Introduction
MicroRNAs (miRNAs) are a class of small, non-coding
RNAs (length, ~22 nt), which regulate gene expression at the
post-transcriptional level. miRNAs are involved in the regulation
of the majority of important biological events, including
differentiation, growth, proliferation, survival, signal
transduction and immune response (1–3).
However, the roles of miRNAs in the activation of human bone
marrow-derived mesenchymal stromal cells (BM-MSCs) remain to be
elucidated.
BM-MSCs are multipotent cells that differentiate
into osteoblasts, adipocytes, chondrocytes and other tissue cells
(4–7). MSCs not only support hematopoiesis
and regulate immunity, but also specifically migrate to sites of
tissue damage, chronic inflammation and tumors (8–11).
For tumor therapy, MSCs can be used as a carrier for tumor
resistance proteins, including interferon-α and -β, and
specifically migrate to tumor sites to inhibit tumor cell growth
(12,13). MSCs can also home to bone marrow,
repair damage in the hematopoietic microenvironment and promote
hematopoietic reconstruction in patients following hematopoietic
stem cell transplantation (14).
MSCs have broad application prospects, however, the mechanism of
MSC migration remains to be fully elucidated. An in-depth
understanding of the mechanisms of MSC migration may enhance
treatment efficiency by improving the ability of MSCs to migrate to
target organs.
Toll-like receptors (TLRs) are an important class of
protein molecules involved in innate immunity and acquired immunity
(15). It has been demonstrated
that TLRs are expressed in MSCs to modulate their proliferation,
cytokine secretion, differentiation, hematopoiesis-supporting
functions and immunosuppressive capacity (16,17). Previous studies have suggested
that the activation of TLR2 inhibits the migration of mice BM-MSCs
(18). In our previous study, it
was demonstrated that TLR2 was expressed on the surface of BM-MSCs
and can suppress their migration (19). Notably, it is well established
that TLRs induce multiple miRNAs, which in turn fine-tunes
TLR-signaling responses at multiple levels. For example, miRNA
(miR)-105, miR-146 and the let-7 miRNA family directly target the
expression of TLR2 and TLR4 (20–23), whereas miR-155 and miR-146b target
numerous TLR downstream signaling proteins (24,25). Regulatory molecules, TLR-induced
transcription factors and the final functional cytokines are also
regulated by miRNAs, including miR-155 (26).
In our previous study, the activation of TLR2
induced the upregulation of miR-27b, miR-146a and miR-155, and the
downregulation of miR-154 in BM-MSCs, indicating that they are
TLR-responsive miRNAs (27). It
was also found that TLR2 was expressed on the surface of BM-MSCs
and that the activation of TLR2 decreased the migration ability of
BM-MSCs (18,19). These findings led to the present
study testing the hypothesis that TLR2-responsive miRNAs are
important in regulating BM-MSC migration ability. The results
showed that miR-155 inhibited the cell migration of BM-MSCs and
provided the first evidence, to the best of our knowledge, that
miR-155 directly targets myosin light chain kinase (MYLK) in
BM-MSCs.
Materials and methods
Cell culture
The present study was approved by the Medical Ethics
Committee of Anhui Medical University (Anhui, China). Written
informed consent was obtained from all participants. The BM-MSCs
were isolated from fresh bone marrow of healthy donors. The
isolation and culture of BM-MSCs have been described previously
(21). In brief, following
density gradient centrifugation for 20 min at 300 × g at room
temperature, mononuclear cells (MNCs) were cultured with high
glucose concentration in Dulbecco's Modified Eagle Medium (HyClone;
GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
fetal calf serum (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) in a 25 cm2 culture flask (Corning
Incorporated, Corning, NY, USA) at a concentration of
1×106 MNCs/ml at 37°C in an atmosphere of 5%
CO2. After 24 h, the non-adherent cells were removed,
and the adherent cells were cultured further. The medium was
replaced twice each week until the cells were ~90% confluent. The
cells were then released by trypsin digestion and passaged into new
culture flasks. Only MSCs in early passages (passage 3–5) were
used. The BM-MSCs were analyzed by flow cytometry following
staining with antibodies against CD90 (cat. no. 555595), CD14 (cat.
no. 555397), CD29 (cat. no. 555443), CD34 (cat. no. 555823), CD166
(cat. no. 559263), CD44 (cat. no. 555478), CD31 (cat. no. 560983),
CD45 (cat. no. 555482), CD13 (cat. no. 560998) and CD105 (cat. no.
561443) were all purchased from all: BD Biosciences, Franklin
Lakes, NJ, USA).
Cell transfection
The miRNAs mimics, miRNAs inhibitor and MYLK small
interfering (si)RNA were purchased from GenePharma Co., Ltd.
(Shanghai, China). The sequences of the miRNAs were as follows:
miR-27b mimics, 5′-AGA GCU UAG CUG AUU GGU GAA C-3′; miR-27b
inhibitor, 5′-GUU CAC CAA UCA GCU AAG CUC U-3′; miR-146a mimics,
5′-UGA GAA CUG AAU UCC AUG GGU U-3′; miR-146a inhibitor, 5′-AAC CCA
UGG AAU UCA GUU CUC A-3′; miR-155 mimics, 5′-UUA AUG CUA AUC GUG
AUA GGG U-3′; miR-155 inhibitor, 5′-ACC CCU AUC ACG AUU AGC AUU
AA-3′; miR-154 mimics, 5′-AAU CAU ACA CGG UUG ACA UAU U-3′; miR-154
inhibitor, 5′-AAU AGG UCA ACC GUG UAU GAU U-3′; MYLK siRNA, 5′-GCC
AAG AUG UUG UGA GCA ATT-3′. At 1 day prior to transfection, the
BM-MSCs were seeded in serum-free medium (Gibco; Thermo Fisher
Scientific, Inc.). The following day, 20 µmol/l miRNA was
transfected using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
cells were treated at 48 h post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). All RT
reactions were performed using 1,000 ng of total RNA according to
the following temperature protocol: 37°C for 60 min and 95°C for 5
min. miRNA quantification was performed by RT-qPCR analysis using
the Step One Real-Time PCR system (Applied Bio systems; Thermo
Fisher Scientific, Inc.) and the SYBR premix Ex Taq II kit (Takara
Biotechnology Co, Ltd., Dalian, China) according to the
manufacturers' protocols. For measurement of the expression of all
miRNA transcripts, U6 was used as the internal reference. Relative
expression of miRNA was evaluated using the 2−ΔΔCq
method (28). Thermo cycling
conditions used for qPCR were as follows: 95°C for 10 min; followed
by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. The primer
sequences were as follows: U6, forward 5′-GCT TCG GCA GCA CAT ATA
CTA AAA T-3′ and reverse 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′;
miR-27b, forward 5′-GGG GAA GAG CTT AGC TGA TTG-3′ and reverse
5′-GTG CGT GTC GTG GAG TCG-3′; miR-146a, forward 5′-GGG TGA GAA CTG
AAT TCC-3′ and reverse 5′-TGC GTG TCG TGG AGT C-3′; miR-154,
forward 5′-GGG GGA ATC ATA CAC GGT TG-3′ and reverse 5′-GTG CGT GTC
GTG GAG TCG-3′; miR-155, forward 5′-GGG GGT AAT GCT AAT CGT GAT-3′
and reverse 5′-GTG CGT GTC GTG GAG TCG-3′.
Cell migration assay
A total of 2×104 BM-MSCs in 100 µl
culture medium (Hyclone; GE Healthcare Life Sciences, Logan, UT,
USA) were seeded in the upper insert of a Transwell with an 8-mm
pore-size membrane (Corning Incorporated, Corning, NY, USA), and
600 µl culture medium was added to the lower chamber.
Following incubation for 24 h at 37°C, each membrane was fixed with
4% paraformaldehyde (Cellchip Biotechnology Co., Ltd., Beijing,
China), and MSCs on the membrane were stained with trypan blue
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The numbers of
migrated BM-MSCs were determined by counting the number of cells
beneath the filter membrane in five fields at a high magnification
under an inverted microscope. The experiments were performed with
three replicates for each condition.
Target gene prediction and Gene Ontology
(GO) analysis
Three online search algorithms, TargetScan version
6.2 (http://www.targetscan.org/vert_60/), miRanda
(http://www.microrna.org/microrna/home.do), and
Microcosm Targets version 5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/)
were used to predict the target genes of miR-155. The overlapping
sections that were identified by these on-line tools were
considered to be the target genes. To confirm the predicted target
genes, the genes were subjected to analysis by the Gene Ontology
project (http://www.geneontology.org). The
ontology covers three domains: Biological Process, Cellular
Component and Molecular Function. Fisher's exact test is used to
determine whether there is more overlap between the differentially
expressed (DE) list and the GO annotation list than would be
expected by chance. The P-value denotes the significance of GO term
enrichment in the DE genes. The lower the P-value, the more
significant the GO term (P≤0.05 is recommended).
Luciferase reporter assay
To confirm whether MYLK is a direct target of
miR-155, a luciferase reporter assay was performed. 293T cells were
seeded in a 48-well plate (Corning Incorporated) at 80% confluence
and co-transfected with miR-155 mimics or inhibitors, and the
psiCHECK-MYLK-3-untranslated region (UTR) or
psiCHECK-MYLK-3-UTR-mutant (mut) vectors (GenePharma Co., Ltd.)
using Lipofectamine 2000. Following incubation for 48 h, the cells
were collected and analyzed using a Dual-Luciferase assay kit
(Promega Corporation, Madison, WI, USA). Each assay was performed
with three replicates for each condition.
Western blot analysis
The cells were harvested 48 h following transfection
with the miR-155 mimics or inhibitor. The cells were pelleted and
lysed in lysing buffer (5 µl protease inhibitor mixture, 5
µl PMSF and 5 µl phosphatase mixture). The lysates
were centrifuged at 16,000 × g for 15 min at 4°C. Supernatants were
subsequently collected and total protein concentration was
determined using the Bio-Rad DC protein assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). An equal quantity of
protein from each cell lysate (20 µg) was separated by 4–12%
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred onto a polyvinylidene fluoride membrane (Kangchen,
Shanghai, China). The membranes were blocked with 5% bovine serum
albumin (Sigma-Aldrich; Merck KGaA) followed by incubation
overnight at 4°C with the following primary antibodies: Rabbit
anti-human MYLK monoclonal antibody (cat. no. ab76092; 1:2,000
dilution; Abcam, Cambridge, MA, USA), rabbit anti-human RhoA
monoclonal antibody (cat. no. ab187027; 1:1,000 dilution; Abcam),
rabbit anti-human Rho-associated, coiled-coil containing protein
kinase (Rock)1 monoclonal antibody (cat. no. ab134181; 1:2,000
dilution; Abcam), rabbit anti-human Rock2 monoclonal antibody (cat.
no. ab125025; 1:10,000 dilution; Abcam) and rabbit anti-human
β-actin monoclonal antibody (cat. no. ab150301; 1:10,000 dilution;
Abcam). Incubation with the corresponding horseradish
peroxidase-conjugated goat anti-rabbit secondary antibodies (cat.
no. 32460; 1:2,000 dilution; Pierce; Thermo Fisher Scientific,
Inc.) was performed for 1 h at 37°C. Following three washes with
TBS, the bound secondary antibody was visualized using enhanced
chemiluminescence solution (Kangchen, Shanghai, China). Images were
captured using the ImageJ system (version 1.50; National Institutes
of Health, Bethesda, MD, USA). Densitometry was performed for
comparison of western blot data (Alpha Innotech, San Leandro, CA,
USA).
Statistical analysis
Data were analyzed using SPSS 13.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Values are presented as
the mean ± standard deviation. For statistical comparisons,
two-tailed Student's t-test (two-group) or one-way analysis of
variance (multi-population) was applied as appropriate. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-155 inhibits the migration of
BM-MSCs
In view of the significant inhibitory effect of TLR2
on the migration of BM-MSCs and the correlation of the TLR2
activation of miR-27b, miR-146a, miR-155 and miR-154, BM-MSCs were
transfected with mimics or inhibitors of miR-27b, miR-146a, miR-155
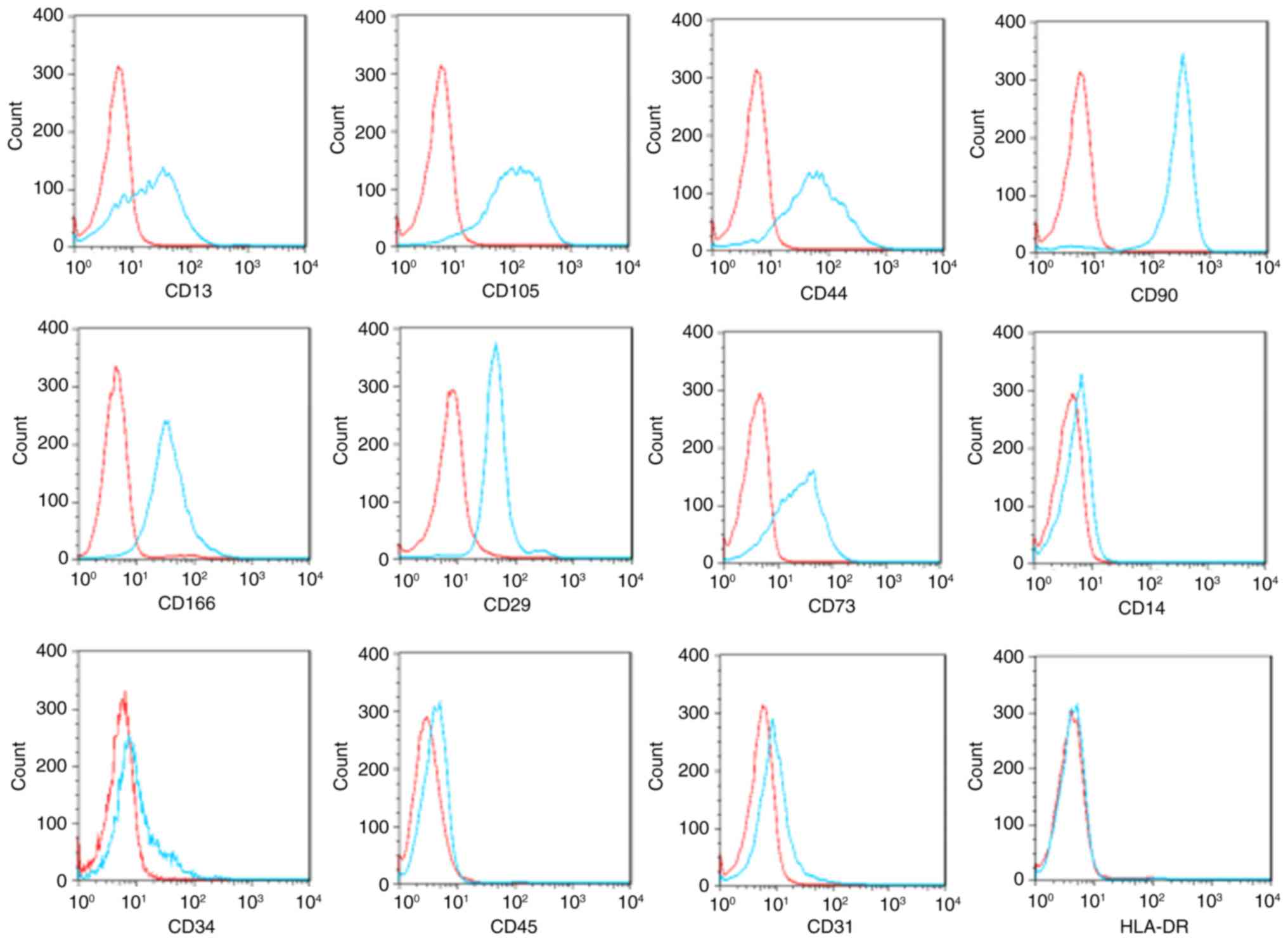
and miR-154. The isolated BM-MSCs were positive for the markers
CD90, CD105, CD166, CD29, CD44, CD13, and CD73, but negative for
hematopoietic and endothelial lineage markers (CD14, CD34, CD31 and
CD45) and HLA-DR (Fig. 1).
RT-qPCR analysis was used to assess the expression of the four
miRNAs in BM-MSCs following transfection. As expected, the
expression levels of miRNAs were significantly upregulated
following transfection with miRNA mimics, compared with levels in
the negative control group. By contrast, the expression levels of
miRNAs were downregulated following transfection with miRNA
inhibitors (Fig. 2).
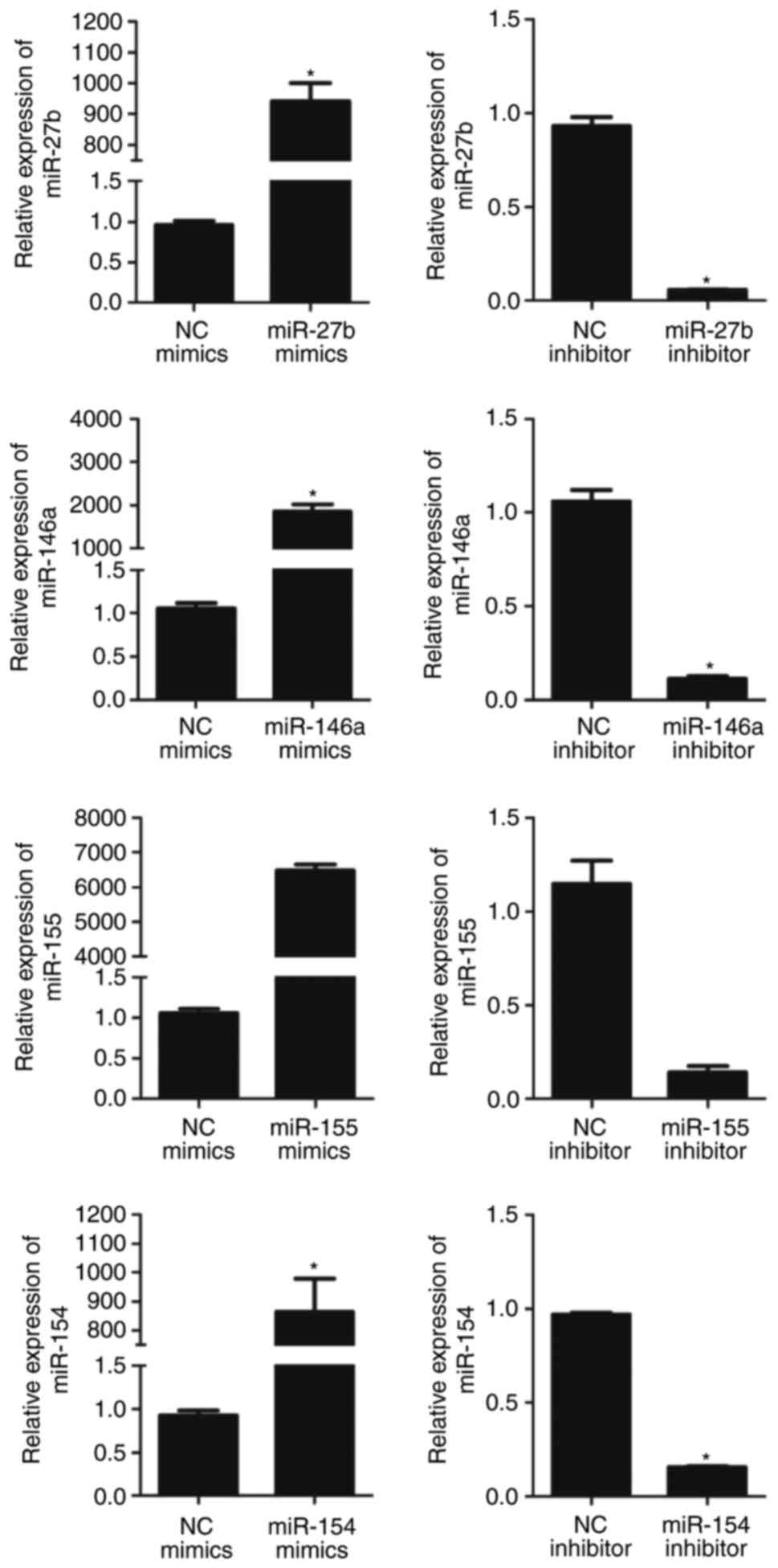 | Figure 2Relative expression of miRNA in
BM-MSCs following transfection with miRNA mimics or inhibitor.
Expression of miRNA was significantly upregulated following
transfection with miRNA mimics and downregulated following
transfection with miRNA inhibitors. The relative expression of
miR-27b, miR-146a, miR-155 and miR-154 in BM-MSCs transfected with
miRNA mimics NC were 0.96±0.05, 1.06±0.06, 1.06±0.06 and 0.92±0.06,
respectively. The relative expression levels of miR-27b, miR-146a,
miR-155 and miR-154 in BM-MSCs transfected with miRNA inhibitor NC
were 0.93±0.05, 1.06±0.07, 1.15±1.12 and 0.97±0.01, respectively.
The relative expression of miR-27b following transfection with
miR-27b mimics and miR-27b inhibitor were 940.43±58.84 (P<0.01)
and 0.06±0.004 (P<0.01), compared with the respective control.
The relative expression of miR-146a following transfection with
miR-146a mimics and miR-146a inhibitor were 1,851.31±167.31
(P<0.01) and 0.11±0.01 (P<0.01) compared with the respective
control. The relative expression of miR-155 following transfection
with miR-155 mimics and miR-155 inhibitor were 6478.00±170.15
(P<0.01) and 0.14±0.04 (P<0.01), compared with the respective
control. The relative expression of miR-154 following transfection
with miR-154 mimics and miR-154 inhibitor were 865.07±114.10
(P<0.01) and 0.15±0.01 (P<0.01), compared with the respective
control. Values are expressed as the mean ± standard deviation.
*P<0.05, compared with the respective control.
BM-MSCs, bone marrow derived-mesenchymal stromal cells; miR,
microRNA; NC, negative control. |
To examine the role of miRNA in the migration of
BM-MSCs, cells transfected with miRNA mimics or inhibitor were
cultured in Transwell chambers. The results of the Transwell assays
showed that miR-155 mimics significantly suppressed the migration
of BM-MSCs, whereas BM-MSC migration was enhanced following
transfection with miR-155 inhibitors. miR-27b, miR-146a and miR-154
had no significant effect on the migration of BM-MSCs (Fig. 3A and B). The present study also
evaluated the effects of the miRNA mimics/inhibitors on the cell
viability of BM-MSCs in vitro. None of these compounds
induced cell death of the BM-MSCs at 48 h post-transfection,
assessed using the trypan blue staining method (data not
shown).
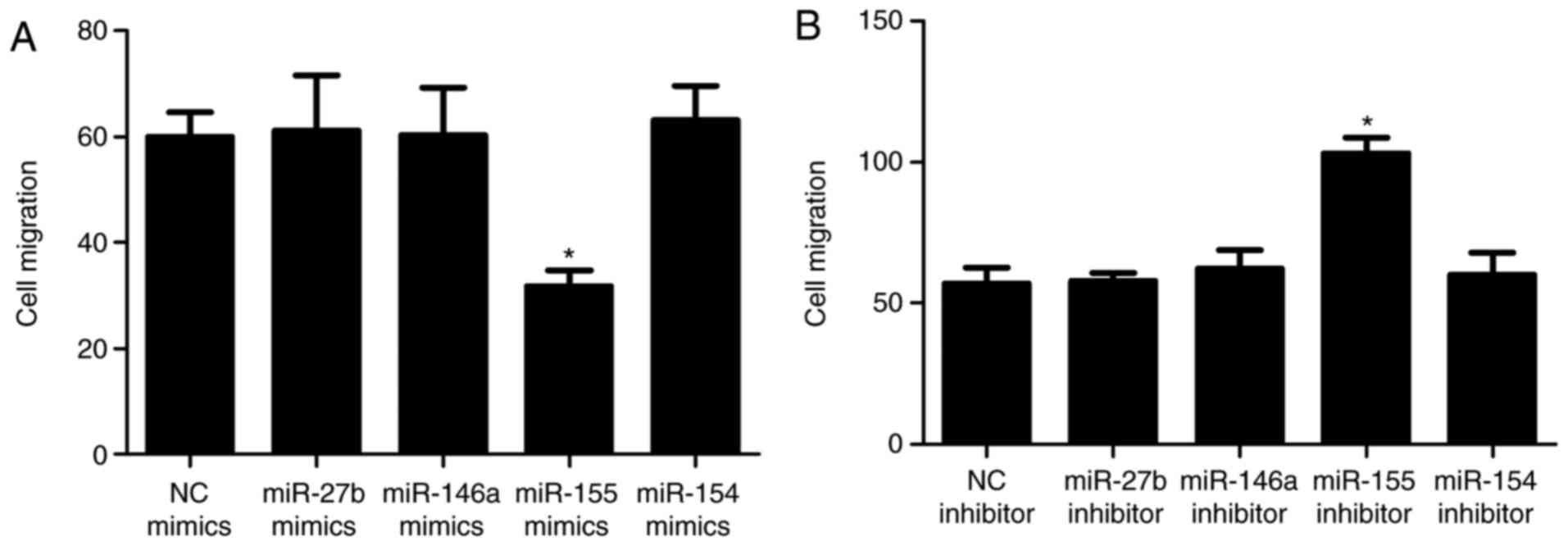 | Figure 3Role of miRNA in the migration of
BM-MSCs. Migration following transfection with (A) miR mimics and
(B) miR inhibitor. The numbers of migrated BM-MSCs following
transfection with miRNA NC and miR-155 mimics were 60.00±4.58 and
31.67±3.06 (P<0.01), respectively; suggesting that miR-155
significantly suppressed the migration of BM-MSCs. Numbers of
migrated BM-MSCs following transfection with miRNA inhibitor NC and
miR-155 inhibitor were 57.00±5.57 and 103.00±5.57 (P<0.01),
respectively, showing BM-MSC migration was enhanced following
transfection with miR-155 inhibitor and confirmed that miR-155
inhibits cell migration. Numbers of migrated BM-MSCs transfected
with miR-27b mimics, miR-146a mimics and miR-154 mimics were
64.33±5.03 (P>0.05), 60.33±8.96 (P>0.05) and 63.00±6.56
(P>0.05) respectively, whereas the numbers of migrated BM-MSCs
following transfection with miR-27b inhibitor, miR-146a inhibitor
and miR-154 inhibitor were 58.00±2.65 (P>0.05), 59.00±5.29
(P>0.05) and 58.00±4.00 (P>0.05), respectively, indicating
miR-27b, miR-146a and miR-154 had no significant effect on
migration. Values are expressed as the mean ± standard deviation.
*P<0.05, compared with the respective control.
BM-MSCs, bone marrow derived-mesenchymal stromal cells; miR,
microRNA; NC, negative control. |
MYLK is a direct target gene of miR-155
in BM-MSCs
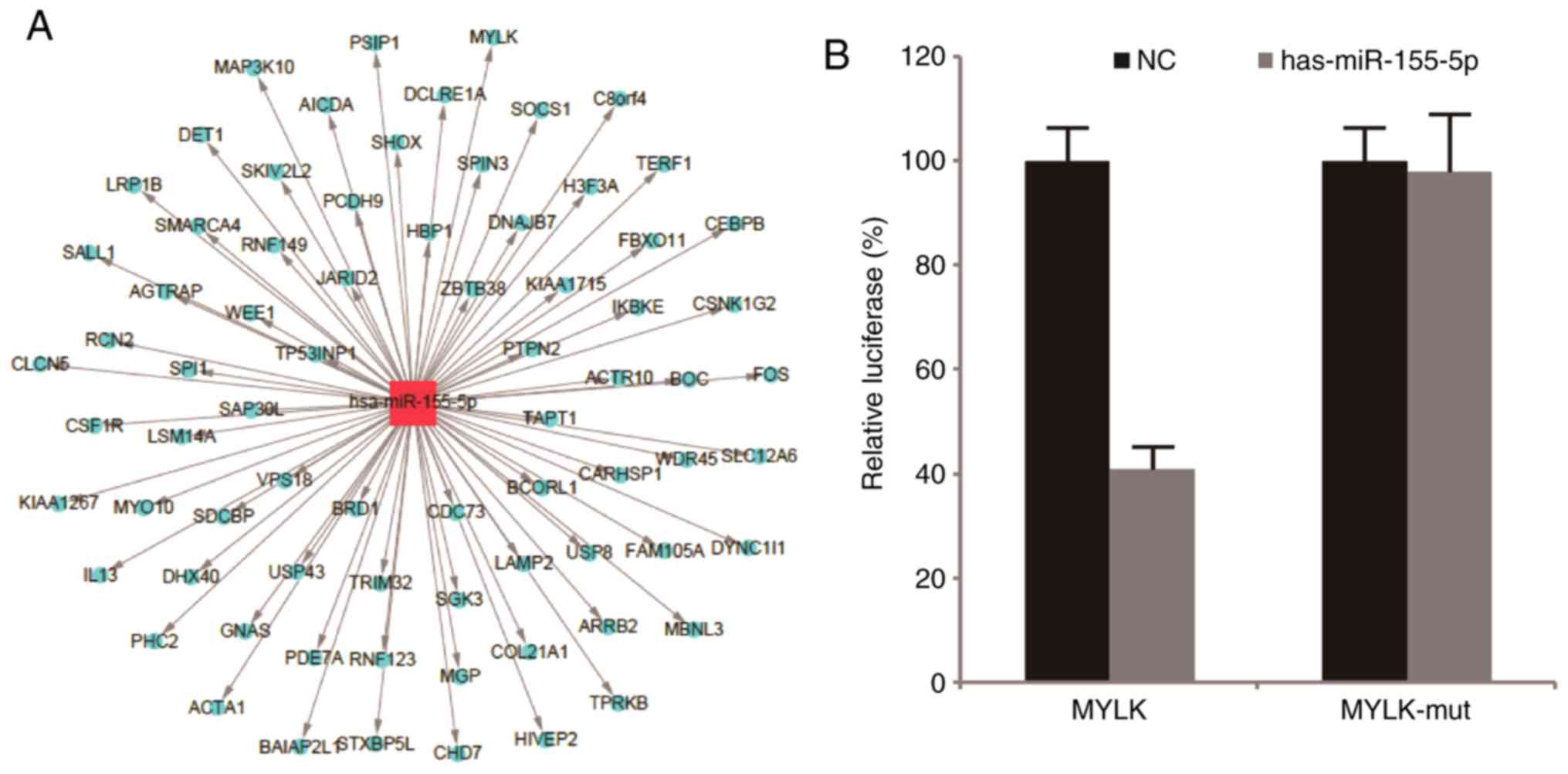
To identify targets of miR-155 in BM-MSCs,
bioinformatics analysis of miR-155 predicted target genes was
performed using three online search algorithms, TargetScan version
6.2, miRanda and Microcosm Targets version 5, which identified 448,
3,387 and 930 potential targets, respectively, with an overlap of
74 genes. The overlapping sections that were identified by these
online tools were considered to be the target genes. As shown in
Fig. 4A, several genes were
identified and subjected to GO analysis. The present study focused
on genes associated with the Biological Process of cell movement.
As a result, MYLK was predicted to be a target of miR-155 as the
3′-UTR of its mRNA contained a region with affinity for miR-155. To
verify whether miR-155 directly targets MYLK, a luciferase reporter
assay was performed. miR-155 was found to significantly inhibit the
luciferase activity of the reporter vector containing the wild-type
sequence of the MYLK 3′UTR targeted by miR-155, whereas the
luciferase activity of the reporter vector containing a mutant
sequence was not affected by miR-155 in the BM-MSCs (Fig. 4B; P<0.05).
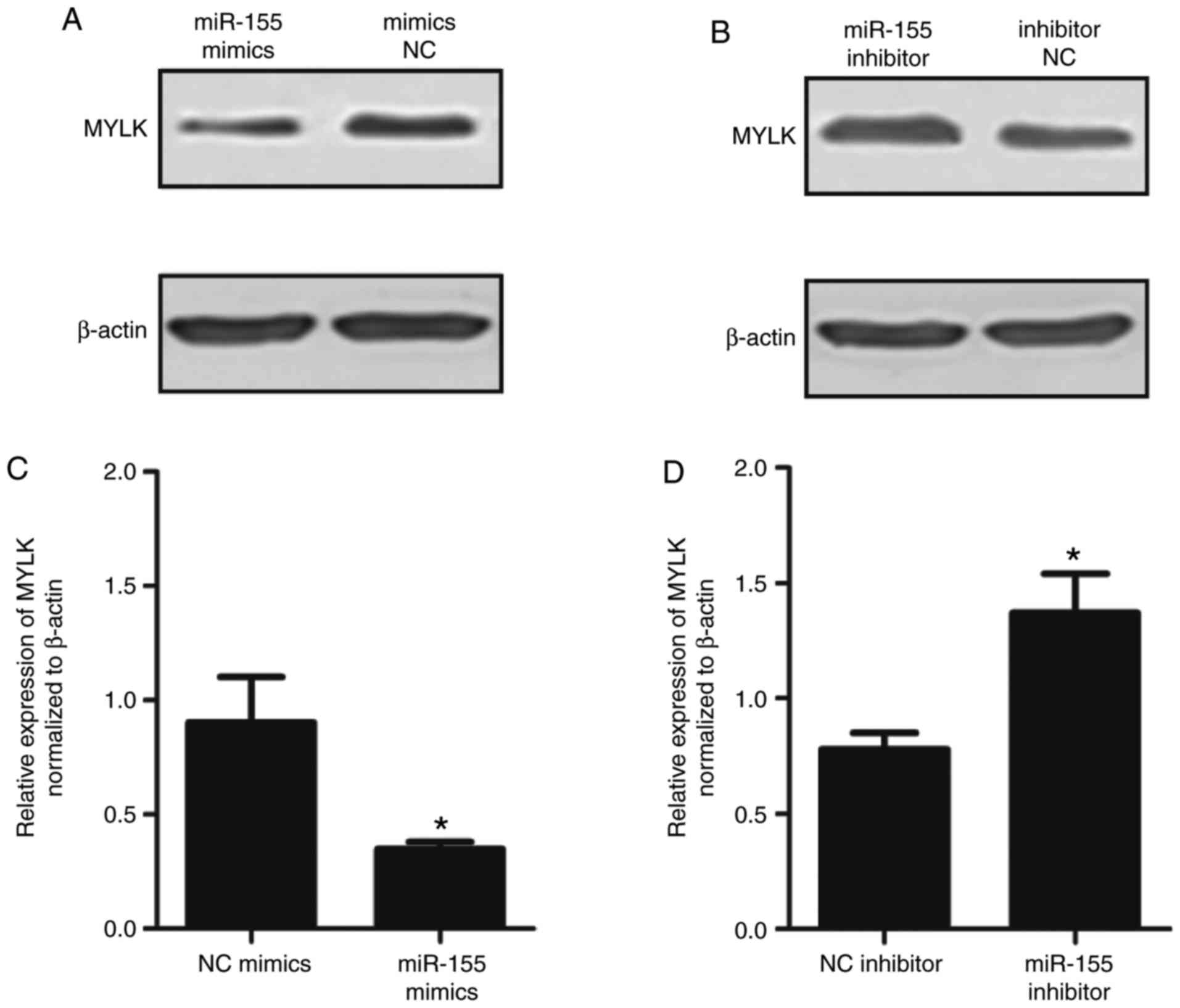
Furthermore, western blot analysis was performed to
determine whether MYLK was decreased following transfection with
miR-155 mimics. As shown in Fig.
5A–D, the protein expression of MYLK was significantly
down-regulated following transfection with miR-155 mimics and
upregulated following transfection with miR-155 inhibitors
(P<0.05). In addition, the upstream proteins, RhoA, Rock1 and
Rock2, were detected. As shown in Fig. 6A–D, no significant differences
between the groups were observed. Therefore, it was hypothesized
that miR-155 targeting MYLK does not affect the RhoA pathway. To
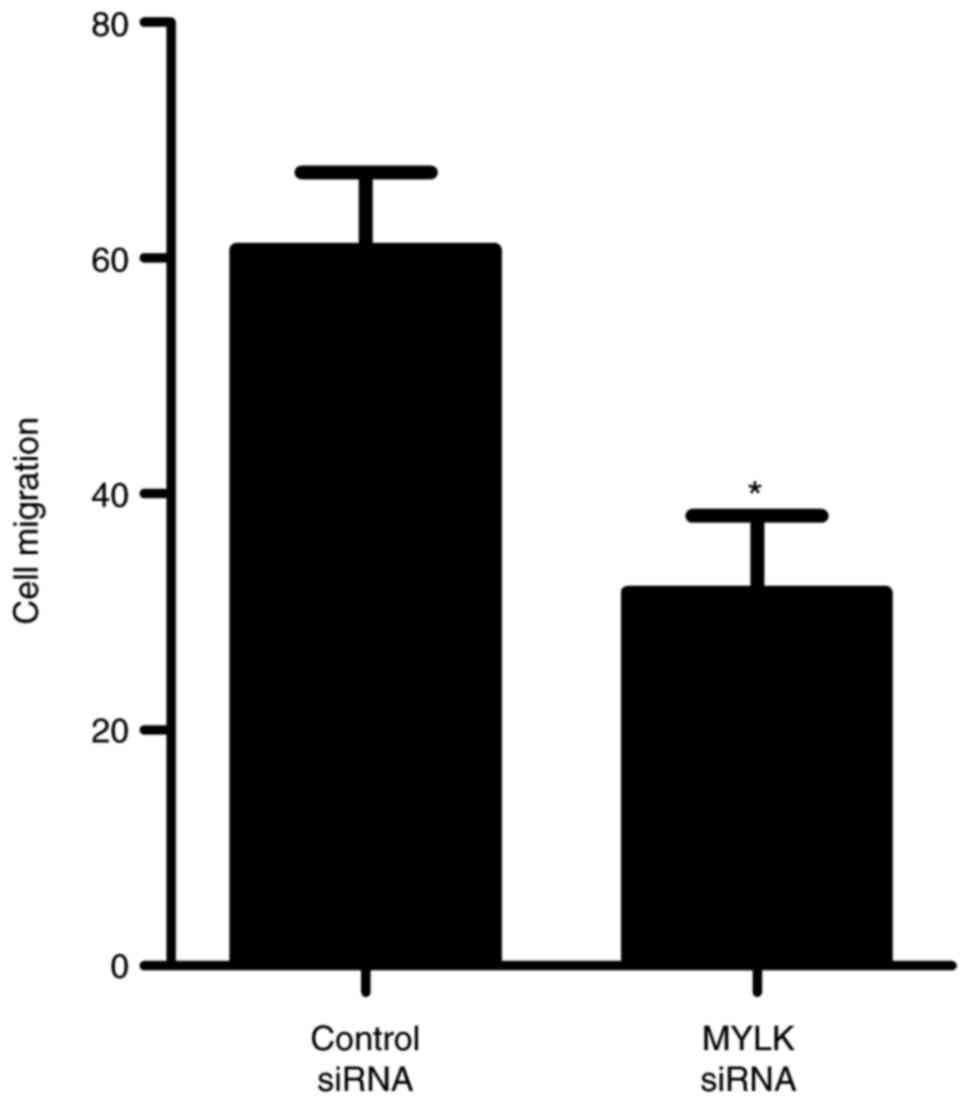
further confirm the role of MYLK in regulating BM-MSC migration,
siRNA was used to knock down MYLK in BM-MSCs. As shown in Fig. 7, the inhibition of MYLK by MYLK
siRNA significantly suppressed the migration of BM-MSCs. Taken
together, these results suggested that miR-155 inhibited BM-MSC
migration via directly targeting MYLK.
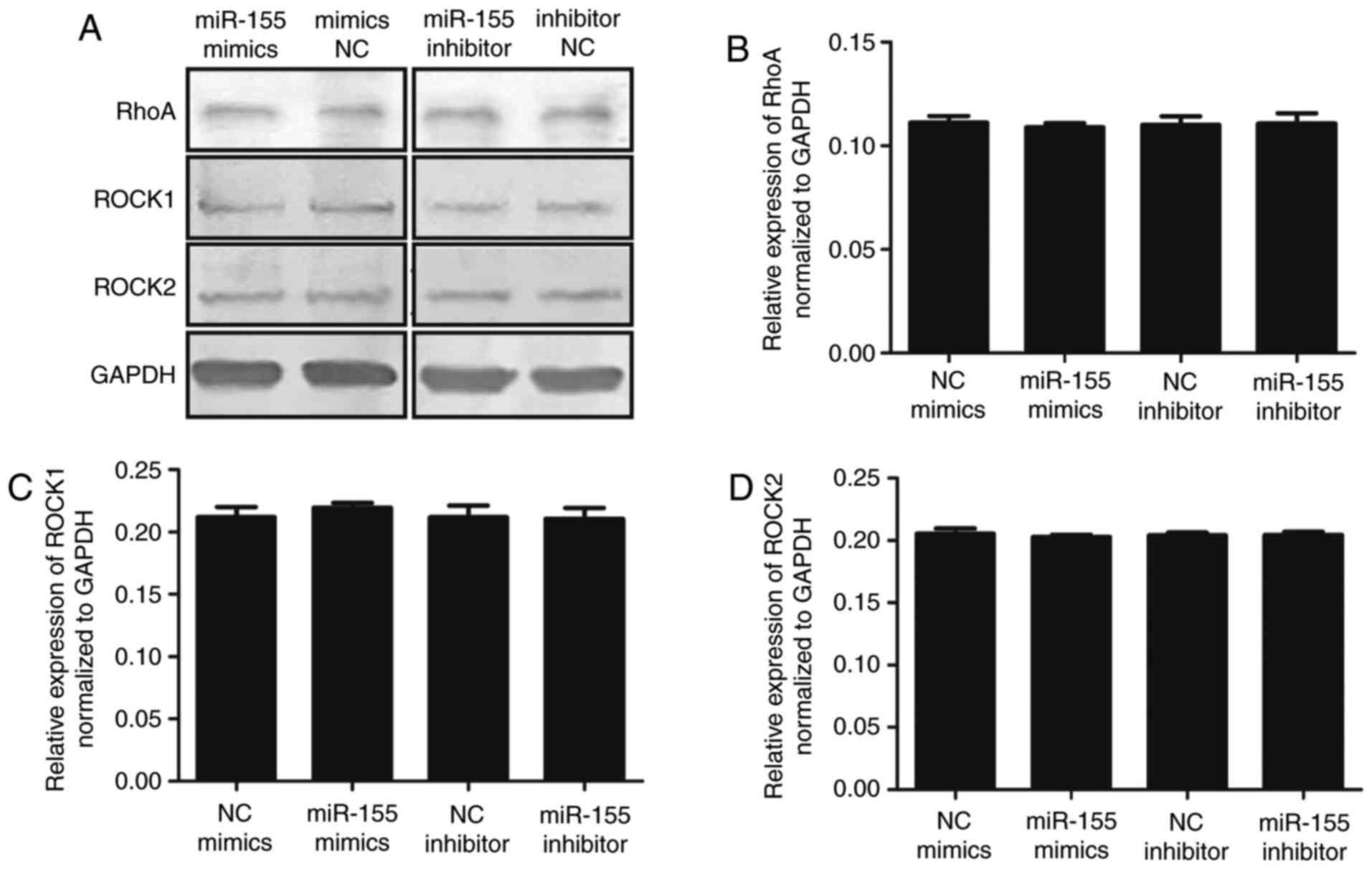 | Figure 6(A) Protein expression of RhoA, ROCK1
and ROCK2 in BM-MSCs transfected with miR-155 mimics or inhibitor.
(B) The relative protein expression of RhoA following transfection
with miRNA mimics NC, miR-155 mimics, miRNA inhibitor NC and
miR-155 inhibitor were 0.111±0.003, 0.108±0.001 (P>0.05),
0.110±0.004 and 0.110±0.005 (P>0.05) respectively. (C) Relative
protein expression of Rock1 following transfection with miRNA
mimics NC, miR-155 mimics, miRNA inhibitor NC and miR-155 inhibitor
were 0.212±0.008, 0.220±0.003 (P>0.05), 0.212±0.009 and
0.211±0.008 (P>0.05), respectively. (D) Relative protein
expression of Rock2 following transfection with miRNA mimics NC,
miR-155 mimics, miRNA inhibitor NC and miR-155 inhibitor were
0.205±0.004, 0.203±0.002 (P>0.05), 0.204±0.002 and 0.204±0.003
(P>0.05) respectively. BM-MSCs, bone marrow derived-mesenchymal
stromal cells; Rock, Rho-associated, coiled-coil containing protein
kinase; miR, microRNA; NC, negative control. |
Discussion
Human miR-155 is encoded by the miR-155 host gene
and is involved in various physiological and pathological
processes. miR-155 has been found to inhibit adipogenesis and
immune regulation of MSCs, however, its effect on migration has not
been reported (29,30). In the present study, the results
showed that miR-155 significantly inhibited the migration of
BM-MSCs. MYLK, a member of the immunoglobulin gene superfamily,
encodes myosin light chain kinase, a calcium/calmodulin-dependent
enzyme (31). MYLK phosphorylates
the N-terminus of the regulatory light chain of the molecular motor
myosin II to produce contractility, which is involved in the
migration of cells (32). Several
studies have shown that MYLK is associated with cell migration.
Weber et al found that MYLK is involved in the migration of
endothelial cells. Their study confirmed that miR155 targets MYLK
to inhibit cell migration (33).
Miao et al found that MYLK-targeted siRNA decreased the
expression of MYLK and cell migration of optic nerve head (ONH)
astrocytes compared with control siRNA (34). This finding indicated that MYLK is
a target in the inhibition of ONH astrocyte migration. Although
MYLK is important in cell migration, its role in the migration of
BM-MSCs has been unclear. Therefore, the present study is the
first, to the best of our knowledge, to show that MYLK and related
targets are involved in BM-MSC migration, and suggests an
additional mechanism for the effect on BM-MSC migration.
The present study is the first, to the best of our
knowledge, to demonstrate that miR-155 inhibits BM-MSC migration by
targeting MYLK. Weber et al reported that miR-155 targeted
MYLK to inhibit the migration of vascular endothelial cells
(33). This issue is important in
future investigations of miRNA, as it is essential for the
application of a target gene of miRNA in one type of cell to other
types of cells. In our previous study, it was found that the
activation of TLR2 significantly inhibited the migration of
BM-MSCs, and the expression of miR-155 was significantly
upregulated following the activation of TLR2 (19,27). The present study confirmed that
miR-155 significantly inhibited the migration of BM-MSCs.
Therefore, the evidence suggests that the mechanism of TLR2
inhibits BM-MSC migration, involving the upregulation of miR-155
and the inhibition of cell migration through miR-155 targeting
MYLK. In addition, miR-27b, miR-146a and miR-154 were examined in
the present study. The results showed that none of these three
miRNAs had a marked effect on cell migration. Of note, a previous
study reported that miR-146a can target stromal cell-derived
factor-1 to inhibit BM-MSC migration (35). By comparison, this previous study
involved counting the numbers of migrated BM-MSCs 12 h following
the beginning of migration, whereas the 24 h time-point was used in
the present study; this indicated a problem for cell migration in
future investigations, as the different migration time may result
in different results. In addition, miR-27b was found to inhibit the
migration of mouse MSCs in a previous study (36). In the present study, miR-27b had
no significant effect on the migration of human BM-MSCs, however,
it is unclear whether this is due to differences between species,
which requires further investigation.
In conclusion, the present study is the first, to
the best of our knowledge, to show that miR-155 inhibited the
migration of BM-MSCs by reducing the expression of MYLK. Through
the identification of novel target genes of miR-155, the present
study enhances current understanding of the mechanisms of BM-MSC
migration. These findings may facilitate the development of the
clinical use of BM-MSCs. Further investigation is required to
identify additional target genes of miR-155 and to assess its
suitability for use in clinical applications of BM-MSCs.
Acknowledgments
Not applicable.
Abbreviations:
|
BM-MSCs
|
bone marrow derived-mesenchymal
stromal cells
|
|
GO
|
Gene Ontology
|
|
miRNAs
|
microRNAs
|
|
MYLK
|
myosin light chain kinase
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TLRs
|
toll-like receptors
|
Funding
This study was supported by the National Natural
Science Foundation (grant no. 81270573), the Science and Technology
key project of Anhui Province (grant no. 1604a0802071) and the
Natural Science Foundation of Anhui Higher Education Institutions
(grant no. KJ2012Z188).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XW designed the study, analyzed the data and
contributed to the writing of the manuscript. XY, JJ and QZ
performed the cell culture, cell transfection, cell migration
assays, Luciferase reporter assays, reverse
transcription-quantitative polymerase chain reaction analysis and
contributed to the writing of the manuscript. BX performed Gene
Ontology analysis. JW and JM performed cell culture and western
blot analyses. XL analyzed the data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Anhui Medical University. Written informed consent was
obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moreno-Moya JM, Vilella F and Simón C:
MicroRNA: Key gene expression regulators. Fertil Steril.
101:1516–1523. 2014. View Article : Google Scholar
|
|
4
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang Y, Jahagirdar BN, Reinhardt RL,
Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund
T, Blackstad M, et al: Pluripotency of mesenchymal stem cells
derived from adult marrow. Nature. 418:41–49. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Charbord P: Bone marrow mesenchymal stem
cells: Historical overview and concepts. Hum Gene Ther.
21:1045–1056. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oreffo RO, Cooper C, Mason C and Clements
M: Mesenchymal stem cells: Lineage, plasticity, and skeletal
therapeutic potential. Stem Cell Rev. 1:169–178. 2005. View Article : Google Scholar
|
|
8
|
Spaeth E, Klopp A, Dembinski J, Andreeff M
and Marini F: Inflammation and tumor microenvironments: Defining
the migratory itinerary of mesenchymal stem cells. Gene Ther.
15:730–738. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chamberlain G, Fox J, Ashton B and
Middleton J: Concise review: Mesenchymal stem cells: Their
phenotype, differentiation capacity, immunological features, and
potential for homing. Stem Cells. 25:2739–2749. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fox JM, Chamberlain G, Ashton BA and
Middleton J: Recent advances into the understanding of mesenchymal
stem cell trafficking. Br J Haematol. 137:491–502. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Warnke PH, Wiltfang J, Springer I, Acil Y,
Bolte H, Kosmahl M, Russo PA, Sherry E, Lützen U, Wolfart S and
Terheyden H: Man as living bioreactor: Fate of an exogenously
prepared customized tissue-engineered mandible. Biomaterials.
27:3163–3167. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ren C, Kumar S, Chanda D, Kallman L, Chen
J, Mountz JD and Ponnazhagan S: Cancer gene therapy using
mesenchymal stem cells expressing interferon-beta in a mouse
prostate cancer lung metastasis model. Gene Ther. 15:1446–1453.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Studeny M, Marini FC, Champlin RE,
Zompetta C, Fidler IJ and Andreeff M: Bone marrow derived
mesenchymal stem cells as vehicles for interferon-beta delivery
into tumors. Cancer Res. 62:3603–3608. 2002.PubMed/NCBI
|
|
14
|
Pontikoglou C, Deschaseaux F, Sensebé L
and Papadaki HA: Bone marrow mesenchymal stem cells: Biological
properties and their role in hematopoiesis and hematopoietic stem
cell transplantation. Stem Cell Rev. 7:569–589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kollmann TR, Levy O, Montgomery RR and
Goriely S: Innate immune function by Toll-like receptors: Distinct
responses in newborns and the elderly. Immunity. 37:771–783. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shirjang S, Mansoori B, Solali S, Hagh MF
and Shamsasenjan K: Toll-like receptors as a key regulator of
mesenchymal stem cell function: An up-to-date review. Cell Immunol.
315:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Cheng Q, Li L, Wang J, Xia L, Xu X
and Sun Z: Toll-like receptors 2 and 4 mediate the capacity of
mesenchymal stromal cells to support the proliferation and
differentiation of CD34+ cells. Exp Cell Res.
318:196–206. 2012. View Article : Google Scholar
|
|
18
|
Lei J, Wang Z, Hui D, Yu W, Zhou D, Xia W,
Chen C, Zhang Q, Wang Z, Zhang Q and Xiang AP: Ligation of TLR2 and
TLR4 on murine bone marrow-derived mesenchymal stem cells triggers
differential effects on their immunosuppressive activity. Cell
Immunol. 271:147–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang ZH, Wang XB, Wang J, Li LL and Zhu
YX: Influence of TLR2 and TLR4 agonists on migration of human bone
marrow mesenchymal stem cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
22:183–186. 2014.In Chinese. PubMed/NCBI
|
|
20
|
Benakanakere MR, Li Q, Eskan MA, Singh AV,
Zhao J, Galicia JC, Stathopoulou P, Knudsen TB and Kinane DF:
Modulation of TLR2 protein expression by miR-105 in human oral
keratinocytes. J Biol Chem. 284:23107–23115. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Quinn EM, Wang JH, O'Callaghan G and
Redmond HP: MicroRNA-146a is upregulated by and negatively
regulates TLR2 signaling. PLoS One. 8:e622322013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
O'Hara SP, Splinter PL, Gajdos GB,
Trussoni CE, Fernandez-Zapico ME, Chen XM and LaRusso NF: NFkappaB
p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated
transcriptional repression of microRNA let-7i following microbial
infection. J Biol Chem. 285:216–225. 2010. View Article : Google Scholar
|
|
23
|
Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu
J, Sun Z and Shen WF: MiR-146a inhibits oxidized low-density
lipoprotein-induced lipid accumulation and inflammatory response
via targeting toll-like receptor 4. FEBS Lett. 585:854–860. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tili E, Michaille JJ, Cimino A, Costinean
S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA and
Croce CM: Modulation of miR-155 and miR-125b levels following
lipo-polysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Curtale G, Mirolo M, Renzi TA, Rossato M,
Bazzoni F and Locati M: Negative regulation of Toll-like receptor 4
signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci USA.
110:11499–11504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ceppi M, Pereira PM, Dunand-Sauthier I,
Barras E, Reith W, Santos MA and Pierre P: MicroRNA-155 modulates
the interleukin-1 signaling pathway in activated human
monocyte-derived dendritic cells. Proc Natl Acad Sci USA.
106:2735–2740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Zhu Y, Xu B, Wang J and Liu X:
Identification of TLR2 and TLR4-induced microRNAs in human
mesenchymal stem cells and their possible roles in regulating TLR
signals. Mol Med Rep. 13:4969–4980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Skårn M, Namløs HM, Noordhuis P, Wang MY,
Meza-Zepeda LA and Myklebost O: Adipocyte differentiation of human
bone marrow-derived stromal cells is modulated by microRNA-155,
microRNA-221, and microRNA-222. Stem Cells Dev. 21:873–883. 2012.
View Article : Google Scholar
|
|
30
|
Xu C, Ren G, Cao G, Chen Q, Shou P, Zheng
C, Du L, Han X, Jiang M, Yang Q, et al: miR-155 regulates immune
modulatory properties of mesenchymal stem cells by targeting
TAK1-binding protein 2. J Biol Chem. 288:11074–11079. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lazar V and Garcia JG: A single human
myosin light chain kinase gene (MLCK; MYLK). Genomics. 57:256–267.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kamm KE and Stull JT: Dedicated myosin
light chain kinases with diverse cellular functions. J Biol Chem.
276:4527–4530. 2001. View Article : Google Scholar
|
|
33
|
Weber M, Kim S, Patterson N, Rooney K and
Searles CD: MiRNA-155 targets myosin light chain kinase and
modulates actin cytoskeleton organization in endothelial cells. Am
J Physiol Heart Circ Physiol. 306:H1192–H1203. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miao H, Crabb AW, Hernandez MR and Lukas
TJ: Modulation of factors affecting optic nerve head astrocyte
migration. Invest Ophthalmol Vis Sci. 51:4096–4103. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hsieh JY, Huang TS, Cheng SM, Lin WS, Tsai
TN, Lee OK and Wang HW: miR-146a-5p circuitry uncouples cell
proliferation and migration, but not differentiation, in human
mesenchymal stem cells. Nucleic Acids Res. 41:9753–9763. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lü MH, Li CZ, Hu CJ, Fan YH, Wang SM, Wu
YY, Liang GP and Yang SM: microRNA-27b suppresses mouse MSC
migration to the liver by targeting SDF-1α in vitro. Biochem
Biophys Res Commun. 421:389–395. 2012. View Article : Google Scholar
|