncorhynchus keta) improves wound healing following
cesarean section in rats. Food Nutr Res. 59:264112015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamamoto T, Uemura K, Sawashi Y, Mitamura
K and Taga A: Optimization of method to extract collagen from
'Emperor' tissue of soft-shelled turtles. J Oleo Sci. 65:169–175.
2016. View Article : Google Scholar
|
|
34
|
Yamamoto T, Nakanishi S, Mitamura K and
Taga A: Shotgun label- free proteomic analysis for identification
of proteins in HaCaT human skin keratinocytes regulated by the
administration of collagen from soft- shelled turtle. J Biomed
Mater Res B Appl Biomater. 2017. View Article : Google Scholar
|
|
35
|
Schägger H: Tricine- SDS- PAGE. Nat
Protoc. 1:16–22. 2006. View Article : Google Scholar
|
|
36
|
Bluemlein K and Ralser M: Monitoring
protein expression in whole-cell extracts by targeted label- and
standard-free LC-MS/MS. Nat Protoc. 6:859–869. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Old WM, Meyer-Arendt K, Aveline-Wolf L,
Pierce KG, Mendoza A, Sevinsky JR, Resing KA and Ahn NG: Comparison
of label-free methods for quantifying human proteins by shotgun
proteomics. Mol Cell Proteomics. 4:1487–1502. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zybailov B, Coleman MK, Florens L and
Washburn MP: Correlation of relative abundance ratios derived from
peptide ion chromatograms and spectrum counting for quantitative
proteomic analysis using stable isotope labeling. Anal Chem.
77:6218–6224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinfor-matics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
41
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
42
|
Parikh P, Bai H, Swartz MF, Alfieris GM
and Dean DA: Identification of differentially regulated genes in
human patent ductus arteriosus. Exp Biol Med (Maywood). 241:2112-
21182016. View Article : Google Scholar
|
|
43
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
44
|
Carbotti G, Nikpoor AR, Vacca P, Gangemi
R, Giordano C, Campelli F, Ferrini S and Fabbi M: IL-27 mediates
HLA class I up-regulation, which can be inhibited by the IL-6
pathway, in HLA-deficient small cell lung cancer cells. J Exp Clin
Cancer Res. 36:1402017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Adnan M, Morton G and Hadi S: Analysis of
rpoS and bolA gene expression under various stress-induced
environments in planktonic and biofilm phase using 2(−ΔΔCT) method.
Mol Cell Biochem. 357:275- 2822011. View Article : Google Scholar
|
|
46
|
Soejima M and Koda Y: TaqMan-based
real-time polymerase chain reaction for detection of FUT2 copy
number variations: Identification of novel Alu-mediated deletion.
Transfusion. 51:762–769. 2011. View Article : Google Scholar
|
|
47
|
Goll DE, Thompson VF, Li H, Wei W and Cong
J: The calpain system. Physiol Rev. 83:731–801. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Momeni HR: Role of calpain in apoptosis.
Cell J. 13:65–72. 2011.PubMed/NCBI
|
|
49
|
Glading A, Lauffenburger DA and Wells A:
Cutting to the chase: Calpain proteases in cell motility. Trends
Cell Biol. 12:46- 542002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sorimachi H, Ishiura S and Suzuki K:
Structure and physiological function of calpains. Biochem J.
328:721- 7321997. View Article : Google Scholar
|
|
51
|
Wu Z, Chen X, Liu F, Chen W, Wu P,
Wieschhaus AJ, Chishti AH, Roche PA, Chen WM and Lin TJ: Calpain-1
contributes to IgE-mediated mast cell activation. J Immunol.
192:5130–5139. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yin G, Zeng Q, Zhao H, Wu P, Cai S, Deng L
and Jiang W: Effect and mechanism of calpains on pediatric lobar
pneumonia. Bioengineered. 8:374- 3822017. View Article : Google Scholar :
|
|
53
|
Kumar V, Everingham S, Hall C, Greer PA
and Craig AW: Calpains promote neutrophil recruitment and bacterial
clearance in an acute bacterial peritonitis model. Eur J Immunol.
44:831–841. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kim H, Kim J, Park J, Kim SH, Uchida Y,
Holleran WM and Cho Y: Water extract of gromwell (Lithospermum
erythrorhizon) enhances migration of human keratinocytes and dermal
fibroblasts with increased lipid synthesis in an in vitro wound
scratch model. Skin Pharmacol Physiol. 25:57–64. 2012. View Article : Google Scholar
|
|
55
|
Amen N, Mathow D, Rabionet M, Sandhoff R,
Langbein L, Gretz N, Jäckel C, Gröne HJ and Jennemann R:
Differentiation of epidermal keratinocytes is dependent on
glucosylceramide: Ceramide processing. Hum Mol Genet. 22:4164-
41792013. View Article : Google Scholar
|
|
56
|
Meckfessel MH and Brandt S: The structure,
function, and importance of ceramides in skin and their use as
therapeutic agents in skin-care products. J Am Acad Dermatol.
71:177–184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gangwar M, Gautam MK, Ghildiyal S, Nath G
and Goel RK: Mallotus philippinensis Muell. Arg fruit glandular
hairs extract promotes wound healing on different wound model in
rats. BMC Complement Altern Med. 15:1232015. View Article : Google Scholar : PubMed/NCBI
|