Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic,
progressive, and usually lethal disorder with an unknown etiology,
a median survival time of 3–5 years and a mortality rate exceeding
those of numerous cancer types (1,2).
Current pathogenetic theories suggest that IPF is a result of an
aberrant wound healing response and characterized by injured
alveolar epithelium, formation of fibroblast/myofibroblast foci and
accumulation of extracellular matrix (ECM) (3). Despite the marked increase in
research efforts, the exact mechanisms involved in the initiation,
maintenance and progression of fibrosis remain largely elusive
(4). In recent years, RNA-based
regulation, including that involving micro (mi)RNA, long non-coding
(nc)RNA or circular (circ)RNA, has been identified in fibrotic
disease (5–7).
circRNA represents a novel class of endogenous
ncRNA, which is observed in a wide variety of organisms (8,9).
Unlike linear RNA, circRNA demonstrates unusual stability due to
its unique covalent loop structure with neither 5′ to 3′ polarity
nor a polyadenylated tail (10).
With the rapid development of high-throughput sequencing
technology, circRNA has been identified to be ubiquitously
expressed in various tissues and cells types (11). Its biogenesis is influenced by cis
elements and transfactors, including Alu repeats, reverse
complementary matches and RNA-binding protein (12). Furthermore, tissue, cell type or
developmental stage-specific expression patterns imply that circRNA
has important biological functions (13,14). Emerging evidence demonstrates that
circRNAs serve as gene regulators in mammals, particularly through
functioning as miRNA sponges or mRNA translation templates,
facilitating transcription of their host genes by directly
associating with RNA polymerase II or forming platforms for protein
interactions (15,16). circRNAs have crucial roles in cell
homeostasis and are closely correlated with the clinical and
pathological features of various human diseases, including
atherosclerosis, neurological disorders, fibrosis and cancer; thus,
circRNAs have potential as novel clinical diagnostic and prognostic
biomarkers (17,18). However, neither the formation
mechanism nor the cellular function of circRNA has been completely
understood in IPF. In the present study, the dysregulated circRNAs
in IPF were identified through microarrays and the potential
circRNA-associated competing endogenous RNA (ceRNA) network was
constructed with the aim to provide novel biomarkers for IPF
diagnosis and pathogenesis.
Materials and methods
Patients and clinical samples
Patients admitted to the Affiliated Hospital of
Binzhou Medical University (Binzhou, China) who were diagnosed with
IPF between October 2015 and June 2017 (n=10) through combined
clinical, radiological and pathological examination based on the
American Thoracic Society/European Respiratory Society consensus
criteria (19) were included in
the present study. Normal controls from healthy volunteers were
matched corresponding to the IPF patients’ sex and age. The healthy
non-smoking volunteers were recruited from individuals undergoing a
physical examination at the hospital over the same time period.
Individuals with respiratory and rheumatic immune diseases were
excluded from the study. Fresh peripheral blood samples were
collected in EDTA-containing tubes and then centrifuged (1,000 × g
for 10 min at room temperature) to isolate the plasma.
RNA extraction and characterization
The total RNA of each sample was isolated using
TRIzol LS reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer’s instructions. The
RNA quantity and quality were determined using a NanoDrop ND-1000
instrument (Thermo Fisher Scientific, Inc.). A 260/280 nm
absorbance ratio between 1.8 and 2.1, or a 260/230 nm absorbance
ratio of >1.8 were acceptable. RNA integrity and genomic DNA
contamination were assessed by standard denaturing agarose gel
electrophoresis after RNA extraction and prior to sample labelling.
DNA contamination was ruled out by the absence of a high
molecular-weight smear or band migration above the 28S ribosomal
RNA band. RNA integrity was confirmed by the absence of RNA
degradation, which was reflected by smearing of ribosomal RNA
bands.
Microarray hybridization
Arraystar Inc. (Rockville, MD, USA) developed the
world’s first commercial circRNA chip for systematically analysing
the expression of circRNA in different physiological and
pathological conditions. The Arraystar Human circRNA Array (8×15 K;
Arraystar Inc.) analysis was performed by Kangchen Corp. (Shanghai,
China) in 3 pairs of IPF samples and normal control samples. In
brief, isolated total RNA was treated with RNase R (Epicenter,
Madison, WI, USA) to remove linear RNA. Subsequently, the samples
were amplified and transcribed into fluorescent complementary
(c)RNA by utilizing a random primer method according to the
Arraystar Super RNA Labeling protocol. Subsequently, the labelled
cRNA was purified using an RNeasy Mini Kit (Qiagen, Hilden,
Germany). The concentration and specific activity of the labelled
cRNA (pmol Cy3/µg cRNA) was measured using a NanoDrop
ND-1000. A total of 1 µg of each labelled cRNA was then
fragmented by adding 5 µl of 10X blocking agent and 1
µl of 25X fragmentation buffer. The mixture was then heated
at 60°C for 30 min, and 25 µl 2X hybridization buffer was
then added to dilute the labelled cRNA. Subsequently, 50 µl
of the hybridization solution was dispensed into the gasket slide
and assembled onto the Arraystar circRNA expression microarray
slide. The slides were incubated for 17 h at 65°C in an Agilent
hybridization oven (Agilent Technologies, Inc., Santa Clara, CA,
USA). The slides were washed and the hybridized arrays were scanned
using an Agilent G2505C scanner (Agilent Technologies, Inc.). The
scanned images were imported into the Agilent feature extraction
software (version 11.0.1.1) to extract raw data (Agilent
Technologies, Inc.).
Microarray data analysis
Quantile normalization of raw data and the
subsequent data processing were performed using the R limma package
(bioconductor. org/packages/release/bioc/html/limma.html). After
quantile normalization of raw data, low-intensity filtering was
performed and circRNA with a minimum of 1 out of 6 samples with
flags in ‘present’ or ‘marginal’ (‘all target values’) were
retained for further analyses. In the comparison of the two groups,
namely disease vs. normal controls, the ‘fold change’ (the ratio of
group average values) among groups was computed for each circRNA.
Statistical significance of the difference was estimated via a
Student’s t-test. circRNA with an absolute value of the fold change
≥1.5 and P-values of ≤0.05 were considered to indicate
significantly different expression. The final outputs were filtered
and the differentially expressed circRNA was ranked based on fold
change and P-value, among others, by using the ‘Data/Sort’ and
‘Filter’ functions of Microsoft Excel2013 (Microsoft Corp.,
Redmond, WA, USA). The dataset was deposited in the Gene Expression
Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) with the accession
no. GSE102660.
Reverse transcription (RT)
cDNA for circRNA and mRNA was generated using the
Super Script™ III First-Strand Synthesis System (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer’s protocol. In
brief, 3 µg total RNA, 1 µl random primer (N9), 4
µl 5X first-strand buffer, 1.6 µl nucleotide mix, 0.3
µl RNase inhibitor, 1 µl dithiothreitol and 0.2
µl Super Script™ III reverse transcriptase were added to the
system, which was then incubated at 50°C for 1 h. The RT reaction
and no-template control were performed simultaneously. RT of miRNAs
was performed using the Bulge-Loop miRNA primer set according to
the manufacturer’s protocol (RT miR-324-5p primer, cat. no.
ssD809230302; RT miR-630 primer, cat. no. ssD809230591; Guangzhou
RiboBio Co., Ltd. (Guangzhou, China).
Quantitative polymerase chain reaction
(qPCR)
The expression levels of circRNA and miRNA were
evaluated via qPCR using 2X PCR master mix (Arraystar Inc.) and
SYBR Green Master Mix (Roche Diagnostics, Basel, Switzerland),
respectively, according to the manufacturer’s protocol. PCR for
circRNA was performed in a 10-µl reaction volume, including
2 µl cDNA, 5 µl 2X Master Mix, 0.5 µl forward
primer (10 µM), 0.5 µl reverse primer (10 µM)
and 2 µl double-distilled water. The reaction was set at
95°C for 10 min for pre-denaturation, followed by 40 cycles of 95°C
for 10 sec and 60°C for 60 sec. GAPDH was used for template
normalization of circRNA (20).
PCR for miR-324-5p and miR-630 entailed initiation at 95°C for 20
sec, followed by 30 cycles at 95°C for 10 sec, 60°C for 20 sec and
72°C for 10 sec. PCR and analyses were performed with a ViiA7
Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The C. elegans miRNA 39-3p was used as a
normalization control. The specific primers for miRNA were
purchased from Guangzhou RiboBio Co., Ltd. and their catalogue
numbers were as follows: miR-324-5p forward, cat. no. ssD809230994;
miR-630 forward, cat. no. ssD090525005; miR-324-5p and miR-630
reverse, cat. no. ssD089261711. Samples for target and reference
gene amplification were prepared in triplicate. The gene expression
levels were calculated using the ΔCt method (20). Divergent primers for circRNA were
designed to amplify the circRNA-specific back-splice junctions and
had the following sequences: hsa_circRNA_100906 forward,
5′-CTGGACAAGGCCACATAGAGT-3′ and reverse,
5′-CAGAGCAGCCAATGAAGACAC-3′; hsa_circRNA_102100 forward,
5′-TCTATTAGGGCATGAGTTTGTCTT-3′ and reverse,
5′-TCCTTGGTTGTGGAGCTGTC-3′; hsa_circRNA_102348 forward,
5′-CCTTTCAGCCCTCCATACTTACT-3′ and reverse,
5′-CCATATTCTTATCAGGCAATCTTGT-3′; hsa_circRNA_101225 forward,
5′-GCACCTGACAGCATCTATTACC-3′ and reverse,
5′-GACAGTAGAAACGCAGTAAGCAA-3′; hsa_circRNA_104780 forward,
5′-ACAGATACCACCGCCGAACT-3′ and reverse, 5′-TCTAGCTCCTTGGCAGGGAT-3′;
hsa_circRNA_101242 forward, 5′-GATGCTGCTCAAATGAGAAATG-3′ and
reverse, 5′-GCAGGAGAAGTATGTGGAGTAATC-3′. GAPDH forward,
5′-GGGAAACTGTGGCGTGAT-3′ and reverse,
5′-GAGTGGGTGTCGCTGTTGA-3.′
Detection of putative miRNA seed
matches
The miRNA response elements on circRNA and mRNA were
scanned using miRNA target prediction software (version 1.0;
Arraystar Inc.) based on miRanda and Target Scan. The
differentially expressed circRNA within all comparisons was
annotated in detail using the circRNA/miRNA interaction
information. mRNA targets were obtained according to the target
scores, which were calculated using the formula (|TargetScan
context + score|+|probabilities of conserved targeting|)/2.
Dual-luciferase reporter assay
Luciferase reporter assays were used to detect
direct binding between circRNA and miRNAs. pMIR-REPORT Luciferase
vector containing firefly luciferase gene (Obio Technology
(Shanghai) Corp., Ltd. (Shanghai, China)) and pRL-cytomegalovirus
Renilla luciferase reporter vector (Promega Corp., Madison,
WI, USA) were applied in this experiment. The full length of the
respective circRNA was obtained by gene synthesis (21). The synthesized circRNA was
verified and inserted (21) into
the pMIR-REPORT Luciferase vector by Obio Technology.
The mutant sequence was amplified from the wild-type
using PCR and mutations in the target site was developed using
primers. The mutant sequence was validated by sequencing following
plasmid construction. The primer sequences used were as follows:
has_circRNA_100906-F1,
5′-ATAGGCCGGCATAGACGCGTTGTCTTCATTGGCTGCTCTG-3′;
hsa_circRNA_100906-R1,5′-AAGGAGCCGTCGTACGGTCATCCTGGGCTGTCAGATTTG
-3′; hsa_circRNA _100906-
F2,5′-TGACCGTACGACGGCTCCTTTACATCTTGCTGCAACTCATG-3′;
hsa_circRNA_100906-R
2,5′-TTCTTACCGTCGTACAAATCACCCTTAAAGGCAGCCACATG-3′;
hsa_circRNA_100906-F3,5′-GATTTGTACGACGGTAAGAAATTAGTGGAAGATGGAGTAATC-3′;
hsa_circRNA_100906-R3,5′AAAGATCCTTTATTAAGCTTCTCTTTGCCACATCACTGGG-3′;
hsa_circRNA_102348-F1,
5′-ATAGGCCGGCATAGACGCGTTTTCAGAAAGTGCTTTCTCTC-3′;
hsa_circRNA_102348-R1,
5′-GTTAACCCGTCGTAGTTTCCAAAGGTGCAACGCTCCGTGG-3′;
hsa_circRNA_102348-F2,
5′-GGAAACTACGACGGGTTAACCCAAGAGAGTGGACTCCAGAGA-3′; and
hsa_circRNA_102348-R2,
5′-AAAGATCCTTTATTAAGCTTAGTTTTTGAACTTCAGGCCACAACCGTCGTAGCAGAAGATGATCCT-3′.
Mutations were introduced to verify the predicted miRNA binding
sites. miRNA mimics and their negative control were obtained from
Gene Pharma (Shanghai, China); they were co-transfected into 293T
cells (Cell Bank of the Chinese Academy of Sciences, Shanghai,
China) with the pMIR-REPORT Luciferase vector with or without the
full-length sequence of circRNA by using
Lipofectamine-2000® (Invitrogen; Thermo Fisher
Scientific, Inc.). After transfection for 48 h, luciferase
activities were detected with the dual-Luciferase Reporter Assay
System (Promega Corp.) according to the manufacturer’s protocols.
Relative light units were determined with a SpectramaxM2 (Molecular
Devices, LLC, Sunnyvale, CA, USA). Firefly luciferase values were
normalized to the corresponding Renilla luciferase values.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Student’s t-test was used for comparison between two
groups. Statistically significant differences from multiple groups
were determined using one-way analysis of variance with the
Student-Newman-Keuls post hoc test. All analyses were performed
using SPSS Statistics software package (version 11.0 for Windows;
SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to indicate
a statistically significant difference.
Results
Identification of differentially
expressed circRNA profiles during IPF development
IPF is clinically characterized by decreased lung
function, increased high-resolution computed tomography evidence of
honeycombing (but not emphysema), as well as significant
microscopic honeycombing and fibroblastic foci on pathology. The
demographic and baseline characteristics of IPF patients and normal
controls are presented in Table
I. No significant differences in the patient number, age and
gender between the normal and IPF groups were present.
 | Table IBaseline characteristics and
physiology of IPF patients and healthy individuals. |
Table I
Baseline characteristics and
physiology of IPF patients and healthy individuals.
| Characteristic | Controls
(n=10) | IPF
patients
(n=10) |
|---|
| Age (years) | 67.5±8.7 | 67.2±9.6 |
| Gender
(male/female) | 6/4 | 6/4 |
| FVC (% of
predicted) | 88.9±10.2 | 59.6±9.2a |
| FEV1/FVC (% of
predicted) | 87.6±4.3 | 85.5±2.4 |
| TLC (% of
predicted) | 88.1±13.2 | 62.5±10.7a |
| DLCO (% of
predicted) | 87.8±4.6 | 53.3±10.6a |
| PaO2
(mmHg) | 86.1±2.7 | 66.3±6.4a |
| PaCO2
(mmHg) | 39.7±3.4 | 35.6±2.7 |
| Smoking history
(%) | 0 | 0 |
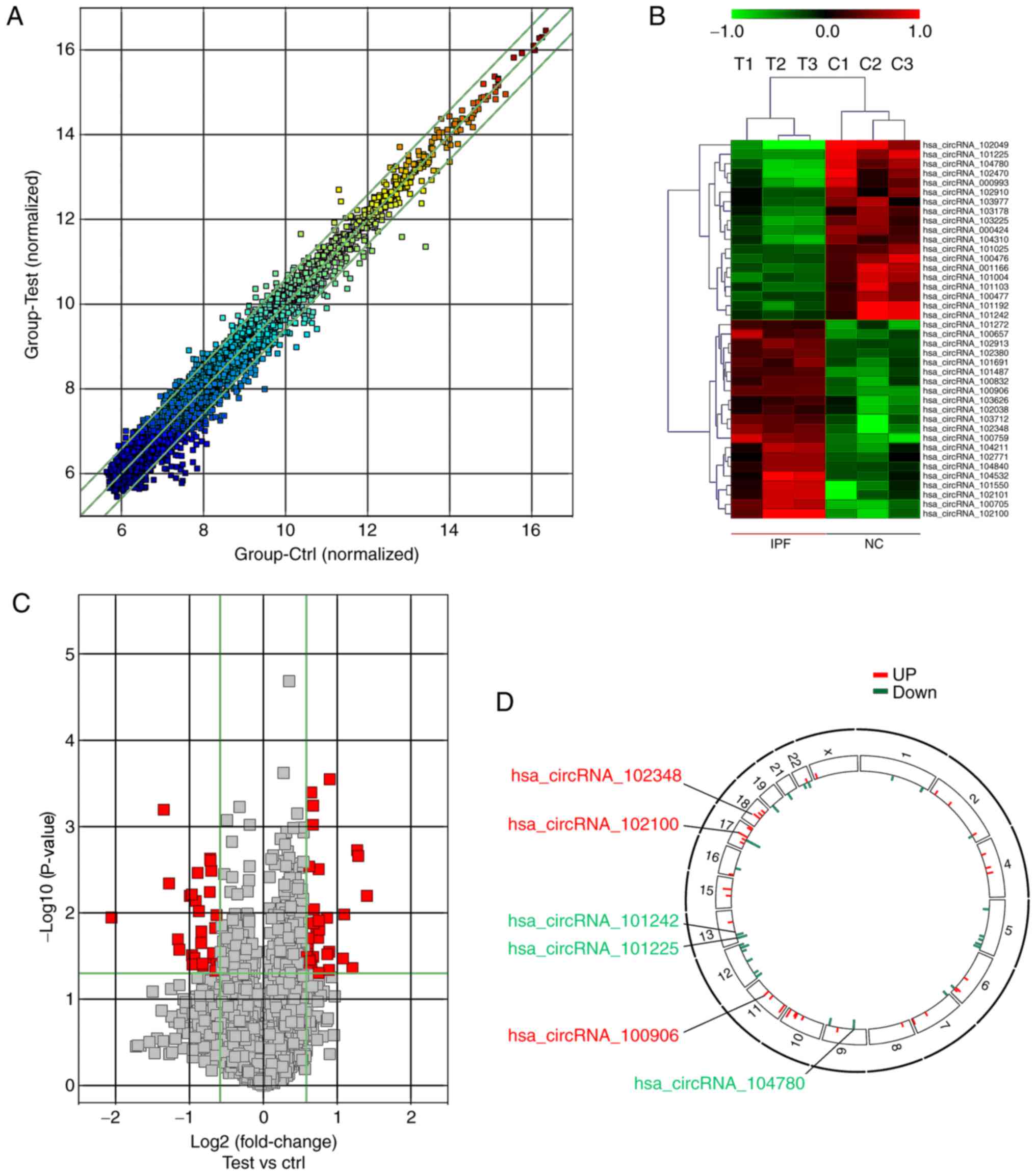
A high-throughput microarray assay was used to
identify circRNAs with abnormal expression in IPF. A total of 4,731
circRNA targets were detected using microarray probes in plasma
samples from IPF patients and healthy individuals. The scatter plot
in Fig. 1A displays the variation
in the circRNA expression ratio between the two groups. The heat
map of circRNA expression in samples ofthe two groups in Fig. 1B displays the regulation of
several circRNA clusters. An upregulated cluster consisting of 38
circRNAs and a down-regulated cluster consisting of 29 circRNAs
were detected in the patients (fold change, ≥1.5; P<0.05). From
the volcano plot in Fig. 1C,
differentially expressed circRNAs with statistical significance
between IPF and control samples were identified. The distribution
of circRNA in human chromosomes is illustrated in Fig. 1D. A total of 21 and 18 of the most
highly upregulated and downregulated circRNAs, respectively,
according to their fold change, are listed in Table II.
 | Table IIMost differentially expressed circRNA
ranked by FC vs. NC group in microarray data. |
Table II
Most differentially expressed circRNA
ranked by FC vs. NC group in microarray data.
| circRNA | P-value | FC (abs) | Change vs. NC | Alias | circRNA_type | Gene symbol |
|---|
|
hsa_circRNA_100906 | 0.00028136 | 1.8654768 | Upregulation |
hsa_circ_0023858 | Exonic | ANKRD42 |
|
hsa_circRNA_102380 | 0.000400941 | 1.5656011 | Upregulation |
hsa_circ_0047841 | Exonic | LIG3 |
|
hsa_circRNA_102913 | 0.000949382 | 1.5911579 | Upregulation |
hsa_circ_0058058 | Exonic | ATIC |
|
hsa_circRNA_100705 | 0.001874312 | 2.4096969 | Upregulation |
hsa_circ_0008898 | Exonic | OAT |
|
hsa_circRNA_100759 | 0.002180791 | 2.4327399 | Upregulation |
hsa_circ_0004099 | Exonic | DENND5A |
|
hsa_circRNA_102100 | 0.006302884 | 2.6442716 | Upregulation |
hsa_circ_0044226 | Exonic | CDC27 |
|
hsa_circRNA_102348 | 0.010380106 | 2.1353111 | Upregulation |
hsa_circ_0007535 | Exonic | ELP2 |
|
hsa_circRNA_102101 | 0.033608302 | 2.1090716 | Upregulation |
hsa_circ_0044234 | Exonic | CDC27 |
|
hsa_circRNA_101550 | 0.043316607 | 2.3059601 | Upregulation |
hsa_circ_0035796 | Exonic | HERC1 |
|
hsa_circRNA_100657 | 0.011363912 | 1.8185249 | Upregulation |
hsa_circ_0006520 | Exonic | MMS19 |
|
hsa_circRNA_103712 | 0.02802823 | 1.8542931 | Upregulation |
hsa_circ_0007540 | Exonic | TBCK |
|
hsa_circRNA_104211 | 0.030050202 | 1.8239174 | Upregulation |
hsa_circ_0007762 | Exonic | STXBP5 |
|
hsa_circRNA_104532 | 0.04568836 | 1.8495971 | Upregulation |
hsa_circ_0001772 | Exonic | RBM33 |
|
hsa_circRNA_102038 | 0.034423041 | 1.541653 | Upregulation |
hsa_circ_0043082 | Exonic | LIG3 |
|
hsa_circRNA_104840 | 0.013111444 | 1.5986307 | Upregulation |
hsa_circ_0002780 | Exonic | CDC14B |
|
hsa_circRNA_101487 | 0.003102427 | 1.6784173 | Upregulation |
hsa_circ_0008926 | Exonic | NUSAP1 |
|
hsa_circRNA_103626 | 0.040907511 | 1.5692823 | Upregulation |
hsa_circ_0007308 | Exonic | PDS5A |
|
hsa_circRNA_102771 | 0.049588502 | 1.5610834 | Upregulation |
hsa_circ_0055377 | Exonic | CTNNA2 |
|
hsa_circRNA_101691 | 0.009068437 | 1.6051068 | Upregulation |
hsa_circ_0007637 | Exonic | CREBBP |
|
hsa_circRNA_100832 | 0.014303719 | 1.6414589 | Upregulation |
hsa_circ_0022378 | Exonic | FADS1 |
|
hsa_circRNA_101272 | 0.012972773 | 1.6474556 | Upregulation |
hsa_circ_0005783 | Exonic | KLF12 |
|
hsa_circRNA_101225 | 0.000635675 | 2.5487789 | Downregulation |
hsa_circ_0029633 | Exonic | ZMYM2 |
|
hsa_circRNA_101025 | 0.002349167 | 1.6468162 | Downregulation |
hsa_circ_0025633 | Exonic | LRMP |
|
hsa_circRNA_104780 | 0.00453198 | 2.4221486 | Downregulation |
hsa_circ_0001861 | Exonic | GRHPR |
|
hsa_circRNA_101004 | 0.006100264 | 1.9569623 | Downregulation |
hsa_circ_0000375 | Exonic | IFFO1 |
|
hsa_circRNA_102049 | 0.011284484 | 4.1574829 | Downregulation |
hsa_circ_0043278 | Exonic | TADA2A |
|
hsa_circRNA_101192 | 0.020134914 | 2.2308587 | Downregulation |
hsa_circ_0005465 | Exonic | CLIP1 |
|
hsa_circRNA_102470 | 0.0264833 | 2.2000704 | Downregulation |
hsa_circ_0049888 | Exonic | EPS15L1 |
|
hsa_circRNA_000993 | 0.030740567 | 1.9544226 | Downregulation |
hsa_circ_0001887 | Intragenic | STRBP |
|
hsa_circRNA_101242 | 0.03320098 | 1.9118807 | Downregulation |
hsa_circ_0029853 | Exonic | PAN3 |
|
hsa_circRNA_100477 | 0.005705005 | 1.6533506 | Downregulation |
hsa_circ_0016867 | Exonic | COG2 |
|
hsa_circRNA_100476 | 0.007246795 | 1.8857807 | Downregulation |
hsa_circ_0016863 | Exonic | COG2 |
|
hsa_circRNA_001166 | 0.006309977 | 1.9935227 | Downregulation |
hsa_circ_0001556 | Intronic | ERGIC1 |
|
hsa_circRNA_103225 | 0.009486195 | 1.8307713 | Downregulation |
hsa_circ_0063331 | Exonic | DDX17 |
|
hsa_circRNA_104310 | 0.016308893 | 1.7897346 | Downregulation |
hsa_circ_0079385 | Exonic | ZDHHC4 |
|
hsa_circRNA_000424 | 0.003435538 | 1.8528501 | Downregulation |
hsa_circ_0001549 | Antisense | FABP6 |
|
hsa_circRNA_103977 | 0.046667432 | 1.5710154 | Downregulation |
hsa_circ_0074362 | Exonic | ARHGAP26 |
|
hsa_circRNA_103178 | 0.029174857 | 1.6035583 | Downregulation |
hsa_circ_0062577 | Exonic | CABIN1 |
|
hsa_circRNA_102910 | 0.041706263 | 1.5010713 | Downregulation |
hsa_circ_0058051 | Exonic | BARD1 |
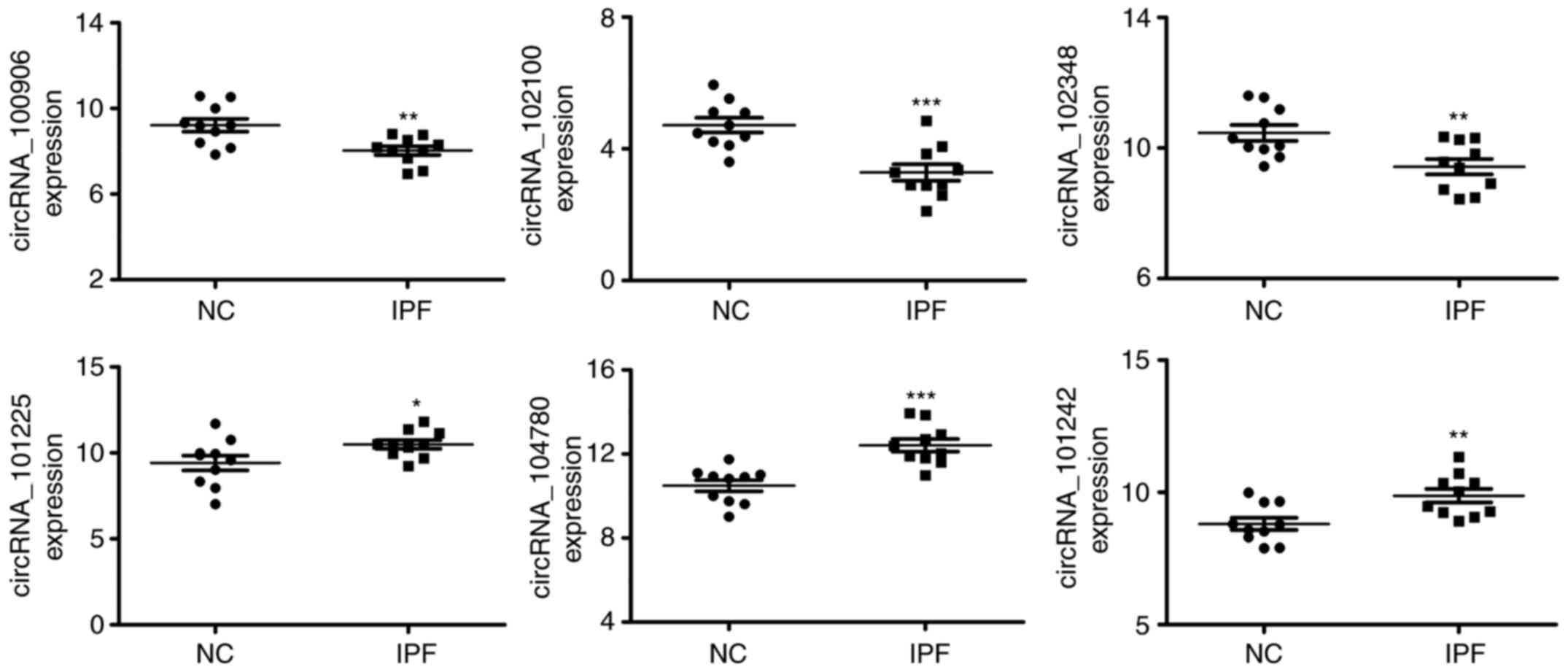
Subsequently, six differentially expressed circRNA
(hsa_circRNA_100906, hsa_circRNA_102100, hsa_circRNA_102348,
hsa_circRNA_101225, hsa_circRNA_104780 and hsa_circRNA_101242) were
selected for further validation and analysis due to their high
fold-changes and low P-values. The expression levels were
quantified via RT-qPCR with circRNA-specific divergent primers
calibrated using standard curves with housekeeping genes as
normalization standards. The RT-qPCR results validated the
differential expression of these circRNAs by using 10 independent
samples. The results indicated that, hsa_circRNA_100906,
hsa_circRNA_102100 and hsa_circRNA_102348 were significantly
upregulated, whereas hsa_circRNA_101225, hsa_circRNA_104780 and
hsa_circRNA_101242 were significantly downregulated in the IPF vs.
normal control samples (Fig. 2).
The changes in the expression of certain circRNAs identified from
the overall array data were therefore in accordance with the
results of theRT-qPCR measurements.
Genomic location of differentially
expressed circRNAs in IPF disease
circRNA is typically generated at the expense of
canonical mRNA isoforms, thereby indicating its possible function
as an important regulator of mRNA production (22). To predict the function of the
circRNAs, their location in the human genome was first detected.
Differentially expressed circRNA was typically derived from
annotated exons (86.6%). Its splice sites typically span 1-11 exons
and overlap the coding exon. Only small fractions of circRNA
originated from introns or were aligned antisense regionsto known
transcripts (Fig. 3A).
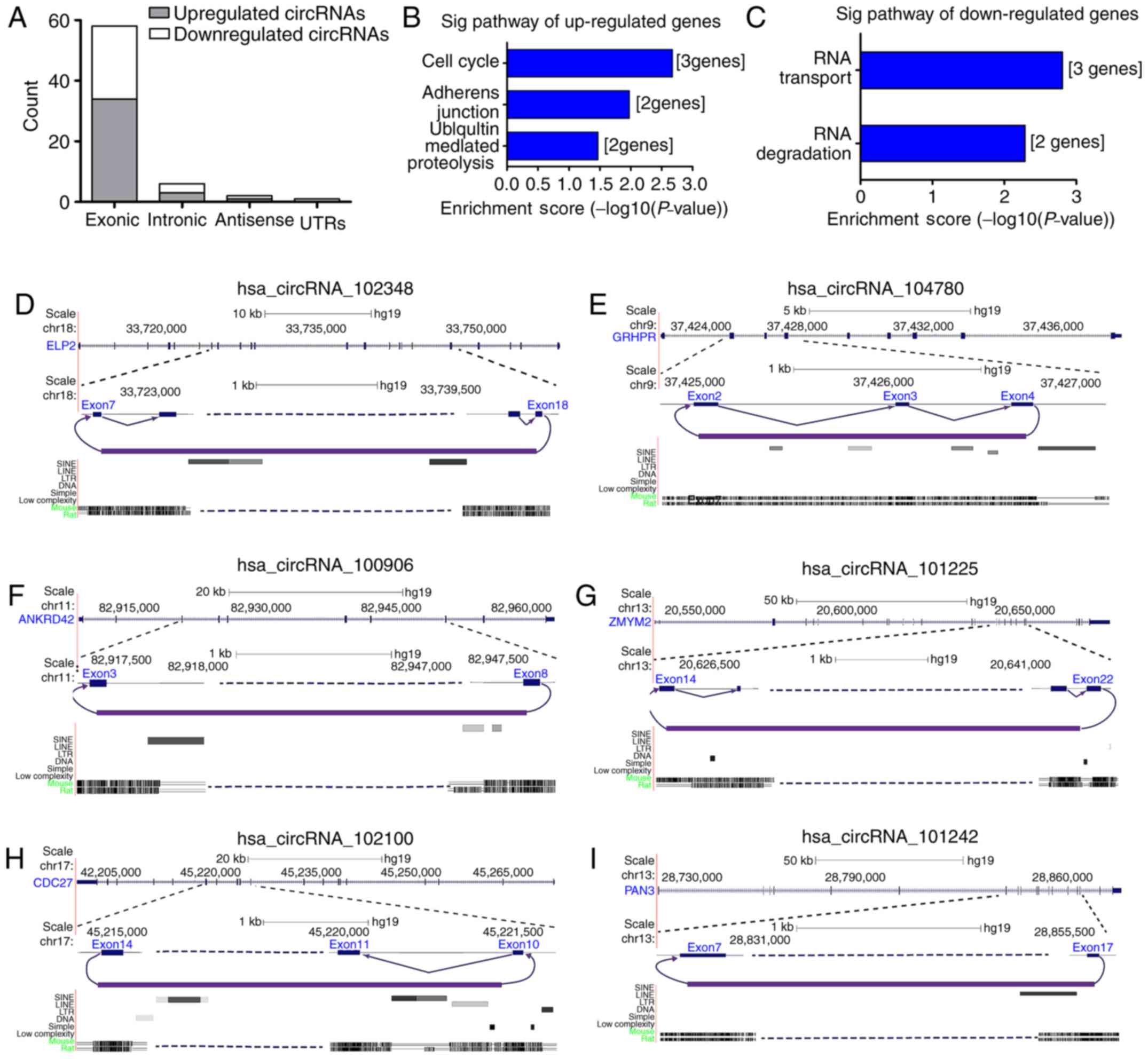 | Figure 3Correlation of differential
expression of circRNAs with their host genes. (A) Bar graph
presenting the circRNA category. (B and C) Results of a Kyoto
Encyclopedia of Genes and Genomes pathway analysis displaying the
participant pathways of host genes. (D-I) Designated orientations
and exon structure of detected six circRNA. For example,
has_circRNA_104780 is formed when the 5′ splice site at the end of
exon 4 is joined to the 3′ splice site at the beginning of exon 2
(purple). circRNA, circular RNA; Chr, chromosome; Sig,
significantly enriched, UTR, untranslated region; hsa, Homo
sapiens. ELP2, elongator acetyltransferase complex subunit 2;
GRHPR, glyoxylate and hydroxypyruvate reductase; ANKRD42, ankyrin
repeat domain 42; ZMYM2, zinc finger MYM-type containing 2; CDC27,
cell division cycle 27; PAN3, poly(A) specific ribonuclease subunit
PAN3. |
Several differentially expressed circRNAs originated
from known protein-coding genes with pivotal roles in fibrosis
[e.g. zinc-finger-DHHC-type with 4 (ZDHHC4)]. Kyoto Encyclopaedia
of Genes and Genomes (KEGG) analysis revealed that genes that
produced dysregulated circRNAs were involved in the pathways of
cell cycle, adherens junctions, ubiquitin-mediated proteolysis, and
RNA transport and degradation (Fig.
3B and C).
As presented in Fig.
3D-I, the tested circRNAs all belonged to exons. In addition,
exonic circRNAs may result from spliceosomal action, e.g. the exon
structure of the human ankyrin repeat domain 42 locus, encompassing
a 797-nt region that includes exons 3-8.
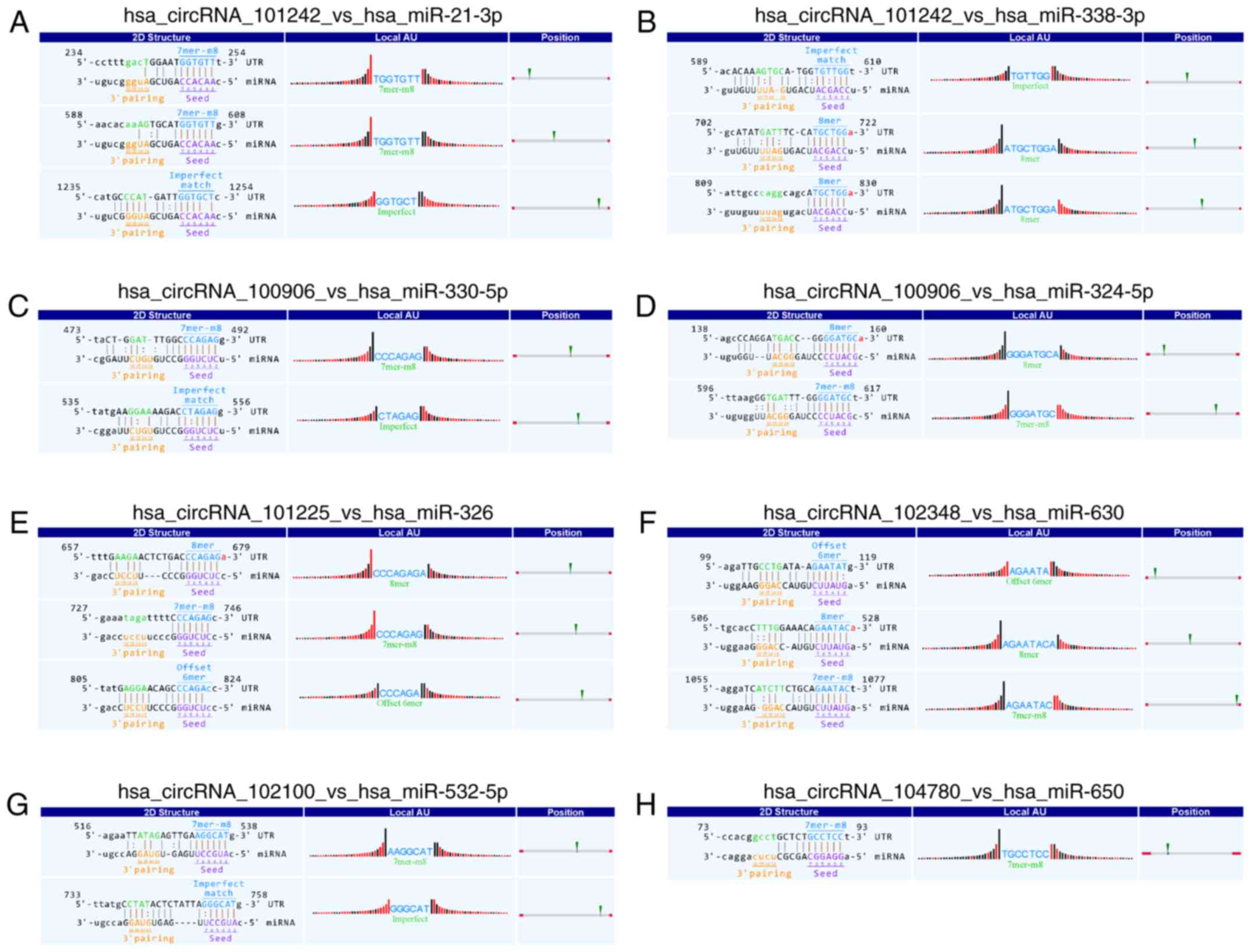
Predicted ceRNA network of
circRNAs/miRNAs/mRNAs in IPF
A considerable number of studies have reported that
circRNAs act as ‘miRNA sponges’ to regulate gene expression. The
present study evaluated the biological role of differentially
expressed circRNAs by drawing and analysing an miRNA-mediated
regulatory network. The number of predicted miRNA binding sites for
all miRNAs (deposited in miR Base version 19) was determined.
Interactions between circRNAs and miRNAs were theoretically
predicted according to their conserved seed-matching sequences. The
analysis revealed that all 67 of the differentially expressed
circRNAs contained response elements for their respective target
miRNAs. The top five miRNAs for each 67 circRNAs were displayed as
a network delineated using Cytoscape software (version 3.1.0;
cytoscape.org/) (Fig. 4A). A sub-network displaying 10
circRNAs and their target miRNAs is displayed in Fig. 4B. Numerous associated miRNAs have
pivotal roles in the progression of fibrotic disease (including
miR-326, -361-5p, -338-3p, -9-5p, -136-5p, -145-5p, -224-5p and
-877-3p). On the basis of the number of binding sites and target
mRNAs, miR-330-5p/miR-324-5p, miR-532-5p, miR-630, miR-326, miR-650
and miR-338-3p/miR-21-3p were selected as potential miRNA targets
for the six selected circRNAs (hsa_circRNA_100906,
hsa_circRNA_102100, hsa_circRNA_102348, hsa_circRNA_101225,
hsa_circRNA_104780 and hsa_circRNA_101242, respectively). The
sequence analysis of the miRNA response elements for the six
selected differentially expressed circRNAs and their potential
complementary binding miRNAs is displayed in Fig. 5. For instance, it was revealed
that in hsa_circRNA_100906, the 154 to 159th and the 611 to 616th
nucleotides starting from the 5′ terminus were completely
complementary to the miR-324-5p seed region in the 8mer or 7mer-m8
binding mode.
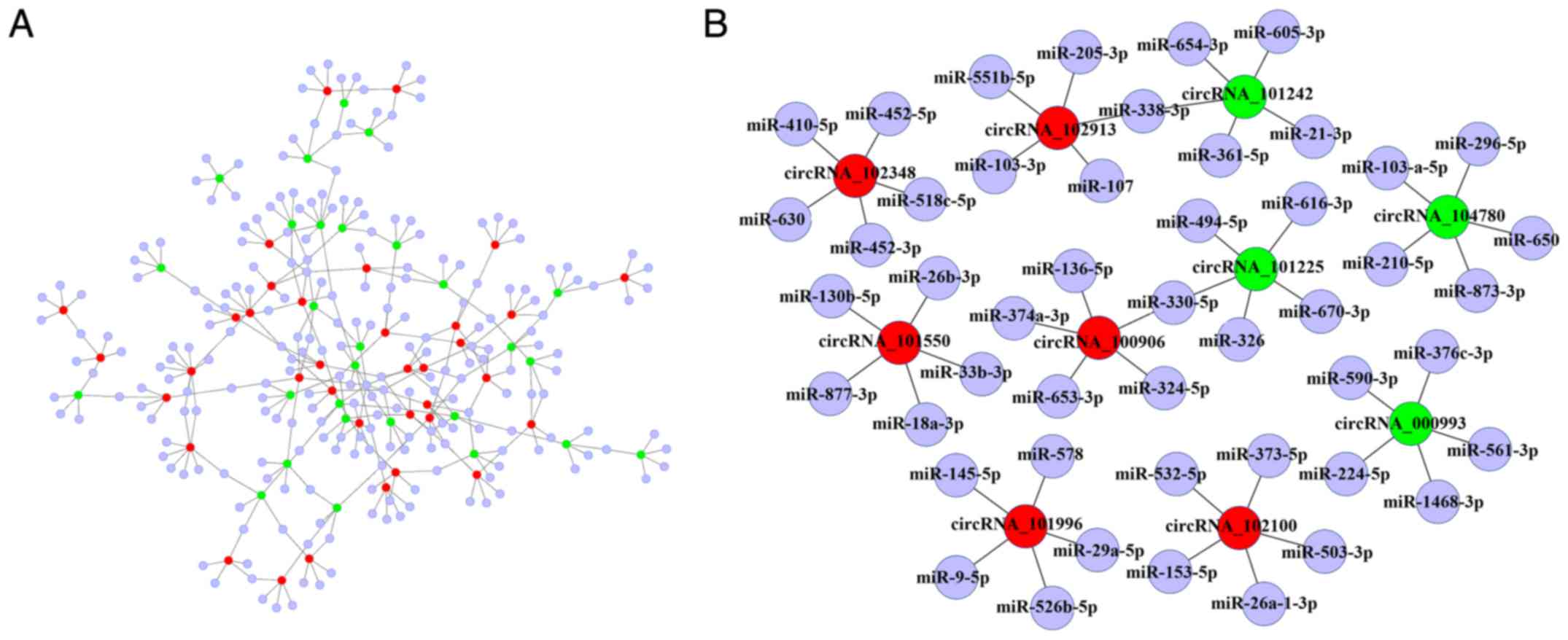 | Figure 4Interaction network of circRNAs and
miRNAs. (A) circRNA-miRNA interaction network consisting of 38
upregulated circRNAs, 29 downregu-lated circRNAs and their target
miRNAs. They were connected by 333 edges based on seed sequence
pairing interactions. (B) Interaction of 10 circRNAs (including 6
verified circRNAs: hsa_circRNA_100906, hsa_circRNA_102100,
hsa_circRNA_102348, hsa_circRNA_101225, hsa_circRNA_104780 and
hsa_circRNA_101242; and 4 circRNAs with high expression and
predicted binding, which serves an important role in lung fibrosis)
and their target miRNAs presented in a magnified network. Red and
green nodes represent upregulated and downregulated circRNAs,
respectively. Purple nodes indicate miRNAs. miR/miRNA, microRNA;
circRNA, circular RNA; hsa, Homo sapiens. |
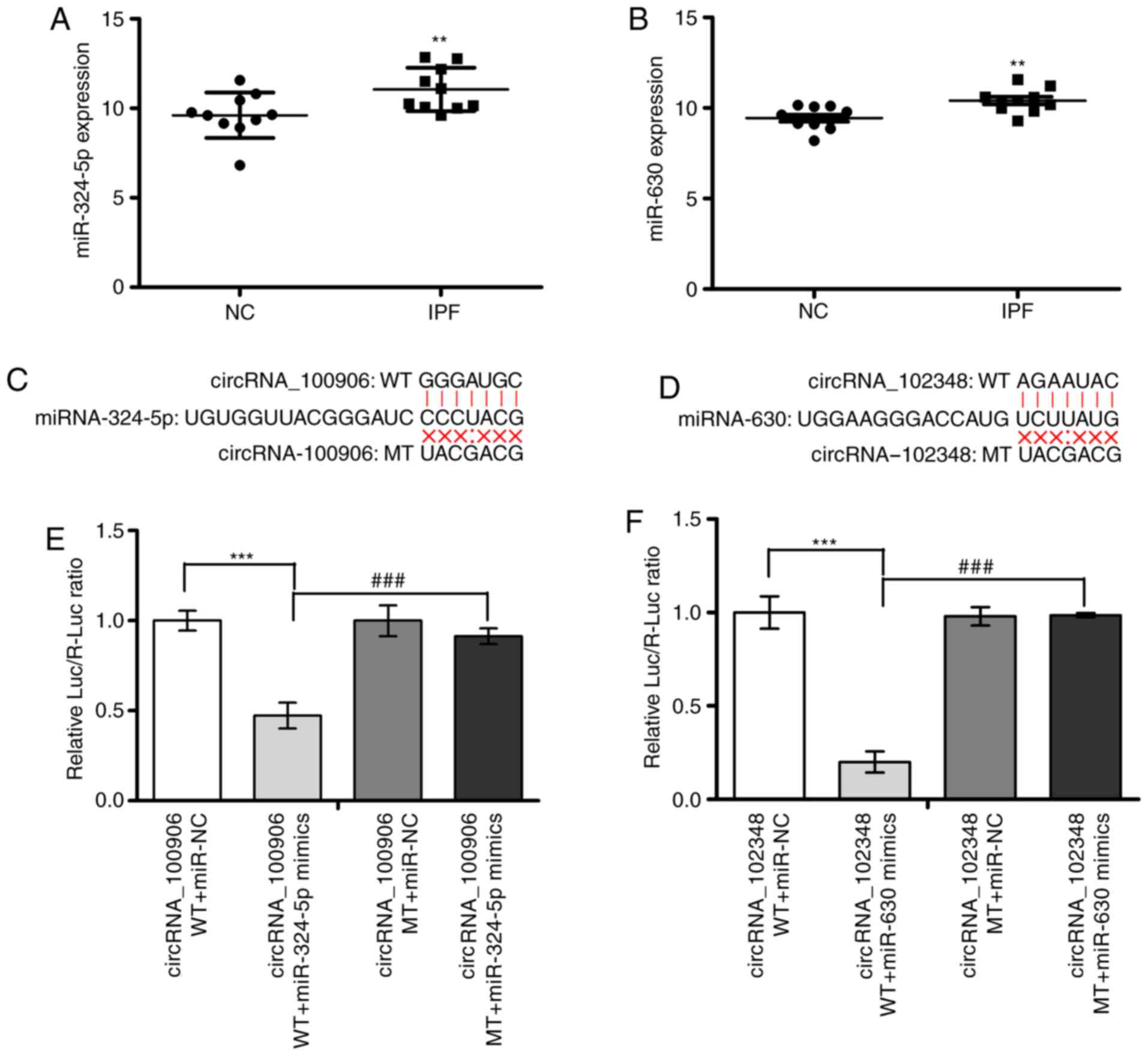
Subsequently, circRNA_100906/miR-324-5p and
circRNA_102348/miR-630 were selected to confirm the predicted
interaction between circRNA and miRNA by lucif-erase reporter
assays. The expression levels of miR-324-5p and miR-630 were
significantly downregulated in IPF patients conversely to those of
their associated circRNAs (Fig. 6A
and B). The luciferase intensity decreased by >50% with the
co-transfection of luciferase reporters containing the
circRNA_100906 or circRNA_102348 full length sequence and mimics of
miR-324-5p or miR-630, respectively. To confirm the direct
interaction, the miRNA response elements (MREs) in the luciferase
reporter were mutated. Co-transfection of miRNA mimics and
luciferase reporter vector with the mutated sequence did not
considerably affect the luciferase activity (Fig. 6C-F).
miR-330-5p/miR-324-5p, miR-532-5p, miR-630, miR-326,
miR-650 and miR-338-3p/miR-21-3p were predicted as target genes and
the regulatory network of circRNAs/miRNAs/mRNAs is presented in
Fig. 7A. Numerous
fibrosis-associated genes [e.g. SMAD3, ECM protein 1 (ECM1),
vimentin, suppressor of cytokine signalling 1 (SOCS1),
rho-associated kinase (ROCK) 1, matrix metalloproteinase (MMP1),
integrin subunit β like 1 (ITGBL1), ADAM metallopeptidase domain 17
and hypoxia-inducible factor 1 subunit α(HIF1A) inhibitor] may be
targeted by the previously mentioned miRNAs. The circRNA/miRNA
regulatory networks act on target genes significantly involved in
transforming growth factor (TGF)-β1, HIF-1, Wnt, Janus kinase
(JAK), ROCK, vascular endothelial growth factor, mitogen-activated
protein kinase, Hedgehog and nuclear factor κB signalling pathways,
which are associated with cell proliferation, migration and
collagen synthesis (Fig. 7B). For
instance, circRNA_100906 may trigger the upregulation of SOCS1,
ROCK1, SP1, nemo-like kinase, MMP1, bromodomain containing 2, ECM1,
ITGBL1, phosphoinositide-3-kinase regulatory subunit 3 and
fibroblast growth factor receptor 2 by sequestering miR-324-5p and
miR-330-5p in the network. The ceRNA network and associated pathway
components may be novel clinical markers and therapeutic targets
for IPF.
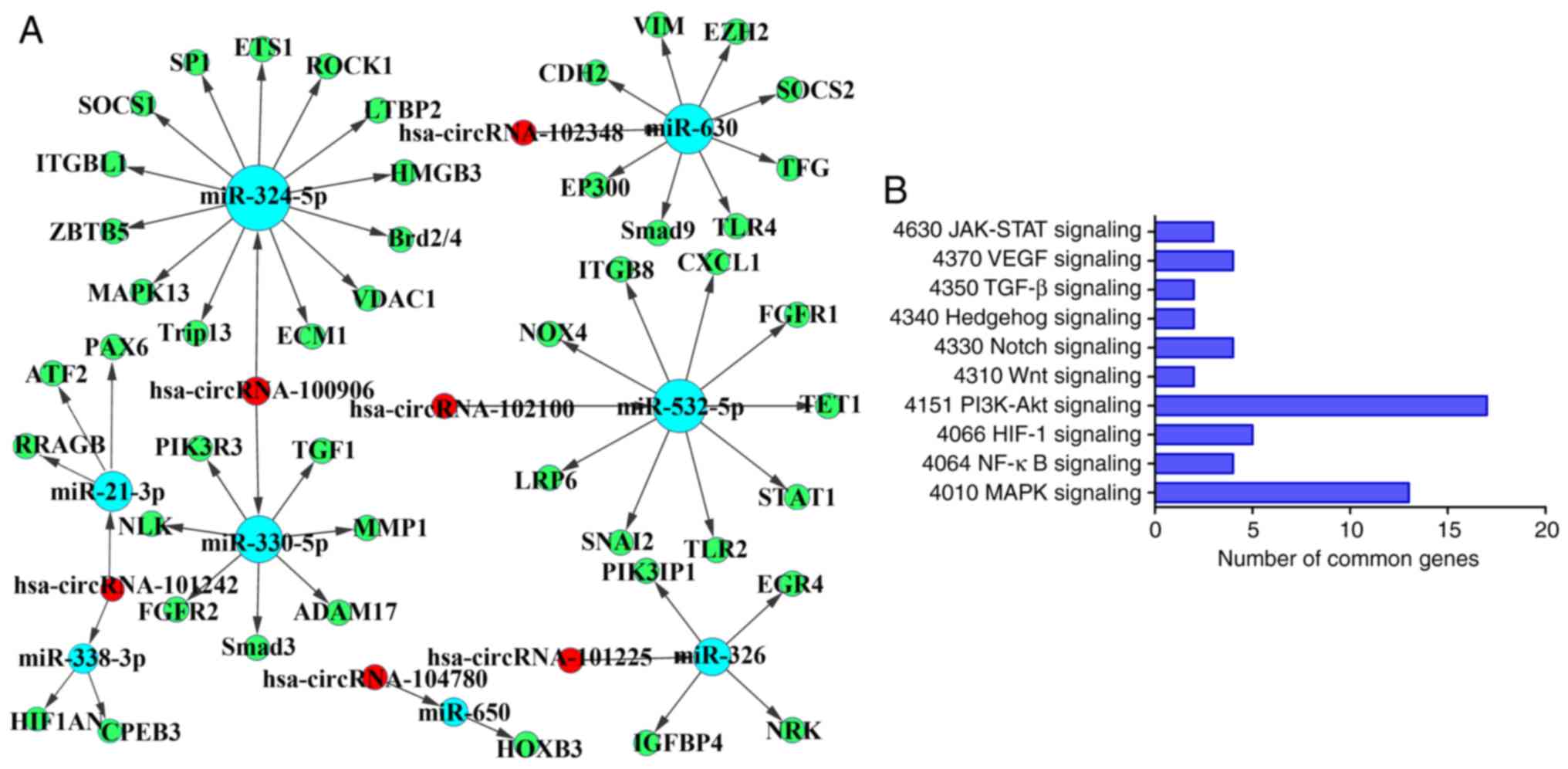 | Figure 7Prediction ofcompeting endogenous RNA
network of circRNAs/miRNAs/mRNAs. (A) Bioinformatics prediction of
the circRNA/miRNA/mRNA network. Red represents circRNA, green
represents mRNA and blue represents miRNA. (B) Kyoto Encyclopedia
of Genes and Genomes analysis of pathways enriched by the targeted
mRNAs. miR/miRNA, microRNA; circRNA, circular RNA. JAK, Janus
kinase; STAT, signal transducer and activator of transcription;
PI3K, phosphoinositide-3 kinase; HIF, hypoxia-inducible factor;
TGF, transforming growth factor; VEGF, vascular endothelial growth
factor; MAPK, mitogen-activated protein kinase. |
Discussion
circRNAs are regarded as novel clinical diagnostic,
prognostic and therapeutic biomarkers that may provide novel
approaches for the treatment of diseases (23). For instance, the expression of
circular ANRIL is correlated with the risk for atherosclerotic
vascular disease (24). However,
whether circRNAs have a role in IPF has remained elusive. In the
present study, circRNA expression profiling was performed in IPF
patients and 67 aberrantly expressed circRNAs were identified.
Furthermore, it was unveiled that certain circRNAs
(hsa_circRNA_100906, hsa_circRNA_102100 and hsa_circRNA_102348)
were upregulated, while others (hsa_circRNA_101225,
hsa_circRNA_104780 and hsa_circRNA_101242) were downregulated in
IPF.
circRNA originates from exons, introns or is aligned
with antisense regions to known transcripts, intergenic sequences
or unannotated regions of the genome (25). Linear splicing and circRNA
production compete against each other for splicing sites and assign
a regulated function to circRNA in their hosted gene (26). Chao et al (27) reported that circRNA originating
from the mouse formin (Fmn) gene functioned as an ‘mRNA trap’ by
leaving a non-coding linear transcript, thereby reducing the
expression levels of the Fmn protein. circRNA to eukaryotic
translation initiation factor 3 subunit J and poly(A) binding
protein interacting protein 2 are predominantly localized in the
nucleus, interacting with U1 small nuclear ribonucleoproteins and
enhancing the transcription of their parental genes in a cisacting
manner (28). In the present
study, numerous parental genes of differentially expressed circRNAs
associated with the biological process of fibrosis were identified.
Yang et al (29) reported
that ZDHHC4, which causes a significant downregulation of
circRNAhsa_circ_104310, is a significant methylation marker to
regulate the expression of large groups of genes in a trans-acting
manner in IPF. André et al (30) demonstrated that BRCA1-associated
RING domain 1, the host gene of hsa_circRNA_102910, regulates lung
epithelial cell damage and fibroblast proliferation by acting as a
mediator of hypoxia and TGF-β may be a novel target for IPF
treatment. hsa_circRNA_102348 is spliced from the elongator
acetyltransferase complex subunit 2 gene, which encodes a general
binding partner or chaperone for the activation and nuclear
translocation of signal transducer and activator of transcription
(STAT)3 (31) and may regulate
the JAK/STAT signalling pathway. hsa_circRNA_102100 and 102101
align with the gene cell division cycle (CDC)27, which encodes a
core component of the anaphase-promoting complex/cyclosome.
Furthermore, aberrant expression of CDC27 may result in chromosomal
aneuploidy integrity and improper cell cycle progression, which may
have roles in cell proliferation in IPF (32). The parental gene of
hsa_circRNA_101225 encodes a zinc finger MYM type 2, which
apparently binds specifically to fibroblast growth factor
receptor-1 or proteins with added small ubiquitin-like modifiers,
including histone deacetylase 1 (33). KEGG analysis supported the concept
of target genes that may regulate crucial biological processes
during the development of IPF.
The highly complex regulator y network of
circRNA/miRNA/mRNA represents another important layer of epigenetic
control over gene expression in health and disease, involving a
variety of cellular processes, including the cell cycle, apoptosis
and adherens junctions (34,35). At present, sponging activity is
the major function of certain circRNAs. A 1.2-kb single-exon
circRNA produced from the mammalian sex determination gene may
function as a miR-138 sponge for 16 target sites (36). The circRNA/miRNA interaction
analysis of the present study indicated that abnormally expressed
circRNAs in IPF patients possess abundant miRNA target sites that
have important roles in fibrotic disease. Zhang et al
(37) reported that the
miR-338-5p/lysophosphatidic acid receptor 1 axis may be regarded as
a target of tectorigenin in bleomycin-induced lung fibrosis. In the
present study, miR-338-5p was matched with hsa_circRNA_102101 and
hsa_circRNA_102100, which originate from the same coding gene
(CDC27). hsa_circRNA_101996 possessed MREs for miR-9 and -145,
which regulated fibrosis by targeting platelet-derived growth
factor receptor β/extracellular signal-regulated kinase signalling
or the TGF-β receptor 2/SMAD3/TGF-β pathway (38,39).
miRNAs including miR-324-5p, miR-330-5p, miR-532-5p,
miR-630, miR-650, miR-21-3p and miR-326 reportedly participate in
cell proliferation, apoptosis, migration, epithelial-mesenchymal
transition (EMT), and cytoskeleton remodelling regarding as tumour
markers (40,41). However, limited information is
available regarding their role in fibrotic disease. Pulmonary
fibrosis is a neoproliferative disorder of the lung that exhibits
several cancer-like pathogenic features, including abnormal
activation, uncontrolled proliferation, resistance to apoptosis and
high migration rates of myofibroblasts. These miRNAs were perfectly
matched with the six circRNAs selected in the present study. MMP1,
Snai2 and vimentin, which are the predicted target genes of
miR-330-5p, 532-5p and 630, respectively, are mostly accepted as
biomarkers for fibrosis. Early growth response 4, which contains
binding sites for miR-326, reportedly counteracts autocrine TGF-β
signalling and abolishes myofibroblast function (42). As a co-receptor for Wnt,
low-density lipoprotein receptor related protein 5 and -6 are
predicted targets of miR-532-5p, transmitting the canonical
Wnt/β-catenin signalling and appearing independently associated
with fibrotic disease progression (43). KEGG analysis further demonstrated
the involvement of the target genes in fibrosis-associated
signalling pathways.
circRNA_000203 is regarded as a ceRNA of collagen
type I alpha 2 chain and connective tissue growth factor by
interacting with miR-26b-5p in cardiac fibrosis (44). Furthermore, Zhou and Yu (45) reported that the profibrotic
function of circRNA_010567 is partly mediated by the miR-141/TGF-β1
pathway. However, differential expression of the two circRNAs and
the associated miRNAs was not observed in the present study.
circRNA_101242, which harbours two binding sites for miR-21-3p, has
been reported to be abnormally expressed in gastric cancer
(46). In the present study,
circRNA_101242 was also verified to be downregulated, whereas
miR-21-3p was significantly upregulated in IPF. The direct binding
and target mRNA require verification in further studies. A previous
study reported that circRNA_100269 prevents gastric cancer
proliferation by sponging miR-630 (47). However, in the present study, no
evident change of circRNA_100269 was noted, whereas miR-630 was
significantly downregulated in IPF patients. In addition,
circRNA_102348, which harbours two MREs for miR-630, was
upregulated and demonstrated to directly interact with miR-630.
Previous studies have only identified a small number of circRNAs or
ceRNAs with multiple binding sites for a particular miRNA; most
circRNAs or ceRNAs were identified to contain only one or two miRNA
binding sites (48). The
predicted number of miRNA binding sites in differentially expressed
circRNAs in the present study was three at the most as determined
by Arraystar’s prediction software. Furthermore, the interaction of
circRNA_100906/miR-324-5p was identified in the present study. As a
predicted target gene of miR-630, E1A binding protein p300was
identified as a co-activator of HIF1A, which is responsible for
lung fibrosis (49). Li et
al (50) reported that the
H19/miR-630/enhancer of zeste homolog 2(EZH2) signalling pathway
has a considerable role in nasopharyngeal carcinoma metastasis. The
activation of ROCK1 and -2, which are targeted by miR-324-5p
according to the present analysis, is involved in actin filament
assembly, actomyosin contraction, cell adhesion and motility,
proliferation and apoptosis, and the remodelling of the ECM in lung
fibrosis (51). miR-324-5p
suppresses ECM degradation in hepatocellular carcinoma by
post-transcriptionally downregulating ETS proto-oncogene 1,
transcription factor and SP1 (52). Furthermore, ITGBL1, the predicted
target gene of miR-324-5p, wasidentified to be significantly
overexpressed in the lung tissue of bleomycin-injured mice and
TGF-β1-treated cells, which inhibited EMT, myofibroblast migration
and collagen synthesis (data not shown). The direct target genes of
miR-630 and miR-324-5p in IPF will be further explored in future
studies by our group.
In conclusion, the present study indicated that
dysregulated circRNAs function as important regulators for pro- or
anti-fibrotic signalling pathways in IPF by sequestering miRNAs or
regulating their host genes. The important roles of circRNAs in IPF
will be considered for further studies in our group. Ongoing
efforts will be made to provide additional fundamental information
for improving the understanding of the regulatory mechanisms in IPF
and to provide novel approaches for the diagnosis, treatment and
prevention of the disease.
Funding
This study was supported by National Natural Science
Foundation of China (81273957, 31300288, 31470415, 81670064,
31670365 and 81530030), Natural Science Foundation of Shandong
Province (2014GSF119014 and ZR2016HP34) and The project of
Traditional Medical Science and Technology for Shandong Province
(2015-271).
Availability of data and materials
The analysed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors’ contributions
RL, YW and XS performed the experiments, analysed
the data and wrote the paper. WS, YL, JZ and QZ performed the
experiments and analysed the data. HL and CM collected the clinical
samples and analysed the data. JZ and CL conceived the experiment,
analysed the data and wrote the paper.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki and the protocol was approved by the
Ethics Committee of Binzhou Medical University (Binzhou, China).
Written informed consent was obtained from the participants of the
present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Selman M and Pardo A: Stochastic
age-related epigenetic drift in the pathogenesis of idiopathic
pulmonary fibrosis. Am J Respir Crit Care Med. 190:1328–1330. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clarke DL, Murray LA, Crestani B and
Sleeman MA: Is personalised medicine the key to heterogeneity in
idiopathic pulmonary fibrosis. Pharmacol Ther. 169:35–46. 2017.
View Article : Google Scholar
|
|
3
|
Tzouvelekis A, Yu G, Lino Cardenas CL,
Herazo-Maya JD, Wang R, Woolard T, Zhang Y, Sakamoto K, Lee H, Yi
JS, et al: SH2 domain-containing phosphatase-2 is a novel
antifibrotic regulator in pulmonary fibrosis. Am J Respir Crit Care
Med. 195:500–514. 2017. View Article : Google Scholar :
|
|
4
|
Richeldi L, Collard HR and Jones MG:
Idiopathic pulmonary fibrosis. Lancet. 389:1941–1952. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mao C, Zhang J, Lin S, Jing L, Xiang J,
Wang M, Wang B, Xu P, Liu W, Song X and Lv C: MiRNA-30a inhibits
AECs-II apoptosis by blocking mitochondrial fission dependent on
Drp-1. J Cell Mol Med. 18:2404–2416. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao G, Zhang J, Wang M, Song X, Liu W, Mao
C and Lv C: Differential expression of long non-coding RNAs in
bleomycin-induced lung fibrosis. Int J Mol Med. 32:355–364. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fang S, Guo H, Cheng Y, Zhou Z, Zhang W,
Han B, Luo W, Wang J, Xie W and Chao J: circ HECTD1 promotes the
silica-induced pulmonary endothelial-mesenchymal transition via
HECTD1. Cell Death Dis. 9:3962018. View Article : Google Scholar
|
|
8
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo Y, Liu S and Yao K: Transcriptome-wide
investigation of mRNA/ circRNA in miR-184 and its r.57c > u
mutant type treatment of human lens epithelial cells. Mol Ther
Nucleic Acids. 7:71–80. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhong Z, Huang M, Lv M, He Y, Duan C,
Zhang L and Chen J: Circular RNA MYLK as a competing endogenous RNA
promotes bladder cancer progression through modulating VEGFA/VEGFR2
signalling pathway. Cancer Lett. 403:305–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aufiero S, van den Hoogenhof MMG, Reckman
YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM and
Creemers EE: Cardiac circRNAs arise mainly from constitutive exons
rather than alternatively spliced exons. RNA. 24:815–827. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou
S, Lin H, Liang X and Cai X: Circular RNA expression is suppressed
by androgen receptor (AR)-regulated adenosine deaminase that acts
on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis.
8:e31712017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA MTO1 acts as the
sponge of miR-9 to suppress hepatocelluar carcinoma progression.
Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wilusz JE and Sharp PA: Molecular biology.
A circuitous route to noncoding RNA. Science. 340:440–441. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
18
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Raghu G, Collard HR, Egan JJ, Martinez FJ,
Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et
al: An official ATS/ERS/JRS/ALAT statement: Idiopathic pulmonary
fibrosis: Evidence-based guidelines for diagnosis and management.
Am J Respir Crit Care Med. 183:788–824. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ausubel FM, Brent R, Kingston RE, Moore
DD, Seidman JG, Smith JA and Struhl K: Short Protocols in Molecular
Biology. 5th edition. Wiley-Blackwell; Hoboken, New Jersey:
2002
|
|
22
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of Intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PloS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y and Wang Z: Efficient backsplicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar :
|
|
27
|
Chao CW, Chan DC, Kuo A and Leder P: The
mouse formin (Fmn) gene: Abundant circular RNA transcripts and
gene-targeted deletion analysis. Mol Med. 4:614–628. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang Li, Bao CZ, Chen C, Lin L, Wang M,
Zhong X, Yu G, Hu B, Dai WL, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang IV, Pedersen BS, Rabinovich E,
Hennessy CE, Davidson EJ, Murphy E, Guardela BJ, Tedrow JR, Zhang
Y, Singh MK, et al: Relationship of DNA methylation and gene
expression in idiopathic pulmonary fibrosis (IPF). Am J Respir Crit
Care Med. 190:1263–1272. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
André PA, Prêle CM, Vierkotten S,
Carnesecchi S, Donati Y, Chambers RC, Pache JC, Crestani B,
Barazzone-Argiroffo C, Königshoff M, et al: BARD1 mediates TGF-β
signaling in pulmonary fibrosis. Respir Res. 16:1182015. View Article : Google Scholar
|
|
31
|
Suaud L, Miller K, Panichelli AE, Randell
RL, Marando CM and Rubenstein RC: 4-Phenylbutyrate stimulates Hsp70
expression through the Elp2 component of elongator and STAT-3 in
cystic fibrosis epithelial cells. J Biol Chem. 286:45083–45092.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Link LA, Howley BV, Hussey GS and Howe PH:
PCBP1/HNRNP E1 protects chromosomal integrity by translational
regulation of CDC27. Mol Cancer Res. 14:634–646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aguilar-Martinez E, Chen X, Webber A,
Mould AP, Seifert A, Hay RT and Sharrocks AD: Screen for
multi-SUMO-binding proteins reveals a multi-SIM -binding mechanism
for recruitment of the transcriptional regulator ZMYM2 to
chromatin. Proc Natl Acad Sci USA. 112:E4854–E4863. 2015.
View Article : Google Scholar
|
|
34
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circlar non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclersis in humans. Nat Commun. 7:124292016.
View Article : Google Scholar
|
|
35
|
Ghosal S, Das S, Sen R and Chakrabarti J:
HumanViCe: Host ceRNA network in virus infected cells in human.
Front Genet. 5:2492014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang H, Liu X, Chen S, Wu J, Ye X, Xu L,
Chen H, Zhang D, Tan R and Wang Y: Tectorigenin inhibits the in
vitro proliferation and enhances miR-338 expression of pulmonary
fibroblasts in rats with idiopathic pulmonary fibrosis. J
Ethnopharmacol. 131:165–173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Ma L, Fan H, Yang Z, Li L and Wang
H: MicroRNA-9 regulates cardiac fibrosis by targeting PDGFR-β in
rats. J Physiol Biochem. 72:213–223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang YS, Li SH, Guo J, Mihic A, Wu J, Sun
L, Davis K, Weisel RD and Li RK: Role of miR-145 in cardiac
myofibroblast differentiation. J Mol Cell Cardiol. 66:94–105. 2014.
View Article : Google Scholar
|
|
40
|
Rupaimoole R, Ivan C, Yang D, Gharpure KM,
Wu SY, Pecot CV, Previs RA, Nagaraja AS, Armaiz-Pena GN, McGuire M,
et al: Hypoxia-upregulated microRNA-630 targets Dicer, leading to
increased tumor progression. Oncogene. 35:4312–4320. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Y, Wang SX, Mu R, Luo X, Liu ZS,
Liang B, Zhuo HL, Hao XP, Wang Q, Fang DF, et al: Dysregulation of
the miR-324 5p-CUEDC2 axis leads to macrophage dysfunction and is
associated with colon cancer. Cell Rep. 7:1982–1993. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kosla J, Dvorakova M, Dvorak M and Cermak
V: Effective myofibroblast dedifferentiation by concomitant
inhibition of TGF-β signaling and perturbation of MAPK signaling.
Eur J Cell Biol. 92:363–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lam AP, Herazo-Maya JD, Sennello JA,
Flozak AS, Russell S, Mutlu GM, Budinger GR, DasGupta R, Varga J,
Kaminski N and Gottardi CJ: Wnt coreceptor Lrp5 is a driver of
idiopathic pulmonary fibrosis. Am J Respir Crit Care Med.
190:185–195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN,
Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al: circRNA_000203
enhances the expression of fibrosis-associated genes by
derepressing targets of miR-26b5p Col1a2 and CTGF, in cardiac
fibroblats. Sci Rep. 7:403422017. View Article : Google Scholar
|
|
45
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J,
Lin H, Liu F and Dai Y: Circular RNA and gene expression profiles
in gastric cancer based on microarray chip technology. Oncol Rep.
37:1804–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: circRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017.
|
|
48
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: circRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kusko RL, Brothers JF II, Tedrow J, Pandit
K, Huleihel L, Perdomo C, Liu G, Juan-Guardela B, Kass D, Zhang S,
et al: Integrated genomics reveals convergent transcriptomic
networks underlying COPD and IPF. Am J Respir Crit Care Med.
194:948–960. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li X, Lin Y, Yang X, Wu X and He X: Long
noncoding RNA H19 regulates EZH2 expression by interacting with
miR-630 and promotes cell invasion in nasopharyngeal carcinoma.
Biochem Biophys Res Commun. 473:913–919. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Knipe RS, Tager AM and Liao JK: The Rho
kinases: Critical mediators of multiple profibrotic processes and
rational targets for new therapies for pulmonary fibrosis.
Pharmacol Rev. 67:103–117. 2015. View Article : Google Scholar :
|
|
52
|
Cao L, Xie B, Yang X, Liang H, Jiang X,
Zhang D, Xue P, Chen D and Shao Z: MiR-324-5p suppresses
hepatocellular carcinoma cell invasion by counteracting ECM
degradation through post-transcriptionally downregulating ETS1 and
SP1. PloS One. 10:e01330742015. View Article : Google Scholar : PubMed/NCBI
|