Introduction
Male fertility is maintained through continuous germ
cell production, which are the only cells able to divide into
haploid cells and carry genetic information generationally
(1). Spermatozoa are produced via
spermatogenesis, which is a complex process involving a series of
mitosis, two stages of meiosis, and post-meiotic differentiation
(2).
In mammals, meiosis is a crucial stage triggered by
the metabolism of retinol and its metabolite, retinoic acid (RA), a
derivative of vitamin A. RA induces the transcription and
expression of stimulated by retinoic acid 8 (Stra8), which commands
the initiation and progression of meiosis (3,4).
Stra8 is a 45-kDa vertebrate-specific gene, without significant
homologous proteins, expressed in the cytoplasm and nuclei of germ
cells between mitosis and meiosis and presents exclusively in the
embryonic and postnatal gonads of mammals. In fetal testis, RA is
temporarily degraded by cytochrome P450 26B1, preventing the
expression of Stra8. Following birth, RA induces the expression of
Stra8 in germ cells, leading to meiotic initiation (4). In vitamin A-deficient (VAD) mice,
the majority of germ cells arrest as undifferentiated spermatogonia
(spg). In VAD rats, some germ cells arrest as preleptotene
spermatocytes. However, when these animals are injected with RA or
vitamin A, the arrested spermatogonia differentiate (5,6).
Stra8 gene-knockout mice are sterile, and accompanied by abnormal
phenotypes, including meiotic cell cycle, chromatin condensation,
homologous recombination, DNA repair and cell apoptosis (7–9).
The testis of Stra8-defecient mice (Stra8−/−mice)
contain a large number of undifferentiated spermatogonia,
preleptotene spermatocytes, and spermatocytes that have features
characteristic of leptonema (i.e., nuclear morphologies) (7–9).
These findings indicate that Stra8 is required for transforming
preleptotene spermatocytes into leptotene spermatocytes (8). According to previous reports, the
phenotypes of meiotic cells are abnormal and spermatogenesis is
interrupted in stra8−/− mice, in which the majority of
spermatogonia undergo apoptosis (8,9);
however, the mechanisms underlying the anti-apoptotic effects of
Stra8 remain to be elucidated.
In the present study, it was found that Stra8
inhibited spermatogenic cell apoptosis in VAD mice,
Stra8−/− mice and Stra8-overexpressing GC1 spg cells.
Compared with the control groups, nine differentially expressed
genes (DEGs) were found to be associated with apoptosis using
Affymetrix GeneChip microarray analysis. Based on these results,
the possible signaling pathway of Stra8 in apoptosis was
discussed.
Materials and methods
Mice
The experiments included two models established in
male mice: i) VAD and vitamin A recovery (VAR), respectively, on a
C57BL/6 genetic background. A total of 20 VAD mice, 18 VAR mice and
36 C57BL/6J wild-type (wT) mice were used during the experiments.
The mice were obtained from the Laboratory Animal Center of
Yangzhou University (Yangzhou, China), and male and female mice
were permitted to mate following being fed a VAD diet for 2 weeks,
with the VAD diet maintained during pregnancy. Neonatal male mice
were fed a VAD diet for 13–14 weeks to obtain VAD mice, and
14-week-old VAD male mice were fed a normal diet containing vitamin
A to obtain VAR mice; ii) Stra8-defecicient mice on a C57BL/6
genetic background. Heterozygous B6.Cg-Stra8tm1Dcp/J mice
(Stra8+/− mice) were purchased from Jackson Laboratories
(Bar Harbor, ME, USA), including one 8-week-old male mouse and two
8-week-old female mice. Stra8-deficient mice (Stra8−/−
mice) were generated by mating heterozygotes (Stra8+/−
mice). Stra8 genotypes were detected by polymerase chain reaction
(PCR) as described previously (10). All mice were housed in the
Laboratory Animal Center of Yangzhou University. They were housed
in a controlled temperature (21–25°C) and light conditions (12 h
dark and 12 h light cycle), and fed a normal diet. The date of
birth was defined as 0 days postpartum (dpp). A total of six 11 dpp
Stra8−/− mice and six C57BL/6J wT mice were used for the
assay and analysis of apoptosis. A total of six adult (7–8-week
old) Stra8−/− mice and six C57BL/6J wT mice were used
for hematoxylin and eosin (H&E) staining. The offspring were
sacrificed at various postnatal ages by decapitation. All
experiments were approved by the Committee on Animal Care of
Yangzhou University.
H&E staining
Following sacrifice of the adult (7–8-week-old) mice
by decapitation, their testes were harvested immediately and fixed
with 4% (w/v) paraformaldehyde for 24 h, and then in chloroform
overnight following dehydrating with a gradient alcohol series. The
tissue was transferred into xylene and then embedded in paraffin. A
microtome was used to cut the tissues into 4-µm sections.
Following dewaxing and hydration, the sections were stained with
hematoxylin for 5 min and eosin for 2 min at room temperature. The
samples were observed under a light microscope (Nikon-70i; Nikon
Corporation, Tokyo, Japan).
Cell cultivation
GC1 spg cells overexpressing Stra8 (Stra8-GC1) and
control GC1 spg (Control-GC1) cells were established at the
laboratory at the Department of Histology and Embryology (School of
Medicine, Yangzhou University) using the following procedure: i)
the entire open reading frame of the Stra8 gene was inserted into
lentivirus-GV34 1 (a lenti-viral expression vector); ii) GV34-Stra8
was transfected into 293T cells (Shanghai Zhong Qiao Xin Zhou
Biotechnology, Co., Ltd., Shanghai, China) together with two helper
vectors; and iii) after 2 days, the lentiviral particles were
collected and used to transfect mouse GC1 spg cells to obtain
Stra8-GC1 spg cells. The lentivirus-GV34 was transfected into mouse
GC1 spg cells to obtain Control-GC1 spg cells.
The Stra8-GC1 and Control-GC1 cells were maintained
in Dulbecco's modified Eagle's medium (wisent, Inc., St. Bruno, QC,
Canada) with 100 U/ml penicillin, 100 mg/ml streptomycin, and 10%
heat-inactivated fetal bovine serum (FBS; Hyclone, GE Healthcare
Life Sciences, Logan, UT, USA), and incubated at 37°C with 5%
CO2. Medium exchange was performed every 48–72 h when
the cell density reached 80–90% confluency. After 24 h, the cells
were harvested and processed for TUNEL, cell apoptosis, microarray,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blot analyses.
TUNEL assay
Apoptosis in the testes and germ cells was evaluated
using an In Situ Cell Death Detection kit, Fluorescein (Roche
Diagnostics, Basel, Switzerland) in accordance with the
manufacturer's protocol. 4′6-diamidino-2-phenylindole technique
(DAPI, Solarbio Science & Technology Co., Ltd., Beijing, China)
was used to visualize the level of DNA fragmentation. Following
dewaxing and rehydration, the 4-µm sections of testes were
incubated with a proteinase K working solution for 15–30 min at
21–37°C. The cells were incubated in a prepared fixation solution
(final concentration: 4% PFA) for 60 min at 15–25°C, and then
incubated with the permeabilization solution (final concentration:
0.1% Triton X-100 with 0.1% sodium citrate) for 2 min on ice
(2–8°C), followed by two washes with PBS. The sections were
incubated in 50 µl TUNEL reaction mixture composed of label
solution and enzyme solution, with label solution as a negative
control. The sections were incubated in a humidified atmosphere for
60 min at 37°C in the dark. Following washing three times with PBS
and then incubating with DAPI, the sections were detected and
analyzed by fluorescence microscopy (Carl Zeiss AG, Oberkochen,
Germany) at 515–565 nm (green light).
Cell apoptosis assay
An Annexin V/Propidium Iodide (PI) Apoptosis
Detection kit (CwBIO, Beijing, China) was used to detect the
apoptosis of Stra8 and control germ cells, according to the
manufacturer's protocol. The cells were cultured in 60 cm dishes
with a density of at least 1×106 cells/dish for 48 h.
The Stra8 and control germ cells were then harvested separately,
and washed twice with PBS. Subsequently, 50 µl of the cell
suspension was mixed with 450 µl Annexin V binding buffer,
and the mixture was incubated with FITC-labeled Annexin V for 5
min, followed by PI for 10 min at room temperature in the dark.
During the final phase of this experiment, the samples were
immediately analyzed via flow cytometry (BD FACSCalibur™, BD
Biosciences, Franklin Lakes, NJ, USA).
Sample processing and microarray
analysis
The microarray analysis was executed using the
Affymetrix GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Inc.,
Santa Clara, CA, USA), and 45,103 probe sets were used to analyze
the expression level of at least 20,000 well-characterized mouse
genes. Total RNA was isolated using the TRIzol method. The quantity
and quality of RNA was determined using a NanoDrop 2000c (Thermo
Fisher Scientific, Inc.) and an Agilent 2100 Bioanalyzer with an
RNA 6000 Nano kit (Agilent Technologies, Inc., Santa Clara, CA,
USA). Samples with an RNA integrity number ≥7.0 and 260/280 nm
ratios between 1.7 and 2.2 were used for detailed analyses. Total
RNA was amplified, labeled and fragmented using the GeneChip 3′IVT
PLUS kit and hybridized for 16 h at 45°C in a GeneChip
Hybridization Oven 645 (Thermo Fisher Scientific, Inc.). The
GeneChips were washed and stained in GeneChip Fluidics Station 450
and scanned with GeneChip Scanner 3000 (Affymetrix, Inc.).
Differentially expressed genes (DEGs) were selected with a fold
change (FC) >1.5 and P<0.01 (one-sample t-test). A functional
analysis based on the Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway database (https://www.kegg.jp/) and Gene Ontology (http://www.geneontology.org/) was accomplished.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
For RT-qPCR analysis, total RNA was isolated from
the Stra8 germ cells and control germ cells using the traditional
TRIzol method and reverse transcribed with the PrimeScript™ RT
reagent kit with gDNA Eraser (Takara Bio, Inc., Otsu, Japan). The
forward and reverse PCR primers were designed by wcgene Biotech
Company (Shanghai, China) in accordance with the PCR conditions
(listed in Table I). The 10
µl RT-qPCR reaction mix contained 5 µl 2X RNA
GoTaq®qPCR Master mix (Promega Corporation, Madison, wI,
USA), 10 pmol of the forward and the reverse primers, and 0.5
µl of the sample/standard. The PCR thermal profile included
40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C for
30 sec, and extension at 72°C for 30 sec. Each sample was analyzed
in triplicate. The mRNA level of GAPDH was used as the internal
control, and relative gene expression was calculated according to
the 2−ΔΔCq method based on the quantification cycle
values (11).
 | Table IPrimers used for microarray data
validation using reverse transcription-quantitative polymerase
chain reaction analysis. |
Table I
Primers used for microarray data
validation using reverse transcription-quantitative polymerase
chain reaction analysis.
| Gene name | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) |
|---|
| GSTP1 |
CTGGAAGGAGGAGGTGGTT |
TTAGATTGGTAAAGGGTGAGGT |
| BIRC6 |
CGTTTGGTTGTGGCAACTGAT |
AAGATATAGCTGTGCCCGTAGTA |
| PDK1 |
GGACTTCGGGTCAGTGAATGC |
TCCTGAGAAGATTGTCGGGGA |
| USP33 |
AACTGGACAAAGACAGGGAC |
TGCTGATAACCGTGGACTAA |
| TCF4 |
CGAAAAGTTCCTCCGGGTTTG |
CGTAGCCGGGCTGATTCAT |
| OSGIN1 |
CCTCCGGTATCTGCCTGTC |
GGAAAGGTACTCTAGGTCCTGG |
| TEAD4 |
GGCACCATTACCTCCAACG |
GCTCATTCCGACCATACATCTT |
| ANG2 |
GCGAAAGTATGATGGTGAAA |
AGTGGTGACCTGGAAGTGAG |
| ANXA10 |
CAAACAGGGAGGAAGGATA |
CAAACAGGGAGGAAGGATA |
| CASP3 |
CTCGCTCTGGTACGGATGTG |
TCCCATAAATGACCCCTTCATCA |
| AKT1 |
ATGAACGACGTAGCCATTGTG |
TTGTAGCCAATAAAGGTGCCAT |
| AKT2 |
ACGTGGTGAATACATCAAGACC |
GGGCCTCTCCTTATACCCAAT |
| AKT3 |
AGGTTGGGTTCAGAAGAGGG |
AGGGGATAAGGTAAGTCCACATC |
| Bcl-2 |
ATGCCTTTGTGGAACTATATGGC |
GGTATGCACCCAGAGTGATGC |
| MAPK3 (ERK2) |
TCCGCCATGAGAATGTTATAGGC |
GGTGGTGTTGATAAGCAGATTGG |
| MAPK14 (P38) |
TGACCCTTATGACCAGTCCTTT |
GTCAGGCTCTTCCACTCATCTAT |
| TRP53 |
CCCCTGTCATCTTTTGTCCCT |
AGCTGGCAGAATAGCTTATTGAG |
| GAPDH |
GACGGCCGCATCTTCTTGT |
ACACCGACCTTCACCATTTTGT |
Western blot analysis
whole proteins from the Stra8-GC1 and Control-GC1
cells were prepared in RIPA buffer (Solarbio Science
&Technology Co., Ltd.) and quantified by using the BCA Protein
Assay kit (CwBIO). The homogenates were centrifuged for 15 min at
10,000 x g at 4°C and the supernatants were collected. Equal
quantities of protein (60 µg/lane) were separated by 10%
sodium dodecyl sulfatepolyacrylamide gel electrophoresis and
transferred onto a polyvinylidene difluo-ride membrane. The
membrane was blocked with Tris-buffered saline (TBS) with 5%
non-fat milk and incubated with the primary detection antibodies:
Phosphatidylinositol-dependent kinase 1 (PDK1; cat. no. sc-293160,
Santa Cruz Biotechnology, Inc., CA, USA; 1:1,000), Phospho-AKT
(cat. no. bs-0876R; BIOSS, Beijing, China, 1:500), B-cell lymphoma
2 (Bcl-2; cat. no. 3498T; Cell Signaling Technology, Inc., Beverly,
MA, USA, 1:1,000), Mitogen-activated protein kinase (MAPK) Family
Antibody Sampler kit for extracellular signal-regulated kinase
(ERK)1/2, c-Jun N-terminal kinase (JNK)1/2, and P38 (cat. no.
9926T; Cell Signaling Technology, Inc., Beverly, MA, USA, 1:1,000),
P53 (cat. no. sc-6243, Santa Cruz Biotechnology, Inc.; 1:1,000),
cleaved caspase 3 (cleaved CASP3; cat. no. 9664T, Cell Signaling
Technology, Inc.; 1:1,000), GAPDH (cat. no. Cw0100, CwBIO; 1:5,000)
at 4°C overnight. Following rinsing three times with TBS/Tween-20,
the membranes were incubated with the following secondary
antibodies: Peroxidase-Conjugated Goat anti-Rabbit IgG (cat no.
ZB-2301; OriGene Technologies, Inc., Rockville, MD, USA; 1:1,000)
and Peroxidase-Conjugated Goat anti-Mouse IgG (cat no. ZB-2305;
OriGene Technologies, Inc.; 1:1,000) at room temperature for 1.5 h.
Protein levels were normalized to GAPDH level and all values were
presented compared with the NC, which was normalized as 1. All
proteins were detected with an electrogenerated chemiluminescence
assay (Tanon-5200; Tanon Science and Technology Co., Ltd. Shanghai,
China) and the Gel Image System, version 4.00 (Tanon Science and
Technology Co., Ltd.).
Statistical analysis
Statistical analysis was performed using Microsoft
Excel 2010 (Microsoft Corporation, Redmond, wA USA). All data are
expressed as the mean ± standard deviation of at least three
independent experiments. To compare between two groups, an unpaired
Student's t-test (one- or two-tailed as indicated) was used.
P<0.05 was considered to indicate a statistically significant
difference.
Results
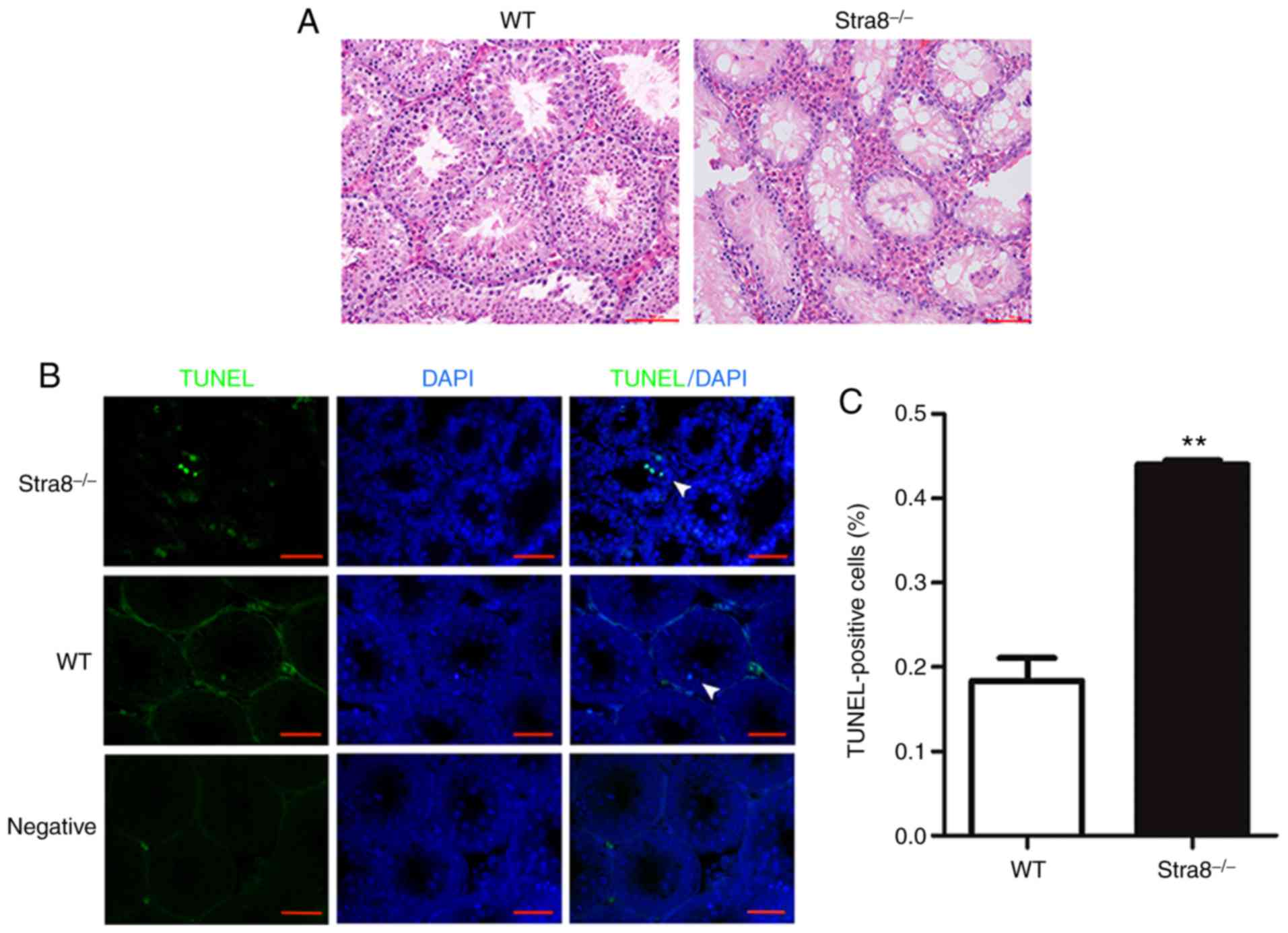
Stra8 exhibits an anti-apoptotic effect
in Stra8−/− male mice
Stra8−/− mice were generated through
mating heterozygotes, and tissue sections were stained with H&E
(Fig. 1A). Compared with the wT
mice, the number of spermatogonia in the semi-niferous tubules of
the Stra8−/− mice began decreasing from 11 dpp;
therefore, the 11 dpp Stra8−/− mice and wT mice testis
were selected for the TUNEL assay to evaluate the number of
apoptotic germ cells in the seminiferous tubules, separately. The
results from the TUNEL assay suggested that the number of apoptotic
germ cells in the Stra8−/− mice was significantly
increased compared with that in the wT mice (Fig. 1B). The apoptotic index was
0.18±0.06 in the wT mice, compared with 0.44±0.01 in the
Stra8−/− mice (Fig.
1C).
Anti-apoptotic effect of Stra8 in VAD and
VAR mice
In addition, the anti-apoptotic effect of Stra8 was
verified in a male VAD and VAR mouse model. Male and female mice
were permitted to mate following being fed a VAD diet for 2 weeks,
with the VAD diet maintained during pregnancy. Neonatal male mice
were fed a VAD diet for 13–14 weeks to obtain VAD mice. According
to morphological observations, there was no phenotypic difference
at 45 days in the VAD mice compared with the wT mice; however, the
number of germ cells was significantly decreased in the testis of
mice at 61 days; and only Sertoli cells, a small number of germ
cells, and a large number of vacuoles in the testes exhibiting
structural disorganization were observed at 93 days. Therefore, 45,
61, and 93 days were selected as the time points to evaluate VAD
and wT mice using a TUNEL assay. The results showed that the
numbers of TUNEL-positive cells in the VAD mice on days 45 and 61
were higher than those in the wT mice (Fig. 2A). The numbers of TUNEL-positive
cells were as follows: 0.69±0.01 in wT mice, 22.22±2.304 in VAD
mice (45 days); 0.62±0.03 in wT mice, 7.81±0.99 in VAD mice (61
days); 0.63±0.05 in wT mice; and 0.97±0.09 in VAD mice (93 days)
(Fig. 2B; Table II).
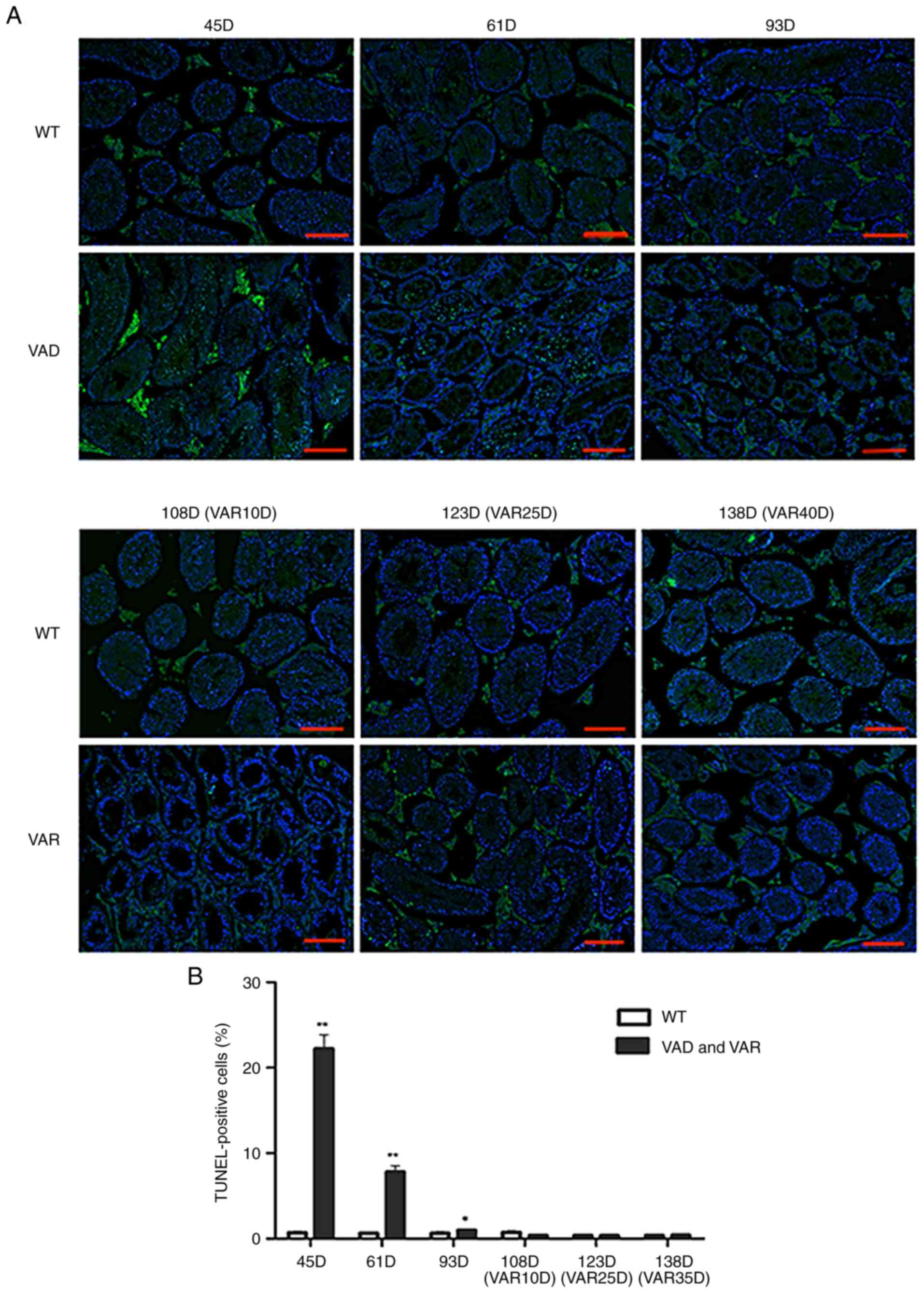 | Figure 2Anti-apoptotic effect of Stra8 in
male VAD and VAR mice. (A) Representative images of TUNEL/DAPI
staining from the seminiferous tubules in the testes of male wT
mice, and VAD and VAR mice at 45, 61, 93, 108 (VAR 10), 123 (VAR
25), and 138 (VAR 40) days. Fluorescence colors show TUNEL in green
and DAPI in blue; scale bar=100 µm. (B) Percentage of
apoptotic cells (TUNEL-positive cells/DAPI-positive cells) among
the seminiferous tubules. N≥6. The data are presented as the mean ±
standard deviation. *P<0.05, **P<0.01
(two-tailed Student's t-test). Stra8, stimulated by retinoic acid
gene 8; wT, wild-type; DAPI, 4′,6-diamidino-2-phenylindole; VAD,
vitamin A deficient; VAR, vitamin A recovery. |
 | Table IIAverage number of TUNEL-positive
cells per tubule from the wT, VAD and VAR mice. |
Table II
Average number of TUNEL-positive
cells per tubule from the wT, VAD and VAR mice.
| Group | 45 D | 61 D | 93 D | 108 D (VAR
10D) | 123 D (VAR
25D) | 138 D (VAR
40D) |
|---|
| wild-type | 0.698±0.010 | 0.626±0.030 | 0.636±0.050 | 0.729±0.050 | 0.401±0.040 | 0.381±0.030 |
| Model | 22.224±2.304 | 7.815±0.999 | 0.979±0.090 | 0.395±0.050 | 0.354±0.030 | 0.415±0.048 |
Subsequently, the 14-week-old VAD male mice were fed
a normal vitamin A diet to obtain VAR mice. According to the
morphological observations, spermatocytes appeared in semi-niferous
tubules in the testis of VAR mice following 10 days on the normal
diets, different types of spermatogenic cells in the seminiferous
tubules had recovered at 25 days, and the seminiferous tubules
exhibited a normal structure at 40 days. Therefore, at 10, 25, and
40 days, VAR mice were selected for further verification using a
TUNEL assay; however, no differences were observed between the VAR
and wT mice (Fig. 2A). The
respective number of TUNEL-positive cells in the wT and VAR mice
were as follows: 0.72±0.05, vs. 0.39±0.05 (10 days); 0.40±0.04, vs.
0.35±0.03 (25 days); and 0.38±0.03, vs. 0.41±0.04 (40 days) group
(Fig. 2B; Table II).
Overexpression of Stra8 inhibits cellular
apoptosis of GC1 spg cells
To further assess the anti-apoptotic effect of Stra8
in vitro, analyses of Stra8-GC1 and Control-GC1 were
performed using an Annexin V/PI apoptosis detection kit via flow
cytometry. These results demonstrated that the proportion of
Stra8-GC1 cells that underwent early apoptosis was lower than that
in the Control-GC1 cells (Fig.
3A). Apoptotic germ cells comprised 29.52±10.53 of the
population in the Control-GC1 cells and 12.35±4.41 in the Stra8-GC1
cells (Fig. 3B).
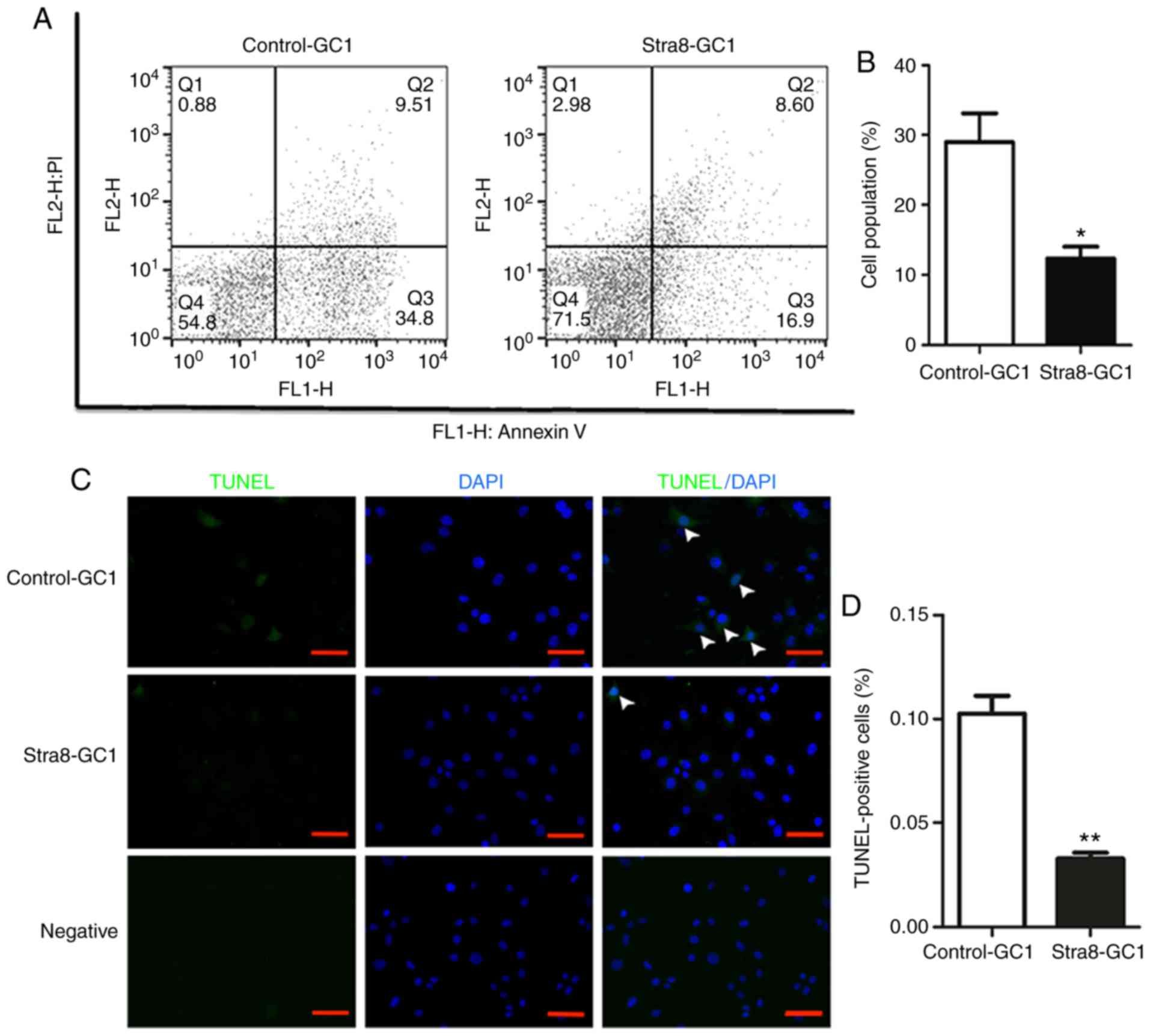 | Figure 3Anti-apoptotic effect of the
overexpression of Stra8 on GC1 spg. (A) Apoptotic cells in the
Stra8-GC1 and Control-GC1 groups were measured by flow cytometry
and the cells were divided into four sections: Q1, mechanical
error; Q2, late apoptosis cells; Q3, early apoptotic cells; and Q4,
living cells. (B) Percentage of cells in early apoptosis in the two
groups (N≥6). Error bars show the mean ± standard deviation.
*P<0.05 (two-tailed Student's t-test). (C) Top and
middle rows show the fluorescence images of TUNEL/DAPI in
Control-GC1 and Stra8-GC1 cells, respectively; bottom row shows the
negative fluorescence images. white arrows indicate TUNEL-positive
cells. Fluorescence colors show TUNEL in green and DAPI in blue;
scale bar=200 µm. (D) Percentage of apoptotic cells
(TUNEL-positive cells/DAPI-positive cells) among the total cells.
n=3/group. Error bars indicate the mean ± standard deviation.
**P<0.01 (two-tailed Student's t-test).). Stra8,
stimulated by retinoic acid gene 8; wT, wild-type; DAPI,
4′,6-diamidino-2-phenylindole; spg, spermatogonia; PI, propidium
iodide. |
Additionally, the results of the TUNEL assay
confirmed the anti-apoptotic effect of Stra8 in germ cells. The
number of apoptotic germ cells was significantly decreased when
Stra8 was overexpressed compared with the control group (Fig. 3C). The apoptotic index was
0.10±0.01 in the Control-GC1 cells compared with 0.03±0.04 in the
Stra8-overexpressing GC1 cells (Fig.
3D).
Anti-apoptotic effect of Stra8 in GC1 spg
cells overexpressing Stra8 according to Affymetrix GeneChip
microarray analysis
To investigate which genes or pathways were involved
in the anti-apoptotic effect of Stra8, an Affymetrix GeneChip
micro-array of the Stra8-GC1 and Control-GC1 cells was completed. A
total of 45,103 probe sets were used to analyze the expression
level of >20,000 well-characterized mouse genes using the
Affymetrix Mouse Genome 430 2.0 Array. DEGs were selected with FC
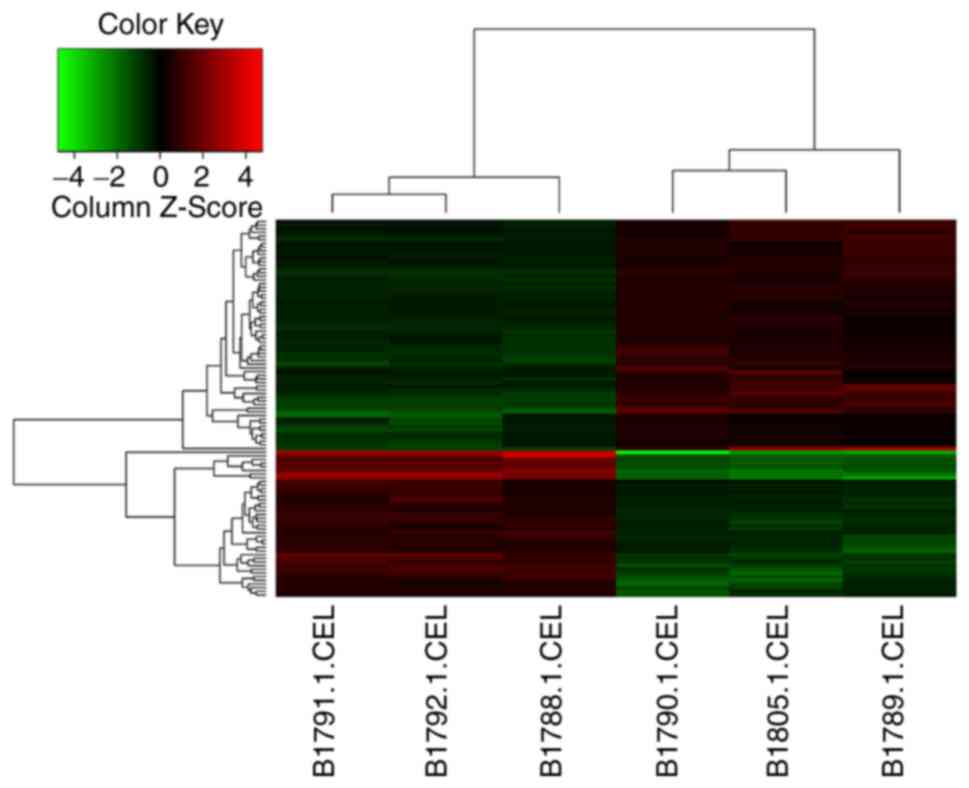
>1.5 and P<0.01 (one-sample t-test). A heat map illustrates
the differential expression patterns between the clustering of the
genes in the Stra8-GC1 cells compared with that of the Control-GC1
cells (Fig. 4).
Based on the analysis of DEGs using the KEGG pathway
database and Gene Ontology, the biological processes involved in
DEGs included the negative regulation of apoptosis, negative
regulation of programmed cell death, system development, structure
development, and the negative regulation of cellular processes. The
top 20 upregulated and down-regulated genes were selected and
displayed according to the P-values (Table III). Among all the DEGs, five
upregulated genes [glutathione S-transferase P91 (GSTP1),
baculoviral inhibitor of apoptosis repeat-containing 6 (BIRC6),
PDK1, ubiquitin-specific protease 33 (USP33), and transcription
factor 4 (TCF4)]; and four downregulated genes [(oxidative stress
induced growth inhibitor 1 (OSGIN1), TEA domain transcription
factor 4 (TEAD4), angiopoietin 2 (ANG2), and Annexin A10 (ANXA10)]
associated with apoptosis were selected according to evidence
obtained from references relating to gene function (Table IV).
 | Table IIITop 20 upregulated and downregulated
genes of the total differentially expressed genes according to the
P-values. |
Table III
Top 20 upregulated and downregulated
genes of the total differentially expressed genes according to the
P-values.
| No | Gene symbol | Description | Fold change | P-value |
|---|
| Upregulated | | | | |
| 1 | BGN | Biglycan | 3.5835900 | 0.012659261 |
| 2 | ANK2 | Ankyrin 2 | 3.2059138 | 6.80039E-06 |
| 3 | GJB5 | Gap junction
protein, β5 | 2.4792826 | 6.03425E-06 |
| 4 | AI256396 | Est ai256396 | 2.3620548 | 0.000472130 |
| 5 | GSTP1 | Glutathione
S-transferase pi 1 | 2.2966921 | 0.003767367 |
| 6 | RBM12B1 | RNA binding motif
protein 12 B1 | 2.2234132 | 0.000175803 |
| 7 | CTH | Cystathionase | 2.2095652 | 0.003464704 |
| 8 | OPLAH | 5-oxoprolinase
(ATP-hydrolysing) | 2.0994134 | 0.016354447 |
| 9 | MBP | Myelin basic
protein | 1.9661545 | 0.001004058 |
| 10 | VAT1L | Vesicle amine
transport protein 1 like | 1.9652303 | 0.033937313 |
| 11 | SLC38A9 | Solute carrier
family 38, member 9 | 1.9384620 | 0.001569600 |
| 12 | ZFP422 | Zinc finger protein
422 | 1.9111286 | 0.000111407 |
| 13 | KLF9 | Kruppel-like factor
9 | 1.8877469 | 0.005244654 |
| 14 | MANBA | Mannosidase, βA,
lysosomal | 1.8711381 | 0.000459721 |
| 15 | CBLN1 | Cerebellin 1
precursor protein | 1.8617241 | 0.000945486 |
| 16 | TRPM3 | Transient receptor
potential cation channel, subfamily M, member 3 | 1.8470881 | 0.015302277 |
| 17 | GNPDA2 |
Glucosamine-6-phosphate deaminase 2 | 1.8342222 | 0.004544168 |
| 18 | ZFP467 | Zinc finger protein
467 | 1.8330021 | 0.028772602 |
| 19 | RNF113A2 | Ring finger protein
113A2 | 1.8240016 | 2.24117E-07 |
| 20 | DMRT2 | Doublesex and mab-3
related transcription factor 2 | 1.8220981 | 0.000109843 |
| Downregulated | | | | |
| 1 | KRT6A | Keratin 6A | −10.943147 | 0.000540374 |
| 2 | ANG2 |
Angiogenin,ribonuclease A family,member
2 | −5.6302330 | 0.000515156 |
| 3 | SERPINI2 | Serine (or
cysteine) peptidase inhibitor, clade I, member 2 | −5.3056600 | 8.79068E-05 |
| 4 | CD302 | CD302 antigen | −4.6146510 | 0.000681903 |
| 5 | IGFBP7 | Insulin-like growth
factor binding protein 7 | −4.0378250 | 8.85925E-05 |
| 6 | EYA2 | EYA transcriptional
coactivator and phosphatase 2 | −3.1538005 | 1.15442E-06 |
| 7 | SMAGP | Small cell adhesion
glycoprotein | −2.9982140 | 2.30374E-06 |
| 8 | TMEM181C-PS | Tmem181c-ps
transmembrane protein 181C, pseudogene | −2.9360726 | 0.000162202 |
| 9 | TMEM181A | Transmembrane
protein 181A | −2.9360726 | 0.000162202 |
| 10 | TMEM181B-PS | Tmem181b-ps
transmembrane protein 181B, pseudogene | −2.9360726 | 0.000162202 |
| 11 | OSGIN1 | Oxidative stress
induced growth inhibitor 1 | −2.5084667 | 0.001495294 |
| 12 | SLC7A2 | Solute carrier
family 7 | −2.3679860 | 0.002371071 |
| 13 | ASS1 | Argininosuccinate
synthetase 1 | −2.2645633 | 0.000348439 |
| 14 | CTSH | Cathepsin H | −2.2382452 | 0.000414636 |
| 15 | CRYAB | Crystallin, αB | −2.1004624 | 0.002057886 |
| 16 | FLT1 | FMS-like tyrosine
kinase 1 | −2.0917470 | 0.009599945 |
| 17 | PTPRG | Protein tyrosine
phosphatase, receptor type, G | −2.0450683 | 0.000267192 |
| 18 | ANXA10 | Annexin A10 | −2.0104885 | 0.000560119 |
| 19 | RANGRF | RAN guanine
nucleotide release factor | −1.8825512 | 0.015963247 |
| 20 | APCDD1 | Adenomatosis
polyposis coli downregulated 1 | −1.8573459 | 6.23783E-05 |
 | Table IVSelected upregulated and
downregulated genes among all differentially expressed genes. |
Table IV
Selected upregulated and
downregulated genes among all differentially expressed genes.
| No | Gene symbol | Description | Fold change | P-value |
|---|
| Upregulated | | | | |
| 1 | GSTP1 | Glutathione
S-transferase pi 1 | 2.2966921 | 0.003767367 |
| 2 | BIRC6 | Baculoviral IAP
repeat-containing 6 | 1.7460216 | 0.000302024 |
| 3 | PDK1 | Pyruvate
dehydrogenase kinase 1 | 1.7768697 | 0.006251959 |
| 4 | USP33 | Ubiquitin specific
peptidase 33 | 1.5531131 | 0.000201205 |
| 5 | TCF4 | Transcription
factor 4 | 1.6565648 | 0.000173130 |
| Downregulated | | | | |
| 1 | OSGIN1 | Oxidative stress
induced growth inhibitor 1 | −2.5084667 | 0.001495294 |
| 2 | EPHA4 | Eph receptor
A4 | −1.5859284 | 0.007054214 |
| 3 | ANG2 | Angiogenin,
ribonuclease A family, member 2 | −5.630233 | 0.000515156 |
| 4 | ANXA10 | Annexin A10 | −2.0104885 | 0.000560119 |
Potential signaling pathways involved in
the anti-apoptotic effect mediated by Stra8
RT-qPCR analysis was performed to verify the
tendency of the nine genes described above, the results of which
were consistent with the GeneChip microarray analysis. The genes in
the caspase family were also examined and it was found that CASP3
was reduced in Stra8-GC1 cells (Fig.
5A). Through a literature review, it was found that PDK1, TCF4,
GSTP1, USP33, ANG2, and OSGIN1 are associated with the AKT
signaling pathway. Among these genes, PDK1 is a key upstream factor
of AKT; therefore, the present study verified the mRNA and protein
expression levels of PDK1 and AKT using RT-qPCR and a western blot
analyses. It was found that the levels of PDK1 and AKT were
increased in Stra8-GC1 cells compared with those in Control-GC1
cells, which is consistent with the hypothesis that Stra8 may exert
an anti-apoptotic effect through the AKT pathway (Fig. 5B-D).
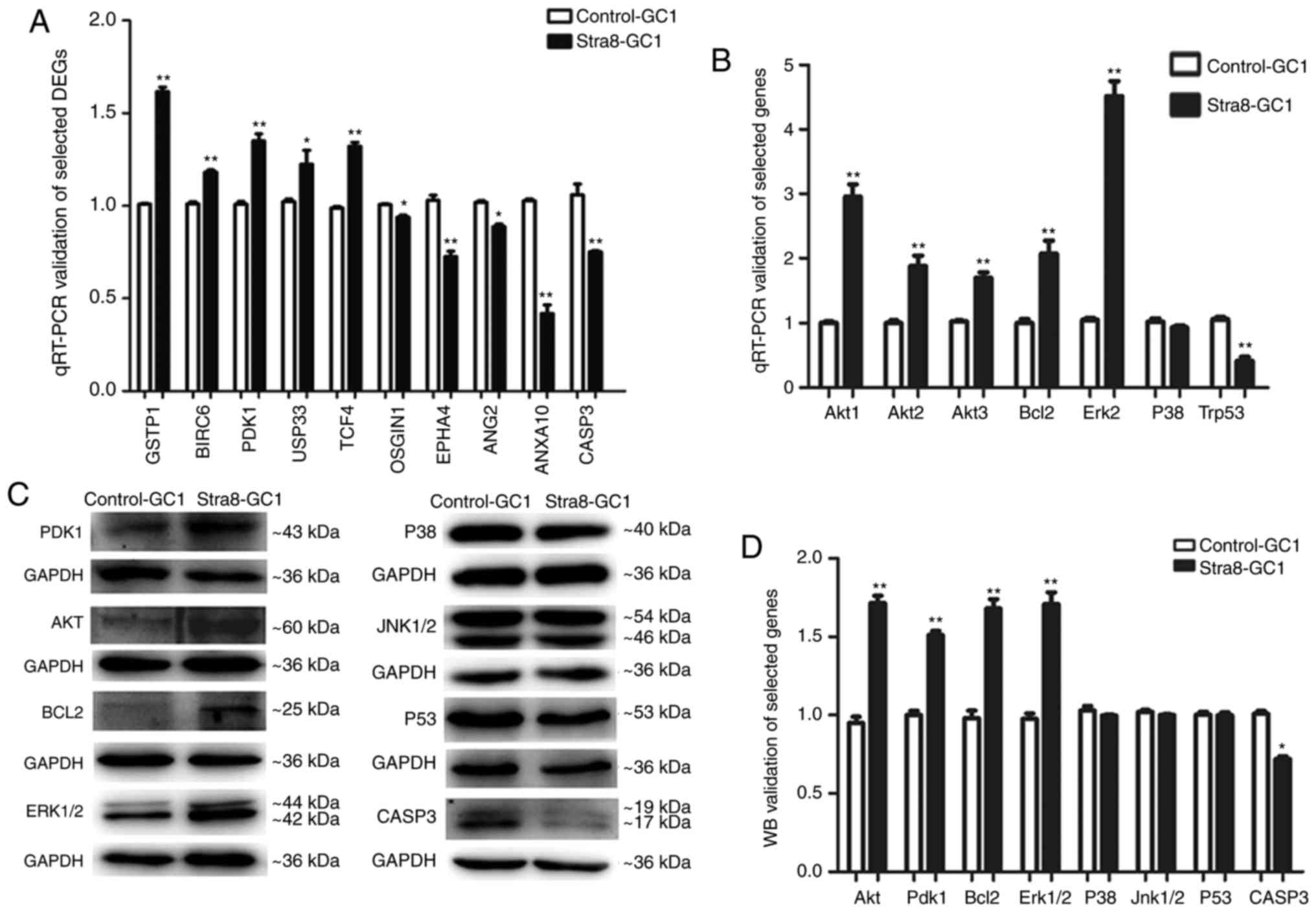 | Figure 5Potential signaling pathways by which
Stra8 mediates an anti-apoptotic effect. (A) Nine DEGs were
associated with apoptosis in Stra8-GC1 cells compared with
Control-GC1 cells, determined through Affymetrix GeneChip
microarray analysis. Among these DEGs, five upregulated genes
(GSTP1, BIRC6, PDK1, USP33, and TCF4) and four downregulated genes
(OSGIN1, TEAD4, ANG2, and ANXA10) were identified through RT-qPCR
analysis. CASP3, one of known executioner caspases, was
downregulated in Stra8-GC1 cells. N=3/group. Error bars show the
mean ± standard deviation, *P<0.05,
**P<0.01 (two-tailed Student's t-test). (B) RT-qPCR
analysis of key genes (AKT1, AKT2, AKT3, Bcl-2, ERK2, P38 and
Trp53) of the AKT pathway in Stra8-GC1 cells compared with
Control-GC1 cells. (C) western blot of key genes (PDK1, AKT, Bcl-2,
ERK1/2, P38, JNK1/2, P53, CASP3) of the AKT pathway in Stra8-GC1
cells compared with Control-GC1 cells. (D) western blot grayscale
value analysis of key genes (PDK1, AKT, Bcl-2, ERK1/2, P38, JNK1/2,
P53, CASP3) of the AKT pathway; N≥3. Error bars show the mean ±
standard deviation *P<0.05, **P<0.01
(two-tailed Student's t-test). Stra8, stimulated by retinoic acid
gene 8; DEGs, differentially expressed genes; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; wB, western
blotting; GSTP1, glutathione S-transferase P1; BIRC6, baculoviral
inhibitor of apoptosis repeat-containing 6; PDK1,
phosphatidylinositol-dependent kinase 1; USP33, ubiquitin-specific
protease 33; TCF4, transcription factor 4; OSGIN1, oxidative stress
induced growth inhibitor 1; TEAD4, TEA domain transcription factor
4; ANG2, angiopoietin 2; ANXA10, Annexin A10; AKT, protein kinase;
Bcl-2, B-cell lymphoma 2; ERK, extracellular signal-regulated
kinase; JNK, c-Jun N-terminal kinase; CASP3, caspase 3. |
ANG2 is a Bcl-2-inhibited gene; TCF4, GSTP1 and
USP33 are MAPK-related genes; and OSGIN1 is related to the P53
pathway. In addition, Bcl-2, p53, ERK (MAPK1/3), JNK (MAPK8/9), and
P38 (MAPK14) are key genes involved in the AKT signaling pathway.
Therefore, the mRNA expression levels of these genes were
determined, which revealed that Bcl-2 and ERK were increased,
whereas P53 was decreased, in the Stra8-GC1 cells compared with the
Control-GC1 cells. In addition, the protein expression levels of
Bcl-2 and ERK were increased, and there was no significant
difference in the expression of P53 in the Stra8-GC1 cells compared
with the control groups. In addition, CASP3, one of the executioner
caspases, was decreased in the Stra8-GC1 cells compared with the
Control-GC1 at the mRNA and protein expression levels (Fig. 5-D). Therefore, it was concluded
that Stra8 may exert an anti-apoptotic effect primarily through the
AKT pathway (Fig. 6).
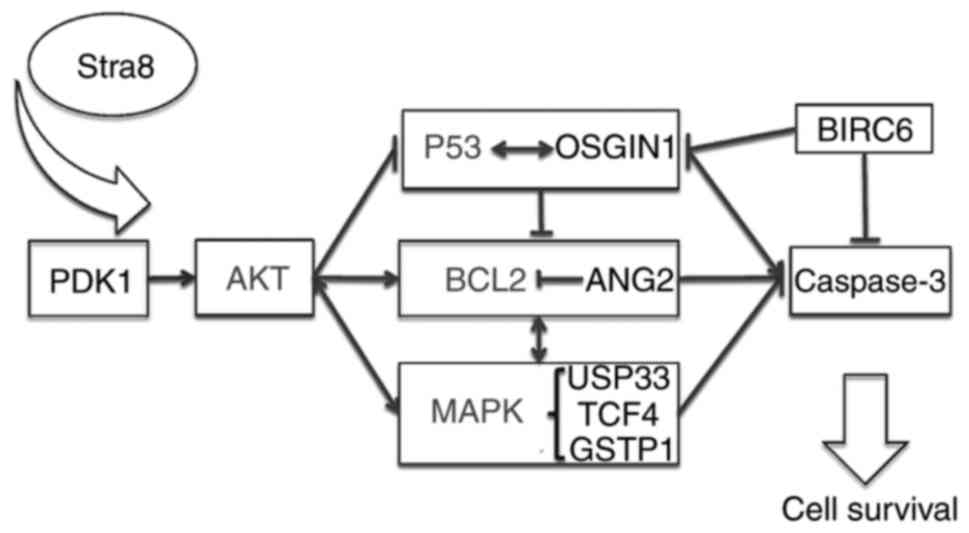 | Figure 6Possible pathway through which Stra8
may exert an anti-apoptotic effect via the AKT signaling pathway.
Stra8, stimulated by retinoic acid gene 8; PDK1,
phosphatidylinositol-dependent kinase 1; AKT, AKT, protein kinase;
OSGIN1, oxidative stress induced growth inhibitor 1; Bcl-2, B-cell
lymphoma 2; ANG2, angiopoietin 2; MAPK, mitogen-activated protein
kinase; USP33, ubiquitin-specific protease 33; TCF4, transcription
factor 4; GSTP1, glutathione S-transferase P1; BIRC6, baculoviral
inhibitor of apoptosis repeat-containing 6. |
Discussion
Previous publications have described Stra8 as an
RA-inducible gene, which is specifically expressed during mouse
embryo-genesis, is involved in the premeiotic phase of
spermatogenesis, and is also required for early meiotic events and
the transformation from mitosis to meiosis (7,12).
In previous experiments, whether Stra8 can affect spermatogenic
cell proliferation was examined using an SRB cell proliferation
assay. The cells were collected and counted at 4 h (day 0), and on
days 1, 2, 3, 4, and 5, and a growth curve was plotted using the
optical density (OD) value every 24 h. No statistical difference in
OD values was found at corresponding time points between the
Stra8-GC1 and Control-GC1 (data not shown). Furthermore, using flow
cytometry, the cell numbers of different stages of the cell cycle
were detected, which revealed no statistically significant
difference between the Stra8-GC1 and Control-GC1 groups (data not
shown). Therefore, it was hypothesized that Stra8 may have no
influence on the proliferation or cell cycle of spermatogenic
cells. In the present study, using a TUNEL assay, a routine and
normal method for visualizing cells undergoing apoptosis, it was
found that Stra8 inhibited spermatogenic cell apoptosis.
Furthermore, this anti-apoptotic effect of Stra8 was verified using
male VAD and VAR mice models. On days 45 and 61, the number of
apoptotic cells in the VAD mice was significantly increased
compared with that in the wT mice on the same days, which was
consistent with expectations. However, no differences were observed
at 93 days between the VAD and wT mice. This may be due to the
reduction in the total number of spermatogenic cells in the
seminiferous tubules at 93 days in the VAD mice, in which severe
vacuolization and disorganized seminiferous tubules were observed.
In addition, a moderate expression of Stra8 remained in the
seminiferous tubules within 10 days in the testis of the VAR mice,
which was sufficient to mediate an anti-apoptotic effect. The
expression level of Stra8 had returned to normal within 40 days in
the VAR mice testis, therefore, no significant difference in the
number of apoptotic cells was observed between the wT and VAR mice.
The present study further verified the anti-apoptotic effect of
Stra8 in vitro using Stra8-overexpressing GC1 spg cells. To
determine the potential genes and possible pathways of the Stra8
anti-apoptotic effects, which have not been previously reported,
GeneChip microarray analysis was performed. A total of 75
upregulated genes and 46 downregulated genes were found through the
GeneChip microarray of the Stra8-GC1 and Control-GC1 cells. Among
these DEGs, five upregulated genes and four downregulated genes
were associated with apoptosis. Additional analyses on the function
and related pathways of these nine genes were performed to obtain
further understanding of the anti-apoptotic effect of Stra8.
Apoptosis is the process of programmed cell death,
which is critical in spermatogenesis and the elimination of germ
cells that carry DNA ns mutations (13). A large number of studies have
indicated that several pathways are involved in apoptosis during
germ development, including the Fas/Fas ligand system (13), MAPK signal pathways, Bcl-2 gene
families (14,15), P53, heat shock protein 70
(16) and AKT signaling pathways
(17). Fas/Fasl are expressed in
Sertoli and spermato-genetic cells, and increase under estrogen
exposure, inducing cell h death (13). In addition, Bcl-2 can prevent germ
cell death during spermatogenesis (14,15). Furthermore, under genotoxic
stress, p53, a sequence specific transcription factor, is involved
in the apoptosis of spermatogonia, spermatocytes and spermatids,
and can regulate the expression of Bcl-2 (16). Abraham et al (18) demonstrated that AKT negatively
regulates levels of p53 by enhancing murine double minute 2. In
addition, previous data show that p53 is involved in promoting
apoptosis by stimulating the release of pro-apoptotic proteins,
including cytochrome c, second mitochondria-derived
activator of caspase, and apoptosis-inducing factor, and the
activation of caspases (Casp3, 6, and 7) (19,20). In addition, MAPK signaling has a
key functional role in numerous male reproductive processes,
including spermatogenesis, capacitation and the acrosome reaction
prior to n fertilization (21).
AKT signaling has an anti-apoptotic effect as a survival factor,
and the inhibition of AKT results in DNA replication damage and
cell cycle arrest, leading to s apoptosis (22,23). Furthermore, phosphoinositide
3-kinase (PI3K)-AKT signaling leads to cellular proliferation,
survival and growth (23).
According to previous studies, RA activates the PI3K-AKT signaling
network to regulate the differentiation of spermatogonia in
neonatal testes (24). In
addition, Busada et al (25) found that mTOR complex 1 is
required to mediate the RA signal to direct spermatogonial
differentiation. Therefore, PI3K-AKT signaling is essential for
male reproduction. Stra8 is one of the genes induced by RA that
shuttles between the cytoplasm and nucleus, depending on the
different dynamics of each cell type, and may exert distinct
functions in various cellular compartments (1). Therefore, the function of Stra8 may
be associated with different cell cycles. Additionally, the
majority of DEGs associated with apoptosis exhibiting an
overexpression of Stra8 GC1 spg are related to AKT signaling,
including PDK1, USP33, TCF4, GSTP1, ANG2, OSGIN1 and CASP3.
Therefore, it was hypothesized that Stra8 may have an apoptotic
role associated with the AKT pathway.
Based on the above results, the present study
further examined the function of AKT-associated genes and pathways.
It has been shown that AKT acts as a survival factor by
phosphorylating pro-apoptotic substrates, including
Bcl-2-associated death promotor, CASP9, forkhead box O, and
apoptosis signal-regulating kinase 1 (22,26,27). According to associated studies in
the literature, AKT can act as a survival factor which balances
pro-survival and pro-death signals, including activating genes in
the Bcl-2 family and interacting with MAPK gene families, which
eventually suppresses escaspases (22,26). There are two major families of
serine/threonine protein kinases, the AGC family and the MAPK
family. The AGC protein kinase family includes PDK1, cyclic
AMP-dependent protein kinase, PKB/AKT and Ca2+-activated
protein e kinase (22,26). Among the DEGs, the present study
found that PDK1, a key upstream gene of AKT, activates ~23 protein
kinases in the AGC family, including PKB/AKT, which is a key
signaling intermediate involved in growth and l survival (27). Therefore, the levels of these
genes were analyzed and it was found that the mRNA and protein
expression levels of AKT and PDK1 were signifi-cantly increased in
the Stra8-GC1 cells compared with those in the Control-GC1 cells.
It has also been found that ANG2 increases cellular apoptosis by
inhibiting the stimulation of AKT/Bcl-2 signaling (28,29). OSGIN1 is a major mediator of
cellular apoptosis under the control of tumor suppressor protein,
p53 (19). In addition, BIRC6
silencing indirectly affects the regulators of p53 or directly
binds and inhibits caspases (Casp3, 6, 7 and 9) through its BIR
domain (30,31). Compared with the Control-GC1
cells, the present study found that Bcl-2 and BIRC6 were
significantly increased; the mRNA levels of ANG2, p53 and OGSIN1
were decreased; and the protein levels of Bcl-2 in Stra8-GC1 cells
was significantly increased. As these genes are all key factors in
the AKT pathway, it was preliminarily hypothesized that the ability
of Stra8 to inhibit apoptosis may be associated with the AKT
pathway.
The other serine/threonine protein kinases family is
the MAPK family, which includes RAF, ERK, MAPK kinase (MEK and
MKK), MAPK kinase kinase, and JNK/p38 (22,26). USP33, TCF4, and GSTP1 are MAPK
signaling-related genes. In addition, USP33 is a gene involved in
ERK signaling, which is important in the regulation of cellular
proliferation, differentiation and s apoptosis (32,33). The phosphorylation of TCF4 at the
serine/threonine proceeds via ERK and p-38 dependent pathways, and
TCF4 knockdown has been shown to induce growth arrest and apoptosis
in human colorectal cancer (34,35). GSTP1, a dimeric protein, binds to
JNK and suppresses downstream JNK g signaling (36). To the best of our knowledge, the
JNK signaling pathway is involved in the processes of cell death
and mediates stress-induced apoptosis via mitochondrial pathways
with the release of cytochrome c-activating caspases (CASP3,
−6, and −7) (36,37). Therefore, the present study
detected the expression of P38, ERK, USP33, TCF4 and GSTP1. It was
found that the mRNA levels of TCF4, USP33 and GSTP1 were
significantly increased. In addition, the mRNA and protein
expression levels of ERK were significantly increased in the
Stra8-GC1 cells, compared with those in the Control-GC1 cells. As
the MAPK pathway is an important AKT interaction pathway, it was
further verified that Stra8 may have an anti-apoptotic effect
through the AKT pathway.
Finally, the present study verified the expression
of the caspase gene families and found that the expression of
CASP3, one of the executioner caspases involved in cellular
degradation during apoptosis, was decreased in the Stra8-GC1 cells
compared with the Control-GC1 cells (38), further supporting the hypothesis
that Stra8 functions to inhibit apoptosis. Therefore, a possible
apoptotic pathway was characterized, in which Stra8 exerts an
anti-apoptotic effect via the AKT signaling pathway. AKT directly
or indirectly inactivates p53 by activating the Bcl-2 family of
genes and interacts with the MAPK family of genes, thereby
inhibiting the caspase gene families and inducing an anti-apoptotic
effect.
In the present study, the anti-apoptotic effects of
Stra8 in the mouse male reproductive system were verified using
in vitro and in vivo methods, and the possible
involvement of the AKT signaling pathway was discussed; however,
the investigations of potential Stra8-related signaling pathways
were primarily based on gene expression analysis, which require
further verification by investigations into the interaction between
genes. Taken together, the findings of the present study provide a
theoretical foundation for future in-depth functional
investigations of Stra8.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 31371174) and the Natural
Science Foundation of Jiangsu Province, China (grant no.
BK20131230).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XS, CN, JG designed and complemented the
experiments; XS wrote the manuscript; MX, JX, and YH collected
literatures and analyzed data; YZ designed the study and all
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Committee on
Animal Care of Yangzhou University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
Stra8
|
stimulated by retinoic acid gene 8
|
|
RA
|
retinoic acid
|
|
DEGs
|
differentially expressed genes
|
|
VAD
|
vitamin A deficient
|
|
VAR
|
vitamin A recovery
|
|
AKT/PKB
|
protein kinase B
|
|
FBS
|
fetal bovine serum
|
|
DAPI
|
4′,6-diamidino-2-phenylindole
|
|
PDK1
|
phosphatidylinositol-dependent kinase
1
|
|
USP33
|
ubiquitin-specific protease 33
|
|
TCF4
|
transcription factor 4
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
GSTP1
|
glutathione S-transferase P1
|
|
ANG2
|
angiopoietin 2
|
|
OSGIN1
|
oxidative stress induced growth
inhibitor 1
|
|
BIRC6
|
baculoviral inhibitor of apoptosis
repeat-containing 6
|
Acknowledgments
Not applicable.
References
|
1
|
Tedesco M, La Sala G, Barbagallo F, De
Felici M and Farini D: STRA8 shuttles between nucleus and cytoplasm
and displays transcriptional activity. J Biol Chem.
284:35781–35793. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ball RL, Fujiwara Y, Sun F, Hu J, Hibbs
MA, Handel MA and Carter GW: Regulatory complexity revealed by
integrated cytological and RNA-seq analyses of meiotic substages in
mouse spermatocytes. BMC Genomics. 17:6282016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
wang S, wang X, Ma L, Lin X, Zhang D, Li
Z, wu Y, Zheng C, Feng X, Liao S, et al: Retinoic acid is
sufficient for the in vitro induction of mouse spermatocytes. Stem
Cell Reports. 7:80–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koubova J, Hu YC, Bhattacharyya T, Soh YQ,
Gill ME, Goodheart ML, Hogarth CA, Griswold MD and Page DC:
Retinoic acid activates two pathways required for meiosis in mice.
PLoS Genet. 10:e10045412014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Endo T, Romer KA, Anderson EL, Baltus AE,
de Rooij DG and Page DC: Periodic retinoic acid-STRA8 signaling
intersects with periodic germ-cell competencies to regulate
spermatogenesis. Proc Natl Acad Sci USA. 112:E2347–E2356. 2015.
View Article : Google Scholar
|
|
6
|
Zhou Q, Nie R, Li Y, Friel P, Mitchell D,
Hess RA, Small C and Griswold MD: Expression of stimulated by
retinoic acid gene 8 (Stra8) in spermatogenic cells induced by
retinoic acid: An in vivo study in vitamin A-sufficient postnatal
murine testes. Biol Reprod. 79:35–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mark M, Jacobs H, Oulad-Abdelghani M,
Dennefeld C, Féret B, Vernet N, Codreanu CA, Chambon P and
Ghyselinck NB: STRA8-deficient spermatocytes initiate, but fail to
complete, meiosis and undergo premature chromosome condensation. J
Cell Sci. 121:3233–3242. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anderson EL, Baltus AE, Roepers-Gajadien
HL, Hassold TJ, de Rooij DG, van Pelt AM and Page DC: Stra8 and its
inducer, retinoic acid, regulate meiotic initiation in both
spermato-genesis and oogenesis in mice. Proc Natl Acad Sci USA.
105:14976–14980. 2008. View Article : Google Scholar
|
|
9
|
Zhang Y, wang Y, Zuo Q, Li D, Zhang W,
wang F, Ji Y, Jin J, Lu Z, wang M, et al: CRISPR/Cas9 mediated
chicken Stra8 gene knockout and inhibition of male germ cell
differentiation. PLoS One. 12:e01722072017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma HT, Niu CM, Xia J, Shen XY, Xia MM, Hu
YQ and Zhen Y: Stimulated by retinoic acid gene 8 (Stra8) plays
important roles in many stages of spermatogenesis. Asian J Androl.
2018.
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−∆∆CT method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
12
|
Miyamoto T, Sengoku K, Takuma N, Hasuike
S, Hayashi H, Yamauchi T, Yamashita T and Ishikawa M: Isolation and
expression analysis of the testis-specific gene, STRA8, stimulated
by retinoic acid gene 8. J Assist Reprod Genet. 19:531–535. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nair R and Shaha C: Diethylstilbestrol
induces rat spermatogenic cell apoptosis in vivo through increased
expression of spermatogenic cell Fas/FasL system. J Biol Chem.
278:6470–6481. 2003. View Article : Google Scholar
|
|
14
|
Liu T, wang L, Chen H, Huang Y, Yang P,
Ahmed N, wang T, Liu Y and Chen Q: Molecular and cellular
mechanisms of apoptosis during dissociated spermatogenesis. Front
Physiol. 8:1882017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ning JZ, Rao T, Cheng F, Yu WM, Ruan Y,
Yuan R, Zhu SM, Du Y and Xiao CC: Effect of varicocelectomy
treatment on spermatogenesis and apoptosis via the induction of
heat shock protein 70 in varicoceleinduced rats. Mol Med Rep.
16:5406–5412. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Chen F, Li C and Chen Y: Quinestrol
induces spermatogenic apoptosis in vivo via increasing
pro-apoptotic proteins in adult male mice. Tissue Cell. 46:318–325.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rogers R, Ouellet G, Brown C, Moyer B,
Rasoulpour T and Hixon M: Cross-talk between the Akt and NF-kappaB
signaling pathways inhibits MEHP-induced germ cell apoptosis.
Toxicol Sci. 106:497–508. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abraham AG and O'Neill E:
PI3K/Akt-mediated regulation of p53 in cancer. Biochem Soc Trans.
42:798–803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brennan MS, Matos MF, Richter KE, Li B and
Scannevin RH: The NRF2 transcriptional target, OSGIN1, contributes
to mono-methyl fumarate-mediated cytoprotection in human
astrocytes. Sci Rep. 7:420542017. View Article : Google Scholar
|
|
20
|
Talos F, Petrenko O, Mena P and Moll UM:
Mitochondrially targeted p53 has tumor suppressor activities in
vivo. Cancer Res. 65:9971–9981. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia X, Xu Y, Wu W, Fan Y, wang G, Zhang T
and Su W: Aroclor1254 disrupts the blood-testis barrier by
promoting endocytosis and degradation of junction proteins via p38
MAPK pathway. Cell Death Dis. 8:e28232017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choudhury R, Bonacci T, wang X, Truong A,
Arceci A, Zhang Y, Mills CA, Kernan JL, Liu P and Emanuele MJ: The
E3 ubiquitin Ligase SCF (Cyclin F) transmits AKT signaling to the
cell-cycle machinery. Cell Rep. 20:3212–3222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matheny RW Jr and Adamo ML: Current
perspectives on Akt Akt-ivation and Akt-ions. Exp Biol Med
(Maywood). 234:1264–1270. 2009. View Article : Google Scholar
|
|
24
|
Busada JT, Chappell VA, Niedenberger BA,
Kaye EP, Keiper BD, Hogarth CA and Geyer CB: Retinoic acid
regulates Kit translation during spermatogonial differentiation in
the mouse. Dev Biol. 397:140–149. 2015. View Article : Google Scholar
|
|
25
|
Busada JT, Niedenberger BA, Velte EK,
Keiper BD and Geyer CB: Mammalian target of rapamycin complex 1
(mTORC1) is required for mouse spermatogonial differentiation in
vivo. Dev Biol. 407:90–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harris TK.PDK1 and PKB/Akt: Ideal targets
for development of new strategies to structure-based drug design.
IUBMB Life. 55:117–126. 2003. View Article : Google Scholar
|
|
27
|
Hurtado E, Cilleros V, Just L, Simó A,
Nadal L, Tomàs M, Garcia N, Lanuza MA and Tomàs J: Synaptic
activity and muscle contraction increases PDK1 and PKCbetaI
phosphorylation in the presynaptic membrane of the neuromuscular
junction. Front Mol Neurosci. 10:2702017. View Article : Google Scholar
|
|
28
|
Shen J, Frye M, Lee BL, Reinardy JL,
McClung JM, Ding K, Kojima SSM, Xia H, Seidel C, Lima e Silva R, et
al: Targeting VE-PTP activates TIE2 and stabilizes the ocular
vasculature. J Clin Invest. 124:4564–4576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Imanishi Y, Hu B, Xiao G, Yao X and Cheng
SY: Angiopoietin-2, an angiogenic regulator, promotes initial
growth and survival of breast cancer metastases to the lung through
the integrin-linked kinase (ILK)-AKT-B cell lymphoma 2 (Bcl-2)
pathway. J Biol Chem. 286:29249–29260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ren J, Shi M, Liu R, Yang QH, Johnson T,
Skarnes WC and Du C: The Birc6 (Bruce) gene regulates p53 and the
mitochondrial pathway of apoptosis and is essential for mouse
embryonic development. Proc Natl Acad Sci USA. 102:565–570. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiu XB and Goldberg AL: The
membrane-associated inhibitor of apoptosis protein, BRUCE/Apollon,
antagonizes both the precursor and mature forms of Smac and
caspase-9. J Biol Chem. 280:174–182. 2005. View Article : Google Scholar
|
|
32
|
wen P, Kong R, Liu J, Zhu L, Chen X, Li X,
Nie Y, wu K and wu JY: USP33, a new player in lung cancer, mediates
Slit-Robo signaling. Protein Cell. 5:704–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu H, Zhang Q, Li K, Gong Z, Liu Z, Xu Y,
Swaney MH, Xiao K and Chen Y: Prognostic significance of USP33 in
advanced colorectal cancer patients: New insights into
β-arrestin-dependent ERK signaling. Oncotarget. 7:81223–81240.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu N, Song Z, Zhang K and Yang X: MAD2B
acts as a negative regulatory partner of TCF4 on proliferation in
human dermal papilla cells. Sci Rep. 7:116872017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jeong JB, Lee J and Lee SH: TCF4 is a
molecular target of resve-ratrol in the prevention of colorectal
cancer. Int J Mol Sci. 16:10411–10425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Domazetovic V, Fontani F, Marcucci G,
Iantomasi T, Brandi ML and Vincenzini MT: Estrogen inhibits
starvation-induced apoptosis in osteocytes by a redox-independent
process involving association of JNK and glutathione S-transferase
P1-1. FEBS Open Bio. 7:705–718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Okamura T, Antoun G, Keir ST, Friedman H,
Bigner DD and Ali-Osman F: Phosphorylation of glutathione
S-transferase P1 (GSTP1) by epidermal growth factor receptor (EGFR)
promotes formation of the GSTP1-c-Jun N-terminal kinase (JNK)
complex and suppresses JNK downstream signaling and apoptosis in
brain tumor cells. J Biol Chem. 290:30866–30878. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Flanagan L, Meyer M, Fay J, Curry S, Bacon
O, Duessmann H, John K, Boland KC, McNamara DA, Kay EW, et al: Low
levels of Caspase-3 predict favourable response to 5FU-based
chemotherapy in advanced colorectal cancer: Caspase-3 inhibition as
a therapeutic approach. Cell Death Dis. 7:e20872016. View Article : Google Scholar : PubMed/NCBI
|