Introduction
Gastric cancer (GC) is the second most common cause
of cancer-related mortality after lung cancer, with 738,000 deaths
and 1 million new diagnoses reported annually worldwide, despite
recent improvements in developed countries (1). In China, 400,000 new cases of GC are
diagnosed annually, accounting for 42% of total cases worldwide
(2). The tumorigenesis of GC is
complex and involves the deregulation of a large number of genes
(3,4). Despite advances in the treatments
available for GC, the prognosis of patients with advanced-stage GC
remains poor. Consequently, further research into GC progression
and the related molecular mechanisms is required.
It has been reported that autophagy, which is
responsible for intracellular degradation and protein recycling,
plays a key role in tumor suppression and anticancer therapy
(5). However, the factors that
regulate the crosstalk between autophagy and apoptosis require
further investigation to elucidate the mechanisms that control cell
survival and death (6).
As a cDNA partially complementing ultraviolet (UV)
radiation sensitivity, UV radiation resistance-associated gene
(UVRAG) was first isolated from xeroderma pigmentosum cells
(7), and there have also been
studies that indicated the important role of UVRAG in mammalian
cell autophagy (8,9). UVRAG, which is a mammalian homolog
of yeast Vps38, activates the Beclin-1/PI3K complex and promotes
the formation of autophagosomes (10). Moreover, UVRAG has been reported
to promote the maturation of autophagosomes via recruiting HOPS
complexes and Rab7 to the late endosome (11,12).
MicroRNAs (miRNAs) are small non-coding RNAs (22-24
nucleotides in length) that play a role in the transcriptional and
post-transcriptional regulation of gene expression (13) by pairing with the 3′-untranslated
region (UTR) of the corresponding mRNAs (14). Since the identification of lin-4
and let-7 in Caenorhabditis elegans (C. elegans),
~2,000 miRNAs have been identified, accounting for 1% of the human
genome (15). miRNAs play a
pivotal role in carcinogenesis; some function as oncogenes, with
their over-expression leading to cancerous growth, while others act
as tumor suppressors, the downregulation of which results in
cancerous growth (16). Mounting
evidence has revealed that miRNAs are crucial for the
proliferation, apoptosis, migration, invasion and metabolism of
tumor cells (17). Reports of
miRNAs in autophagy have improved our understanding of the
pathogenesis of human diseases (18), indicating that miRNAs may regulate
autophagy-related genes and affect the progression of cancer.
The aim of the present study was to explore miRNAs
that target UVRAG and participate in the regulation of autophagy
and apoptosis in GC.
Materials and methods
Cell culture
The GC cell line MKN28 was purchased from The
Institute of Biochemistry and Cell Biology at The Chinese Academy
of Sciences (Shanghai, China) and grown in RPMI-1640 medium
containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with antibiotics
at 37°C in a humidified atmosphere containing 5% CO2.
Cells at passages 5-20 were used in the present study. To induce
autophagy, MKN28 cells were rinsed with phosphate-buffered saline
(PBS) twice and incubated with Hank's Balanced Salt solution (HBSS;
Cellgro; Corning Incorporated, Corning, NY, USA) for 4 h at 37°C in
a humidified atmosphere containing 5% CO2, as previously
described (19).
Transfection of miRNAs
miR-NC mimics, miR-183 mimics, miR-NC inhibitors or
miR-183 inhibitors (Ambion; Thermo Fisher Scientific, Inc.) were
transfected into MKN28 cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) and OPTI-MEM I
Reduced-Serum Medium (Invitrogen; Thermo Fisher Scientific, Inc.)
in accordance with the manufacturer's instructions. The cell
culture medium was changed at 24 h post-transfection. The following
experiments were performed at 48 h post-transfection in
triplicate.
Cell viability assay
MKN28 cells were seeded on 96-well plates at a
density of 3x104 cells/well, grown in RPMI-1640 medium
containing 10% fetal bovine serum (FBS), and transiently
transfected with pre-miR-183 and miR-control (Ambion; Thermo Fisher
Scientific, Inc.; cat. no. AM17110). At 5 days post-transfection,
cell viability was detected using Cell Counting Kit-8 (CCK-8;
Dojindo Molecular Technologies, Inc., Kumamoto, Japan). CCK-8
reagent (10 µl) was added into each well and incubated at 37°C for
3 h. Absorbance was measured at 450 and 600 nm using a
spectrophotometer. Cell viability was evaluated using optical
density (OD) as follows: Cell viability = OD450 nm -
OD600 nm. Each experiment was performed at least in
triplicate. For the detection of cell viability in the control,
starvation and starvation + miR-183 mimic/inhibitor groups, cells
were treated as follows: Control, cells incubated with RPMI-1640
medium containing 10% FBS; starvation, cells incubated with HBSS;
starvation + miR-183 mimic/inhibitor, cells incubated with HBSS,
which were transfected with miR-183 mimic/inhibitor.
Flow cytometry assay
MKN28 cells were seeded in 12-well plates and
incubated for 48 h at 37°C. A total of 1x105 MKN28 cells
were collected by centrifugation at 1,000 x g for 5 min at room
temperature and washed three times with PBS. Thereafter, MKN cells
were fixed by pre-cooled 70% ethanol at -20°C overnight. On the
next day, MKN28 cells were re-suspended in RPMI-1640 medium with
10% FBS, mixed with Annexin V-enhanced fluorescent dye and
prop-idium iodide (PI) and incubated for 20 min in the dark at room
temperature. Assay results were evaluated using flow cytometry; the
results were measured by Cell Quest software (BD Biosciences,
Franklin Lakes, NJ, USA). For the detection of cell apoptosis in
the control, starvation and starvation + miR-183 mimic/inhibitor
groups, cells were treated as follows: Control, cells incubated
with RPMI-1640 medium containing 10% FBS; starvation, cells
incubated with HBSS; starvation + miR-183 mimic/inhibitor, cells
incubated with HBSS, which were transfected with miR-183
mimic/inhibitor.
Dual luciferase reporter assay
The potential binding site of UVRAG 3′-UTR with
miRNA was predicted using TargetScan (www.targetscan.org). miR-183 was predicted to bind
with UVRAG 3′-UTR. Wild-type (wt) and mutant (mut) UVRAG 3′-UTR
were synthesized and cloned into the XbaI site (downstream
of the luciferase stop codon) of pGL3 vectors (Promega Corp.,
Madison, WI, USA). The resulting vectors were named pGL3-wt-UVRAG
and pGL3-mut-UVRAG, respectively. 293 cells were cultured in
24-well plates and transfected with 0.4 mg pGL3-Control,
pGL3-wt-UVRAG or pGL3-mut-UVRAG with pRL-TK luciferase reporter (25
ng/well) and 20 nm pcDNA-miR-183 or pcDNA-miR-NC using
Lipofectamine® 2000. At 48 h post-transfection, a
Dual-Luciferase Reporter Assay was performed to assess luciferase
activity (Promega Corp.).
Western blotting
MKN28 cells were lysed using RIPA buffer containing
complete protease inhibitor cocktail (Roche Molecular Diagnostics,
Pleasanton, CA, USA). Protein concentrations in each sample were
determined using a BCA kit (Beyotime Institute of Biotechnology,
Haimen, China) in accordance with the manufacturer's instructions.
Denatured proteins (15 µg) were separated by 12% SDS-polyacrylamide
gels and transferred to nitrocellulose membranes. Membranes were
blocked in 5% skimmed milk for 2 h at room temperature, followed by
incubation with primary antibodies in PBS containing 0.5% Tween-20
(PBST) at 4°C overnight. The primary antibodies were as listed:
UVRAG (ABS1600; 1:1,000; Millipore, Darmstadt, Germany), BAX
(50599; 1:1,000; ProteinTech Group, Inc., Chicago, IL, USA), Bcl-2
(12789; 1:1,000; ProteinTech Group, Inc.), LC3B-I/II (L8918;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and GAPDH (5174;
1:1,000, Cell Signaling Technology, Inc.). Subsequently, the
membranes were washed three times with PBST for 10 min, followed by
incubation with horseradish peroxidase (HRP)-conjugated secondary
antibody (ab97080; 1:5,000; Abcam, Cambridge, MA, USA) at room
temperature for 1 h. Finally, the nitrocellulose membranes were
washed three times with PBST for 10 min and the bands were
visualized using enhanced chemiluminescence (GE Healthcare Life
Science, Little Chalfont, UK) in accordance with the manufacturer's
instructions. For the detection of UVRAG protein level in the
control and starvation groups, the cells were treated as follows:
Control, cells incubated with RPMI-1640 medium containing 10% FBS;
starvation, cells incubated with HBSS.
For the detection of UVRAG protein level in the
control, miR-NC mimic/inhibitor and miR-183 mimic/inhibitor groups,
the cells were treated as follows: Control, cells incubated with
RPMI-1640 medium containing 10% FBS; miR-NC mimic/inhibitor, cells
incubated with RPMI-1640 medium containing 10% FBS, which were
transfected with miR-NC mimic/inhibitor; miR-183 mimic/inhibitor,
cells incubated with RPMI-1640 medium containing 10% FBS, which
were transfected with miR-183 mimic/inhibitor. For the detection of
BAX, Bcl-2, LC3B-I/II and UVRAG in the control, starvation and
starvation + miR-183 mimic/inhibitor groups, the cells were treated
as follows: Control, cells incubated with RPMI-1640 medium
containing 10% FBS; starvation, cells incubated with HBSS;
starvation + miR-183 mimic/inhibitor, cells incubated with HBSS,
which were transfected with miR-183 mimic/inhibitor.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was isolated from cultured MKN28 cells
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). The
integrity, quantity and purity of total RNA were determined using a
Nano-Drop 8000 Spectrophotometer (Thermo Fisher Scientific, Inc.).
miR-183 was reverse-transcribed and cDNA was synthesized from total
RNA (10 ng) using a TaqMan® Small RNA Assay (Thermo
Fisher Scientific, Inc.) in accordance with the manufacturer's
instructions. Relative mRNA and miRNA levels were quantified using
a mirVanaqRT-PCR miRNA Detection kit and SYBR Green I (Applied
Biosystems; Thermo Fisher Scientific, Inc.). RT-qPCR was performed
using an ABI Prism 7900 Sequence Detection System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Subsequently, the
quantification cycle (Cq) of each sample was determined. The
relative mRNA levels and miRNA levels were calculated based on the
Cq values and 2-∆∆Cq method (20) and normalized to GAPDH or U6 level
in each sample, respectively. For the detection of miR-183 and
UVRAG in the control and starvation groups, cells were treated as
follows: Control, cells incubated with RPMI-1640 medium containing
10% FBS; starvation, cells incubated with HBSS. For the detection
of UVRAG mRNA level and miR-183 in the control, miR-NC
mimic/inhibitor, and miR-183 mimic/inhibitor groups, cells were
treated as follows: Control, cells incubated with RPMI-1640 medium
containing 10% FBS; miR-NC mimic/inhibitor, cells incubated with
RPMI-1640 medium containing 10% FBS, which were transfected with
miR-NC mimic/inhibitor; miR-183 mimic/inhibitor, cells incubated
with RPMI-1640 medium containing 10% FBS, which were transfected
with miR-183 mimic/inhibitor.
Immunofluorescence staining
MKN28 cell were seeded on glass slides in 24-well
plates and incubated for 24 h at 37°C, then rinsed with PBS three
times. Thereafter, MKN28 cells were fixed in 4% paraformaldehyde
for 30 min and permeabilized with 0.5% Triton X-100 for 1 h at 4°C.
After blocking with 3% bovine serum albumin (Solarbio Life
Sciences, Beijing, China) for 1 h at room temperature, the cells
were incubated with rabbit anti-LC3B antibody (Sigma-Aldrich; Merck
KGaA) overnight at room temperature and goat anti-rabbit Alexa
Fluo488 (Invitrogen; Thermo Fisher Scientific, Inc.) for 2 h at
room temperature. The cells were then mounted onto coverslips using
Prolong Gold anti-fade reagent with 4'-6-diamidino-2-phenylindole
(Invitrogen; Thermo Fisher Scientific, Inc.) for 2 h at room
temperature. Images of immunofluorescence staining in each group
were captured using an AV300-ASW confocal microscope (Olympus
Corp., Tokyo, Japan). For the detection of LC3 dot formation in the
control, starvation and starvation + miR-183 mimic/inhibitor
groups, the cells were treated as follows: Control, cells incubated
with RPMI-1640 medium containing 10% FBS; starvation, cells
incubated with HBSS; starvation + miR-183 mimic/inhibitor, cells
incubated with HBSS, which were transfected with miR-183
mimic/inhibitor.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean. GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla,
CA, USA) was used for statistical analysis. Student's t-test was
used for comparisons between two groups, while multiple group
comparisons were performed using one-way analysis of variance
followed by Bonferroni's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
UVRAG and miR-183 expression following
induction of autophagy
Recent studies have revealed several
autophagy-related microRNAs in cancer, including miR-183 (21,22). miR-183 has clinical implications
in cancer patients; for example, it was reported to be a biomarker
for bladder cancer (23). To
explore the potential association between miR-183 and autophagy in
GC, the level of miR-183 expression was evaluated under baseline
conditions and after the induction of autophagy.
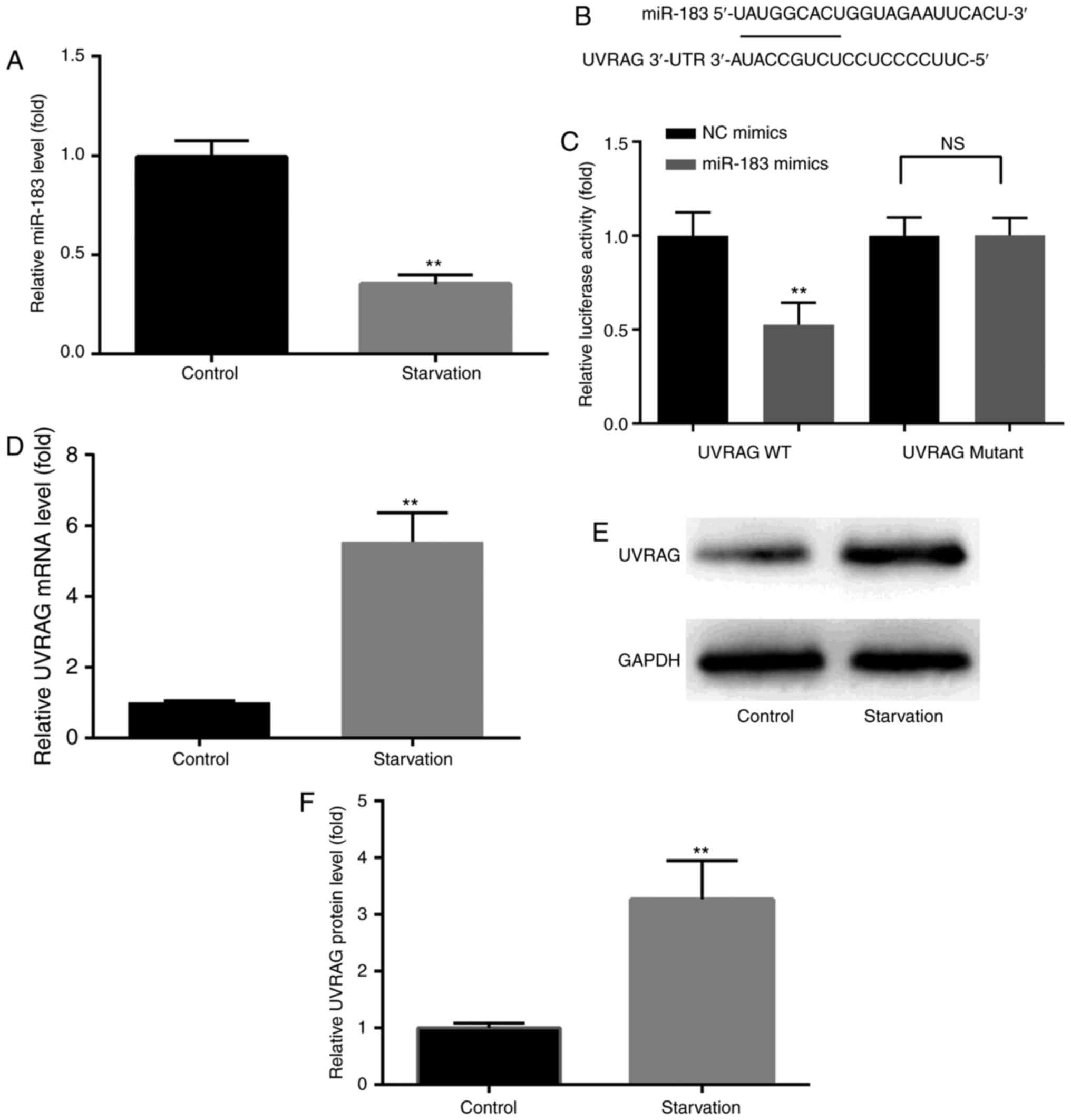
As shown in Fig.
1A, RT-qPCR analysis revealed that serum starvation resulted in
a distinct downregulation of miR-183 in MKN28 cells. In addition,
UVRAG was identified as an mRNA target for miR-183 (Fig. 1B), and the dual luciferase
reporter assay revealed that the 3′UTR of UVRAG was targeted by
miR-183 (Fig. 1C). UVRAG protein
and mRNA expression were increased following induction of autophagy
in MKN28 cells, as demonstrated by western blotting and RT-qPCR,
respectively (Fig. 1D-F). These
results indicate a potential physiological function of miR-183 in
GC autophagy, prompting us to further explore its precise
function.
miR-183 mimics and inhibitors regulate
the expression of miR-183 and UVRAG
RT-qPCR was performed to assess the effects of
miR-183 mimics and inhibitors on the expression of miR-183 and
UVRAG in MKN28 cells. It was observed that, compared with the
control and miR-NC groups, miR-183 mimics significantly upregulated
the expression of miR-183, while miR-183 inhibitor significantly
downregulated the expression of miR-183 (Fig. 2A and B). Compared with the control
and miR-NC groups, miR-183 mimics significantly downregulated the
expression of UVRAG mRNA, while miR-183 inhibitors had the opposite
effect (Fig. 2C and D). The
effects of miR-183 mimics and inhibitors on the expression of UVRAG
protein was similar to that observed for mRNA (Fig. 2E and F). These findings provide a
new insight into the exact effects of miR-183 mimics and inhibitors
on miR-183 and UVRAG expression. Transfection with miR-NC mimics or
inhibitors resulted in no significant changes in miR-183 or UVRAG
expression.
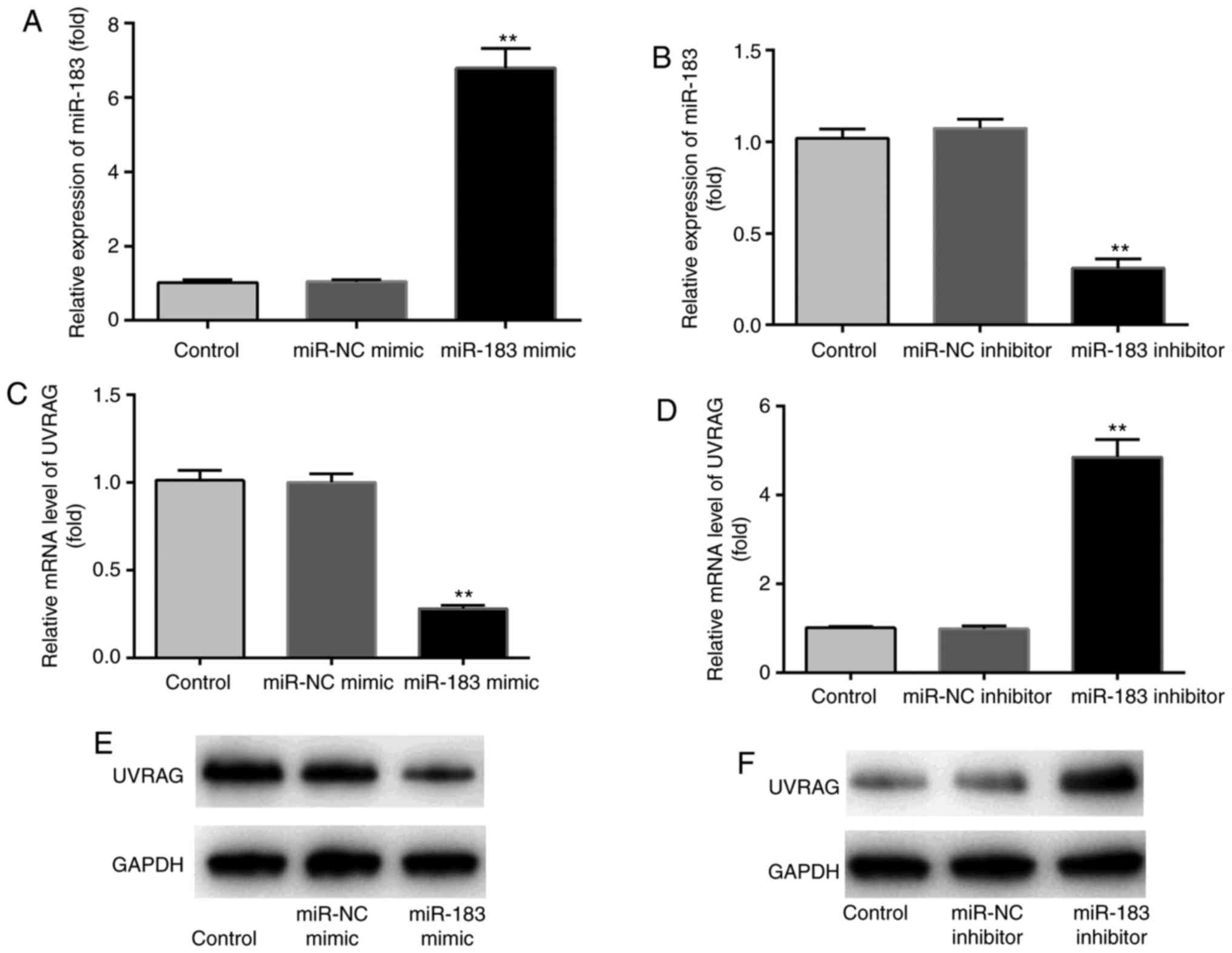 | Figure 2Effects of miR-183 mimics and
inhibitors on miR-183 and UVRAG expression. Compared with the
control and miR-NC groups, the expression of miR-183 was (A)
significantly upregulated by miR-183 mimics and (B) downregulated
by miR-183 inhibitors. Compared with the control and miR-NC groups,
the expression of UVRAG mRNA was (C) significantly downregulated by
miR-183 mimics and (D) significantly upregulated by miR-183
inhibitors. (E and F) miR-183 mimics and inhibitors exerted similar
effects on the expression of UVRAG protein. n=3.
**P<0.01 miR-183 mimic vs. miR-NC mimic or miR-183
inhibitor vs. miR-NC inhibitor. n.s., no significant difference.
Control, cells incubated with RPMI-1640 medium containing 10% FBS;
miR-NC mimic/inhibitor, cells incubated with RPMI-1640 medium
containing 10% FBS, which were transfected with miR-NC
mimic/inhibitor; miR-183 mimic/inhibitor, cells incubated with
RPMI-1640 medium containing 10% FBS, which were transfected with
miR-183 mimic/inhibitor. NC, negative control; UVRAG, ultraviolet
radiation resistance-associated gene; FBS, fetal bovine serum. |
miR-183 regulates starvation-induced
changes in cell viability and apoptosis
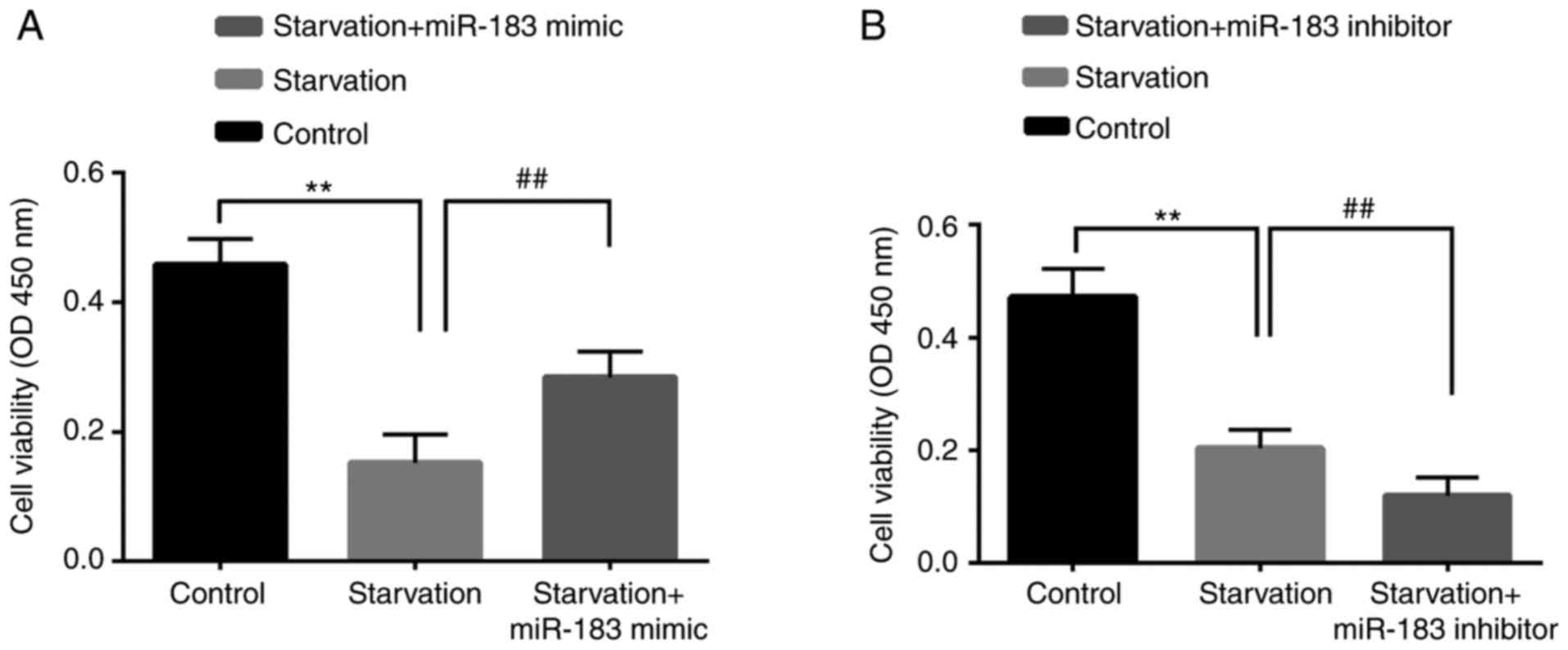
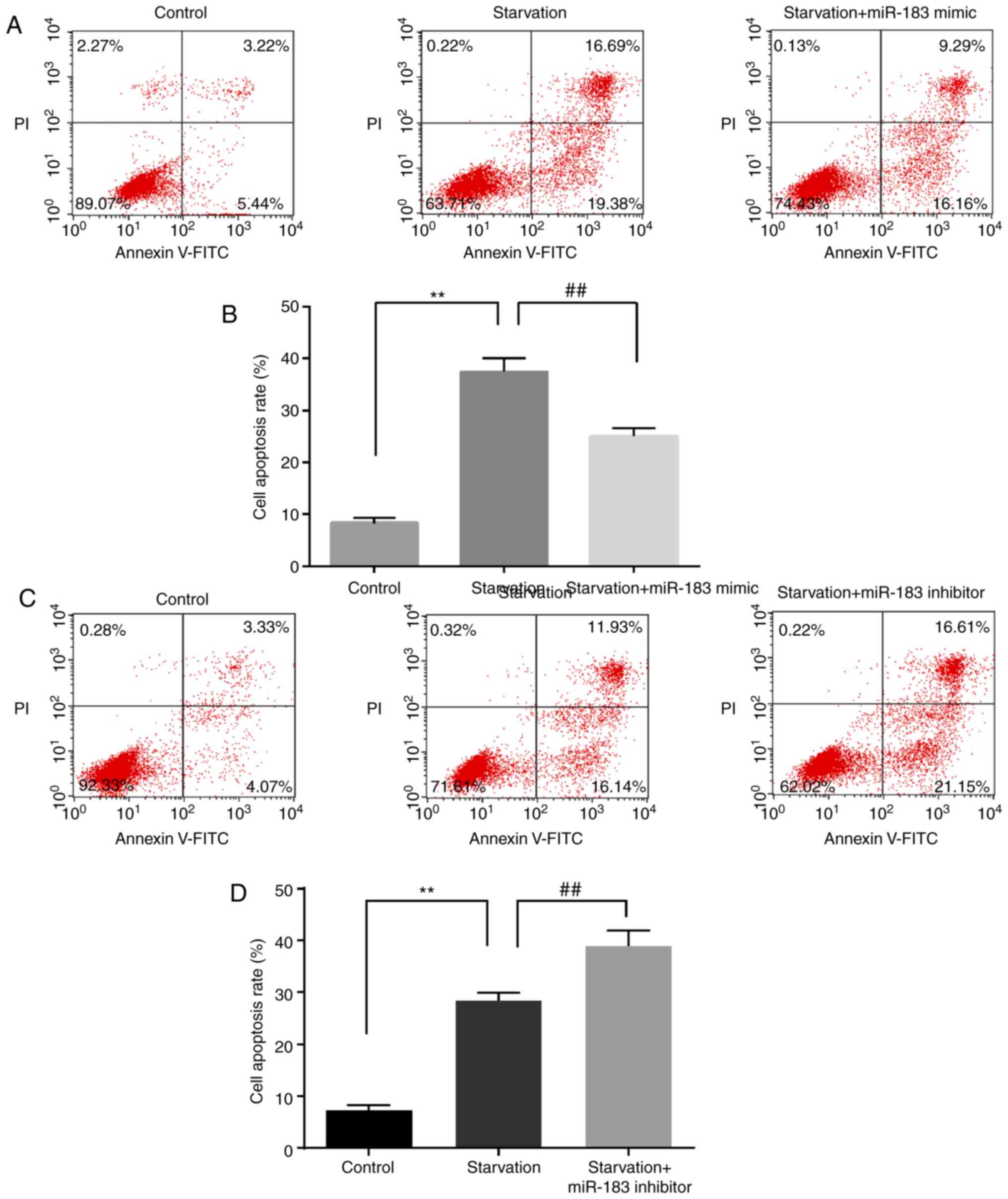
The effect of miR-183 on starvation-mediated cell
death was assessed using CCK-8 and flow cytometry assays. Compared
with the control group, starvation induction significantly
downregulated cell viability; this effect was inhibited by miR-183
mimics (Fig. 3A) and enhanced by
miR-183 inhibitors (Fig. 3B).
Compared with the control group, starvation induction significantly
upregulated cell apoptosis; this effect was reversed by miR-183
mimics (Fig. 4A and B) and
augmented by miR-183 inhibitors (Fig.
4C and D). These findings may provide novel insight into the
exact role of miR-183 mimics and inhibitors in starvation-induced
cell viability and cell apoptosis. However, the molecules that are
involved in cell apoptosis remain to be investigated.
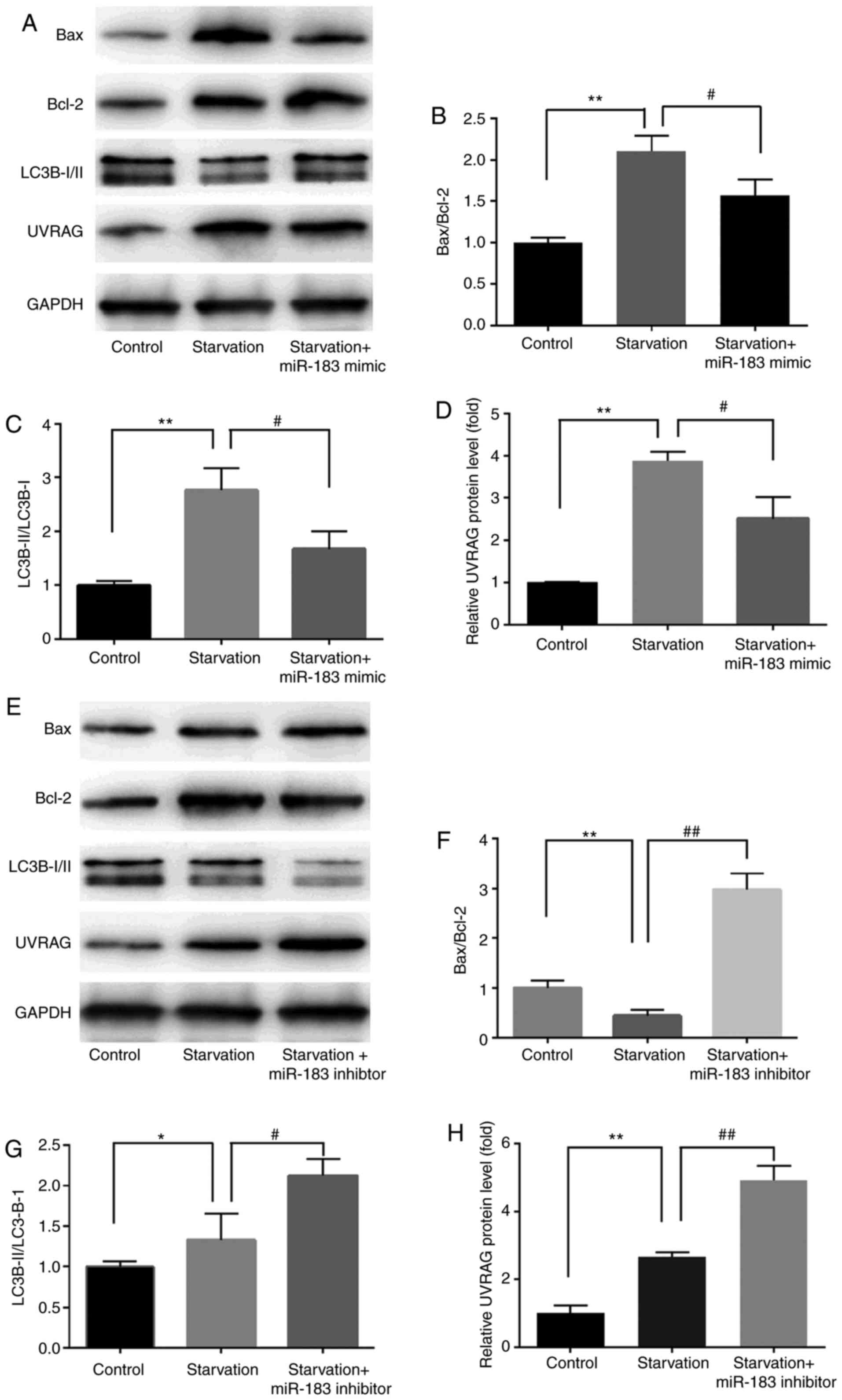
miR-183 regulates starvation-induced
changes in Bax/Bcl-2, LC3B-II/LC3B-I and UVRAG expression
We next examined the effect of miR-183 on cell
death-related proteins in MKN28 cells following serum starvation.
Treatment with miR-183 mimics significantly enhanced cell survival
and desensitized cells to starvation-induced apoptosis, as
evidenced by decreased Bax/Bcl-2 and LC3B-II/LC3B-I ratios.
Treatment with miR-183 mimics significantly reduced the
starvation-induced upregulation of UVRAG (Fig. 5A-D), while treatment with miR-183
inhibitors exerted the opposite effects (Fig. 5E-H). These findings may provide
novel insight into the exact role of miR-183 mimics and inhibitors
in starvation-induced Bax/Bcl-2, LC3B-II/LC3B-I and UVRAG
expression. However, the role of miR-183 in starvation-induced
autophagy remains to be investigated.
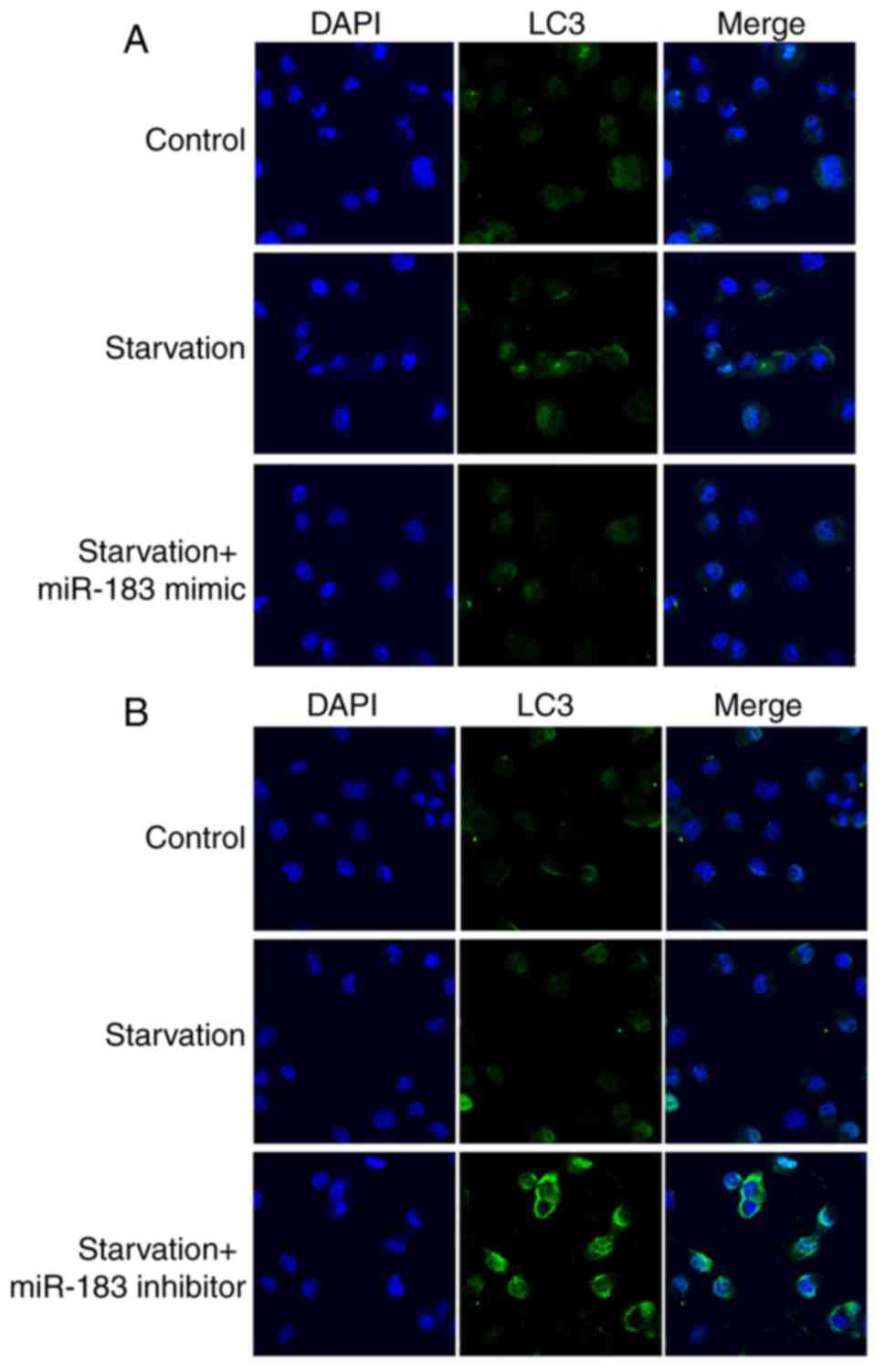
miR-183 regulates starvation-induced
autophagy
The effects of miR-183 on autophagy were assessed
using immunofluorescence staining. Starvation-induced LC3 dot
formation (Fig. 6A and B) was
reversed by miR-183 mimics (Fig.
6A) and promoted by miR-183 inhibitors in MKN28 cells (Fig. 6B). In summary, miR-183 inhibited
starvation-induced autophagy.
Discussion
Apoptosis is established as the major mechanism of
programmed cell death (PCD) and has been widely investigated
(24). However, apoptosis is not
the sole type of PCD; autophagy (ʻself-eatingʼ) plays a role in
self-destructive cell progression and is intimately associated with
the pathogenesis of a number of human diseases, including cell
death in GC (25). Autophagy and
apoptosis commonly occur in the same cell, and autophagy precedes
apoptosis (26). Autophagy
triggers necrosis and stimulates cell death by apoptosis (27); however, the final cell death is
achieved by autophagy but not apoptosis (28). It remains to be determined how
autophagosome accumulation triggers apoptosis (29).
The role of miRNAs in autophagy may improve our
understanding of the pathogenesis of multiple types of human
diseases (18), as miRNAs
regulate autophagy-related genes and affect the progression of
cancer. For example, miR-183 has been reported to modulate
autophagy and apoptosis in colorectal cancer (19). miR-183 has clinical implications
in cancer patients; for example, it was reported to be a biomarker
for bladder cancer (23).
Furthermore, miR-183 inhibits GC cell proliferation and invasion by
targeting Bmi-1 (30), while
miR-183-5p is negatively correlated with the overall survival of GC
patients (31). These findings
suggest the potential role of miR-183 in GC and autophagy. In
colorectal cancer, autophagy is an important mechanism affecting
the interaction between genetic susceptibility and metabolic
dysregulation, which contributes to the pathogenesis of the disease
(32). Consequently,
investigating the mechanism by which miR-183 and autophagy pathways
are modulated may improve our understanding of GC pathogenesis.
UVRAG has recently been identified as a risk factor
for colorectal cancer (33).
Genotoxic and metabolic stress may result in UVRAG upregulation and
apoptosis of human tumor cells (34). Surprisingly, UVRAG binds to Bax in
the cytosol and prevents translocation of Bax to the mitochondria
(35), suggesting that UVRAG may
interact with other Bcl-2 family members to induce autophagy and
trigger cell death.
LC3 is one of the biomarkers of autophagy. Once
autophagy is initiated, cytosolic LC3-I converts to membrane-bound
LC3-II, which is indispensable for the formation of autophagosomes
(36).
In summary, the results of the present study
demonstrated that miR-183 inhibits starvation-induced autophagy
(LC3) and apoptosis (BAX/Bcl-2) in the human GC cell line MKN28
[identified as a derivative of the MKN74 cell line (https://web.expasy.org/cellosaurus/CVCL_1416)
(37), suggesting that miR-183
acts as an autophagy-related oncogene; these findings were
consistent with previous studies (19,34-36). Understanding the newly identified
functions of miR-183 may allow for the development of promising
therapeutic strategies for improving the clinical outcomes of
cancer patients.
There were certain limitations to the present study.
For example, only one cell line was used, and we plan on using at
least one more cell line in our future research. Furthermore, the
molecular mechanisms underlying the effects of miR-183 remain
unclear and, thus, require further investigation in future
studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data presented and analyzed are available from
the corresponding author on special request.
Authors' contributions
The study was designed and data were acquired by YY,
YZ, LH, SS and YS. Results interpretation was performed by YY and
YZ. YY and YS wrote the manuscript. The study was supervised by YS.
All authors read and approved the final manuscript
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang L: Incidence and mortality of gastric
cancer in China. World J Gastroenterol. 12:17–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Houghton J, Stoicov C, Nomura S, Rogers
AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR and Wang TC:
Gastric cancer originating from bone marrow-derived cells. Science.
306:1568–1571. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng L, Wang L, Ajani J and Xie K:
Molecular basis of gastric cancer development and progression.
Gastric Cancer. 7:61–77. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rubinsztein DC, Codogno P and Levine B:
Autophagy modulation as a potential therapeutic target for diverse
diseases. Nat Rev Drug Discov. 11:709–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kroemer G and Levine B: Autophagic cell
death: The story of a misnomer. Nat Rev Mol Cell Biol. 9:1004–1010.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perelman B, Dafni N, Naiman T, Eli D,
Yaakov M, Feng TL, Sinha S, Weber G, Khodaei S, Sancar A, et al:
Molecular cloning of a novel human gene encoding a 63-kDa protein
and its sublocalization within the 11q13 locus. Genomics.
41:397–405. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang Y, Quach C and Liang C: Autophagy
modulator plays a part in UV protection. Autophagy. 12:1677–1678.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahashi Y, Coppola D, Matsushita N,
Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, et
al: Bif-1 interacts with Beclin 1 through UVRAG and regulates
autophagy and tumorigenesis. Nat Cell Biol. 9:1142–1151. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liang C, Feng P, Ku B, Oh BH and Jung JU:
UVRAG: A new player in autophagy and tumor cell growth. Autophagy.
3:69–71. 2007. View Article : Google Scholar
|
|
11
|
McKnight NC, Zhong Y, Wold MS, Gong S,
Phillips GR, Dou Z, Zhao Y, Heintz N, Zong WX and Yue Z: Beclin 1
is required for neuron viability and regulates endosome pathways
via the UVRAG-VPS34 complex. PLoS Genet. 10:e10046262014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang C, Lee JS, Inn KS, Gack MU, Li Q,
Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C and Jung JU:
Beclin1-binding UVRAG targets the class C Vps complex to coordinate
autopha-gosome maturation and endocytic trafficking. Nat Cell Biol.
10:776–787. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lawrie CH: MicroRNAs and haematology:
Small molecules, big function. Br J Haematol. 137:503–512. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frankel LB and Lund AH: MicroRNA
regulation of autophagy. Carcinogenesis. 33:2018–2025. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huangfu L, Liang H, Wang G, Su X, Li L, Du
Z, Hu M, Dong Y, Bai X, Liu T, et al: miR-183 regulates autophagy
and apoptosis in colorectal cancer through targeting of UVRAG.
Oncotarget. 7:4735–4745. 2016. View Article : Google Scholar :
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Pan B, Yi J and Song H: MicroRNA-mediated
autophagic signaling networks and cancer chemoresistance. Cancer
Biother Radiopharm. 28:573–578. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abraham D, Jackson N, Gundara JS, Zhao J,
Gill AJ, Delbridge L, Robinson BG and Sidhu SB: MicroRNA profiling
of sporadic and hereditary medullary thyroid cancer identifies
predictors of nodal metastasis, prognosis, and potential
therapeutic targets. Clin Cancer Res. 17:4772–4781. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nagata M, Muto S and Horie S: Molecular
biomarkers in bladder cancer: Novel potential indicators of
prognosis and treatment outcomes. Dis Markers. 2016:82058362016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu G, Pei F, Yang F, Li L, Amin AD, Liu
S, Buchan JR and Cho WC: Role of autophagy and apoptosis in
non-small-cell lung cancer. Int J Mol Sci. 18:E3672017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qian HR and Yang Y: Functional role of
autophagy in gastric cancer. Oncotarget. 7:17641–17651. 2016.
View Article : Google Scholar :
|
|
26
|
Maiuri MC, Zalckvar E, Kimchi A and
Kroemer G: Self-eating and self-killing: Crosstalk between
autophagy and apoptosis. Nat Rev Mol Cell Biol. 8:741–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Galluzzi L, Vitale I, Abrams JM, Alnemri
ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry
WS, Fulda S, et al: Molecular definitions of cell death
subroutines: Recommendations of the nomenclature committee on cell
death 2012. Cell Death Differ. 19:107–120. 2012. View Article : Google Scholar :
|
|
28
|
Shen HM and Codogno P: Autophagic cell
death: Loch Ness monster or endangered species? Autophagy.
7:457–465. 2011. View Article : Google Scholar
|
|
29
|
Ma X, Liu H, Foyil SR, Godar RJ,
Weinheimer CJ, Hill JA and Diwan A: Impaired autophagosome
clearance contributes to cardiomyocyte death in
ischemia/reperfusion injury. Circulation. 125:3170–3181. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu L, Li Y, Yan D, He J and Liu D:
MicroRNA-183 inhibits gastric cancer proliferation and invasion via
directly targeting Bmi-1. Oncol Lett. 8:2345–2351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Identification
and functional characterization of microRNAs reveal a potential
role in gastric cancer progression. Clin Transl Oncol. 19:162–172.
2017. View Article : Google Scholar
|
|
32
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goi T, Kawasaki M, Yamazaki T, Koneri K,
Katayama K, Hirose K and Yamaguchi A: Ascending colon cancer with
hepatic metastasis and cholecystolithiasis in a patient with situs
inversus totalis without any expression of UVRAG mRNA: Report of a
case. Surg Today. 33:702–706. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao Z, Ni D, Ghozalli I, Pirooz SD, Ma B
and Liang C: UVRAG: At the crossroad of autophagy and genomic
stability. Autophagy. 8:1392–1393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yin X, Cao L, Kang R, Yang M, Wang Z, Peng
Y, Tan Y, Liu L, Xie M, Zhao Y, et al: UV irradiation
resistance-associated gene suppresses apoptosis by interfering with
BAX activation. EMBO Rep. 12:727–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee J, Giordano S and Zhang J: Autophagy,
mitochondria and oxidative stress: Cross-talk and redox signalling.
Biochem J. 441:523–540. 2012. View Article : Google Scholar :
|
|
37
|
Motoyama T, Hojo H and Watanabe H:
Comparison of seven cell lines derived from human gastric
carcinomas. Acta Pathol Jpn. 36:65–83. 1986.PubMed/NCBI
|