Introduction
Gastric cancer is one of the most common types of
malignant tumor and is the leading causes of cancer-associated
mortality worldwide (1,2). The risk factors for this disease
include diet, Helicobacter pylori infection and genetic
alterations (3-5). It is reported that >95% of
malignancies of the stomach are adenocarcinomas (6). The aggressiveness of human gastric
cancer is associated with the activation of oncogenes, inactivation
of tumor suppressor genes, and perturbation of growth factors and
their receptors (7,8). However, the mechanisms controlling
its level of aggression remain to be fully elucidated.
Human epidermal growth factor receptor 2 (HER2) is a
proto-oncogene, which is encoded by ERBB2 on chromosome 17. Its
amplification is detected in >15% of gastric cancer cases and is
associated with poor clinical outcomes (9-14).
Lapatinib (Tykerb; GlaxoSmithKline, Brentford, UK), a potent
ATP-competitive inhibitor, is a small, orally active molecule,
which inhibits the tyrosine kinases of HER2 and epidermal growth
factor receptor type 1 (EGFR1) (15). Several studies have shown that the
activation of receptor tyrosine kinases can mediate resistance to
HER-targeted therapy (11).
Fibroblast growth factor receptor 2 (FGFR2), a receptor tyrosine
kinase, has been shown to be activated in several types of cancer
through a variety of mechanisms, including gene amplification,
translocations and point mutations (16). The expression of FGFR2 is
increased in tumor tissues and positively correlated with
clinicopathological factors; it promotes the invasion and migration
of human gastric cancer cells (17,18).
Previous reports have shown that testican-1-mediated
epithelial-mesenchymal transition signaling confers acquired
resistance to lapatinib in HER2-positive gastric cancer (19). In addition, the expression of
phosphorylated (p-)MET, phosphorylated signal transducer and
activator of transcription 3 (p-Stat3) and p-HER3 have been
suggested as markers positively associated with resistance to
lapatinib (19). Although these
markers have been recognized, their regulatory mechanisms remain to
be fully elucidated. The aim of the present study was to explore
the effects of miR-494 and FGFR2 in regulation of cancer-initiating
cell phenotypes and therapeutic efficiency of lapatinib in
HER2-positive gastric cancer.
Materials and methods
Clinical specimens
The Ethics Committees of Qilu Hospital of Shandong
University (Jinan, China) approved the present study. Patient
consent was obtained prior to tissue collection. In total, six
gastric cancer samples and matched control tissue samples were
frozen in liquid nitrogen immediately following surgical resection
and were stored at −80°C until protein extraction. The surgical
samples were obtained at Qilu Hospital of Shandong University
between January 2015 and January 2017. The patients, 4 males and 2
females, ranged in age from 35-70 years, with a mean age of 61
years. According to American Joint Committee on Cancer clinical
cancer stage (20), 2 patients
were stage I/II, and 4 patients were stage III/IV. None of the
patients received preoperative treatment, for example radiation or
chemotherapy.
Cell culture
The YCC1 gastric cancer cell line (HER2-positive
gastric cancer cells) and YCC1-F (HER2-positive, FGFR2
overexpressing and lapatinib-resistant gastric cancer cells) were
obtained from Tiangen Biotech Co., Ltd. (Beijing, China). They were
grown in RPMI-1640 medium (Sigma; Merck Millipore, Darmstadt,
Germany) containing 10% fetal bovine serum (FBS; Shanghai ExCell
Biology, Shanghai, China) and 100 mg/ml penicillin and streptomycin
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
incubated at 37°C in a humidified atmosphere of 5%
CO2.
Pre-miR-494/control miR and
transfection
The pre-miR-494 and control miR were purchased from
Ambion; Thermo Fisher Scientific, Inc. The cells were seeded at a
density of 1.5×105 per well in 6-well plates or 60-mm
dishes in 2 ml complete medium containing 10% FBS for 24 h.
Transfections were performed using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). For each well, the
pre-miR-494 or negative control precursor miRNA (mock) was
transfected into cells (Ambion; Thermo Fisher Scientific, Inc.).
The mixture of Lipofectamine and miRNA (50 nM miR-494) was then
administered to cells at 37°C in the presence of serum-free medium
for up to 72 h.
Western blot analysis
Western blot analysis was performed as described
previously (21). Total protein
was prepared using extraction buffer comprising NaCl/Pi
containing 0.5% Triton X-100, 1 mM EDTA, 1 mM phenylmethyl sulfonyl
fluoride, and complete protease inhibitors (Roche Diagnostics,
Basel, Switzerland). The concentration of each protein lysate was
determined using a BCA™ protein assay kit (Thermo Fisher
Scientific, Inc.). Equal quantities (20 µg) of total protein
were subjected to 12% SDS-PAGE. The samples were then transferred
onto nitrocellulose membranes and blocked for 60 min at room
temperature in 5% skim milk powder in NaCl/Pi. The
membranes were immunoblotted using antibodies against human FGFR2
(ab10648; 1:500), CD44 (ab157107; 1:500; Abcam), testican-1
(ab229935; 1:500; Abcam), MET (ab51067; 1:500; Abcam), HER2
(ab16901, 1:500; Abcam), Stat3 (ab119352; 1:500; Abcam), HER3
(ab32121; 1:500; Abcam), p-MET (ab5662, 1:500; Abcam), p-Stat3
(ab76315; 1:500; Abcam), p-HER3 (ab101407; 1:500; Abcam), c-myc
(ab32072; 1:500; Abcam), insulin-like growth factor 1 receptor
(IGF1R; ab39398; 1:500; Abcam), or β-actin (ab5694; 1:500; all from
Abcam, Cambridge, MA, USA) overnight at 4°C. IRDye®
800-conjugated anti-rabbit secondary antibody (ab191866; 1:10,000;
Abcam) was used for incubation at room temperature for 30 min. The
specific proteins were visualized using an Odyssey™ infrared
imaging system (Gene Company, Ltd., Lincoln, NE, USA). The
expression of β-actin was used as an internal control to ensure
equal loading of the protein samples.
Sphere growth
The measurement of sphere growth was performed as
described previously (22). Cells
(103/ml) in serum-free RPMI-1640/1 mM Na-pyruvate were
seeded on 0.5% agar precoated 6-well plates. After 10 days, half
the medium was exchanged every third day. Single spheres were
picked and counted. The sphere forming ability of the cells was
recorded for the next 7-14 days. The size and number of spheres
were assessed under a routine Leica microscope (Leica Microsystems
GmbH, Wetzlar, Germany).
MTT assay
To monitor resistance to Lapatinib, YCC1 and YCC1-F
cells were treated with 1 µM Lapatinib for 24 h and then MTT
assay was performed as described previously (19). Data were analyzed with Origin
software version 7.5 (OriginLab, Northampton, MA, USA) to fit a
sigmoidal curve. IC50 is the Lapatinib concentration
that reduces proliferating cells by 50%.
Bioinformatics analysis
The analysis of potential miRNA target sites was
performed using miRanda (http://www.microrna.org/microrna/home.do).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the cells by
homogenizing cells in TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA (500 ng) was quantitated at 260 nm and reverse-transcribed into
cDNA using the PrimeScript RT reagent kit (Takara Biotechnology,
Co., Ltd., Dalian, China) according to the manufacturer's protocol,
at 37°C for 15 min and 85°C for 30 sec.
For miRNA qPCR, reverse transcription was performed
using the QuantMir RT kit (System Biosciences, Mountain View, CA,
USA). cDNA was quantitated based on the absorption at 260 nm and
served as the template for SYBR real-time PCR using Power
SYBR-Green PCR Master mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.). All reactions were run in triplicate on the
iCycler iQ Multicolor Real-Time PCR detection system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) using miR-494-specifc
primers (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
following primers were used: miR-494, forward
5′-TGGTGATGGGATTTGAAACATACACGGGAAAC-3′, and reverse
5′-AGATAGACGG-TGTCGCTGTTGAAGTCAG-3′; U6: Forward,
5′-GCTTCGGCAGCACATATACTAA-3′; and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The amplification profile was as
follows: Denaturation at 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 15 sec, annealing at 60°C for 30 sec, and
extension at 72°C for 1 min. The comparative cycle quantification
(Cq) method was used to quantify the miRNA expression levels. The
relative quantity of miR-494 to small nuclear U6 RNA was calculated
using the 2−ΔCq equation, where ΔCq =
(CqmiR-494 − CqU6 RNA). The fold change of
gene expression was calculated using the 2−ΔΔCq method
(23). U6 small nuclear RNA was
used as the internal standard.
For mRNA qPCR, analysis was performed as described
above. cDNA was quantitated based on the absorption at 260 nm and
served as the template for qPCR. The thermal cycle profile was as
follows: Denaturation for 30 sec at 95°C, annealing for 45 sec at
52-58°C depending on the primers used, and extension for 45 sec at
72°C. Each PCR reaction was performed for 28-32 cycles. PCR
products were visualized on 2% agarose gels stained with ethidium
bromide under UV transillumination. RT-qPCR was performed using a
StepOne™ real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Fast SYBR®−Green Master Mix was also
obtained from Applied Biosystems. Data are shown as a relative
expression level after normalization to glyceraldehyde-3-phosphate
dehydrogenase (GAPDH). The following primers were used: FGFR2,
forward 5′-GGTCGTTTCATCTGCCTGGT-3′ and reverse
5′-CCTTCCCGTTTTTCAGCCAC-3′; CD44, forward
5′-CAGCAACCCTACTGATGATGACG-3′ and reverse
5′-GCCAAGAGGGATGCCAAGATGA-3′; GAPDH, forward
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Statistical analysis
The results were analyzed using SAS software
(version 9.4; SAS Institute, Inc., Cary, NC, USA) (24). Data are presented as the mean ±
standard error of the mean of separate experiments (n=3).
Statistical significance was determined using Student's t-test
(two-tailed). P<0.05 was considered to indicate a statistically
significant difference.
Results
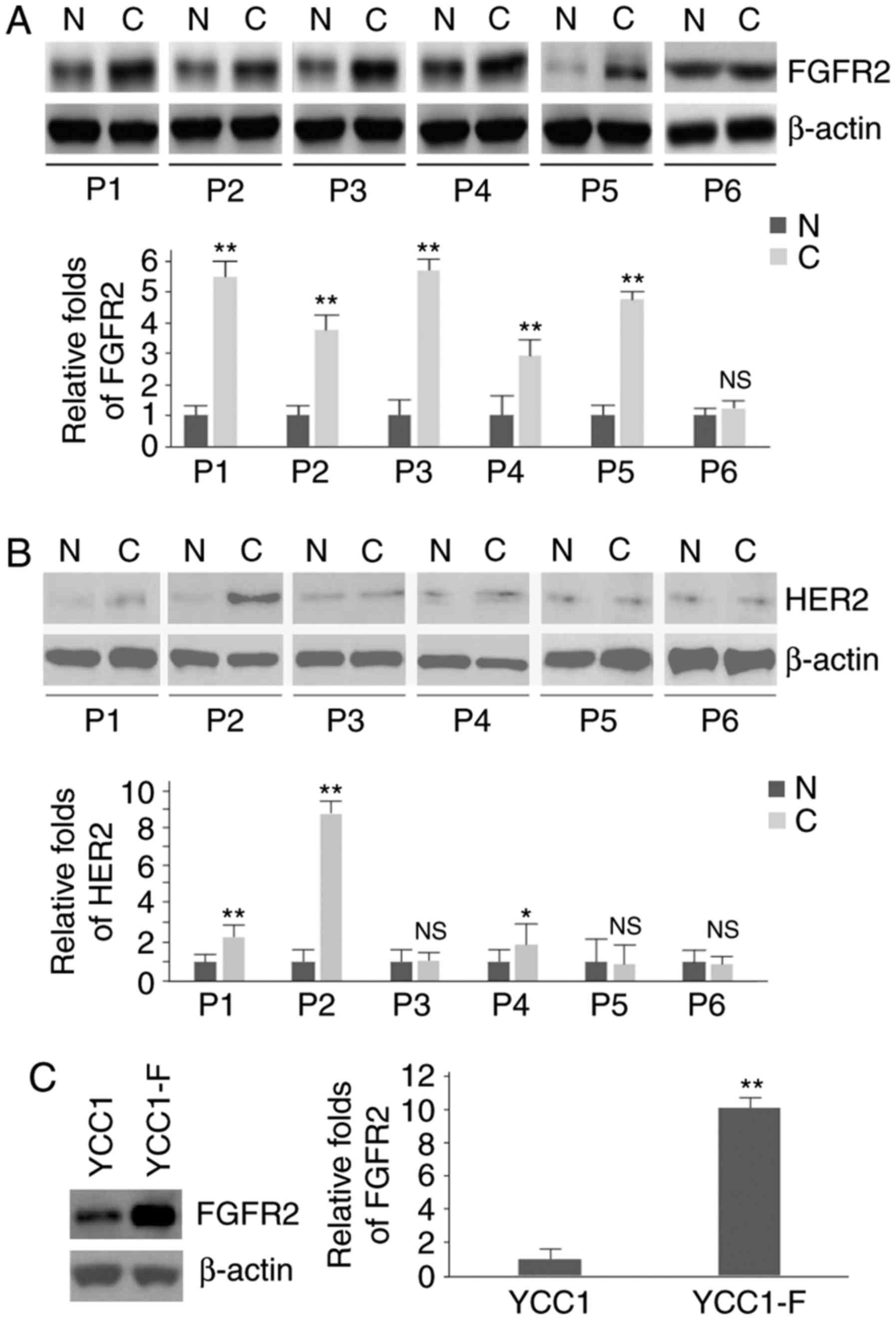
FGFR2 protein is increased in gastric
cancer tissues and YCC1-F cells
To identify the protein expression FGFR2 and HER2 in
gastric cancer tissues and their adjacent normal tissues, western
blot analysis was performed in six pairs of gastric cancer tissues
and their adjacent normal tissues. It was observed that the protein
expression of FGFR2 was significantly increased in five tumor
tissues (patients 1-5; Fig.
1A).
Statistically significant differences in the protein
expression of HER2 were observed between the gastric cancer tissues
and their adjacent normal tissues in patient 1, patient 2 and
patient 4 (Fig. 1B). In addition,
western blot analysis was performed to determine the protein
expression of FGFR2 in YCC1 cells (HER2-positive gastric cancer
cells) and YCC1-F cells (HER2-positive and FGFR2-overexpressing
gastric cancer cells). The results showed that the protein level of
FGFR2 was upregulated in the YCC1-F cells (Fig. 1C).
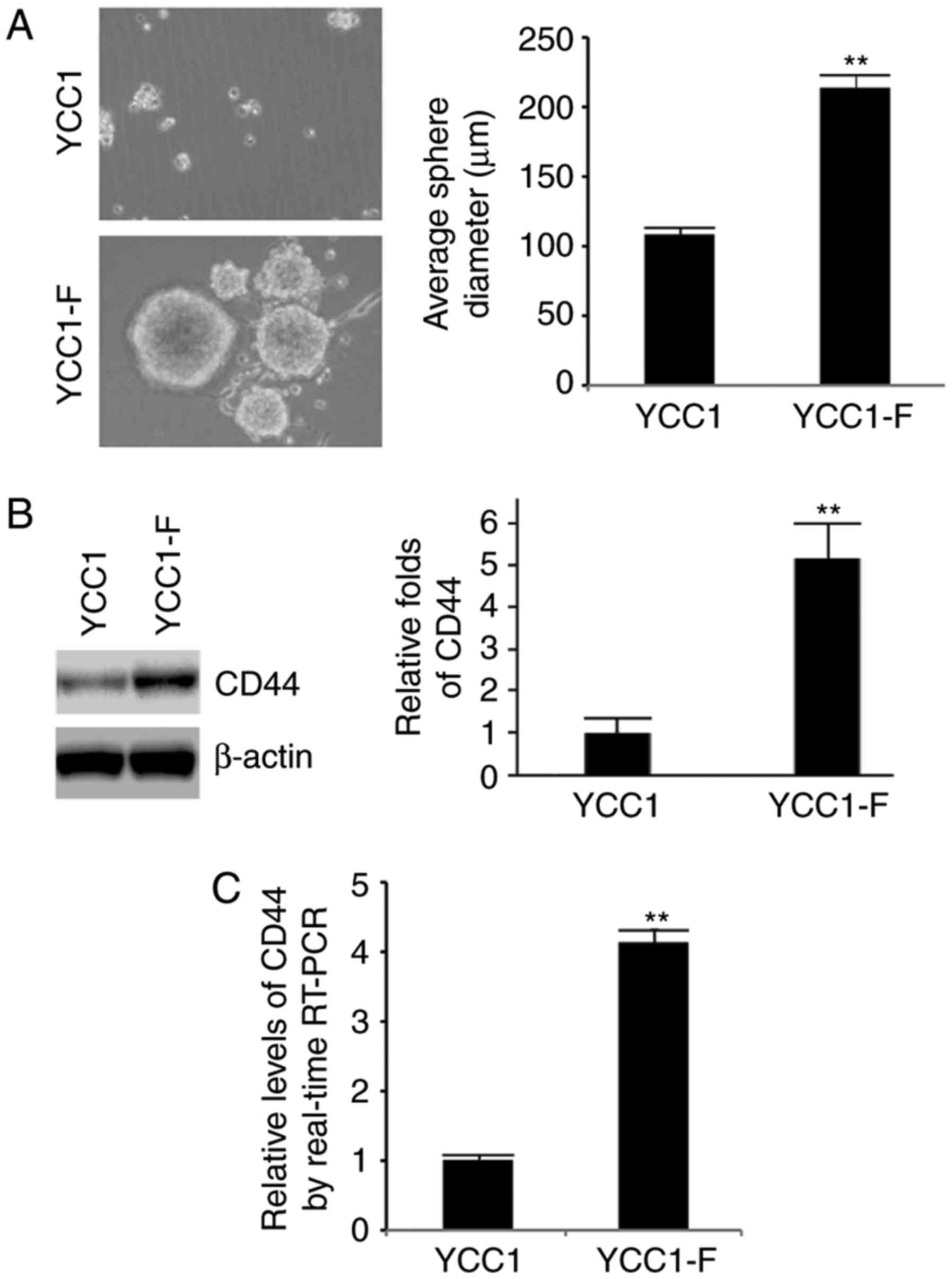
FGFR2 induces the formation of
cancer-initiating cell (CIC) phenotypes
To determine whether FGFR2 affects CICs, a
sphere-forming assay was performed to assess the formation of stem
cell-like populations. It was found that the formation of spheres
was increased in the YCC1-F cells (Fig. 2A). CD44 is a robust marker and is
of functional importance for CICs (25). In order to detect whether the
expression of CD44 can be affected by FGFR2, western blot and
RT-qPCR analyses were performed. It was observed that the protein
and mRNA expression levels of CD44 were upregulated in the YCC1-F
cells (Fig. 2B and C).
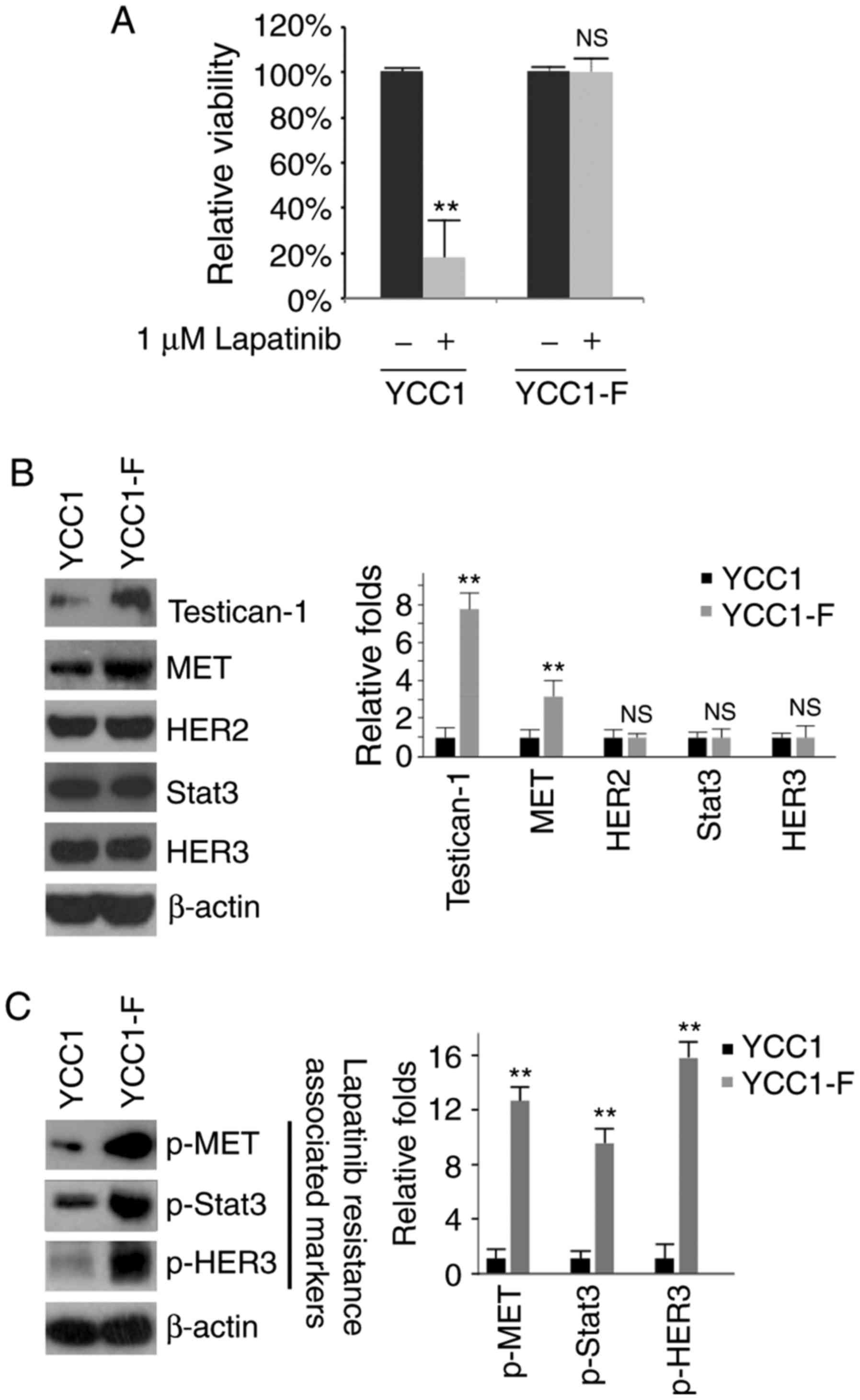
Overexpression of FGFR2 promotes
resistance to lapatinib
In order to determine whether FGFR2 can affect the
efficacy of lapatinib, an MTT assay was performed in the treated
YCC1 and YCC1-F cells (Fig. 3A).
The results showed that the overexpression of FGFR2 promoted
resistance to lapatinib (Fig.
3A). The expression of testican-1, HER3, p-HER3, MET, p-MET and
p-Stat3 has been suggested as markers positively associated with
lapatinib resistance in HER2-positive gastric cancer (19). Therefore, the present study
performed western blot analysis to examine the protein expression
levels of testican-1, MET, HER2, Stat3, HER3, p-MET, p-Stat3 and
p-HER3 in the YCC1 and YCC1-F cells. It was observed that the
protein expression levels of testican-1, p-MET, p-Stat3 and p-HER3
were increased by FGFR2 (Fig. 3B and
C).
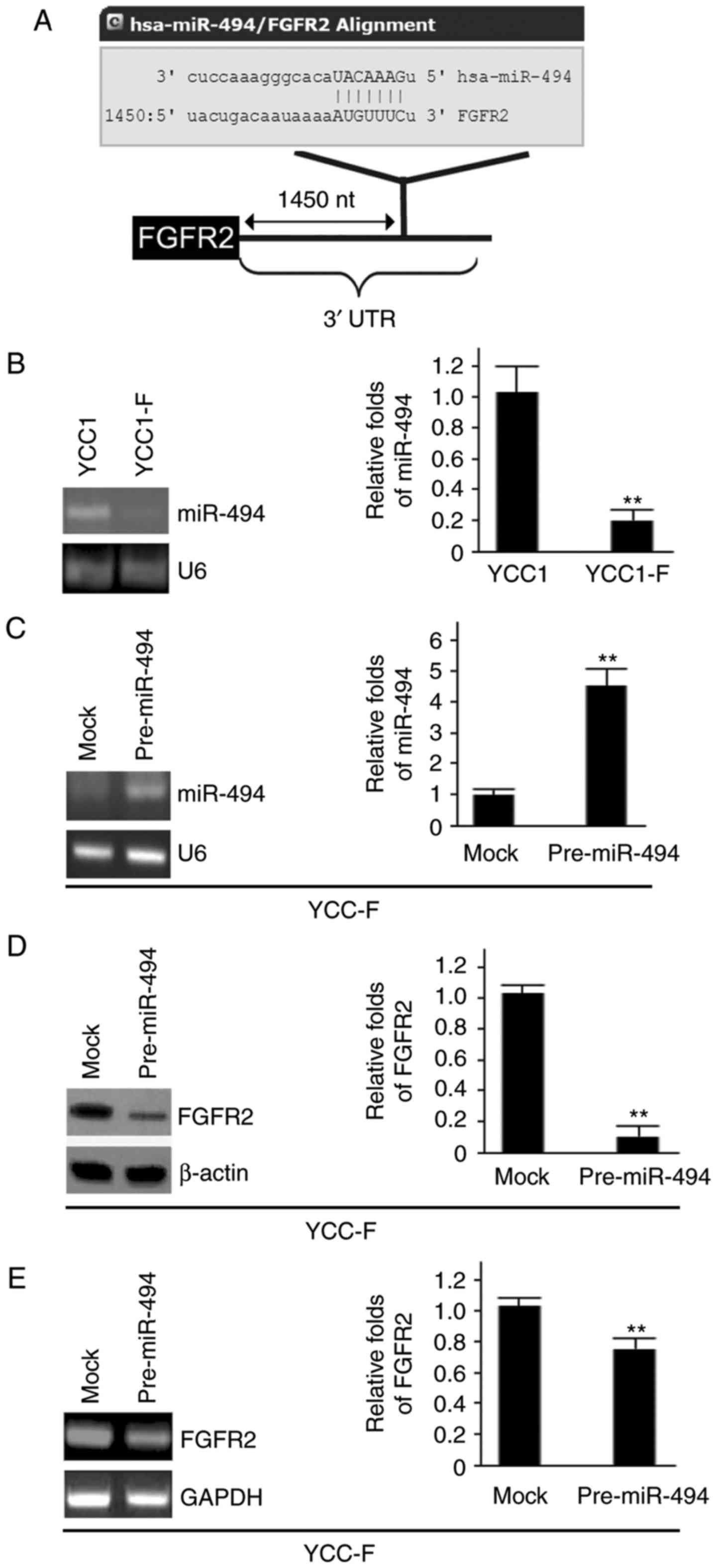
miR-494 inhibits the expression of FGFR2
in YCC1-F cells
The present study also aimed to investigate the
molecular mechanism regulating the expression of FGFR2 in YCC1-F
cells. miRNAs are an emerging class of small, non-coding,
single-stranded RNAs, which serve as important regulators of gene
expression by binding to the 3' untranslated region (UTR) of target
mRNAs, leading to their translational repression and/or degradation
(26-28). To examine whether FGFR2 can be
regulated by miRNAs, the present study used miRanda, a commonly
used prediction algorithm (http://www.microrna.org/microrna/home.do) to analyze
the 3′UTR of FGFR2. In total, 39 miRNAs were found using the
algorithm: miR-494; miR-374b; miR-374a; miR-590-3p; miR-217;
miR-381; miR-300; miR-103; miR-107; miR-15b; miR-424; miR-15a;
miR-497; miR-16; miR-195; miR-223; miR-410; miR-543; miR-153;
miR-431; miR-485-5p; miR-194; miR-544; miR-382; miR-33a; miR-33b;
miR-22; miR-125a-5p; miR-125b; miR-542-3p; miR-330-5p; miR-326;
miR-429; miR-200b; miR-200c; miR-145; miR-340; miR-218 and
miR-296-3p. However, the present study focused on miR-494, as
miR-494 is a candidate tumor suppressor gene (29). It was hypothesized that miR-494
downregulates the protein expression of FGFR2 by targeting its
3′UTR in YCC1-F cells. The target sites on the 3′UTR of FGFR2 are
shown in Fig. 4A.
To examine the expression of miR-494 in the YCC1 and
YCC1-F cells, RT-qPCR analysis was performed. It was found that
miR-494 was decreased in the YCC1-F cells (Fig. 4B). To examine the effect of
miR-494, YCC1-F cells were transfected with pre-miR-494 and control
miR, and the expression of miR-494 was examined using RT-qPCR
analysis. It was found that miR-494 was increased by pre-miR-494 in
the YCC1-F cells (Fig. 4C). To
determine whether miR-494 regulates the protein expression of
FGFR2, western blot analysis was performed to examine the protein
expression of FGFR2 in YCC1-F cells transfected with pre-miR-494
and control miR. It was observed that FGFR2 protein was
significantly inhibited by miR-494 (Fig. 4D). RT-qPCR analysis was then
performed to examine the mRNA expression of FGFR2 in the YCC1-F
cells transfected with pre-miR-494 and control miR. It was found
that the mRNA level of FGFR2 was inhibited by miR-494 (Fig. 4E).
miR-494 inhibits the formation of CIC
phenotypes in YCC1-F cells
To determine whether miR-494 can affect the
formation of CICs, a sphere-forming assay was performed to assess
the formation of stem cell-like populations. It was found that the
formation of spheres was decreased in the YCC1-F cells transfected
with pre-miR-494 (Fig. 5A). To
determine whether the expression of CD44 was affected by miR-494,
western blot and RT-qPCR analyses were performed to examine its
expression. The results showed that the protein and mRNA levels of
CD44 were downregulated in the YCC1-F cells transfected with
pre-miR-494 (Fig. 5B and C).
miR-494 reverses lapatinib-resistance in
YCC1-F cells
To determine whether miR-494 affects lapatinib
efficacy, an MTT assay was performed of the YCC1-F cells following
treatment (Fig. 6A). The results
showed that the overexpression of miR-494 reversed resistance to
lapatinib (Fig. 6A). Western blot
analysis was also performed to detect the protein expression levels
of testican-1, MET, HER2, Stat3, HER3, p-MET, p-Stat3 and p-HER3 in
the YCC1-F cells. The results showed that the protein expression
levels of testican-1, p-MET, p-Stat3 and p-HER3 in the YCC1-F cells
were inhibited by miR-494 (Fig. 6B
and C). To investigate whether the expression of c-myc and
IGF1R were regulated by miR-494 in YCC1-F cells, western blot
analysis was performed to determine the protein expression of c-myc
and IGF1R in cells transfected with pre-miR-494 or control miR. It
was observed that the overexpression of miR-494 significantly
inhibited the protein expression of c-myc and IGF1R in the YCC1-F
cells (Fig. 6D and E).
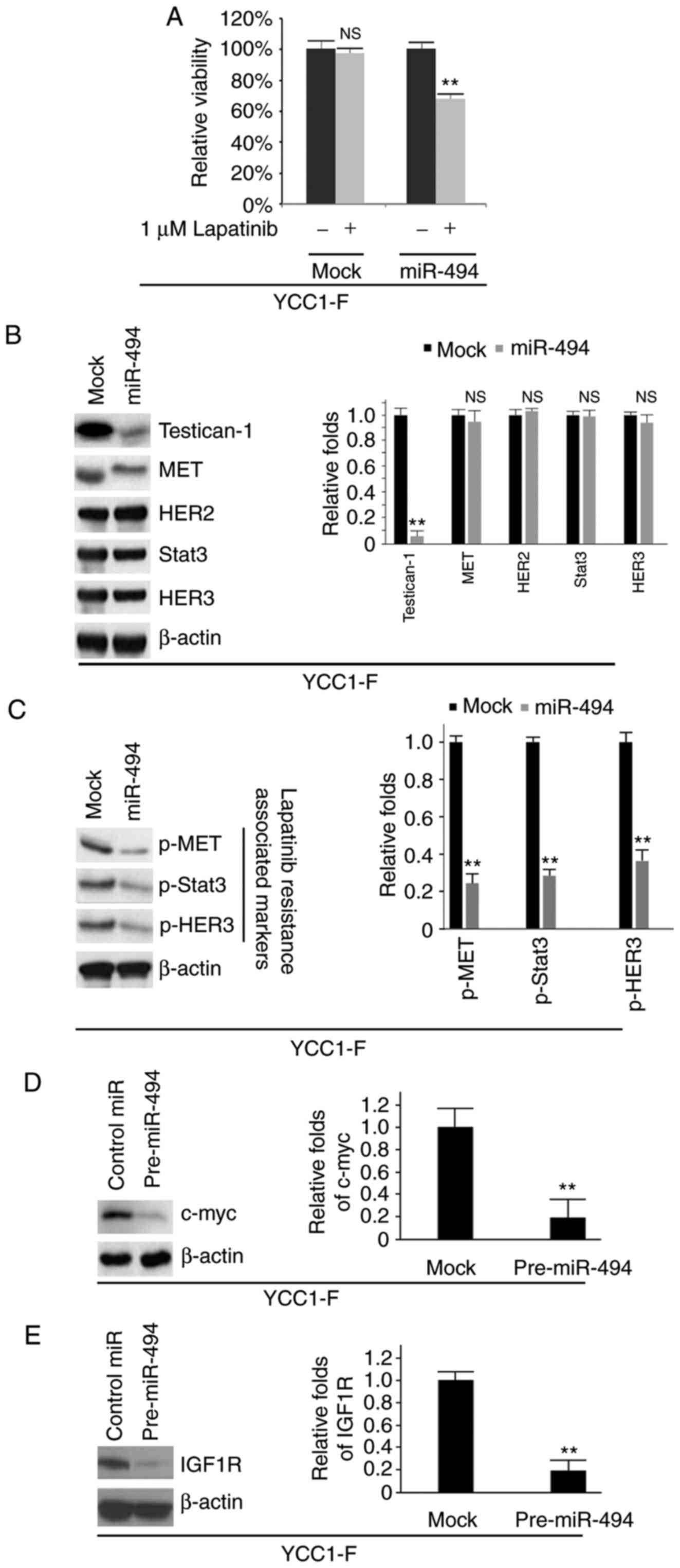 | Figure 6miR-494 reverses resistance to
lapatinib in YCC1-F cells. (A) MTT assay of YCC1-F cells. YCC1-F
cells transfected with pre-miR-494 and control miR (mock) were
untreated or treated with lapatinib (n=3). (B) Western blot
analysis of testican-1, MET, HER2, Stat3 and HER3 proteins in
YCC1-F cells transfected with pre-miR-494 or control (mock) miR
(n=3). Bar chart shows the quantified data for the blots. (C)
Western blot analysis of p-MET, p-Stat3 and p-HER3 proteins in
YCC1-F cells transfected with pre-miR-494 or control (mock) miR
(n=3). Bar chart shows the quantified data for the blots. (D)
Western blot analysis of c-myc protein in YCC1-F cells transfected
with pre-miR-494 and control miR (n=3). Bar chart shows the
quantified data for the blots. (E) Western blot analysis of IGF1R
protein in YCC1-F cells transfected with pre-miR-494 and control
miR (n=3). Bar chart shows the quantified data for the blots.
**P<0.01 vs. control. NS, no significant difference;
miR, microRNA; HER, human epidermal growth factor receptor; Stat3,
signal transducer and activator of transcription 3; p-,
phosphorylated; IGF1R, insulin-like growth factor 1 receptor. |
Discussion
The frequency of overexpression was reported as
11.8% for HER2 and 31.1% for FGFR2 in a large cohort of patients
with gastric cancer (30).
HER2-positive gastric cancer exhibits more differentiated tumor
types (papillary and tubular adenocarcinoma) and is more frequently
associated with venous invasion and regional lymph node metastasis,
compared with HER2-negative cancer (30). Similar to HER2-positive gastric
cancer, FGFR2-positive gastric cancer is more frequently associated
with vascular invasion and a more advanced tumor stage, compared
with FGFR2-negative gastric cancer (30). In the present study, it was
observed that the protein expression of FGFR2 was significantly
increased in five of six tumor tissues examined, and statistically
significant differences in the protein expression of HER2 were
observed between gastric cancer tissues and their adjacent normal
tissues in three of the six patients.
Small molecule inhibitors of HER2, including
lapatinib, are clinically active in women with advanced
HER2-positive gastric cancer (11). However, the effectiveness of this
class of agent is limited by either primary resistance or acquired
resistance. The molecular mechanisms underlying the resistance of
HER2-positive gastric cancer cells to lapatinib remain to be fully
elucidated. The activation of receptor tyrosine kinases can
contribute to lapatinib resistance in HER2 positive gastric cancer
(11). FGFR2 is a receptor
tyrosine kinase (31), and its
overexpression promotes the expression of CD44 and accelerates
tumor growth in mice, and can maintain stemness in gastric cancer
(32). In the present study, it
was observed that FGFR2 was increased in gastric cancer and this
overexpression promoted the formation of CICs, consistent with the
findings of a previous report (32). This increased formation of CICs
can lead to drug resistance (33-35). In accordance with previous reports
(33-35), the present study found that the
overexpression of FGFR2 contributed to lapatinib resistance in
gastric cancer.
Testican-1 can confer acquired resistance to
lapatinib (19). The results of
the present study showed that testican-1 was upregulated by FGFR2
protein in the HER2-positive gastric cancer cells. The expression
levels of p-MET, p-Stat3, p-HER3 are positively associated with
resistance to lapatinib (19).
The present study found that FGFR2 promoted the protein expression
of p-MET, p-Stat3 and p-HER3 in the HER2-positive gastric cancer
cells. These results suggested that FGFR2 contributes to lapatinib
resistance by regulating the protein expression levels of
testican-1, p-MET, p-Stat3 and p-HER3 in HER2-positive gastric
cancer cells. Lapatinib, a potent ATP-competitive inhibitor, is a
small, orally active molecule, which inhibits HER2 and EGFR
(15). One of limitations of the
present study was the lack of HER2 and EGFR antibody
expression.
The expression of miR-494 is decreased in gastric
cancer and acts as an anti-oncogene (36). The present study found that
overexpressing miR-494 downregulated the expression of FGFR2. In
contrast to the role of FGFR2, it was shown that miR-494 inhibited
the formation of CICs. In addition, it was observed that
overexpressing miR-494 reversed resistance to lapatinib. It was
hypothesized that miR-494 regulates the expression of FGFR2 by
targeting its 3′UTR in gastric cancer, and that the downregulation
of miR-494 may contribute to the upregulation of FGFR2, promoting
resistance to lapatinib in HER2-positive gastric cancer. miR-494
acts as a tumor suppressor gene in gastric cancer by targeting
c-myc and IGF1R (36,37). Consistent with previous reports
(36,37), the present study confirmed that
the c-myc and IGF1R proteins were inhibited by miR-494. These
results suggested that the decrease of miR-494 may be causal in the
upregulated protein expression of FGFR2.
Taken together, the results of the present study
provide novel insights for understanding the stemness phenotype and
resistance to lapatinib developed by HER2-positive gastric cancer
cells, which may contribute to the future development of novel
anti-gastric cancer strategies.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yuan E: Taiwan area: Death rate of ten
leading sites of malignant neoplasms. Taiwan: Department of Health,
Executive Yuan; pp. 160–173. 2006
|
|
2
|
Terry MB, Gaudet MM and Gammon MD: The
epidemiology of gastric cancer. Semin Radiat Oncol. 12:111–127.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ushijima T and Sasako M: Focus on gastric
cancer. Cancer Cell. 5:121–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
González CA, Sala N and Capellá G: Genetic
susceptibility and gastric cancer risk. Int J Cancer. 100:249–260.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zheng L, Wang L, Ajani J and Xie K:
Molecular basis of gastric cancer development and progression.
Gastric Cancer. 7:61–77. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smith MG, Hold GL, Tahara E and El-Omar
EM: Cellular and molecular aspects of gastric cancer. 12:2979–2990.
2006.
|
|
7
|
El-Rifai W and Powell SM: Molecular
biology of gastric cancer. Semin Radiat Oncology. 12:128–140. 2002.
View Article : Google Scholar
|
|
8
|
Watson SA, Grabowska AM, El-Zaatari M and
Takhar A: Gastrin-active participant or bystander in gastric
carcinogenesis? Nat Rev Cancer. 6:936–946. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Resende C, Ristimäki A and Machado JC:
Genetic and epigenetic alteration in gastric carcinogenesis.
Helicobacter. 15(Suppl 1): S34–S39. 2010. View Article : Google Scholar
|
|
10
|
Nakajima M, Sawada H, Yamada Y, Watanabe
A, Tatsumi M, Yamashita J, Matsuda M, Sakaguchi T, Hirao T and
Nakano H: The prognostic significance of amplification and
overexpression of c-MET and c-erb B-2 in human gastric carcinomas.
Cancer. 85:1894–1902. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen CT, Kim H, Liska D, Gao S,
Christensen JG and Weiser MR: MET activation mediates resistance to
lapatinib inhibition of HER2-amplified gastric cancer cells. Mol
Cancer Ther. 11:660–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park DI, Yun JW, Park JH, Oh SJ, Kim HJ,
Cho YK, Sohn CI, Jeon WK, Kim BI, Yoo CH, et al: HER-2/neu
amplification is an independent prognostic factor in gastric
cancer. Dig Dis Sci. 51:1371–1379. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang XL, Yang YS, Xu DP, Qu JH, Guo MZ,
Gong Y and Huang J: Comparative study on overexpression of HER2/neu
and HER3 in gastric cancer. World J Surg. 33:2112–2118. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
De Vita F, Giuliani F, Silvestris N,
Catalano G, Ciardiello F and Orditura M: Human epidermal growth
factor receptor 2 (HER2) in gastric cancer: A new therapeutic
target. Cancer Treat Rev. 36(Suppl 3): S11–S15. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Geyer CE, Forster J, Lindquist D, Chan S,
Romieu CG, Pienkowski T, Jagiello-Gruszfeld A, Crown J, Chan A,
Kaufman B, et al: Lapatinib plus capecitabine for HER2-positive
advanced breast cancer. N Engl J Med. 355:2733–2743. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hierro C, Rodon J and Tabernero J:
Fibroblast growth factor (FGF) receptor/FGF inhibitors: Novel
targets and strategies for optimization of response of solid
tumors. Semin Oncol. 42:801–819. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deng N, Goh LK, Wang H, Das K, Tao J, Tan
IB, Zhang S, Lee M, Wu J, Lim KH, et al: A comprehensive survey of
genomic alterations in gastric cancer reveals systematic patterns
of molecular exclusivity and co-occurrence among distinct
therapeutic targets. Gut. 61:673–684. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang T, Wang L, Liu D, Li P, Xiong H,
Zhuang L, Sun L, Yuan X and Qiu H: FGF7/FGFR2 signal promotes
invasion and migration in human gastric cancer through upregulation
of thrombospondin-1. Int J Oncol. 50:1501–1512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim HP, Han SW, Song SH, Jeong EG, Lee MY,
Hwang D, Im SA, Bang YJ and Kim TY: Testican-1-mediated
epithelial-mesenchymal transition signaling confers acquired
resistance to lapatinib in HER2-positive gastric cancer. Oncogene.
33:3334–3341. 2014. View Article : Google Scholar
|
|
20
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar
|
|
22
|
Ghosh RD, Ghuwalewala S, Das P, Mandloi S,
Alam SK, Chakraborty J, Sarkar S, Chakrabarti S, Panda CK and
Roychoudhury S: MicroRNA profiling of cisplatin-resistant oral
squamous cell carcinoma cell lines enriched with
cancer-stem-cell-like and epithelial-mesenchymal transition-type
features. Sci Rep. 6:239322016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Lu DL, Sookthai D, Le Cornet C, Katzke VA,
Johnson TS, Kaaks R and Fortner RT: Reproducibility of serum
oxysterols and lanosterol among postmenopausal women: Results from
EPIC-Heidelberg. Clin Biochem. 52:117–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zöller M: CD44: Can a cancer-initiating
cell profit from an abundantly expressed molecule? Nat Rev Cancer.
11:254–267. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshikawa K, Noguchi K, Nakano Y, Yamamura
M, Takaoka K, Hashimoto-Tamaoki T and Kishimoto H: The Hippo
pathway transcriptional co-activator, YAP, confers resistance to
cisplatin in human oral squamous cell carcinoma. Int J Oncol.
46:2364–2370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heter-ochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luqmani YA, Graham M and Coombes RC:
Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in
normal and malignant human breast, and comparison with other normal
tissues. Br J Cancer. 66:2731992. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nagatsuma AK, Aizawa M, Kuwata T, Doi T,
Ohtsu A, Fujii H and Ochiai A: Expression profiles of HER2, EGFR,
MET and FGFR2 in a large cohort of patients with gastric
adenocarcinoma. Gastric Cancer. 18:227–238. 2015. View Article : Google Scholar
|
|
31
|
Wesche J, Haglund K and Haugsten EM:
Fibroblast growth factors and their receptors in cancer. Biochem J.
437:199–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park J, Kim SY, Kim HJ, Kim KM, Choi EY
and Kang MS: A reciprocal regulatory circuit between CD44 and FGFR2
via c-myc controls gastric cancer cell growth. Oncotarget.
7:28670–28683. 2016.PubMed/NCBI
|
|
33
|
Moriyama T, Ohuchida K, Mizumoto K, Cui L,
Ikenaga N, Sato N and Tanaka M: Enhanced cell migration and
invasion of CD133+ pancreatic cancer cells cocultured
with pancreatic stromal cells. Cancer. 116:3357–3368. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fang XJ, Jiang H, Zhu YQ, Zhang LY, Fan QH
and Tian Y: Doxorubicin induces drug resistance and expression of
the novel CD44st via NF-κB in human breast cancer MCF-7 cells.
Oncol Rep. 31:2735–2742. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kinugasa Y, Matsui T and Takakura N: CD44
expressed on cancer-associated fibroblasts is a functional molecule
supporting the stemness and drug resistance of malignant cancer
cells in the tumor microenvironment. Stem Cells. 32:145–156. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He W, Li Y, Chen X, Lu L, Tang B, Wang Z,
Pan Y, Cai S, He Y and Ke Z: miR-494 acts as an anti-oncogene in
gastric carcinoma by targeting c-myc. J Gastroenterol Hepatol.
29:1427–1434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao XQ, Liang TJ and Fu JW: miR-494
inhibits invasion and proliferation of gastric cancer by targeting
IGF-1R. Eur Rev Med Pharmacol Sci. 20:3818–3824. 2016.PubMed/NCBI
|